
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 23 June 2022
Sec. Animal Nutrition and Metabolism
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.902001
 Yan Li1†
Yan Li1† Ning Ma1†
Ning Ma1† Liyuan Ren2
Liyuan Ren2 Meimei Wang2
Meimei Wang2 Linqi Hu2
Linqi Hu2 Yizhao Shen2
Yizhao Shen2 Yufeng Cao2,3
Yufeng Cao2,3 Qiufeng Li2,3
Qiufeng Li2,3 Jianguo Li2,3,4*
Jianguo Li2,3,4* Yanxia Gao2,3,4*
Yanxia Gao2,3,4*The objective of the present study was to investigate the effects of N-carbamylglutamate (NCG) supplementation on metabolic profile and microbiota in ruminal content and feces of lactating dairy cows under heat stress (HS). Forty-eight lactating Holstein cows (154 ± 13.6 days in milk) were assigned randomly to four treatments (n = 12), to receive 0, 15, 20, or 25 g/day of commercial NCG (proportion: 97.7%) for the period of 60 days. The recorded ambient temperature–humidity index (THI) suggested that the cows were exposed to HS for almost the entire experimental period (average THI: 80.6). Samples of ruminal content and feces were collected at the end of the trial (day 60) to determine the biological effects of NCG supplementation on metabolome and microbiota using mass spectrometry-based metabolomics and 16S rRNA gene sequencing techniques, respectively. Results showed that NCG supplementation enhanced the levels of ruminal microbial protein, total volatile fatty acids (VFAs), and the molar proportion of propionate in the rumen, but lowered the ruminal pH, ammonia nitrogen (NH3-N), and the ratio of acetate to propionate. NCG at doses of 20 and 25 g/day reduced the community richness and diversity of ruminal microbiota with the decrease of Shannon and Simpson diversity. Compositions of ruminal and fecal microbiotas were altered by NCG, and the PICRUSt results revealed that metabolic pathways of the bacteria, such as amino acid metabolism, energy metabolism, and pyruvate metabolism, were enriched in NCG groups. Distinct changes in the metabolomic profile of ruminal fluid were observed between the control and NCG groups. Changes of 26 metabolites mainly involved in arginine metabolism, glutamate metabolism, and nitrogen metabolism were observed associated with NCG supplementation. These results provided new insights into the effects of NCG on metabolomic profile and microbiota in ruminal content and feces, and the optimal dose of NCG supplemented to dairy cows was 20 g/hd/day, which contributed to understanding the effects of NCG on HS in lactating dairy cows.
The gut microbiota plays a key role in nutrition acquisition, and gut microbiota improvement can increase the efficiency of ruminal fermentation and feed utilization in ruminant production (1). The structure and abundance of the gut microbiota are affected by many factors such as antibiotics, age, and dietary structure (2). It is well known that ruminal content and feces are important biological samples to investigate the changes in gut microbiota in dairy cows. A previous study found that the diversity and composition of fecal microbiota in dairy cows under summer heat stress (HS) were changed, and these changes were related to cows' health (3). Moreover, the studies found that the microbial community composition, bacterial diversity, and richness are different in feces and ruminal content (4). Gut microbiota also affects the host metabolism, immune response, and other physiological processes through the production of bioactive metabolites such as vitamins, amino acids, and volatile fatty acids (VFAs) (5). Recent studies have proved that microbial metabolites can be present in the plasma and other body fluids, and in this way, the gut microbiota can make interactions with the host (6, 7). Thus, the integrative analysis of gut microbiota and metabolite profiling can help us to improve animal production performance by increasing the understanding of interactions between the host and microbiota (8).
Dairy cows are sensitive to the high-temperature environment. With the aggravation of the greenhouse effect, HS causes more and more harm to dairy production, reproductive performance, and economic loss in the dairy industry (9, 10). HS can change the ruminal function and shift the composition and community structure of ruminal microbiota. Previous studies have found that ruminal pH and acetate concentration were decreased in cows under HS, whereas lactate concentration and energy-related bacteria such as Streptococcus ruminobacter and Treponema were increased (11). HS can also change the fecal microbial population, microbiota diversity, and composition, resulting in functional disorders in the immune system and lipid metabolism (3, 12).
Arginine is one of the essential amino acids for dairy cows. In addition to being a building block of protein synthesis, arginine is also used to convert into biological molecules such as nitric oxide, polyamines, and creatine, which play important roles in animal development and health. Dou et al. (13) reported that arginine could reduce the damage in intestinal epithelial cells of dairy cows caused by HS by lowering inflammation and oxidant stress. However, the short biological half time, ruminal degradation, and the competition in the transportation of alkaline amino acids limit the application of arginine as a feed additive in dairy farming (14–16). N-carbamylglutamate (NCG) can increase endogenous arginine synthesis through the activation of carbamyl phosphate synthetase-1 (17). Currently, a growing number of studies have indicated that NCG can be used as a promising feed additive in the livestock industry because NCG treatment can increase the growth rate, muscle protein synthesis, reproductive efficiency, and fetal development in mammals (18–20). However, as a feed additive enhancing arginine synthesis, the effects of NCG on HS in dairy cows remain unknown.
In dairy studies, NCG has been reported to improve production performance and milk quality (21). Our previous study found that NCG supplementation had no effects on dry matter intake and regulated plasma metabolomics profile in dairy cows under HS. Meanwhile, NCG increased milk yield, milk protein percentage, serum glucose concentration, and improved antioxidant capacity and immune function in dairy cows, which indicated the improving effect of NCG in HS (22). However, little information is known concerning the effects on metabolite profiles of ruminal fluid and gut microbiota associated with NCG treatment in dairy cows under HS. In conjunction with our previous study, it is necessary to further investigate the action mechanism of NCG on HS by ruminal fluid and gut microbiota.
16S rRNA gene sequencing technology has served as an important tool for the analysis of the structure and relative abundance of microbial communities. Metabolomics based on liquid chromatography mass spectrometry (LC-MS) is widely used to obtain metabolic information in biological systems (23). Notably, 16S rRNA gene sequencing coupled with metabolomics has been widely applied to investigate the effects of food type in dairy cows on the changes of microbiota and key metabolic pathways through the comprehensive analysis of biological samples such as milk, plasma, ruminal fluid, and urine (24–26). In this study, we applied LC-MS-based metabolomics to characterize ruminal metabolites and 16S rRNA gene sequencing technology to determine the changes in the ruminal and fecal microbial population in dairy cows treated by NCG under HS. The comprehensive analysis of the metabolites and microbiota offered important insights into metabolic processes and the microbial variations in the rumen and feces, which help to understand the mechanism of improved efficiency by NCG supplementation in dairy cows.
Forty-eight lactating Holstein cows (154 ± 13.6 days in milk, 1–3 parity and similar body condition score, BCS) from Hong Da dairy farm (Baoding, Hebei) were used in a randomized complete design and randomly assigned to four treatments (n = 12). The measurement of BCS was performed according to previous reports (27). In each treatment, 0 (control), 15 (low dose, LNCG), 20 (medium dose, MNCG), and 25 g/hd/day (high dose, HNCG) of NCG was precisely supplemented by mixing with total mixed rations (TMR) in the morning, respectively (20). The dairy cows were grouped to receive NCG supplementation treatment and free access to freshwater. Commercial NCG used in this study was purchased from Beijing Animore Sci. & Tech. Co., Ltd. (Beijing, China), the proportion of NCG was 97.7% and the rest were water and impurities. All cows were fed a standard TMR diet, the compositions and nutrient levels of TMR are listed in Supplementary Table S1. The experiment was conducted for 60 days. Ambient temperature and relative humidity of each day were measured to calculate the temperature-humidity index (THI) (28). With the use of a thermo-hygrometer probe (Model: AR837, XiMa Smart Sensor Co., Ltd., Hongkong, China), the temperature and relative humidity of cowsheds were measured in the morning (06:30), midday (14:00), and night (20:00), three times a day. As shown in Supplementary Figure S1, the THI was >72, indicating that the cows were under HS during the entire experiment.
Five cows from each group were selected randomly for ruminal collection on day 60. Ruminal samples were collected using an oral stomach tube for 4 h after morning feeding, and the ruminal fluid samples were harvested by filtering ruminal contents with sterile four-layer cheesecloth (29). The oral stomach tube was inserted to a depth of 180 cm for ruminal fluid collection. In order to avoid ruminal fluid contamination from saliva or mucus in the mouth and esophagus, the first 150 ml of ruminal fluid was discarded, and then 150 ml of ruminal fluid was collected (29). The collected 150 ml ruminal contents were mixed evenly, divided into three 10 ml sterilization tubes, and quickly stored in liquid nitrogen. After being returned to the laboratory, they were stored at −80°C for ruminal microbiota analysis. The fecal samples were collected from the rectum using sterile gloves on day 60 (one pair of gloves per cow). The obtained fecal samples were divided into three 5 ml sterilized frozen tubes and put into liquid nitrogen immediately, and then the samples were stored in a refrigerator at −80°C for microbiota analysis.
The pH of the ruminal fluid was immediately measured using a digital pH meter (DENVER UB-7, Denver Instrument, Denver, United States). Two parts of the ruminal fluid samples (10 ml) were transferred to the refrigerator at −80°C within 2 h for metabolomic and VFAs analysis. In VFAs analysis, the ruminal fluid samples were acidified with 25% metaphosphoric acid, followed by standing for 30 min, and then centrifuged (10,000 g, 15 min). The supernatant was collected for VFA analysis using gas chromatography [7890A, Agilent Technologies, Chandler, United States, (30)]. An aliquot of ruminal fluid (10 ml) was acidified with 6 mol/L hydrochloric acids and stored at −20°C for the analysis of NH3-N concentrations (31). Ruminal microbial protein (MCP) was determined according to the previous study (32).
Microbial DNA was extracted from ruminal contents using PowerFecal™ DNA Isolation kit (MO BIO Laboratories, Carlsbad, CA, United States), and the concentration and purity of DNA samples were determined by Nanodrop spectrophotometer (Thermo Scientific, Wilmington, NC, United States). 515f/806r primer set targeted the V4 region was used as PCR primer for 16S rDNA amplicon libraries. The PCR reactions were performed using Phusion High-fidelity PCR Master Mix (New England Biolabs LTD., Shanghai, China), and the amplified products were detected and purified by Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany). Following amplification, sequencing libraries were constructed and then sequenced on an Illumina HiSeq 2500 platform at Novogene Technology Co., Ltd. (Tianjin, China) according to standard protocols.
Based on the unique barcode and primer sequence, the paired-end reads were assigned to the samples. Then, with the use of FLASH (Version 1.2.7), the paired-end reads were merged to obtain raw tags (33). For high-quality clean tags, raw tags were filtered by QIIME (Version 1.7.0) with the main parameters as follows: minimum quality score = 19, minimum/maximum length = 200/1,000, and error-correcting Golay 12 nucleotide barcodes (34). Tags were clustered to operational taxonomic units (OTUs) using UPARSE (Version 7.1) with a 97% similarity cutoff. Chimeras were filtered out using UCHIME (Version 4.2.40). The indices of Alpha diversity including Shannon, Chao1, Simpson, and ACE were calculated using MOTHUR (version v.1.30.1). Beta diversity was calculated with QIIME (Version 2.0) and displayed with R software (Version 2.15.3). Cluster analysis was performed by principal coordinate analysis (PCoA) to exhibit the differences of the samples using the WGCNA package, stat packages, and ggplot2 package in R software. Phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) analysis was used to assess the metabolic changes of the microbiota in ruminal content and feces (35).
Prior to analysis, the ruminal fluid samples were thawed on ice. Next, 200 μl of the sample was placed into a centrifuge tube, and 400 μl of methanol/water (4:1, v/v) was added. After vortex-mixing for 30 s, the mixture was incubated (1 h at −20°C) and centrifuged (14,000 g, 20 min at 4°C). And then, the supernatant of the ruminal fluid sample was further collected and dried. The obtained lyophilized powder was reconstituted (60% methanol, 100 μl), vortex-mixed (1 min), and centrifuged (14,000 g, 15 min at 4°C) for metabolomic analysis. Quality control (QC) samples were prepared by pooling equal volumes of supernatant from each sample and placed in the analysis sequence for system evaluation.
Metabolomic data acquisition was carried out by Vanquish ultra high-performance liquid chromatography (UHPLC) system coupled to a Q ExactiveTM HF-X mass spectrometer (Thermo Fisher, Waltham, MA, United States). Metabolomic data acquisition and analysis were performed according to previous studies (25). The detailed parameters of the UHPLC system, mass spectrometer, and data processing method are provided in Supplementary Material S1.
With exacted molecular weight and MS/MS fragments, online databases such as HMDB (www.hmdb.ca/), METILN (metlin.scripps.edu), and Mass Bank (www.massbank.jp/) were used to identify the chemical structures of metabolites. For further biological interpretation, pathway analysis and biochemical reactions of the metabolites were conducted by MetaboAnalyst 4.0 software (www.metaboanalyst.ca/) and KEGG (www.kegg.jp/).
Data were analyzed by Proc Mixed of SAS 9.4 (SAS Institute Inc., Cary, NC) for a randomized complete design. In the MIXED model, NCG supplementation dose was a fixed effect and dairy cows were random variables. The linear, quadratic, and cubic NCG dose responses were determined by using specific pre-planned contrasts. The liner, quadratic, and cubic NCG dose responses were determined by using specific pre-planned contrasts. Data were reported as mean and standard error of mean (SEM). Significant differences were declared at P ≤ 0.05, and the tendency for a difference was declared at 0.05 < P ≤ 0.10.
Table 1 shows the effects of NCG supplementation on pH, NH3-N, MCP, and VFAs. The ruminal pH decreased linearly with the increase of NCG (P = 0.02). Compared with the control group, ruminal concentrations of NH3-N in the rumen in NCG-treated groups also decreased linearly (P < 0.01) with increasing NCG doses. The concentration of MCP cubically (P = 0.02) increased with NCG doses, where the medium dosage of NCG showed the best effect on MCP. There were quadratic (P = 0.01) and cubic (P < 0.01) effects of NCG levels on propionate and total VFAs, with the highest propionate and total VFAs at 20 g/hd/day NCG. The molar proportion of acetate tended to decrease linearly (P = 0.01). NCG dosage tended to have cubic effects on the molar proportion of butyrate (P = 0.01), and the ratio of acetate/propionate reduced quadratically with the increase of NCG (P = 0.01).
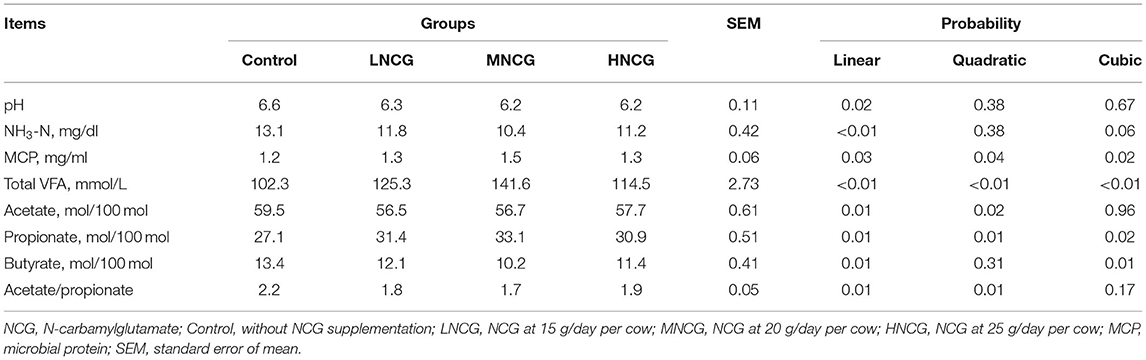
Table 1. Effects of NCG supplementation on ruminal fermentation patterns of lactating dairy cows under heat stress.
Effects of NCG on ruminal and fecal microbiota were evaluated by 16S RNA gene sequencing. A total of 19 ruminal contents and 20 fecal samples were used for sequencing. A summary of the sequencing data is provided in Supplementary Table S2. In ruminal and fecal samples, 2,159 and 2,272 OTUs were identified, respectively. Meanwhile, the results of rarefaction analysis indicated that the sequencing depth of the samples was sufficient to reflect the abundance and diversity of the microbiota (Supplementary Figure S2). Alpha diversity and beta diversity were used to evaluate the changes in microbial communities. Table 2 shows the summary of the Alpha diversity of the bacterial community. For ruminal content, no effect of NCG levels was observed on Simpson (P > 0.05), but Shannon (P = 0.01), Chao1 (P = 0.01), and ACE (P = 0.02) were reduced linearly with NCG dosage. In regard to the feces, the response of Shannon (P = 0.05) and Simpson (P = 0.03) to the increased dosage of NCG was cubic. Plots of PCoA revealed the clustering of ruminal and fecal samples in different groups, indicating the difference in the bacterial community structure (Figures 1A,B). At the phylum level, the results of OTU analysis of the ruminal contents are shown in Figure 1C. The relative abundance of ruminal microbiota at the phylum level was not affected by NCG treatment (Supplementary Table S3). In the feces, Bacteroidetes and Firmicutes were abundant in all groups (Figure 1D). The relative abundance of Firmicutes (P = 0.05) and Proteobacteria (P = 0.04) was reduced cubically with increased NCG (Supplementary Table S3). The P-value for quadratic contrast on Tenericutes was significant (P = 0.04), with the cows supplied NCG at 15 g/day having a lower abundance than cows supplied 20 or 25 g/hd/day of NCG. Linear discriminant analysis effect size (LEfSe) analysis was performed to identify the bacteria that were significantly different among the control and three NCG treatment groups. Supplementary Figure S3 displays the bacterial taxa represented in ruminal content and feces. In ruminal content, four taxa were overrepresented in the control, while five bacterial taxa were abundant in LNCG (Supplementary Figure S3A). No significant differences in the abundance of bacterial taxa in ruminal content were found between MNCG and HNCG groups. As to the feces, 22 bacterial taxa were detected in the LNCG and MNCG, and 19 distinct taxa in the control (Supplementary Figure S3B). Based on the relative abundance and statistical difference, seven genera in ruminal content and eight genera in feces were identified (Supplementary Table S4). Among them, unidentified Prevotellaceae in feces was increased cubically with increasing NCG (P = 0.03, Supplementary Table S4), while Acetitomaculum, Marvinbryantia, and Unidentified Christensenellaceae were reduced cubically (Supplementary Table S4). Meanwhile, the relative abundance Mogibacterium in ruminal content was reduced linearly with raising the dosage of NCG (P < 0.01).
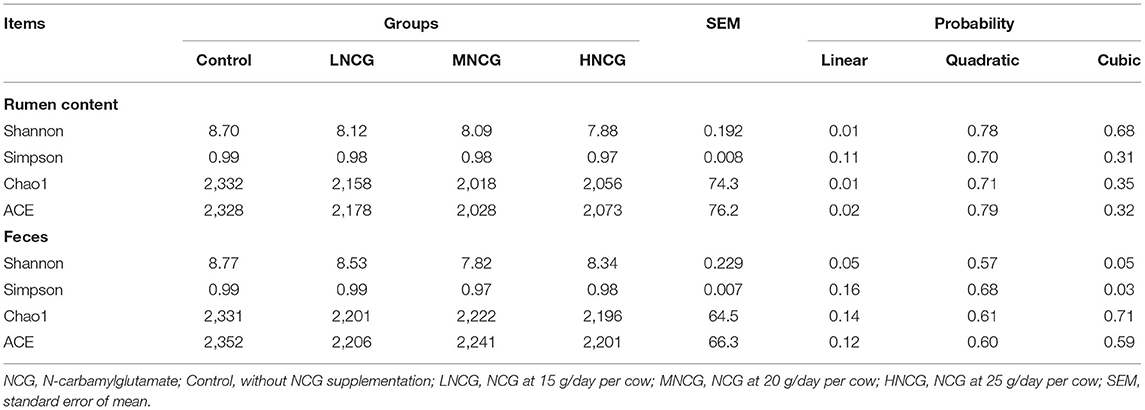
Table 2. Summary of the Alpha diversity of the bacterial community in ruminal and fecal microbiota in different groups.
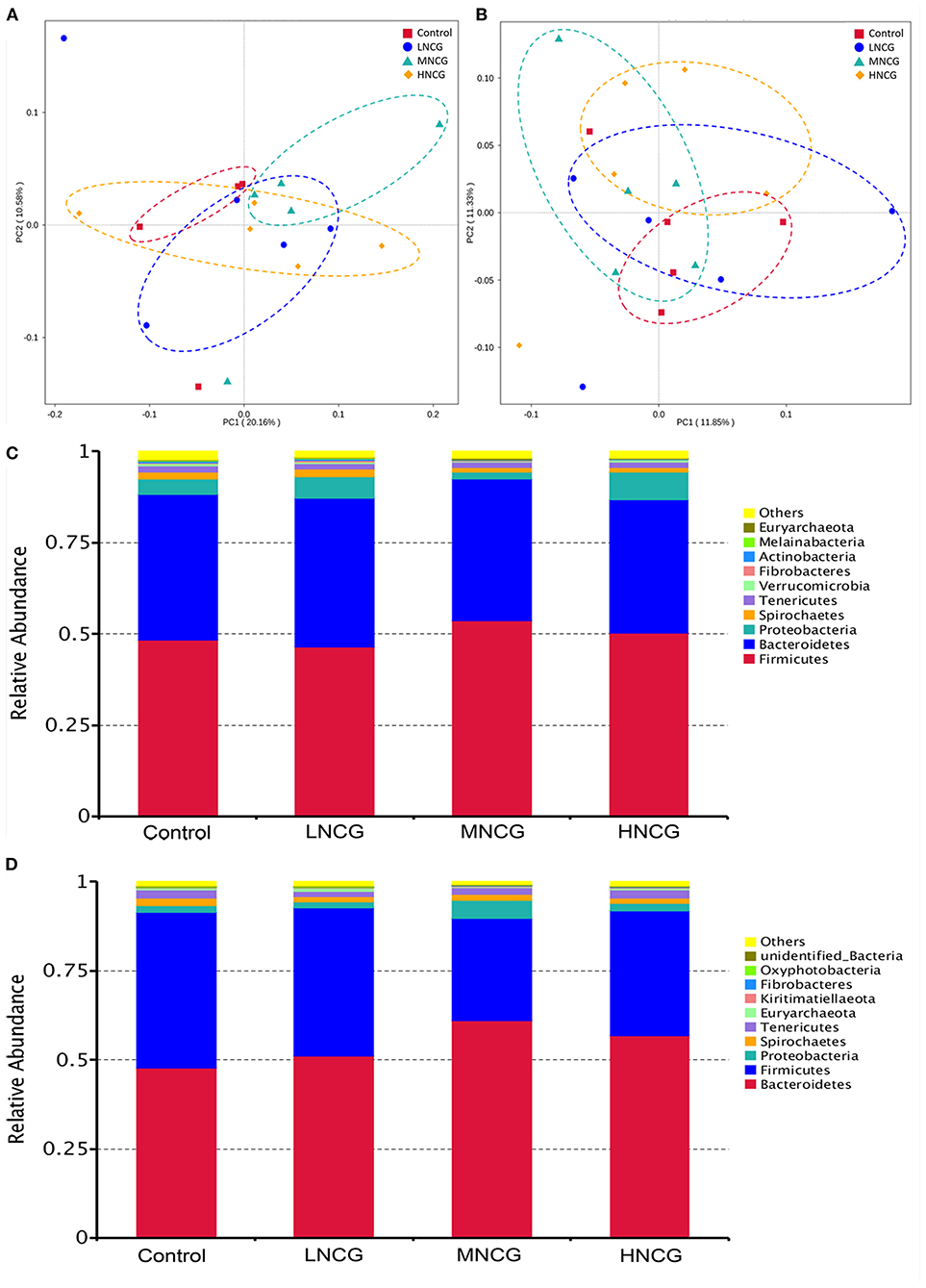
Figure 1. Effects of NCG supplementation on microbiota structure and composition in ruminal content and feces. NCG, N-carbamylglutamate; Control, without NCG supplementation; LNCG, NCG at 15 g/day per cow; MNCG, NCG at 20 g/day per cow; HNCG, NCG at 25 g/day per cow. (A,B) Principal coordinate analysis (PCoA) of bacterial community structures in ruminal content (A) and feces (B). PCoA shows the difference in the bacterial communities among control and NCG groups. (C,D) The composition of the ruminal (C) and fecal (D) microbial community at the phylum level.
The functional capacity of the microbiota in ruminal content and feces was inferred by the PICRUSt analysis. Heatmap of PICRUSt results (level 3) showed that the control group and NCG groups clustered differently, and differences in KEGG metabolic pathways were also observed in various groups (Figure 2). In the ruminal content, oxidative phosphorylation, purine metabolism, phenylalanine, tyrosine, and tryptophan biosynthesis, and alanine, aspartate, and glutamate metabolism were up-regulated in the LNCG compared with the control. Meanwhile, methane metabolism, energy metabolism, pyruvate metabolism, and glycolysis/gluconeogenesis were up-regulated in the MNCG but not in LNCG (Figure 2A). Interestingly, pyrimidine metabolism and arginine and proline metabolism were down-regulated in the HNCG. In feces, pathways of purine metabolism, oxidative phosphorylation, and pyrimidine metabolism were up-regulated in the MNCG, and the adverse effects were observed in the LNCG (Figure 2B).
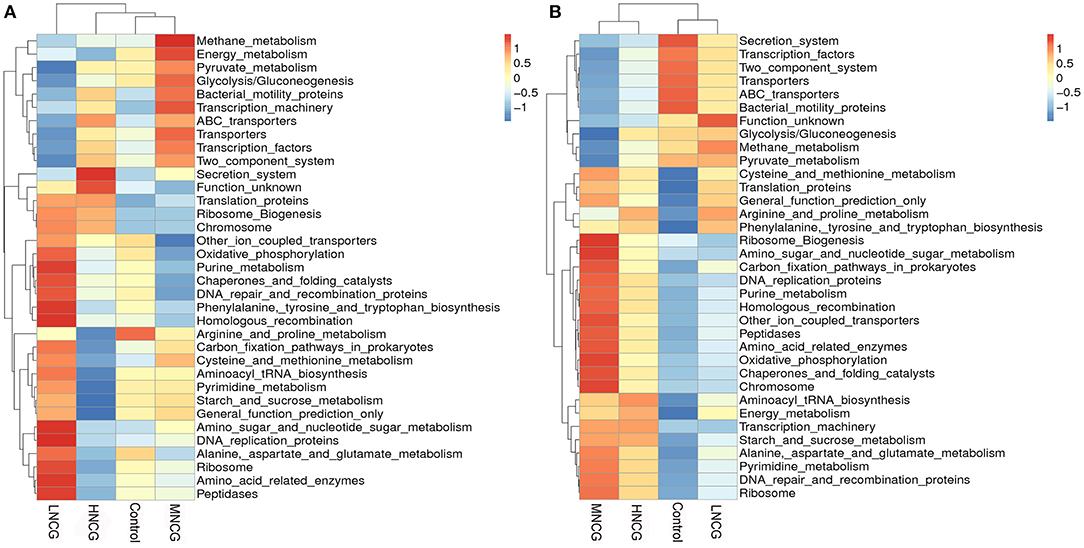
Figure 2. Heatmap of bacterial metabolic pathways obtained from PICRUSt analysis. NCG, N-carbamylglutamate; Control, without NCG supplementation; LNCG, NCG at 15 g/day per cow; MNCG, NCG at 20 g/day per cow; HNCG, NCG at 25 g/day per cow. PICRUSt analysis, Phylogenetic investigation of communities by reconstruction of unobserved states analysis. Microbiota functions in ruminal content (A) and feces (B) were analyzed by PICRUST. The ruminal and fecal microbiota function in NCG treatment groups were differed from those in the control that metabolism of energy, amino acid, pyruvate, glycolysis, and carbon fixation were enhanced in NCG groups.
In order to validate the repeatability and stability of the method, pooled QC sample was applied in the ESI positive and negative analysis sequence, respectively. Principal component analysis (PCA) was first performed on all the samples in the study to find out the metabolic distinction. The distribution of the QC samples is shown in Figures 3A,B. Three QC samples were tightly clustered, which suggested that the method was robust with good suitability in the metabolomic experiment. Meanwhile, the control group and NCG-treated groups showed a slight but not significant separation trend in the PCA score plots in both ESI modes (Figures 3A,B).
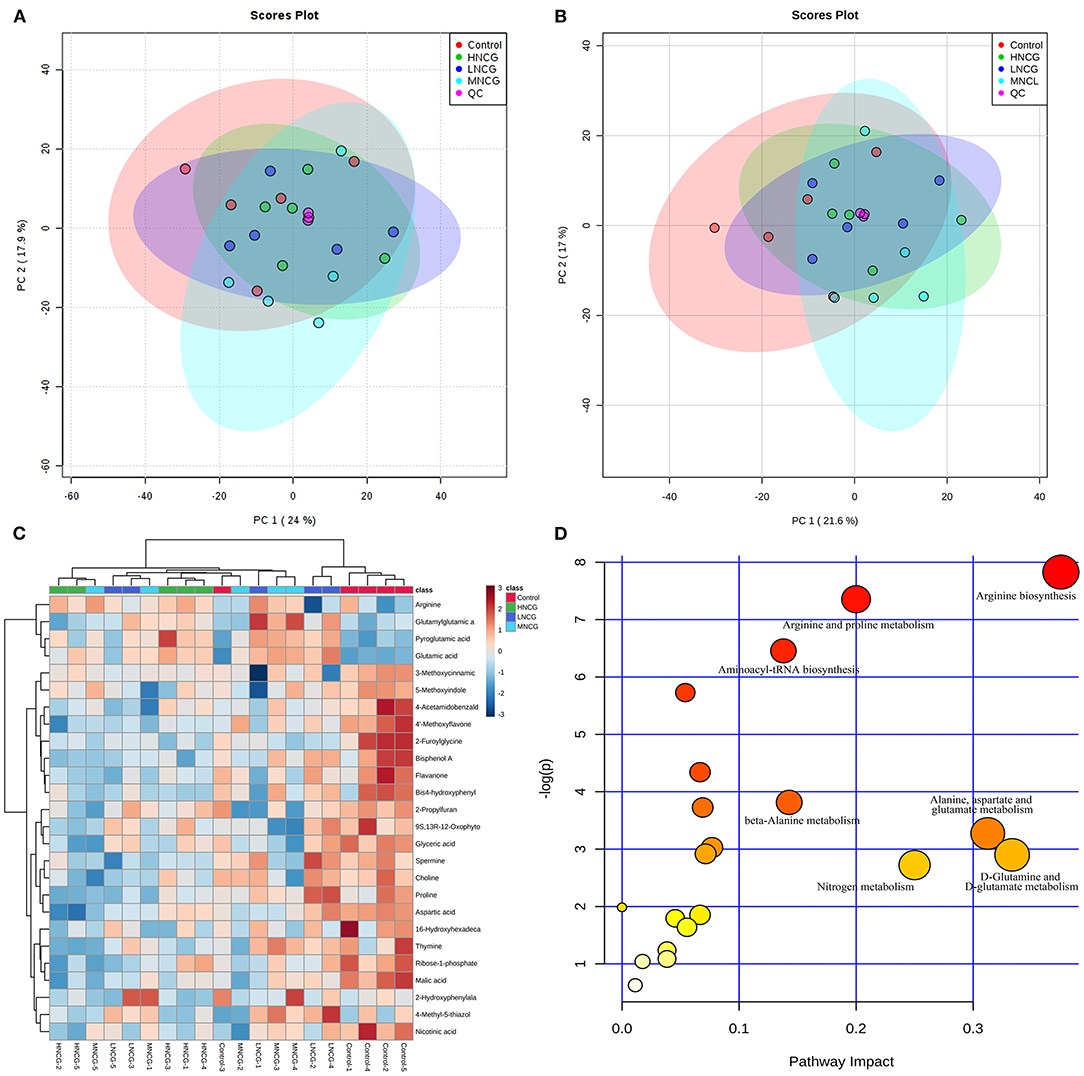
Figure 3. Effects of NCG supplementation on metabolomic profile in ruminal fluid. NCG, N-carbamylglutamate; Control, without NCG supplementation; LNCG, NCG at 15 g/day per cow; MNCG, NCG at 20 g/day per cow; HNCG, NCG at 25 g/day per cow. (A,B) PCA score plots of the ruminal fluid from the control and NCG groups analyzed by UPLC-Q-TOF/MS analysis in positive (A) and negative modes (B). PCA, principal component analysis; QC, quality control samples. (C) Hierarchical clustering heatmap of the identified metabolites. The relative intensity of the metabolites was indicated by the color of the heatmap. (D) Pathway analysis of the metabolites obtained from ruminal fluid in response to NCG supplementation.
The partial least squares discriminant analysis (PLS-DA) model was built to further identify the molecular features differentiated among the groups. Clear separation of the samples between control and NCG-treated groups could be observed in the PLS-DA score plots in both ESI positive and negative modes (Supplementary Figure S4), indicating there existed significant discrimination in ruminal fluid metabolic profile. Variables with VIP >1 and P-value <0.05 were selected as potential metabolites. As a result, 26 metabolites were identified as potential biomarkers related to NCG treatment (Supplementary Table S5). Compared with the control, 20 metabolites were reduced significantly, and 5 metabolites showed a remarkable increase in concentration in the NCG-treated groups. According to the metabolite abundance, the hierarchical clustering analysis was carried out to display the relationships and differences of the ruminal liquid metabolites. As shown in Figure 3C, metabolites in the same metabolic pathways or similar abundance pattern were clustered together. Heatmap also revealed that these metabolites significantly differed between the control and NCG-treated groups in ruminal liquid.
In order to visualize the metabolic pathways in response to NCG treatment, pathways analysis of the potential metabolites was performed by MetaboAnalyst. In this study, Bos taurus (cow) pathway library was selected and the pathway impact-value threshold was set to 0.10. Figure 3D showed that the potential metabolites were mainly involved in seven metabolic pathways, including arginine biosynthesis, arginine, and proline metabolism, aminoacyl-tRNA biosynthesis, nitrogen metabolism, D-glutamine, and D-glutamate metabolism, beta-alanine metabolism, and alanine, aspartate and glutamate metabolism.
Potential links among the ruminal metabolites and altered microbiota genera in ruminal content and feces were identified by Pearson correlation analysis. A clear correction with the metabolites was found for the ruminal and fecal microbiota at the genus level. As shown in Figure 4, positive correlations between metabolites and genera were indicated by the red color, and the negative correlations by blue. As shown in Figure 4, glutamylglutamic acid, pyroglutamic acid, spermine, choline, thymine, 2-hydroxyphenylalanine, and glutamic acid displayed negative corrections with the identified seven genera in the ruminal content. Three genera including Marvinbryantia, Acetitomaculum, and Intestinimonas in feces showed positive corrections with glyceric acid, spermine, choline, and aspartic acid, and negative corrections with 5-methoxyindole and 2-hydroxyphenylalanine. The association analysis between fermentation parameters and microbiota genera in ruminal content is shown in Supplementary Table S6. At the genus level, it was found that the seven genera such as Pseudobutyrivibrio, Acetitomaculum, Schwartzia, and Saccharofermentans were positively related to the changes of pH, NH3-N, acetate, butyrate, and the ratio of acetate/propionate, and negatively related to MCP, VFAs, and propionate.
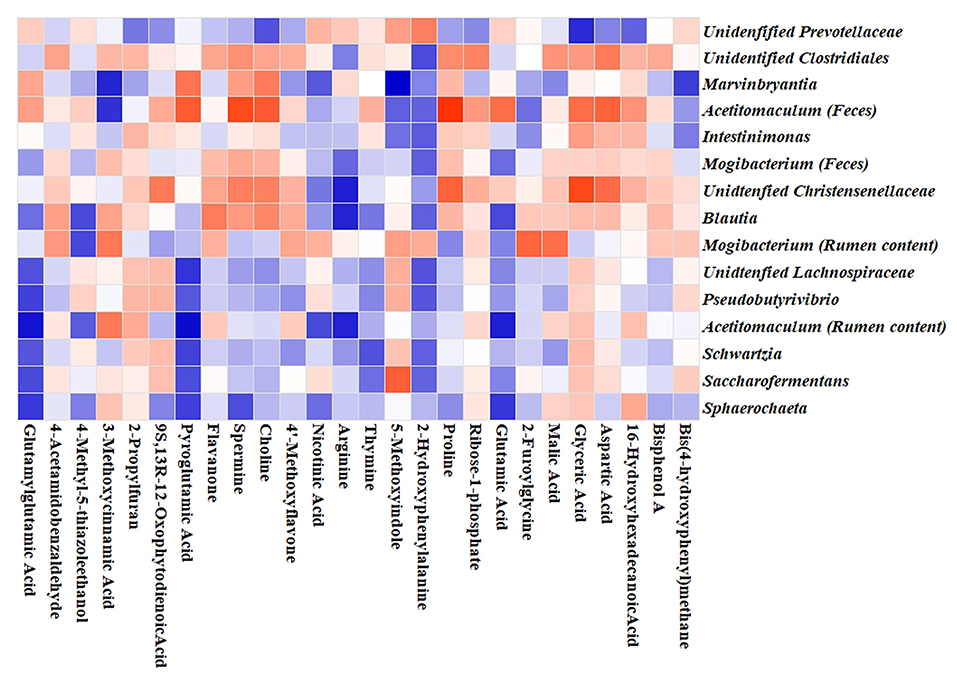
Figure 4. Correlation heatmap between microbiota genera and the altered metabolites. Pearson correlation between metabolites and microbiota genera in ruminal content and feces affected by NCG supplementation. Blue squares indicate negative correlations, and red squares indicate positive correlations.
Heat stress affects ruminal fermentation, metabolic profile as well as gut microbiota. As a cost-effective way, feed additive is used to ameliorate the HS in dairy cows to improve their health and production performance (28, 36). Effects of supplementation with NCG in ruminal content and feces of dairy cows under HS were studied for the first time in this study. The results found that dietary administration of NCG altered ruminal fermentation pattern, ruminal metabolic profile, and microbiota in ruminal content and feces of dairy cows exposed to HS. These findings provided evidence to illustrate the possible action mechanism of NCG in the amelioration of HS and promote the application of NCG in dairy farming. Moreover, this study integrates metabolomics and gut microbiota as an effective approach in ruminant nutritional research.
Previous studies found that HS could decrease the level of total VFAs and elevate the ratio of acetate to propionate (37, 38). In this study, the molar proportion of propionate and VFAs concentrations were increased by the NCG supplementation, while butyrate and the ratio of acetate to propionate were reduced. These results indicated that NCG could improve ruminal fermentation and have positive effects on energy production. In this study, with increasing NCG supplementation, the molar proportion of acetate and butyrate decreased. Reduced pH is conducive to the absorption of VFAs in the rumen epithelium (39). It was found NCG linearly reduced the pH in the rumen, which might be the reason for the decrease of acetate and butyrate. Propionate was increased with the addition of NCG, which was consistent with the results reported by Chacher et al. (40). As a pre-cursor of propionate, fumarate can promote propionate production in the rumen (41). In the process of gluconeogenesis, NCG is metabolized and converted to fumarate through the α-ketoglutaric acid, which may be responsible for the increase of propionate. It was noticed that ruminal pH decreased in the NCG groups, which might be resulted from the increase of the VFAs that dissociated more hydrogen ions in the rumen (42). In addition, NCG could promote nutrient digestion to increase the accumulation of organic acids in the rumen, leading to a decrease in pH (40). Oba et al. (43) reported that NCG could activate carbamyl phosphate synthetase-1 to increase the utilization of NH3-N in the rumen epithelial and duodenal mucosal cells, which may explain the reduced concentration of NH3-N in NCG groups in this study. MCP is an index to reflect microorganism growth and digestibility, and MCP synthesis is closely related to energy release. The addition of 20 g NCG/day to cows increased MCP levels in the rumen, suggesting NCG might improve the ruminal microbial community to enhance the nutrient availability for dairy cows under HS.
In our previous study, it was found that the levels of blood glucose and blood urea nitrogen were respectively increased and decreased in the dairy cows with NCG supplementation under HS (22). Propionate is the pre-cursor for glucose production. In the present study, the molar proportion of propionate was linearly increased by NCG, which might result in an increase in blood glucose. There is a linear correlation between blood urea nitrogen and rumen NH3-N concentration (44). It was noticed that both blood urea nitrogen and rumen NH3-N were reduced, which were consistent with each other and beneficial to improving the utilization rate of nitrogen.
In the present study, both Alpha diversity and PCoA indicated that NCG supplementation altered ruminal and fecal microbial community, such as the reduction of community richness and diversity. In addition, the microbial composition at the phylum and genus levels was changed by NCG. The abundance of Proteobacteria and Actinobacteria was reduced in the ruminal content in NCG groups. Proteobacteria have been reported to increase by HS with the presence of lipopolysaccharide in many inflammation-associated diseases (45, 46). Thus, the reduction of Proteobacteria caused by NCG may inhibiting inflammation to maintain the health of dairy cows. Li et al. (3) reported that phyla Firmicutes and Bacteroidetes were increased and reduced in the fecal samples of dairy cows under seasonal HS, respectively. Interestingly, a decrease in Firmicutes and an increase in Bacteroidetes were found in the feces of the dairy cows after NCG supplementation, indicating NCG reversed the dysbiosis of fecal microbiota caused by HS. At the genus level, Mogibacterium, Sphaerochaeta, Marvinbryantia, Acetitomaculum, and Unidentified Christensenellaceae were reduced, while Unidentified Prevotellaceae was enriched in NCG groups. As a pathogen, Mogibacterium was isolated from adult human patients with periodontal disease (47). Marvinbryantia plays an important role in butyrate production and energy metabolism of intestinal epithelial cells, which is associated with intestinal diseases such as colorectal cancer and ulcerative colitis (48, 49). NCG had the effect of inhibiting the level of Mogibacterium and Marvinbryantia in ruminal content and feces, indicating NCG might have the ability to protect the dairy cows from the diseases caused by potentially pathogenic bacteria.
Microbial functions of microbiota such as carbohydrate metabolism, amino acid metabolism, and energy metabolism were reduced in rumen and feces in the dairy cows or goats under HS (3). In the present study, the function of microbiota in ruminal content and feces was revealed by PICRUSt analysis, which found that the microbiota functions in NCG treatment groups differed from those in the control. Ruminal and fecal microbiota in MNCG group had an enhanced capacity to the metabolism of energy, amino acid, pyruvate, glycolysis, and carbon fixation. These results indicated that NCG could improve metabolic disorders caused by HS through the regulation of the bacterial functions in ruminal content and feces.
Metabolomic analysis in the ruminal fluid was conducted to further explore the response of rumen to NCG supplementation. The results of the score plots indicated that there were significant differences in metabolic profiles of ruminal fluid between the control and NCG groups. Twenty-six metabolites involved in arginine and proline metabolism, glutathione metabolism, citric acid cycle, glycerophospholipid metabolism, and purine metabolism were found to be associated with NCG supplementation. Moreover, the metabolomic results of ruminal fluid were partly in agreement with the findings in ruminal and fecal microbiota.
Metabolites involved in arginine metabolisms, such as arginine, aspartic acid, proline, and spermine, were affected by the NCG supplementation. Previous studies have demonstrated that NCG could increase the concentration of arginine by the activation of carbamyl phosphate synthetase-1 (19). Consistent with the previous results, the levels of arginine in the ruminal fluid were higher in NCG groups than in the control, indicating that NCG promoted the endogenous synthesis of arginine. As a toxic biogenic amine, spermine is related to subacute ruminal acidosis and can be oxidized into aldehyde and hydrogen to cause oxidative stress in ruminants (50). In the present study, NCG supplementation showed favorable inhibition of spermine concentration in the rumen, which may release less spermine into the blood to reduce oxidative stress. Proline and aspartic acid are building blocks of proteins. Chacher et al. (21) reported that dietary NCG supplementation increases aspartic acid in the plasma of the high-yield dairy cows. Inconsistent with this observation, the levels of proline and aspartic acid were reduced in the rumen of the dairy cows supplemented with NCG. The possible explanation for this discrepancy is that NCG might have different effects on the concentrations of proline and aspartic acid in plasma and ruminal content. Glutamic acid is an important nitrogen donor, it is produced from the incorporation of carbon skeleton and ammonia in the ruminal bacteria (51). In the present study, levels of glutamic acid and its derivatives such as glutamylglutamic acid and pyroglutamic acid were significantly higher in the NCG groups than in the control group, suggesting that NCG might improve the nitrogen utilization in the dairy cows under HS. As an organic acid, a high level of glyceric acid leads to general metabolic acidosis in the rumen (52). Glyceric acid was decreased in the ruminal fluid in NCG groups, which is potentially beneficial to maintaining rumen health. It was also noticed that some metabolites such as malic acid, nicotinic acid, and choline were also affected by NCG supplementation. The precise roles of these metabolites in the rumen are not clear, but the increased levels of these metabolites by NCG might be important for ruminal functions.
A great number of studies have confirmed there is a close relationship between the metabolites and gut microbiota in dairy cows (53). In the present study, correlation analysis was conducted to explore the connections between them. Our analysis revealed that a number of metabolites in ruminal fluid and feces were positively or negatively correlated with the bacterial taxa. These corrections might be important for NCG's effects on dairy cow health. Blautia species have been reported to be associated with glutamic acid and the transformation of host bile acids (54, 55). Consistent with the previous results, corrections among Blautia, choline, and glutamic acid were observed in this study, which might suggest that the reduction of Blautia in the feces of NCG groups had effects on choline transportation and glutamic acid production. Many ruminal metabolites were correlated with the abundance of the bacterial taxa. However, the sophisticated relationships among NCG, metabolites, and the bacterial community are poorly characterized, and further work are needed to elucidate their interactions.
In the present study, an integrative approach through the combination of LC-MS-based metabolomics and 16S rRNA gene sequencing was utilized to assess the effects of NCG supplementation on microbiota and metabolites in ruminal content and feces of the dairy cows under HS. The results showed that NCG altered the ruminal fermentation pattern, including the increase of acetate, propionate, butyrate, and total VFAs and the decrease of ruminal pH and NH3-N. Meanwhile, NCG changed the diversity and composition of the microbiota and ameliorated the functional metabolic pathways such as amino acid metabolism, energy metabolism, and pyruvate metabolism. Metabolomic analysis revealed that 26 metabolites mainly involved in arginine metabolism, glutamate metabolism, and nitrogen metabolism were regulated by NCG. Furthermore, potential links between bacterial genera and metabolites were also revealed by correlation analysis. Combining the results of ruminal fermentation, metabolomics, and gut microbiota, the optimal dose of NCG for dairy cows under HS was 20 g/hd/day. These findings provided new insights into the effects of NCG on metabolites and microbiota in ruminal content and feces, suggesting NCG might be considered a potential dietary feed additive for the improvement of HS in dairy cows. Further research are required to investigate the underlying interactions among NCG, metabolites, and microbiota.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA811687; https://www.ncbi.nlm.nih.gov/, PRJNA817393.
The animal study was reviewed and approved by the Institutional Animal Care and Use Committee of Hebei Agricultural University and carried out in accordance with the Guidelines of the Care and Use of Laboratory Animals of China.
YG and JL: conceptualization, methodology, supervision, and funding acquisition. YL, LR, NM, MW, and LH: investigation and data curation. YC, QL, and YS: resources. LH, NM, and YL: writing and editing. All authors read and approved the final manuscript.
This research was financially supported by the earmarked fund for China Agriculture Research System (CARS-36, Beijing, China), Hebei Dairy Cattle Innovation Team of Modern Agro-industry Technology Research System (HBCT2018120203, Shijiazhuang, China), Key Research and Development Project of Hebei (20326606D, Shijiazhuang, China), and Precision Animal Husbandry Discipline Group Construction Project of Hebei Agricultural University (2020, Hebei, China).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank Prof. Fengqi Zhao (University of Vermont) for its linguistic assistance during the preparation of this manuscript. The authors thank the entire staff of Hongda Dairy Farm (Baoding, China) for their assistance in animal care and sample collection.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.902001/full#supplementary-material
1. Gomez DE, Arroyo LG, Costa MC, Viel L, Weese JS. Characterization of the fecal bacterial microbiota of healthy and diarrheic dairy calves. J Vet Intern Med. (2017) 31:928–39. doi: 10.1111/jvim.14695
2. McCann JC, Elolimy AA, Loor JJ. Rumen microbiome, probiotics, and fermentation additives. Vet Clin North Am Food Anim Pract. (2017) 33:539–53. doi: 10.1016/j.cvfa.2017.06.009
3. Li H, Li R, Chen H, Gao J, Wang Y, Zhang Y, et al. Effect of different seasons (spring vs. summer) on the microbiota diversity in the feces of dairy cows. Int J Biometeorol. (2020) 64:345–54. doi: 10.1007/s00484-019-01812-z
4. Azad E, Derakhshani H, Forster RJ, Gruninger RJ, Acharya S, McAllister TA, et al. Characterization of the rumen and fecal microbiome in bloated and non-bloated cattle grazing alfalfa pastures and subjected to bloat prevention strategies. Sci Rep. (2019) 9:4272. doi: 10.1038/s41598-019-41017-3
5. Steele MA, Penner GB, Chaucheyras-Durand F, Guan LL. Development and physiology of the rumen and the lower gut: targets for improving gut health. J Dairy Sci. (2016) 99:4955–66. doi: 10.3168/jds.2015-10351
6. Liu C, Wu H, Liu S, Chai S, Meng Q, Zhou Z. Dynamic alterations in yak rumen bacteria community and metabolome characteristics in response to feed type. Front Microbiol. (2019) 10:1116. doi: 10.3389/fmicb.2019.01116
7. Xue MY, Sun HZ, Wu XH, Liu JX, Guan LL. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome. (2020) 8:64. doi: 10.1186/s40168-020-00819-8
8. Denman SE, Morgavi DP, McSweeney CS. Review: the application of omics to rumen microbiota function. Animal. (2018) 12:s233–45. doi: 10.1017/S175173111800229X
9. Allen JD, Hall LW, Collier RJ, Smith JF. Effect of core body temperature, time of day, and climate conditions on behavioral patterns of lactating dairy cows experiencing mild to moderate heat stress. J Dairy Sci. (2015) 98:118–27. doi: 10.3168/jds.2013-7704
10. Wheelock JB, Rhoads RP, Vanbaale MJ, Sanders SR, Baumgard LH. Effects of heat stress on energetic metabolism in lactating Holstein cows. J Dairy Sci. (2010) 93:644–55. doi: 10.3168/jds.2009-2295
11. Zhao S, Min L, Zheng N, Wang J. Effect of heat stress on bacterial composition and metabolism in the rumen of lactating dairy cows. Animals. (2019) 9:925. doi: 10.3390/ani9110925
12. Chen S, Wang J, Peng D, Li G, Chen J, Gu X. Exposure to heat-stress environment affects the physiology, circulation levels of cytokines, and microbiome in dairy cows. Sci Rep. (2018) 8:14606. doi: 10.1038/s41598-018-32886-1
13. Dou M, Zhang Y, Shao Q, Zhu J, Li W, Wang X, et al. L-arginine reduces injury from heat stress to bovine intestinal epithelial cells by improving antioxidant and inflammatory response. Anim Biotechnol. (2021):1–9. doi: 10.1080/10495398.2021.2009491
14. Wu G, Knabe DA, Kim SW. Arginine nutrition in neonatal pigs. J Nutr. (2004) 134:2783S−90S, 2796S−97S. doi: 10.1093/jn/134.10.2783S
15. Wu G, Bazer FW, Cudd TA, Jobgen WS, Kim SW, Lassala A, et al. Pharmacokinetics and safety of arginine supplementation in animals. J Nutr. (2007) 137:1673S−80S. doi: 10.1093/jn/137.6.1673S
16. Sun L, Zhang H, Fan Y, Guo Y, Zhang G, Nie H, et al. Metabolomic profiling in umbilical venous plasma reveals effects of dietary rumen-protected arginine or N-carbamylglutamate supplementation in nutrient-restricted Hu sheep during pregnancy. Reprod Domest Anim. (2017) 52:376–88. doi: 10.1111/rda.12919
17. Sugiyama Y, Shimura M, Ogawa-Tominaga M, Ebihara T, Kinouchi Y, Isozaki K, et al. Therapeutic effect of N-carbamylglutamate in CPS1 deficiency. Mol Genet Metab Rep. (2020) 24:100622. doi: 10.1016/j.ymgmr.2020.100622
18. Palencia JYP, Saraiva A, Abreu MLT, Zangeronimo MG, Schinckel AP, Pospissil Garbossa CA. Effectiveness of citrulline and N-carbamoyl glutamate as arginine pre-cursors on reproductive performance in mammals: a systematic review. PLoS ONE. (2018) 13:e209569. doi: 10.1371/journal.pone.0209569
19. Hu Y, Shao D, Wang Q, Xiao Y, Zhao X, Shen Y, et al. Effects of dietary N-carbamylglutamate supplementation on growth performance, tissue development and blood parameters of yellow-feather broilers. Poult Sci. (2019) 98:2241–9. doi: 10.3382/ps/pey591
20. Gu FF, Liang SL, Wei ZH, Wang CP, Liu HY, Liu JX, et al. Short communication: effects of dietary addition of N-carbamoylglutamate on milk composition in mid-lactating dairy cows. J Dairy Sci. (2018) 101:10985–90. doi: 10.3168/jds.2018-15162
21. Chacher B, Zhu W, Ye JA, Wang DM, Liu JX. Effect of dietary N-carbamoylglutamate on milk production and nitrogen utilization in high-yielding dairy cows. J Dairy Sci. (2014) 97:2338–45. doi: 10.3168/jds.2013-7330
22. Ma N, Li Y, Ren L, Hu L, Xu R, Shen Y, et al. Effects of dietary N-carbamylglutamate supplementation on milk production performance, nutrient digestibility and blood metabolomics of lactating Holstein cows under heat stress. Anim Feed Sci Tech. (2021) 273:114797. doi: 10.1016/j.anifeedsci.2020.114797
23. Wilson ID, Plumb R, Granger J, Major H, Williams R, Lenz EM. HPLC-MS-based methods for the study of metabonomics. J Chromatogr B. (2005) 817:67–76. doi: 10.1016/j.jchromb.2004.07.045
24. Humer E, Kröger I, Neubauer V, Schedle K, Reisinger N, Zebeli Q. Supplementing phytogenic compounds or autolyzed yeast modulates ruminal biogenic amines and plasma metabolome in dry cows experiencing subacute ruminal acidosis. J Dairy Sci. (2018) 101:9559–74. doi: 10.3168/jds.2018-14744
25. Yang Y, Dong G, Wang Z, Liu J, Chen J, Zhang Z. Treatment of corn with lactic acid or hydrochloric acid modulates the rumen and plasma metabolic profiles as well as inflammatory responses in beef steers. BMC Vet Res. (2018) 14:408. doi: 10.1186/s12917-018-1734-3
26. Elolimy A, Alharthi A, Zeineldin M, Parys C, Helmbrecht A, Loor JJ. Supply of methionine during late-pregnancy alters fecal microbiota and metabolome in neonatal dairy calves without changes in daily feed intake. Front Microbiol. (2019) 10:2159. doi: 10.3389/fmicb.2019.02159
27. Edmonson AJ, Lean IJ, Weaver LD, Farver T, Webster G. A body condition scoring chart for Holstein dairy cows. J Dairy Sci. (1989) 72:68–78. doi: 10.3168/jds.S0022-0302(89)79081-0
28. Perdomo MC, Marsola RS, Favoreto MG, Adesogan A, Staples CR, Santos J. Effects of feeding live yeast at 2 dosages on performance and feeding behavior of dairy cows under heat stress. J Dairy Sci. (2020) 103:325–39. doi: 10.3168/jds.2019-17303
29. Shen JS, Chai Z, Song LJ, Liu JX, Wu YM. Insertion depth of oral stomach tubes may affect the fermentation parameters of ruminal fluid collected in dairy cows. J Dairy Sci. (2012) 95:5978–84. doi: 10.3168/jds.2012-5499
30. Shen YZ, Ran T, Saleem AM, Wang HR, Yang WZ. Short communication: Ground corn steeped in citric acid modulates in vitro gas production kinetics, fermentation patterns and dry matter digestibility. Anim Feed Sci Tech. (2019) 247:9–14. doi: 10.1016/j.anifeedsci.2018.10.014
31. Broderick GA, Kang JH. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci. (1980) 63:64–75. doi: 10.3168/jds.S0022-0302(80)82888-8
32. Cotta MA, Russell JB. Effect of peptides and amino acids on efficiency of rumen bacterial protein synthesis in continuous culture. J Dairy Sci. (1982) 65:226–34. doi: 10.3168/jds.S0022-0302(82)82181-4
33. Magoč T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. (2011) 27:2957–63. doi: 10.1093/bioinformatics/btr507
34. Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. (2011) 10:10–7. doi: 10.1002/0471250953.bi1007s36
35. Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. (2013) 31:814–21. doi: 10.1038/nbt.2676
36. Sun Y, Liu J, Ye G, Gan F, Hamid M, Liao S, et al. Protective effects of zymosan on heat stress-induced immunosuppression and apoptosis in dairy cows and peripheral blood mononuclear cells. Cell Stress Chaperones. (2018) 23:1069–78. doi: 10.1007/s12192-018-0916-z
37. Freestone P, Lyte M. Stress and microbial endocrinology: prospects for ruminant nutrition. Animal. (2010) 4:1248–57. doi: 10.1017/S1751731110000674
38. Zhong S, Ding Y, Wang Y, Zhou G, Guo H, Chen Y, et al. Temperature and humidity index (THI)-induced rumen bacterial community changes in goats. Appl Microbiol Biotechnol. (2019) 103:3193–203. doi: 10.1007/s00253-019-09673-7
39. Aschenbach JR, Penner GB, Stumpff F, Gäbel G. Ruminant nutrition symposium: role of fermentation acid absorption in the regulation of ruminal pH. J Anim Sci. (2011) 89:1092–107. doi: 10.2527/jas.2010-3301
40. Chacher B, Wang D, Liu H, Liu J. Degradation of L-arginine and N-carbamoyl glutamate and their effect on rumen fermentation in vitro. Ital J Anim Sci. (2012) 11:e68. doi: 10.4081/ijas.2012.e68
41. Gadhia MM, Maliszewski AM, O'Meara MC, Thorn SR, Lavezzi JR, Limesand SW, et al. Increased amino acid supply potentiates glucose-stimulated insulin secretion but does not increase β-cell mass in fetal sheep. Am J Physiol Endocrinol Metab. (2013) 304:E352–62. doi: 10.1152/ajpendo.00377.2012
42. Aikman PC, Henning PH, Humphries DJ, Horn CH. Rumen pH and fermentation characteristics in dairy cows supplemented with Megasphaera elsdenii NCIMB 41125 in early lactation. J Dairy Sci. (2011) 94:2840–9. doi: 10.3168/jds.2010-3783
43. Oba M, Baldwin RL, Owens SL, Bequette BJ. Metabolic fates of ammonia-N in ruminal epithelial and duodenal mucosal cells isolated from growing sheep. J Dairy Sci. (2005) 88:3963–70. doi: 10.3168/jds.S0022-0302(05)73082-4
44. Whitelaw FG, Milne JS, Chen XB. The effect of a rumen microbial fermentation on urea and nitrogen metabolism of sheep nourished by intragastric infusion. Exp Physiol. (1991) 76:91–101. doi: 10.1113/expphysiol.1991.sp003484
45. Shi D, Bai L, Qu Q, Zhou S, Yang M, Guo S, et al. Impact of gut microbiota structure in heat-stressed broilers. Poult Sci. (2019) 98:2405–13. doi: 10.3382/ps/pez026
46. Rizzatti G, Lopetuso LR, Gibiino G, Binda C, Gasbarrini A. Proteobacteria: a common factor in human diseases. Biomed Res Int. (2017) 2017:9351507. doi: 10.1155/2017/9351507
47. Sousa V, Nibali L, Spratt D, Dopico J, Mardas N, Petrie A, et al. Peri-implant and periodontal microbiome diversity in aggressive periodontitis patients: a pilot study. Clin Oral Implants Res. (2017) 28:558–70. doi: 10.1111/clr.12834
48. Rajilić-Stojanović M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm Bowel Dis. (2013) 19:481–8. doi: 10.1097/MIB.0b013e31827fec6d
49. Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, et al. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. (2012) 6:320–9. doi: 10.1038/ismej.2011.109
50. Humer E, Kröger I, Neubauer V, Reisinger N, Zebeli Q. Supplementation of a clay mineral-based product modulates plasma metabolomic profile and liver enzymes in cattle fed grain-rich diets. Animal. (2019) 13:1214–23. doi: 10.1017/S1751731118002665
51. Pengpeng W, Tan Z. Ammonia assimilation in rumen bacteria: a review. Anim Biotechnol. (2013) 24:107–28. doi: 10.1080/10495398.2012.756402
52. Alarcon P, Hidalgo AI, Manosalva C, Cristi R, Teuber S, Hidalgo MA, et al. Metabolic disturbances in synovial fluid are involved in the onset of synovitis in heifers with acute ruminal acidosis. Sci Rep. (2019) 9:5452. doi: 10.1038/s41598-019-42007-1
53. Wang Y, Nan X, Zhao Y, Wang H, Wang M, Jiang L, et al. Coupling 16S rDNA sequencing and untargeted mass spectrometry for milk microbial composition and metabolites from dairy cows with clinical and subclinical mastitis. J Agric Food Chem. (2020) 68:8496–8508. doi: 10.1021/acs.jafc.0c03738
54. Ottosson F, Brunkwall L, Ericson U, Nilsson PM, Almgren P, Fernandez C, et al. Connection between BMI-related plasma metabolite profile and gut microbiota. J Clin Endocrinol Metab. (2018) 103:1491–501. doi: 10.1210/jc.2017-02114
Keywords: cow, metabonomics, microbiota, N-carbamylglutamate, rumen
Citation: Li Y, Ma N, Ren L, Wang M, Hu L, Shen Y, Cao Y, Li Q, Li J and Gao Y (2022) Microbiome-Metabolome Responses in Ruminal Content and Feces of Lactating Dairy Cows With N-Carbamylglutamate Supplementation Under Heat Stress. Front. Vet. Sci. 9:902001. doi: 10.3389/fvets.2022.902001
Received: 22 March 2022; Accepted: 30 May 2022;
Published: 23 June 2022.
Edited by:
Tugay Ayasan, Osmaniye Korkut Ata University, TurkeyReviewed by:
Pongsatorn Gunun, Rajamangala University of Technology Isan, ThailandCopyright © 2022 Li, Ma, Ren, Wang, Hu, Shen, Cao, Li, Li and Gao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianguo Li, MTE4MTkzNTA5NEBxcS5jb20=; Yanxia Gao, eWFueGlhLmdhb0BoZWJhdS5lZHUuY24=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.