
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 31 May 2022
Sec. Livestock Genomics
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.892815
This article is part of the Research Topic Genetic Analysis of Reproductive Traits in Livestock View all 14 articles
 Jiayuan Mo1
Jiayuan Mo1 Yujie Lu1
Yujie Lu1 Siran Zhu1
Siran Zhu1 Lingli Feng1
Lingli Feng1 Wenjing Qi1
Wenjing Qi1 Xingfa Chen1
Xingfa Chen1 Bingkun Xie1,2
Bingkun Xie1,2 Baojian Chen2
Baojian Chen2 Ganqiu Lan1
Ganqiu Lan1 Jing Liang1*
Jing Liang1*Litter size and teat number are economically important traits in the porcine industry. However, the genetic mechanisms influencing these traits remain unknown. In this study, we analyzed the genetic basis of litter size and teat number in Bama Xiang pigs and evaluated the genomic inbreeding coefficients of this breed. We conducted a genome-wide association study to identify runs of homozygosity (ROH), and copy number variation (CNV) using the novel Illumina PorcineSNP50 BeadChip array in Bama Xiang pigs and annotated the related genes in significant single nucleotide polymorphisms and common copy number variation region (CCNVR). We calculated the ROH-based genomic inbreeding coefficients (FROH) and the Spearman coefficient between FROH and reproduction traits. We completed a mixed linear model association analysis to identify the effect of high-frequency copy number variation (HCNVR; over 5%) on Bama Xiang pig reproductive traits using TASSEL software. Across eight chromosomes, we identified 29 significant single nucleotide polymorphisms, and 12 genes were considered important candidates for litter-size traits based on their vital roles in sperm structure, spermatogenesis, sperm function, ovarian or follicular function, and male/female infertility. We identified 9,322 ROHs; the litter-size traits had a significant negative correlation to FROH. A total of 3,317 CNVs, 24 CCNVR, and 50 HCNVR were identified using cnvPartition and PennCNV. Eleven genes related to reproduction were identified in CCNVRs, including seven genes related to the testis and sperm function in CCNVR1 (chr1 from 311585283 to 315307620). Two candidate genes (NEURL1 and SH3PXD2A) related to reproduction traits were identified in HCNVR34. The result suggests that these genes may improve the litter size of Bama Xiang by marker-assisted selection. However, attention should be paid to deter inbreeding in Bama Xiang pigs to conserve their genetic diversity.
Bama Xiang pigs, an indigenous Chinese pig breed, are famous for their excellent meat quality and early maturation (1). Owing to their small size, Bama Xiang pigs are easy to handle. Further, Bama Xiang pigs have anatomical and physiological traits similar to humans. Therefore, these pigs could be used in human medical research, such as hypertrophic scarring and diabetes (2, 3). However, genetic diversity in Bama Xiang pigs is declining due to historical inbreeding. Runs of homozygosity (ROH) arise when the same haplotypes are inherited from parents (4), especially in inbreeding Bama Xiang pigs. Bama Xiang boars sexually mature in 76 days and often mate with their mothers resulting in inbreeding (5). The increased inbreeding and declining genetic diversity may hamper the sustainable production of the Bama Xiang pig. Moreover, replacing traditional breeding with intensive pig farming has been beneficial to Bama Xiang pig breeding. The ROH analysis and evaluation of inbreeding rates are important for conserving Bama Xiang pig resources.
Copy number variations (CNVs) are a subtype of genomic structural variation ranging from 50 bp to several Mb in length. The copy number variation region (CNVR) is the area adjacent to the copy number with overlapping regions (6). The CNVR, owing to its length, has a higher probability of changing gene structure and gene dosage and is known to affect several traits in pigs. Qiu et al. demonstrated that nine CNVRs were associated with average daily gain and days to 100 kg in Duroc (7). Zheng et al. (8) reported that the copy number of the AHR had a positive effect on reproduction traits. The copy number variation in GPER1 might be related to the litter size in the Large White pig breed (9). Bovo et al. (10) showed that the CNVR in MSRB3 may be associated with the ear size. Thus, analyzing CNVR function has become an important part of porcine genetics and breeding.
Single nucleotide polymorphism (SNP) arrays can genotype hundreds of thousands of SNPs distributed throughout the genome (11). Based on the density, the porcine SNP arrays were split into 50K (12), 60K (13), and 80K (14). The beadchips have been widely used for genomic selection (15), selection signature research (14), and genome-wide association studies (16). The array can be used to complete population genetics research (17) and CNV detection (18).
Litter size and teat number, which are the base index, are typically associated with economic benefits and production ability in the porcine industry. While the teat number has a medium level of heritability, the litter-size heritability is low. During breeding, the teat number trait increases with an increase in the litter-size trait (19, 20). Litter size represents productivity levels per sow per year in pigs (21). Teat number is a proxy for lactation ability and, thus, is related to piglet mortality rates (22). Numerous researchers have focused on identifying SNPs, quantitative trait loci (QTLs), and candidate genes associated with litter size and teat number (23–25). However, the studies of quantitative traits based on CNVs have rarely been completed, especially on litter size and teat number. Therefore, a study on CNV associated with litter size and teat number in pigs is required.
In this study, we completed the genome-wide association study (GWAS) and the ROH analysis to evaluate the genomic inbreeding coefficients (FROH) of Bama Xiang pigs using SNP array data. We also performed CNV detection and CNVR-based association analysis of litter size and teat number of Bama Xiang pigs. We identified the genes in significant SNPs, ROH, and CNVR and provided the candidate genes associated with litter size and the teat number of Bama Xiang pigs. The information on the Bama Xiang pig can provide the development of the molecular mechanisms of litter size and teat number. Understanding the genetic basis of litter size and teat number in Bama Xiang pigs should help us improve their reproductive capacity.
This study collected ear tissues of 403 Bama Xiang sows from the Agriculture and Animal Husbandry Co. Ltd. (Guangxi, China) for genomic DNA extraction. Parity and teat number data from 297 sows (2,199 dens) were also obtained to calculate phenotype statistics of 14 litter-size traits and five teat-number traits (Supplementary Table S1).
Using a tissue DNA isolation mini kit (Vazyme, Nanjing, Jiangsu, China, Cat. #DC112-01), the genomic DNA was isolated from the ear tissue. Porcine SNP50 BeadChip (Illumina, Inc.) containing 51,315 SNPs was used to genotype the genomic DNA. The raw data were called using the GenomeStudio 2.0 software. Single nucleotide polymorphism arrays with call rates less than 0.9 or with minor allele frequencies of <0.05 were removed in PLINK, version 1.90. Single nucleotide polymorphism arrays without location information were also deleted. After quality control, 403 sows and 24,123 autosomal SNPs were used for analysis. Beagle, version 5.0, was used to impute missing alleles.
For each trait, we implemented GWAS using a univariate linear mixed model in GEMMA. The GWAS model was as follows (26):
where y is an n-vector of reproduction traits, W is a matrix of fixed effects including three principal constituents and the parity effect, x is the SNP genotype, α and β are the corresponding coefficients, u is the random effect, ε is the random error, τ−1 is the variance of residual errors, λ is the ratio between the two variance components, K is the kinship matrix, In is the n × n identity matrix, and MVNn denotes the n-dimensional multivariate normal distribution. Thresholds for Bonferroni-adjusted genome-wide significance and suggestive significance were defined as –log10(p) = 5.68 (0.05/24,123) and –log10(p) = 4.38 (1/24,123), respectively (27–29). We defined the 200 kb regions upstream and downstream of the saliency marker as significant for identifying genes in the Biomart program (Sus scrofa 10.2).
The ROH was identified using detectRUNS packages with the following parameters: Window size was 15; the threshold was 0.05; the minimum number of homozygous/heterozygous SNP in the window was 30; the maximum number of SNP with opposite genotype was 1; the maximum number of missing genotypes was 1; the maximum gap between consecutive SNPs was 250,000; minimum length of the run was 1,000,000; and number of SNPs every kilo-basepairs was 1/100. The ROH length was classified into four classes: 0–6, 6–12, 12–24, and 24–48 Mb. The FROH for Bama Xiang pigs was estimated using the following formula (30):
where LROH is the total length of ROH on autosomes and LAUTO is the total length of the autosomes.
The relationship between FROH and Bama Xiang pig reproduction traits was calculated using the Spearman coefficient. The SPSS 19.0 was used to calculate the general lines model.
The CNVs were detected using cnvPartition 3.2.0 and PennCNV 1.0.5 software. As a plug-in software in GenomeStudio 2.0, cnvPartition detected different copy numbers using Gaussian distribution by LogR ratio and B allele frequency values. To increase the accuracy of CNVs, the cnvPartition was completed with the following parameters: Confidence threshold, 35; minimum homozygous region size, 1,000,000; and minimum probe count, 3. PennCNV used the LogR ratio and B allele frequency values to identify the copy number base on the hidden Markov model with parameters: numSNP, 3; length, 1 K. The CNVs were detected using cnvPartition, and PennCNV was merged into CNVRs using bedtools software. According to the types of CNVRs, we merged the same type of CNVR from two software into CCNVR. The CCNVR of Bama Xiang pigs was annotated using the BioMart program and David database. The Pig QTL database (https://www.animalgenome.org/cgi-bin/QTLdb/SS/index) was used to annotate the traits related to the CCNVR.
According to the CNVR frequency detected by cnvPartition and PennCNV, we defined the CNVR frequency over 5% as the HCNVR. To identify the effect of HCNVR on Bama Xiang pigs' reproductive traits, we completed a mixed linear model association analysis with the Q + K method using TASSEL software, where Q is the principal component matrix and K is the kinship matrix. The Q and K matrices were calculated by SNP array data using Tassel software. According to the Bonferroni thresholds of GWAS, we defined p = 0.001 (0.05/50) and p = 0.02 (1/50) as the significance thresholds and suggestive significance thresholds, respectively (27–29). The significance and suggestive significance HCNVRs were annotated using the BioMart program.
We found that 29 SNPs exceeded the suggestive significance threshold in SSC 1, 3, 4, 5, 7, 14, 16, and 17. Seven litter-size traits (Figure 1) and four teat-number traits (Supplementary Figure S1; Supplementary Table S2) were covered, including average birth number (ABN, one SNP), birth number–second (BN2, two SNPs), birth number–fourth (BN4, one SNPs), birth number–fifth (BN5, 10 SNPs), birth number–sixth (BN6, four SNPs), birth number–ninth (BN9, two SNPs), the maximum number of births (MAXBN, three SNPs), teat numbers on the left side (LTN, one SNP), teat numbers on the right side (RTN, one SNP), the total number of teats (TTN, three SNPs), and minimum teat number (MINTN, one SNP). The SNP rs80960023 on SSC4 was significant in ABN. The SNP rs80967544 was a significant SNP in BN5 and MAXBN. The SNP rs81450533 on SSC14 was significant in LTN, TTN, and MINTN. Forty-seven genes were significantly associated with litter-size traits (Supplementary Table S3). Notably, 12 genes were identified as related to reproduction, including CIB4, DRC1, HADHA, DMC1, DDX17, RAB23, HMGA1, SLC26A8, MAPK14, ARRDC4, CDH6, and BMP7.
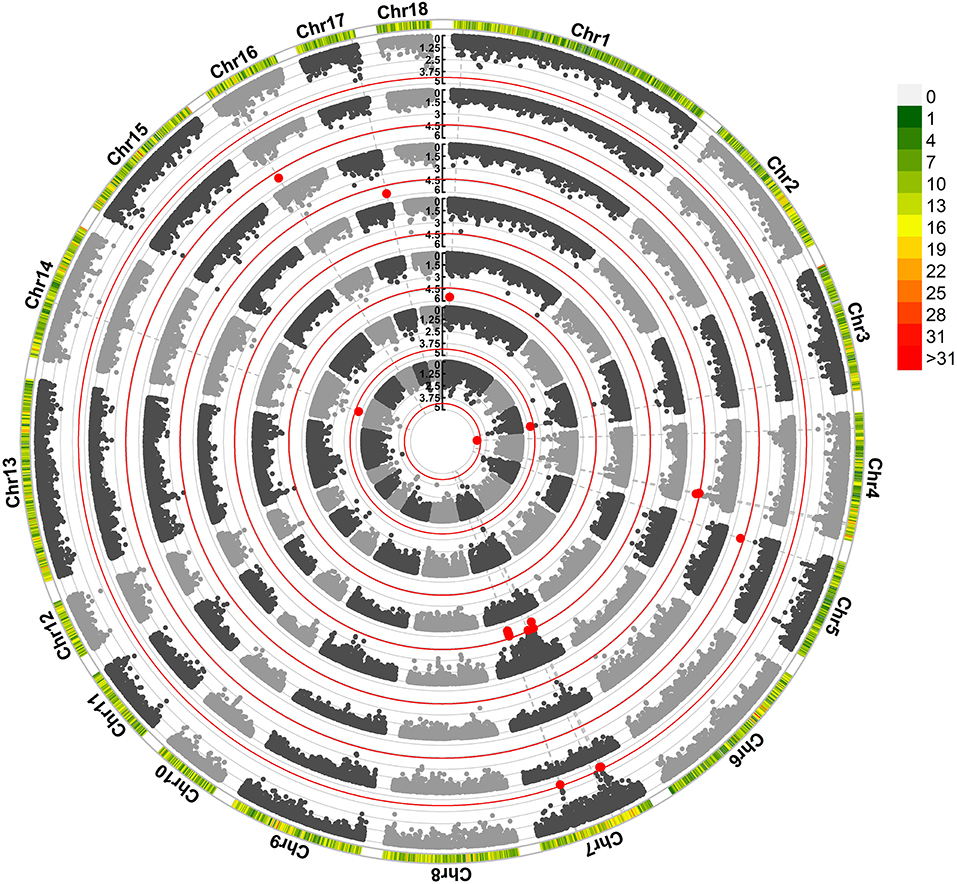
Figure 1. Manhattan plots obtained in the GWAS of litter-size traits. The red line identifies the cut-off for suggestive significance. Red spots identify SNPs with suggestive significance. Traits, from the inner to outer lanes are average birth number (ABN), birth number–second (BN2), birth number–fourth (BN4), birth number–fifth (BN5), birth number–ninth (BN9), and maximum number of births.
We detected 9,322 ROHs, with an average length of 88.78 Mb, in 403 Bama Xiang pigs. The number of ROH per animal ranged from 1 to 136, with the average number of ROH being 23.14 ± 13.74 (mean ± SD). The number of ROH for 0–6, 6–12, 12–24, and 24–48 Mb was 8,369 (89.77%), 831 (8.91%), 110 (1.18%), and 12 (0.13%), respectively. The genomic inbreeding coefficients per sow ranged from 0.0013 to 0.24, with the average genomic inbreeding coefficient as 0.036 ± 0.024 (mean ± SD). A total of 12 litter-size traits had a significantly negative relation with FROH (p < 0.05), including ABN (−0.28), BN1 (−0.19), BN2 (−0.19), BN3 (−0.25), BN4 (−0.15), BN5 (−0.20), BN6 (−0.23), BN8 (−0.17), BN9 (−0.22), MAXBN (−0.22), and the minimum numbers of birth (MINBN) (−0.25) (Supplementary Table S4). The general linear model about ABN and FROH was ABN = 10.359–15.875 *FROH, with R2 was 0.62, and p < 0.05.
We identified 2,920 and 397 CNVs using cnvPartition and PennCNV software, respectively. After the merge, we removed the length of CNVRs over 4 Mb. Finally, we got 197 CNVRs ranging from 0.036 to 3.72 Mb, including 159 deletions, 10 duplications, and 28 mixed in cnvPartition (Figure 2A). We got 45 CNVRs ranging from 0.093 to 2.97 Mb, including 26 deletions, six duplications, and 13 mixed in PennCNV (Figure 2B). We merged the CNVRs from cnvPartition and PennCNV based on their type. Finally, we got 24 CCNVR, including 11 deletions, one duplication, and 12 mixed (Figure 2C; Supplementary Table S5). A total of 64 genes were located in CCNVRs. The genes were significantly enriched (p < 0.05) in the cilium movement and outer dynein arm assembly pathway. We located 637 QTLs in CCNVRs, including 26 average backfat thickness QTLs, 21 backfat at last rib QTLs, and 17 loin muscle area QTLs (Supplementary Figure S2).
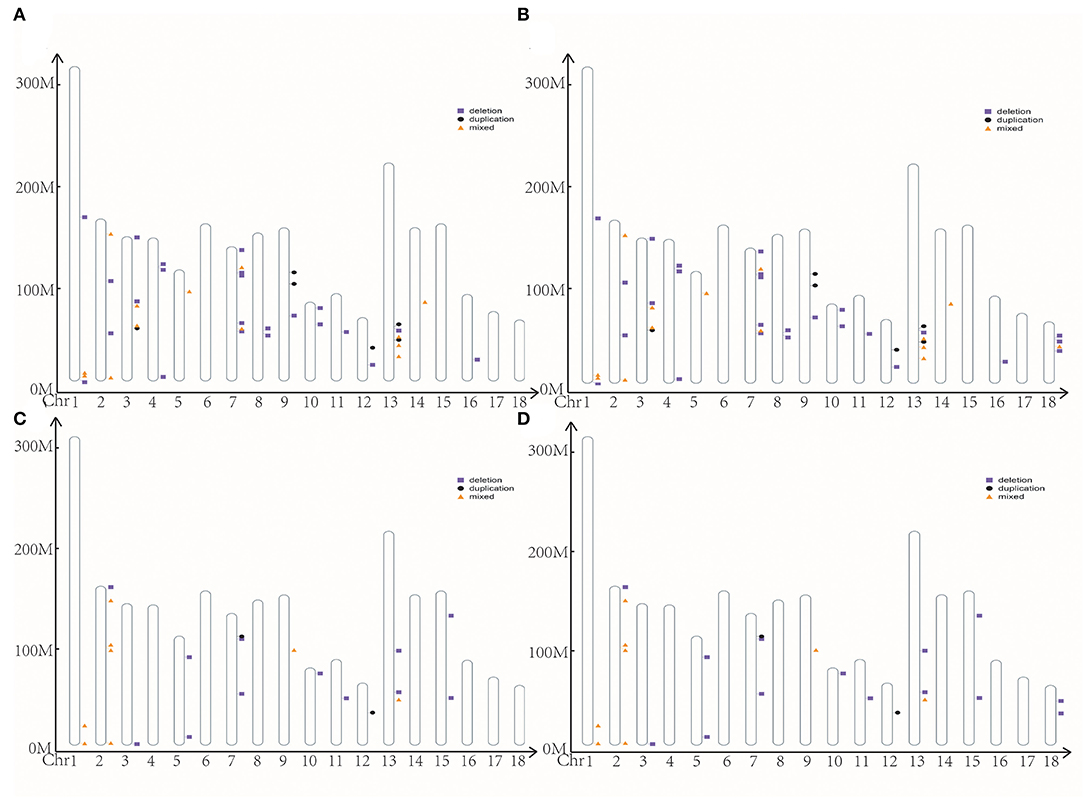
Figure 2. The position of CNVR. (A) CNVR from cnvPartition; (B) CNVR from PennCNV; (C) the common CNVR; (D) the high frequency CNVR.
With cnvPartition and PennCNV software, we got 242 CNVRs in Bama Xiang pigs, including 50 HCNVR (Figure 2D; Supplementary Table S6). After execution of the association analysis, four HCNVRs were over the significance thresholds, and nine HCNVRs were over the suggestive significance thresholds. Interestingly, HCNVR10 (chr2 from 152455383 to 152748172) was over the suggestive significance thresholds in ABN and BN4, BN5, and MINBN (Supplementary Table S7). Additionally, HCNVR34 (chr14 from 124353052 to 124685417) was over the suggestive significance thresholds in ABN and MAXBN, the maximum number of teats (MAXTN), TTN, and LTN (Supplementary Table S8). We executed a t-test with reproduction traits between different CNVR types in HCNVR10 and HCNVR34 (Supplementary Table S8). The deletions were lower than normal in ABN, BN4, BN7, MINBN, and other reproductive traits such as HCNVR10. In ABN, MAXBN, MAXTN, TTN, and LTN, the phenotype of deletions was lower than normal in HCNVR34. After annotation, we got two genes (NEURL1 and SH3PXD2A) related to reproduction traits located in HCNVR34.
We identified 18 genes by using GWAS. Of these, MAPK14 was the only gene that was not associated with a specific sex; MAPK14 promotes phosphorylation and is therefore involved in both male and female reproduction (31). It is critical for the heat-induced proliferation of spermatogenic cells. Moreover, MAPK14 is mainly localized in granulosa and follicle cells in the ovaries, implying its importance during follicular development (32).
Most of the remaining genes were related to sperm structure, spermatogenesis, sperm function, and male infertility. Gene CIB4, which is strongly expressed in mouse and human testes, plays an essential role in the spermatid head. Gene CIB4 knockout (KO) mice are sterile because haploid differentiation becomes impaired (33). Knocking out the DRC1 gene in mice completely disorders the axoneme structure of sperm flagella, impairing sperm motility (34). Research in bulls shows that HADHA proteins are significantly more abundant in the immotile sperm of low-fertility males than in the sperm from high-fertility males (35). Gene DMC1 plays a vital role in repairing DNA double-strand breaks, and disruption of this repair process is linked to male infertility (36). Gene DDX17 increases during the transition from spermatocytes to sperm (37). Research in a Duroc × Erhualian F2 population identified RAB23 as a candidate gene for pubertal reproductive failure (38). In humans, HMGA1 is a stage-specific marker gene for germ cells, and in mice, it is essential for sperm production (39). Gene SLC26A8 is a sperm-specific member of the SLC26 family, and its heterozygous missense mutations are highly associated (power of >95%) with asthenozoospermia (40). Gene ARRDC4 mediates extracellular vesicle biogenesis, which appears to be required for sperm function, given that ARRDC4 KO mice have impaired sperm (41). Two of the 18 genes were related to ovaries, follicles, and female infertility. Gen CDH6 regulates endometrial adhesion and implantation. Studies show that the CDH6 gene is dysregulated in the endometrium of women with infertility (42). Gene BMP7 regulates steroidogenesis, granulosa cell states, and follicular development. A study on Yorkshire pigs found that it is a candidate gene for litter size (43, 44).
In this study, we successfully identified 9,322 ROHs. Similar to Laiwu pig (45), Diannan xiaoer pig (46) and Large White (47), the majority ROHs in Bama Xiang pigs were short segments. The significant Spearman correlation coefficient of FROH and litter size ranged from −0.14 to −0.28. No significant correlation was noticed between the FROH and teat number. Our result, same as proposed by Tao et al. (48), illustrated that the inbreeding had a significant effect on the litter size but had no effect on the teat number in Bama Xiang pigs. According to the general linear model, the ABN decreased to 0.16 when the FROH was increased to 0.01. Thus, the average number of births decreased to 0.57 ± 0.38 in Bama Xiang pig. We established that decreased FROH improves the litter size in Bama Xiang pigs.
We identified 3,317 CNVs, 242 CNVRs, and 13 CCNVRs in Bama Xiang pigs. A total of 11 genes related to reproduction, TUBB4B (49), STPG3 (50), SNRPA1 (51), NRARP (52), NEUROD4 (53), MGAT1 (54), LRGUK (55), LCN8 (56), LCN12 (57), LCN10 (58), and CCDC183 (58), were identified in CCNVR. Bama Xiang pigs are famous for their excellent meat quality and early maturation; however, their lower growth rate and higher backfat content make them less desirable. Thus, the result that the top 10 QTLs in CCNVRs are major related to backfat, meat quality, growth, and teat number is reasonable. However, reproduction-related QTLs were not found in CCNVR1. After executing association analysis, we discovered that HCNVR10 and HCNVR34 were associated with the reproduction traits in Bama Xiang pigs, including litter size and teat number. The phenotype of deletions of HCNVR10 and HCNVR34 were lower than normal in litter size and teat number. NEURL1 and SH3PXD2A were located in HCNVR34. However, NEURL1 and SH3PXD2A, possibly related to reproduction traits, have not been reported. The HCNVR34 (chr14 from 124353052 to 124685417) contained NEURL1 and SH3PXD2A gene may be the candidate CNVR and candidate genes, which is related to litter size and teat number in Bama Xiang pigs.
In GWAS, we found that nine genes were related to testis and sperm, including CIB4, DRC1, HADHA, DMC1, DDX17, RAB23, HMGA1, SLC26A8, and ARRDC4. In CNV detection, we found that TUBB4B, STPG3, NRARP, LCN8, LCN12, LCN10, and CCDC183 were related to the testis and sperm. This result indicated that the sperm of Bama Xiang pigs were different than those of other pigs. Our previous study found that the sperm from Duroc improved the litter size of Bama Xiang pig sows compared to the sperm from Bama Xiang boars (59). We speculate that the reduced litter size in Bama Xiang is due to inbreeding; however, the quality of sperm should be studied further to ascertain the cause of the reduced litter size. Several genes related to testis and sperm were identified from litter-size traits, illustrating that the marks play an important role in Bama Xiang pigs. Bama Xiang boars' sperm quality could be enhanced to improve the litter size of Bama Xiang sows. However, inbreeding in parent and offspring should be deterred to improve the fertility of Bama Xiang pigs.
In this study, we executed the GWAS, ROH analysis, and CNV detection using porcine 50K Beadchip in Bama Xiang pigs. A total of 29 candidate SNPs for seven litter-size traits and four teat-number traits were identified in Bama Xiang pigs using GWAS. Twelve candidate genes were identified in litter-size traits. A total of 9,322 ROHs were found, and the litter-size traits had a significant negative correlation with FROH. A total of 3,317 CNVs were identified, of which 11 genes may be the candidates for reproduction traits. Sixteen genes related to the testis and sperm function were identified. Our results confirm that by using marker-assisted selection on 16 genes, we can improve the litter size of Bama Xiang boars.
The original contributions presented in the study are publicly available. This data can be found here: https://doi.org/10.6084/m9.figshare.19564270.v1.
The animal study was reviewed and approved by Guangxi University.
JM and YL designed the study, conducted the experiments, and drafted original manuscripts. SZ, LF, WQ, and XC conducted parts of experiments. BX, BC, GL, and JL revised the manuscripts. All authors have read and agreed to the published version of the manuscript.
This research was funded by the National Modern Agricultural Industrial Technology System (nycytxgxcxtd-15-01), the National Natural Science Foundation of China (81860150), and the Science and Technology Major Project of Guangxi (Guike-AA17292002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
We thank the National Natural Science Foundation of China for funding this research.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.892815/full#supplementary-material
Supplementary Figure S1. The Manhattan plots obtained in the GWAS with teat number traits. The red line identifies the cut-off for suggestive significance. Red spots identify SNPs with suggestive significance. Traits, from the inner to outer lanes are the teat numbers on the left side (LTN), the teat numbers on the right side (RTN), total number of teats (TTN), and the minimum number of teats (MINTN).
Supplementary Figure S2. The top 10 QTLs in common CNVRs.
1. Yang Y, Adeola AC, Xie HB, Zhang YP. Genomic and transcriptomic analyses reveal selection of genes for puberty in Bama Xiang pigs. Zool Res. (2018) 39:424–30. doi: 10.24272/j.issn.2095-8137.2018.068
2. Ning X, Yang K, Shi W, Xu C. Comparison of hypertrophic scarring on a red Duroc pig and a Guangxi Mini Bama pig. Scars Burn Heal. (2020) 6:2059513120930903. doi: 10.1177/2059513120930903
3. Zhang L, Huang Y, Wang M, Guo Y, Liang J, Yang X, et al. Development and genome sequencing of a laboratory-inbred miniature pig facilitates study of human diabetic disease. iScience. (2019) 19:162–76. doi: 10.1016/j.isci.2019.07.025
4. Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. (2018) 19:220–34. doi: 10.1038/nrg.2017.109
5. Wang LY, Lin AG, Wang LX, Li K, Yang GS, He RG, et al. Animal Genetic Resources in China Pigs. Beijing: China Agriculture Press (2011), p. 237.
6. Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. (2006) 444:444–54. doi: 10.1038/nature05329
7. Qiu Y, Ding R, Zhuang Z, Wu J, Yang M, Zhou S, et al. Genome-wide detection of CNV regions and their potential association with growth and fatness traits in Duroc pigs. BMC Genomics. (2021) 22:332. doi: 10.1186/s12864-021-07654-7
8. Zheng X, Zhao P, Yang K, Ning C, Wang H, Zhou L, et al. CNV analysis of Meishan pig by next-generation sequencing and effects of AHR gene CNV on pig reproductive traits. J Anim Sci Biotechnol. (2020) 11:42. doi: 10.1186/s40104-020-00442-5
9. Wang Y, Zhang T, Wang C. Detection and analysis of genome-wide copy number variation in the pig genome using an 80 K SNP Beadchip. J Anim Breed Genet. (2020) 137:166–76. doi: 10.1111/jbg.12435
10. Bovo S, Ribani A, Muñoz M, Alves E, Araujo JP, Bozzi R, et al. Genome-wide detection of copy number variants in European autochthonous and commercial pig breeds by whole-genome sequencing of DNA pools identified breed-characterising copy number states. Anim Genet. (2020) 51:541–56. doi: 10.1111/age.12954
11. Ramos AM, Crooijmans RP, Affara NA, Amaral AJ, Archibald AL, Beever JE, et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE. (2009) 4:e6524. doi: 10.1371/journal.pone.0006524
12. Zhang Z, Zhang Z, Oyelami FO, Sun H, Xu Z, Ma P, et al. Identification of genes related to intramuscular fat independent of backfat thickness in Duroc pigs using single-step genome-wide association. Anim Genet. (2021) 52:108–13. doi: 10.1111/age.13012
13. Salek Ardestani S, Jafarikia M, Sargolzaei M, Sullivan B, Miar Y. Genomic prediction of average daily gain, back-fat thickness, and loin muscle depth using different genomic tools in Canadian swine populations. Front Genet. (2021) 12:665344. doi: 10.3389/fgene.2021.665344
14. Wang X, Zhang H, Huang M, Tang J, Yang L, Yu Z, et al. Whole-genome SNP markers reveal conservation status, signatures of selection, and introgression in Chinese Laiwu pigs. Evol Appl. (2020) 14:383–98. doi: 10.1111/eva.13124
15. Melnikova E, Kabanov A, Nikitin S, Somova M, Kharitonov S, Otradnov P, et al. Application of genomic data for reliability improvement of pig breeding value estimates. Animals. (2021) 11:1557. doi: 10.3390/ani11061557
16. Xue Y, Li C, Duan D, Wang M, Han X, Wang K, et al. Genome-wide association studies for growth-related traits in a crossbreed pig population. Anim Genet. (2021) 52:217–22. doi: 10.1111/age.13032
17. Valluzzi C, Rando A, Macciotta NPP, Gaspa G, Di Gregorio P. The Nero Lucano pig breed: recovery and variability. Animals. (2021) 11:1331. doi: 10.3390/ani11051331
18. Long Y, Su Y, Ai H, Zhang Z, Yang B, Ruan G, et al. A genome-wide association study of copy number variations with umbilical hernia in swine. Anim Genet. (2016) 47:298–305. doi: 10.1111/age.12402
19. Martínez-Giner M, Noguera JL, Balcells I, Alves E, Varona L, Pena RN. Expression study on the porcine PTHLH gene and its relationship with sow teat number. J Anim Breed Genet. (2011) 128:344–53. doi: 10.1111/j.1439-0388.2011.00925.x
20. Merks JW, Mathur PK, Knol EF. New phenotypes for new breeding goals in pigs. Animal. (2012) 6:535–43. doi: 10.1017/S1751731111002266
21. Leman AD. Optimizing farrowing rate and litter size and minimizing nonproductive sow days. Vet Clin North Am Food Anim Pract. (1992) 8:609–21. doi: 10.1016/S0749-0720(15)30707-6
22. Chalkias H, Jonas E, Andersson LS, Jacobson M, de Koning DJ, Lundeheim N, et al. Identification of novel candidate genes for the inverted teat defect in sows using a genome-wide marker panel. J Appl Genet. (2017) 58:249–59. doi: 10.1007/s13353-016-0382-1
23. Li Y, Pu L, Shi L, Gao H, Zhang P, Wang L, et al. Revealing new candidate genes for teat number relevant traits in Duroc pigs using genome-wide association studies. Animals (Basel). (2021) 11:806. doi: 10.3390/ani11030806
24. Bovo S, Ballan M, Schiavo G, Ribani A, Tinarelli S, Utzeri VJ, et al. Single-marker and haplotype-based genome-wide association studies for the number of teats in two heavy pig breeds. Anim Genet. (2021) 52:440–50. doi: 10.1111/age.13095
25. Vashi Y, Magotra A, Kalita D, Banik S, Sahoo NR, Gupta SK, et al. Evaluation of candidate genes related to litter traits in Indian pig breeds. Reprod Domest Anim. (2021) 56:577–85. doi: 10.1111/rda.13895
26. Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. (2012) 44:821–4. doi: 10.1038/ng.2310
27. Singh A, Kumar A, Gondro C, Pandey AK, Dutt T, Mishra BP. Genome wide scan to identify potential genomic regions associated with milk protein and minerals in vrindavani cattle. Front Vet Sci. (2022) 9:760364. doi: 10.3389/fvets.2022.760364
28. Zhou S, Ding R, Zhuang Z, Zeng H, Wen S, Ruan D, et al. Genome-wide association analysis reveals genetic loci and candidate genes for chest, abdominal, and waist circumferences in two duroc pig populations. Front Vet Sci. (2022) 8:807003. doi: 10.3389/fvets.2021.807003
29. Huang J, Wang C, Ouyang J, Tang H, Zheng S, Xiong Y, et al. Identification of key candidate genes for beak length phenotype by whole-genome resequencing in geese. Front Vet Sci. (2022) 9:847481. doi: 10.3389/fvets.2022.847481
30. Hidalgo J, Cesarani A, Garcia A, Sumreddee P, Larios N, Mancin E, et al. Genetic background and inbreeding depression in Romosinuano cattle breed in Mexico. Animals. (2021) 11:321. doi: 10.3390/ani11020321
31. Zou L, Cheng G, Xu C, Liu H, Wang Y, Li N, et al. The role of miR-128-3p through MAPK14 activation in the apoptosis of GC2 spermatocyte cell line following heat stress. Andrology. (2021) 9:665–72. doi: 10.1111/andr.12923
32. Hu S, Rao M, Lei H, Wu Y, Wang Y, Ke D, et al. Expression patterns of p38αMAPK during follicular development in the ovaries of neonatal rats. Acta Histochem. (2017) 119:538–42. doi: 10.1016/j.acthis.2017.05.007
33. Xu Z, Miyata H, Kaneda Y, Castaneda JM, Lu Y, Morohoshi A, et al. CIB4 is essential for the haploid phase of spermatogenesis in mice. Biol Reprod. (2020) 103:235–43. doi: 10.1093/biolre/ioaa059
34. Zhang J, He X, Wu H, Zhang X, Yang S, Liu C, et al. Loss of DRC1 function leads to multiple morphological abnormalities of the sperm flagella and male infertility in human and mouse. Hum Mol Genet. (2021) 30:1996–2011. doi: 10.1093/hmg/ddab171
35. D'Amours O, Calvo É, Bourassa S, Vincent P, Blondin P, Sullivan R. Proteomic markers of low and high fertility bovine spermatozoa separated by Percoll gradient. Mol Reprod Dev. (2019) 86:999–1012. doi: 10.1002/mrd.23174
36. Shang Y, Huang T, Liu H, Liu Y, Liang H, Yu X, et al. MEIOK21: a new component of meiotic recombination bridges required for spermatogenesis. Nucleic Acids Res. (2020) 48:6624–39. doi: 10.1093/nar/gkaa406
37. Pandey A, Yadav SK, Vishvkarma R, Singh B, Maikhuri JP, Rajender S, et al. The dynamics of gene expression during and post meiosis sets the sperm agenda. Mol Reprod Dev. (2019) 86:1921–39. doi: 10.1002/mrd.23278
38. Xin WS, Zhang F, Yan GR, Xu WW, Xiao SJ, Zhang ZY, et al. A whole genome sequence association study for puberty in a large Duroc × Erhualian F2 population. Anim Genet. (2018) 49:29–35. doi: 10.1111/age.12623
39. Wang M, Liu X, Chang G, Chen Y, An G, Yan L, et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. (2018) 23:599–614.e4. doi: 10.1016/j.stem.2018.08.007
40. Dirami T, Rode B, Jollivet M, Da Silva N, Escalier D, Gaitch N, et al. Missense mutations in SLC26A8, encoding a sperm-specific activator of CFTR, are associated with human asthenozoospermia. Am J Hum Genet. (2013) 92:760–6. doi: 10.1016/j.ajhg.2013.03.016
41. Foot NJ, Gonzalez MB, Gembus K, Fonseka P, Sandow JJ, Nguyen TT, et al. Arrdc4-dependent extracellular vesicle biogenesis is required for sperm maturation. J Extracell Vesicles. (2021) 10:e12113. doi: 10.1002/jev2.12113
42. Zhou W, Santos L, Dimitriadis E. Characterization of the role for cadherin 6 in the regulation of human endometrial receptivity. Reprod Biol Endocrinol. (2020) 18:66. doi: 10.1186/s12958-020-00624-w
43. Du X, Yin H, Pan Z, Wu W, Shang P, Chamba Y, et al. BMP7 is a candidate gene for reproductive traits in Yorkshire sows. Anim Reprod Sci. (2020) 221:106598. doi: 10.1016/j.anireprosci.2020.106598
44. Li X, Ye J, Han X, Qiao R, Li X, Lv G, et al. Whole-genome sequencing identifies potential candidate genes for reproductive traits in pigs. Genomics. (2020) 112:199–206. doi: 10.1016/j.ygeno.2019.01.014
45. Fang Y, Hao X, Xu Z, Sun H, Zhao Q, Cao R, et al. Genome-wide detection of runs of homozygosity in Laiwu pigs revealed by sequencing data. Front Genet. (2021) 12:629966. doi: 10.3389/fgene.2021.629966
46. Wu F, Sun H, Lu S, Gou X, Yan D, Xu Z, et al. Genetic diversity and selection signatures within Diannan small-ear pigs revealed by next-generation sequencing. Front Genet. (2020) 11:733. doi: 10.3389/fgene.2020.00733
47. Shi L, Wang L, Liu J, Deng T, Yan H, Zhang L, et al. Estimation of inbreeding and identification of regions under heavy selection based on runs of homozygosity in a Large White pig population. J Anim Sci Biotechnol. (2020) 11:46. doi: 10.1186/s40104-020-00447-0
48. Tao L, He X, Wang X, Di R, Chu M. Litter size of sheep (Ovis aries): inbreeding depression and homozygous regions. Genes. (2021) 12:109. doi: 10.3390/genes12010109
49. Sarkar H, Arya S, Rai U, Majumdar SS. A study of differential expression of testicular genes in various reproductive phases of Hemidactylus flaviviridis (Wall Lizard) to derive their association with onset of spermatogenesis and its relevance to mammals. PLoS ONE. (2016) 11:e0151150. doi: 10.1371/journal.pone.0151150
50. Park S, Shimada K, Fujihara Y, Xu Z, Shimada K, Larasati T, et al. CRISPR/Cas9-mediated genome-edited mice reveal 10 testis-enriched genes are dispensable for male fecundity. Biol Reprod. (2020) 103:195–204. doi: 10.1093/biolre/ioaa084
51. Wu H, Sun L, Wen Y, Liu Y, Yu J, Mao F, et al. Major spliceosome defects cause male infertility and are associated with nonobstructive azoospermia in humans. Proc Natl Acad Sci USA. (2016) 113:4134–9. doi: 10.1073/pnas.1513682113
52. Murta D, Batista M, Silva E, Trindade A, Henrique D, Duarte A, et al. Dynamics of Notch pathway expression during mouse testis post-natal development and along the spermatogenic cycle. PLoS ONE. (2013) 8:e72767. doi: 10.1371/journal.pone.0072767
53. Kottmann JS, Jørgensen MGP, Bertolini F, Loh A, Tomkiewicz J. Differential impacts of carp and salmon pituitary extracts on induced oogenesis, egg quality, molecular ontogeny and embryonic developmental competence in European eel. PLoS ONE. (2020) 15:e0235617. doi: 10.1371/journal.pone.0235617
54. Biswas B, Batista F, Akintayo A, Aguilan J, Stanley P. Transgenic rescue of spermatogenesis in males with Mgat1 deleted in germ cells. Front Cell Dev Biol. (2020) 8:212. doi: 10.3389/fcell.2020.00212
55. Okuda H, DeBoer K, O'Connor AE, Merriner DJ, Jamsai D, O'Bryan MK. LRGUK1 is part of a multiprotein complex required for manchette function and male fertility. FASEB J. (2017) 31:1141–52. doi: 10.1096/fj.201600909R
56. Wen Z, Liu D, Zhu H, Sun X, Xiao Y, Lin Z, et al. Deficiency for Lcn8 causes epididymal sperm maturation defects in mice. Biochem Biophys Res Commun. (2021) 548:7–13. doi: 10.1016/j.bbrc.2021.02.052
57. Suzuki K, Lareyre JJ, Sánchez D, Gutierrez G, Araki Y, Matusik RJ, et al. Molecular evolution of epididymal lipocalin genes localized on mouse chromosome 2. Gene. (2004) 339:49–59. doi: 10.1016/j.gene.2004.06.027
58. Vandenbrouck Y, Pineau C, Lane L. The Functionally Unannotated Proteome of Human male tissues: a shared resource to uncover new protein functions associated with reproductive biology. J Proteome Res. (2020) 19:4782–94. doi: 10.1021/acs.jproteome.0c00516
Keywords: GWAS, ROH, CNV, litter size, teat number, candidate genes, Bama Xiang pig
Citation: Mo J, Lu Y, Zhu S, Feng L, Qi W, Chen X, Xie B, Chen B, Lan G and Liang J (2022) Genome-Wide Association Studies, Runs of Homozygosity Analysis, and Copy Number Variation Detection to Identify Reproduction-Related Genes in Bama Xiang Pigs. Front. Vet. Sci. 9:892815. doi: 10.3389/fvets.2022.892815
Received: 09 March 2022; Accepted: 22 April 2022;
Published: 31 May 2022.
Edited by:
Yang Zhou, Huazhong Agricultural University, ChinaReviewed by:
Kejun Wang, Henan Agricultural University, ChinaCopyright © 2022 Mo, Lu, Zhu, Feng, Qi, Chen, Xie, Chen, Lan and Liang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jing Liang, bGlhbmdqaW5nQGd4dS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.