
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 30 June 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.883416
This article is part of the Research TopicRespiratory Diseases in Veterinary Medicine: Time for Some Fresh AirView all 6 articles
 Bingbing Zong1
Bingbing Zong1 Yongwei Zhu2
Yongwei Zhu2 Manli Liu3
Manli Liu3 Xiangru Wang2,4,5,6
Xiangru Wang2,4,5,6 Huanchun Chen2,4,5,6
Huanchun Chen2,4,5,6 Yanyan Zhang1*
Yanyan Zhang1* Chen Tan2,4,5,6*
Chen Tan2,4,5,6*Mycoplasma hyopneumoniae is the primary pathogen of swine enzootic pneumonia and causes great economic losses to the swine industry worldwide. In China, M. hyopneumoniae seriously hinders the healthy development of the native black pigs. To prevent and treat porcine respiratory disease caused by M. hyopneumoniae, the characteristics of M. hyopneumoniae strain ES-2 isolated from Chinese native black pig lungs with gross lesions at post-mortem were studied for the first time in this study. Strain ES-2 cell was round or oval cells and most sensitive to kanamycin. The diameters of most strain ES-2 cells ranged from 0.4 to 1.0 μm with maximum viability of 1010 CCU/ml. Experimental challenge of animals with strain ES-2 showed respiratory disease could be reproduced, with pneumonic lung lesions evident. Comparative genomics analysis identified that 2 genes are specific to pathogenic M. hyopneumoniae strains, which may be predicted to be a molecular marker. These findings suggest that the study on the characteristics of M. hyopneumoniae strain ES-2 will guide the rapid and accurate drug use in the clinic, and develop a theoretical foundation for accurately diagnosing and treating the infection caused by pathogenic M. hyopneumoniae.
Mycoplasma hyopneumoniae (M. hyopneumoniae) is a host-specific microorganism that only infects swine (1, 2). M. hyopneumoniae does not have a cell wall, and its genome consists of 893–920 kilobase pairs which encode only a few biosynthetic pathways (3, 4). Due to the limited metabolism, M. hyopneumoniae needs to obtain essential metabolites from the host (5). In addition, M. hyopneumoniae is very susceptible to the change in environmental conditions since it lacks a cell wall, thus it is difficult to cultivate M. hyopneumoniae in the medium without cells (6). Although the cultivation of M. hyopneumoniae in vitro has been improved since the 1970s (7), the successful isolation of M. hyopneumoniae is still complicated (8). M. hyopneumoniae mainly infects pig lung tissue, once M. hyopneumoniae invades and colonizes the respiratory tract of pigs, the pathogen will persist in the host (9, 10). M. hyopneumoniae first adheres to, and then colonizes the ciliated epithelial cells of the respiratory tract (1), resulting in ciliostasis, clumping, and finally loss (11). Once the ciliary activity of the respiratory tract is impaired, mucosal immunity and clearance is damaged (12). In addition, M. hyopneumoniae is capable of modulating both innate and adaptive immune systems (13), leading to the damage of the host defense system and the chronic colonization of this pathogen in the respiratory tract (4). Clinically, M. hyopneumoniae is able to infect suckling piglets, weaned piglets, and fattening pigs. To date, M. hyopneumoniae has mostly been detected in intensive swine production farms and herds all over the world (14, 15). Therefore, M. hyopneumoniae, as a widespread respiratory pathogen, is the primary etiological agent responsible for enzootic pneumonia (EP) which is a chronic respiratory disease causing huge economic losses to the pig industry world (16).
About 100 years ago, the chronic and prevalent EP were described (17). In 1963, M. hyopneumoniae strain J was isolated (18). Subsequently, M. hyopneumoniae was identified as the causative agent of EP in 1965 (7). In 1975, Friis et al. (7) reported that the Friis medium can be used to isolate and cultivate M. hyopneumoniae. Although it is very complicated and time-consuming to isolate M. hyopneumoniae from lung tissue of pigs, several strains of M. hyopneumoniae J (GenBank assembly accession: GCA_000008205.1), M. hyopneumoniae 7422 (GenBank assembly accession: GCA_000427215.1), M. hyopneumoniae 7448 (GenBank assembly accession: GCA_000008225.1), M. hyopneumoniae 168 (GenBank assembly accession: GCA_000183185.1), M. hyopneumoniae 232 (GenBank assembly accession: GCA_000008405.1), M. hyopneumoniae KM014 (GenBank assembly accession: GCA_002257505.1), M. hyopneumoniae TB1(GenBank assembly accession: GCA_002213485.1), and M. hyopneumoniae 11 (GenBank assembly accession: GCA_002193015.1) have been reported (8, 19–24). And their genomes are available in GenBank. Enshi black pigs, a typical Chinese native black breed, are best known for cold-wet tolerance and their fat storage ability, which is mainly raised in mountainous regions with an average altitude of 800 m above sea level in southwest China. However, they suffer from swine EP for a long time. So far, M. hyopneumoniae which causes Enshi black pig EP has not been studied. To effectively prevent and treat the EP in Enshi black pigs, M. hyopneumoniae from the lungs of Enshi black pigs will be isolated and cultured in this study.
In addition, with the rapid development of the pig industry, porcine respiratory diseases have become more and more complicated (25–27). In clinical cases, respiratory disease caused by only one pathogen is very rare. In most cases, the respiratory disease is complicated, which is caused by multiple pathogens, namely, M. hyopneumoniae, porcine circovirus type 2 (PCV-2), porcine reproductive and respiratory syndrome virus (PRRSV), and so on (28, 29). Of which M. hyopneumoniae is one of the main pathogens (30). To effectively prevent and control complex porcine respiratory diseases, the characteristics of M. hyopneumoniae were studied by many countries. Moreover, a variety of methods that can accurately detect M. hyopneumoniae in pig lung lesions were developed, and the sensitivity of the isolates to different antibiotics was determined (25–27, 29). Simultaneously, as more and more M. hyopneumoniae genomes were available, many genes that play important roles in the growth or pathogenicity of M. hyopneumoniae were identified by comparative proteomics, transcriptomic, and comparative genomics analyses (23, 24, 31–34). These findings provide a lot of reliable theoretical basis for the local treatment and prevention of M. hyopneumoniae infection. Therefore, the characteristics of the M. hyopneumoniae strain isolated from Enshi black pig lungs will be studied, which will lay a theoretical foundation for the development of treatment for infections caused by M. hyopneumoniae.
In this study, M. hyopneumoniae strain ES-2 was isolated from Chinese native pig lungs with gross lesions at post-mortem and determined the growth characteristics and the pathogenicity of isolates. The susceptibility of M. hyopneumoniae strain ES-2 to different antibiotics was determined. Based on the comparative genomics analyses, the genes which are present in pathogenic M. hyopneumoniae strains and absent in nonpathogenic M. hyopneumoniae strains will be identified. This study will provide a theoretical foundation to effectively prevent the EP.
Mycoplasma hyopneumoniae (M. hyopneumoniae) ES-2 strain is a China field isolate from the lung with gross lesions of Enshi Black pig. And the lung was from the slaughterhouse. M. hyopneumoniae strain ES-2 was isolated by using Friis liquid medium modified Friis liquid medium and modified Friis solid medium as previously described (19).
To improve the isolation rate of M. hyopneumoniae, the lung is required to be harvested from 3–4-month-old, chronic coughing, and skinny pigs with obvious lesions of M. hyopneumoniae pneumonia in the apical and cardiac lobes of the lung. The lungs harvested from the diseased pig were immediately put into the clean bench. A total of 9 lung tissue samples from the different sites of the same lung were placed in a centrifuge tube with sterile phosphate-buffered saline (PBS), and then cut the lung tissue in the centrifuge tube into small pieces with scissors and grind it with a tissue grinder. Subsequently, the supernatants were collected by centrifugation at 5,000 rpm for 10 min, and bacteria in the supernatants were removed by 0.45 μm filtration. The supernatants were transferred into a fresh Friis medium. The mixtures were incubated in 5% CO2 at 37°C for 6 days. On day 6 of culture, M. hyopneumoniae in Friis medium was identified by PCR amplifying the highly conserved 16S rRNA region from M. hyopneumoniae (19). The samples identified correctly by PCR were continued to subculture. If PCR identification of the culture at passage 4 was still correct, the positive samples were placed on the modified Friis solid medium. After culture for 9 days, a single colony on the solid medium was cultured in the modified Friis liquid medium. After three times of purification on the modified Friis solid medium, the purified colony was identified by PCR and sequencing.
Minimal inhibitory concentrations (MICs) of kanamycin for planktonic cultures of M. hyopneumoniae ES-2 were determined using a broth microdilution method previously described (35).
For the purpose of the visualization of M. hyopneumoniae ES-2 strain, scanning electron microscopy (SEM) and transmission electron microscopy (TEM) were performed as previously described with some modifications (36). The insoluble impurities in the fresh-modified Friis medium were removed by centrifugation at 10,000 rpm for 20 min and then centrifuged fresh-modified Friis medium was used to culture the ES-2 strain. Two days after culture, 100 ml of cultured ES-2 was centrifuged at 12,000 rpm for 15 min at 4°C, and the pellets were washed three times with PBS. The pellets were fixed with 2.5% glutaraldehyde in PBS at 4°C for 2 h. And then samples were sent to Servicebio for further processing. ES-2 was visualized using an HT7700 TEM and a SU8010 SEM. The adobe photoshop CC was used to determine the diameter of ES-2.
More than 20 continuous passages of M. hyopneumoniae ES-2 were performed. And the colonies of the ES-2 strain were cultured to confirm that there are no other microorganisms except M. hyopneumoniae ES-2 strain in the medium. And then ES-2 was inoculated into fresh-modified Friis medium. The mixture was cultured in 5% CO2 at 37°C for 6 days. Meanwhile, the growth of ES-2 was examined every 12 h, between 0 and 144 h with the metabolic activity recorded by the color-changing units (CCU) which was indicated by the phenol red as previously described (37).
To observe the growth characteristic of ES-2 strain on a solid culture medium, the modified Friis solid medium was prepared as previously described (19). ES-2 at the exponential growth phase with five-fold serial dilutions in fresh-modified Friis medium was spotted (200 ml) onto the modified Friis solid medium. After ES-2 was cultured in 5% CO2 at 37°C for 9 days, ES-2 strain colonies were observed by Inverted Microscope (Olympus, Japan).
Three-week-old piglets were purchased from a commercial herd. Before M. hyopneumoniae inoculation, blood samples were collected from each piglet. Anti-M. hyopneumoniae IgG antibody in sera, PCV-2 and PRRSV in blood were separately detected by the test kit (IDEXXCo.) and PCR was used to confirm that no piglets were exposed to M. hyopneumoniae, PCV-2, or PRRSV. On the 28th day, 4-week-old pigs were weighed and randomly allocated to two different groups. Different groups with 5 pigs in each group were housed in different rooms to avoid the spread of M. hyopneumoniae between groups. All pigs were inoculated and then euthanized as described previously (38) with minor modifications. Experimental group was inoculated intratracheally with 7 ml inoculum containing 7×107 CCU of M. hyopneumoniae ES-2 strain. The control group was inoculated with 7 ml modified Friis liquid medium without ES-2.
On day 38 after infection, all pigs were euthanized and necropsied as previously described (38). For histopathological analysis, the parts of the lungs were fixed in 4% paraformaldehyde. And then, the fixed lungs were processed, stained with H&E, and visualized under a light microscope. To observe the ciliated epithelial cells and microvilli of the tracheal surface of the infected lungs, the lung bronchi were gently washed three times with 0.1 M PBS, and then were immediately put into fixative (G1102, Servicebio) at 4°C for 2–4 h for TEM. The remaining lungs were stored at −20°C, which were used to identify M. hyopneumoniae by PCR and isolate the ES-2 strain.
After recovery of the frozen M. hyopneumoniae ES-2 strain, the liquid culture with ES-2 strain was placed on a modified Friis solid medium. After culture for 9 days, the colony of M. hyopneumoniae ES-2 strain was cultured in 150 ml modified Friis liquid medium at 37°C. And then the M. hyopneumoniae ES-2 strain was harvested by centrifugation at 14,000 g for 15 min. Afterward, the DNA of the ES-2 strain was extracted by using a TIANamp Genomic DNA Kit (Q5502, Tiangen, China). The complete genome of the M. hyopneumoniae ES-2 strain was sequenced using the PacBio method. The raw reads (approximately 906,783,884 bp polymerase read bases) were obtained by the third-generation sequencing. Polymerase read quality was 0.876. The reads were assembled de novo using the HGAP2 protocol on the SMRT Portal v2.3.0 (39). A circular chromosome was obtained with 751× average base coverage. The annotation and analysis of the sequences of the genome were performed using Glimmer3 v3.02 (40) with the default parameters.
The complete sequences of a total of 162 mycoplasma strains were collected from the genome assembly database. A phylogenetic tree including 59 human isolates, 35 bovine isolates, 17 porcine isolates, 18 chicken isolates, 13 sheep isolates, and 20 other isolates (unknown host) was generated based on CDS existence information in 162 mycoplasma strains using amino acid multiple sequence alignment in Roary v3.11.2 (http://sanger-pathogens.github.io/Roary/) with default parameters (41). To analyze the genes which are present in pathogenic M. hyopneumoniae strains and absent in nonpathogenic M. hyopneumoniae strains, BlastP with a similarity cutoff (0.75, 0.80, 0.85, 0.90, 0.95, and 0.99) (42) was performed to distinguish homologous protein-coding regions from nonhomologous ones. These diagrams were developed by VENNY.
After the first generation was cultured for a week, 8 out of 9 samples were observed to be positive by PCR (Figure 1A). The 8 positive samples were separately transferred into a fresh Friis liquid medium (1:4). After one week's culture, 5 out of 8 samples were found to be positive by PCR (Figure 1B). These 5 positive samples were separately transferred into fresh liquid Friis medium (1:4). After one week's culture, 3 out of 5 samples were detected to be positive by PCR (Figure 1C). In this continuous passage culture, 2 samples of the fourth generation remained positive by PCR (Figure 1D). Subsequently, the serial passage culture of these 2 positive samples was performed in the modified Friis liquid medium. After 20 passage cultures, M. hyopneumoniae was still identified by PCR (Figure 1E).
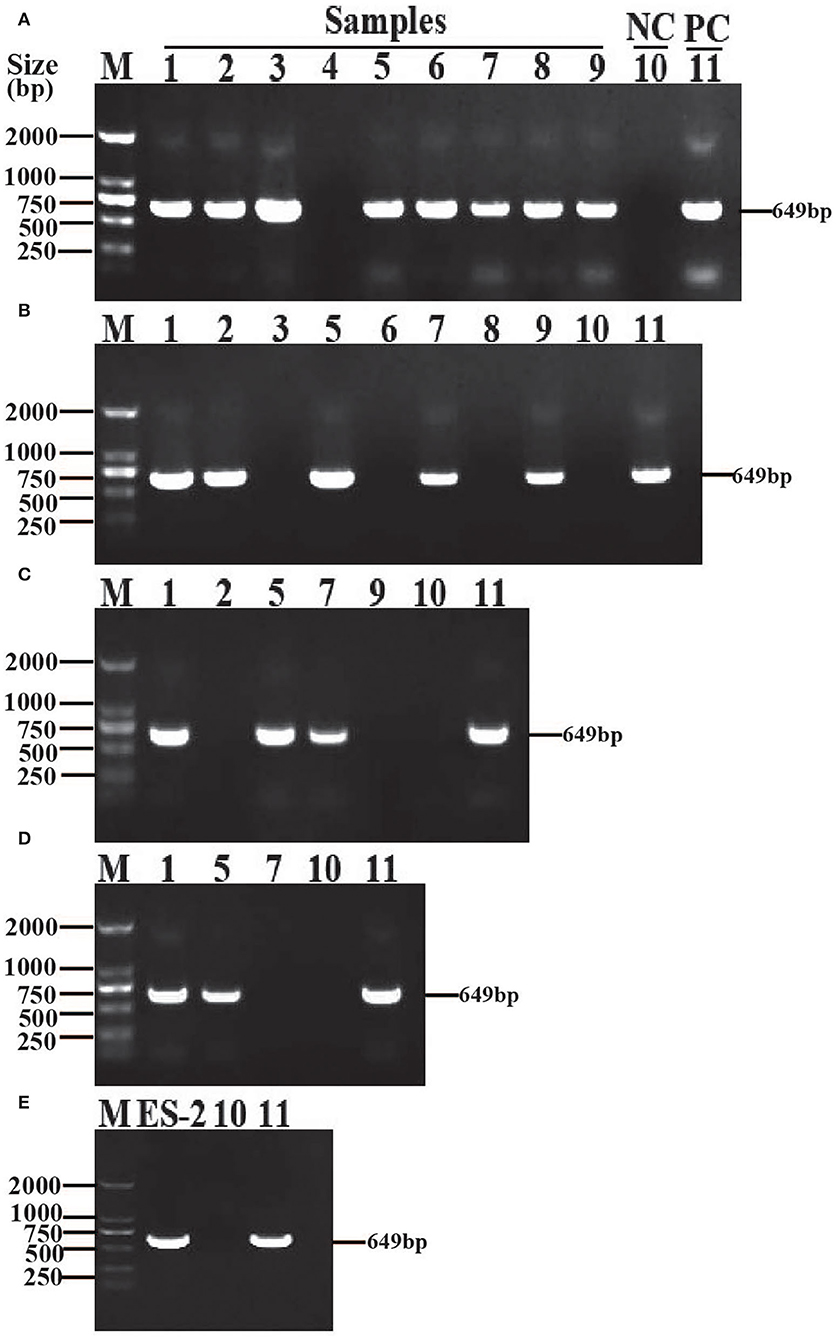
Figure 1. Isolation and identification of M. hyopneumoniae strain ES-2. The samples (1–9) were harvested from different parts of the same lung with gross lesions of EP, M. hyopneumoniae strain ES-2 was isolated by in vitro continuous passage and was confirmed by amplifying the highly conserved 16S rRNA region through PCR with the forward primer 5′-GAGCCTTCAAGCTTCACCAAG A-3′and the reverse primer 5′-TGTGTTAGTGACTTTTGCCACC-3′. (A) The first generation. (B) The second generation. (C) The third generation. (D) The fourth generation. (E) The twentieth generation. M, marker; NC, negative control; PC, positive control.
To get pure M. hyopneumoniae strain, the liquid cultures were placed on the modified Friis solid medium. And then a single colony was cultured in the modified Friis liquid medium. After three times of purification, sequencing results showed that the microorganism purified in liquid medium is M. hyopneumoniae strain (GenBank accession number GCA_004768725.1). Ultimately, a field M. hyopneumoniae strain was successfully isolated and purified from the lung of Enshi black pigs suffering from typical swine mycoplasma pneumonia. And we named this newly isolated strain M. hyopneumoniae strain ES-2. Meanwhile, this study found that the minimum inhibitory concentration of M. hyopneumoniae ES-2 strain to kanamycin was 2 μg/ml.
To study the colony morphology of ES-2 strain on solid media, after 9 days of culture, the colonies from the M. hyopneumoniae ES-2 strain on solid medium were observed through microscopic in this study. The colonies were found to be very small and granular with irregular margins with their central parts burrowed into the medium, which exhibited a typical “fried egg” appearance (Figures 2A,B). To further observe the morphology of cells, M. hyopneumoniae ES-2 strain was collected by centrifugation. ES-2 strain cells were found to be spherical or elliptic and they differed in sizes under SEM (Figures 2C,D). Meanwhile, TEM results exhibited that the morphology of most cells was spherical and a very small number of them was irregular in morphology (Figures 2E,F). The diameters of ES-2 strain cells ranged from 0.2 to 1.6 μm and the diameters of approximately 80% of cells were between 0.4 and 1.0 μm under the electron microscope (Figure 2G).
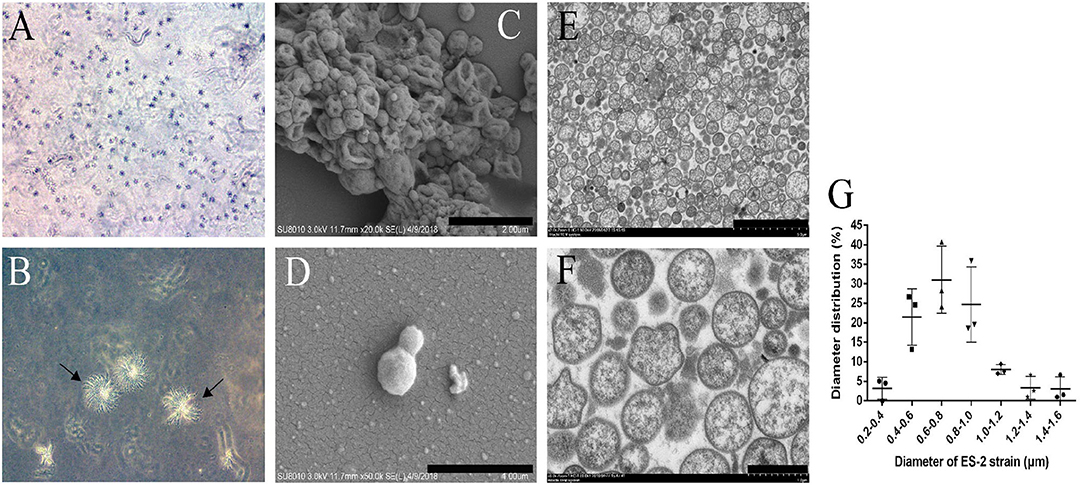
Figure 2. Colony of M. hyopneumoniae strain ES-2 on modified Friis solid media and the ES-2 cell morphology viewed under the electron microscope. A, B The colony of ES-2 strain were viewed by the inverted microscope, (A) 5× and (B) 10×. (C,D) The morphology of strain ES-2 was viewed by scanning electron microscope. (C) Bars = 2.0 μm, (D) Bars = 1.0 μm. (E,F) The cytoplasmic structure of strain ES-2 was viewed by transmission electron microscope. (E) Bars = 5.0 μm, (F) Bars = 1.0 μm. (G) The diameter of the ES-2 strain (μm).
Color-changing units (CCU) of liquid medium were commonly used to determine the viability of M. hyopneumoniae. After M. hyopneumoniae ES-2 strain culture was transferred into a fresh medium, the results showed that the number of ES-2 strain cells was approximate 107 CCU/ml on day 0 (Figure 3A). From day 0 to day 2, the number of ES-2 strain cells was constantly increasing. However, the pH of the color shift continued to decline (pH 7.55–6.37) during the whole incubation process. And the viability in cultures reached a maximum (1010 CCU/ml) approximately on day 2 postincubation with the corresponding pH of 7.24. After 2 days of culture, the viability in cultures continuously declined. On day 8, only 10 CCU/ml of ES-2 strains viability was detected with the corresponding color shift pH being 6.37. On day 7, color shift pH was minimum (6.37) and stopped declining (Figures 3A,B).
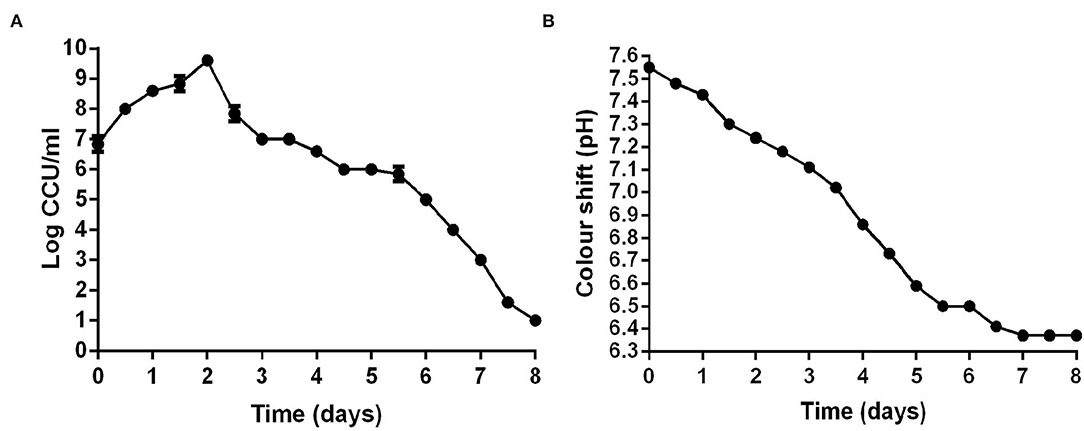
Figure 3. CCU of M. hyopneumoniae and pH of corresponding the modified Friis broth from day 0 to day 8. (A) CCU of M. hyopneumoniae was recorded every 12 h. (B) pH of corresponding the modified Friis broth was measured every 12 h. The growth curve of three repeating experiments were developed.
Some studies reported that M. hyopneumoniae could infect pigs of various ages resulting in typical lesions of EP (43). In this study, the 28-day-old pigs infected with M. hyopneumoniae ES-2 strain for 38 days showed typical pneumonia in the accessory lobes, apical lobes, cardiac lobes, and the cranial portion of the caudal lobes of the lungs (Figure 4F). However, typical lesions of EP were not discovered in the lungs of pigs inoculated with fresh medium without ES-2 strain in the control group (Figures 4A–D). In the pathogenesis of M. hyopneumoniae infections, it is crucial for pathogenic M. hyopneumoniae to damage the ciliated epithelial cells of the trachea, bronchi, and bronchioles (1, 2). SEM results showed that the cilia of the bronchi in the lungs of the pigs infected with the ES-2 strain were tangled, clumped, split, and lost, compared to those of the healthy lungs (Figures 4G–I). In addition, the histological examination results of the lungs of pigs infected with M. hyopneumoniae ES-2 strain showed that much less air was present in the infected lung due to the massive infiltration of inflammatory and lymphocytes cells (Figure 4J), compared to that in the healthy lung (Figure 4E). Therefore, it could be concluded that the M. hyopneumoniae ES-2 strain isolate is a pathogenic strain.
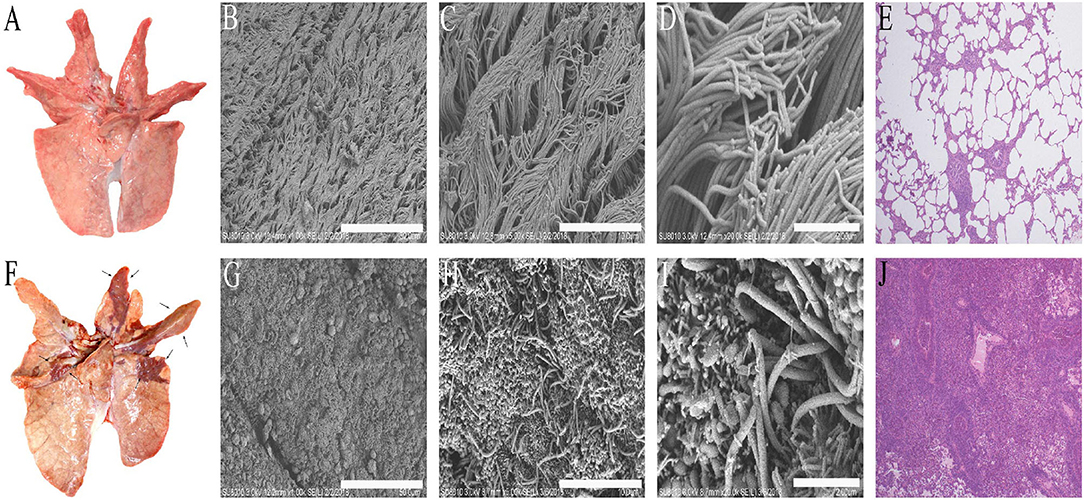
Figure 4. The typical lesions caused by M. hyopneumoniae occurred in the lungs of pigs infected with strain ES-2. The lung tissues were observed on day 38 after infection. (A) The lung tissues of pigs inoculated with modified Friis liquid medium served as blank group. (B) (Bars = 50 μm), (C) (Bars = 10 μm), and (D) (Bars = 2 μm). The tracheal surface (ciliated epithelial cells and microvilli) was observed by scanning electron microscope. (E) (40×) HE staining of lung tissues of blank group is shown. (F) Typical macroscopic lesions caused by M. hyopneumoniae were observed in the lungs of pigs infected with 7 × 107 CCU of M. hyopneumoniae strain ES-2. (G) (Bars = 50 μm), (H) (Bars = 10 μm), and (I) (Bars = 2 μm). The tracheal surfaces of the lungs of pigs infected with strain ES-2 (almost all the fluff is loss) were observed by scanning electron microscope. (J) (40×) The hyperplasia of peribronchiolar accumulation of mononuclear cells and the epithelial cells was observed from the lungs of pigs infected with ES-2 by HE staining.
The genomes of 9 M. hyopneumoniae strains were available from the National Center for Biotechnology Information (NCBI) database. Of these 9 genomes of M. hyopneumoniae strains, the assembly level of 6 genomes (J, 7422, 7448, 168, 168-L, 232, and KM014) was complete. The other 2 genomes (11 and TB1) were incomplete. The genome size of M. hyopneumoniae strains ranged from 0.8 to 1.0 Mb (Supplementary Table S1). In this study, the complete genome of M. hyopneumoniae ES-2 strain (GenBank accession number GCA_004768725.1), which was sequenced using the PacBio method, contained a 956,514 bp (GC content 28.36%) single circular chromosome. The total number of genes predicted was 733, and the average gene length was 1,164 bp (Supplementary Table S1). Subsequently, 25 virulence factors of mycoplasma reported (http://www.mgc.ac.cn/cgi-bin/VFs/compvfs.cgi?Genus=Mycoplasma) were collected from the virulence factor database to investigate the distribution of virulence-related factors among 9 M. hyopneumoniae strains. By aligning the proteins (identity ≥ 80% and coverage ≥ 80%) from each strain against the reported 25 virulence factors, a total of 11 virulence factors were identified in these genomes. In total, 10 out of 11 virulence factors were found to be present in each of 9 M. hyopneumoniae strains, and some virulence factors had multiple copies in these genomes. The cytoadherence protein (P200) (44) of Mycoplasma pneumoniae which functions in cytoadherence and gliding motility was found to be present only in the genome of the ES-2 strain and absent in other strains (Supplementary Table S1). So putative protein P200 has existed in M. hyopneumoniae ES-2.
In addition, the above experiments show that M. hyopneumoniae strain ES-2 is pathogenic for pigs. Among the M. hyopneumoniae strains which have been reported, M. hyopneumoniae strains 7422, 7448, 168, 168-L, and 232 were also reported to be pathogenic for pigs. Only M. hyopneumoniae strain J was nonpathogenic. The pathogenicity of the remaining 3 strains (KM014, TB1, and 11) was unknown.
To understand the phylogenetic relationship between different mycoplasma species, all 162 complete genomes of different species of mycoplasma strains were collected from the NCBI database. However, no core gene was found in the 162 mycoplasma strains. Therefore, the phylogenetic tree was first constructed based on CDS existence information in 162 mycoplasma strains. The phylogenetic tree showed that 15 porcine mycoplasmas (8 M. hyopneumoniae, 1 M. flocculare, and 6 M. hyorhinis), 2 M. dispar, 1 M. bovoculi, 3 other mycoplasmas form a group; 53 human mycoplasmas form another group; 10 sheep mycoplasmas and 14 bovine mycoplasmas form the third group; 12 chicken mycoplasmas, 2 human mycoplasmas, 2 porcine mycoplasmas, 1 bovine mycoplasma, and 7 other mycoplasmas form the fourth group; and 17 bovine mycoplasmas, 6 chicken mycoplasmas, 53 human mycoplasmas, and 13 other mycoplasmas form the fifth group (Figure 5).
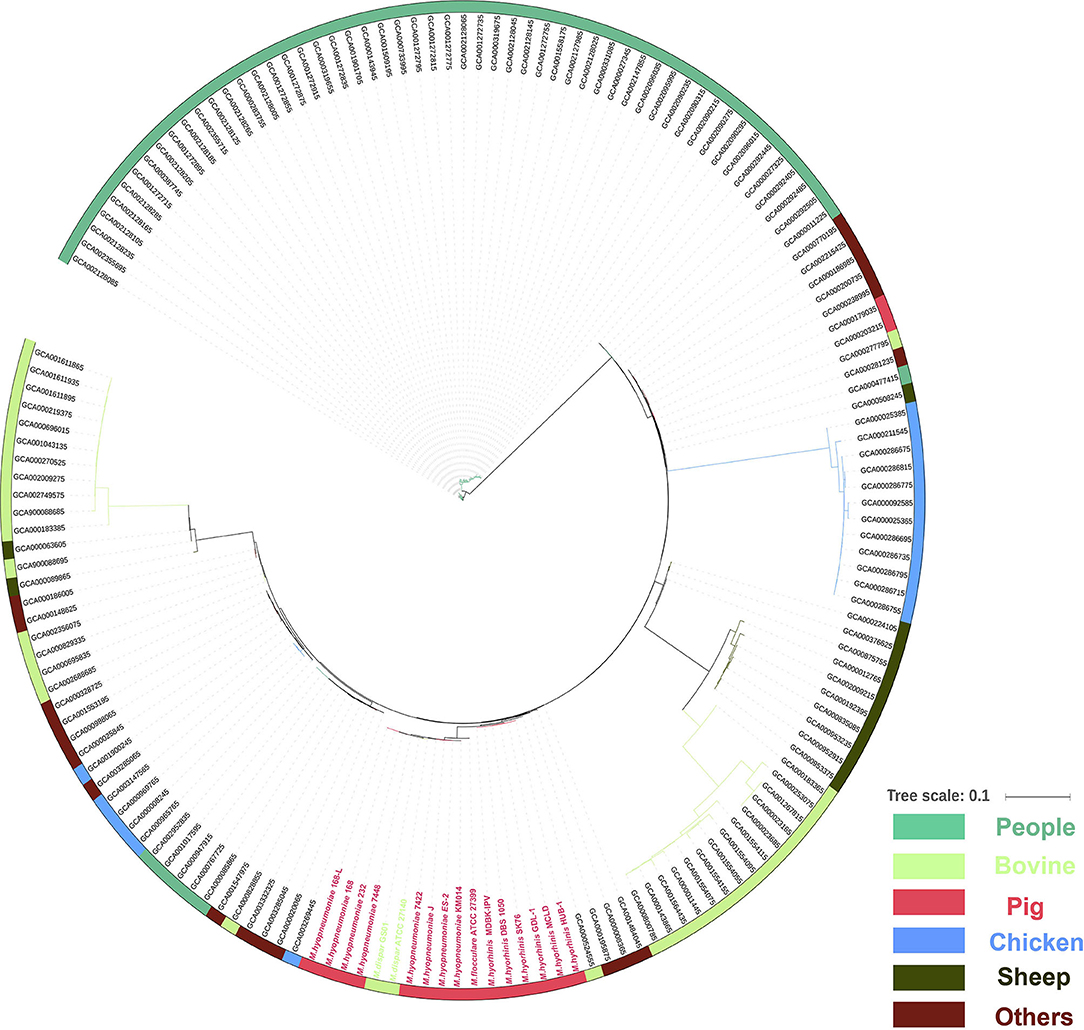
Figure 5. Phylogenetic tree of the whole genome sequences of 162 mycoplasma strains isolated from human (green), bovine (light green), pig (red), chicken (blue), sheep (blackish green), and others species (purple) based on amino acid alignments of all genes.
To detect the genes which are present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strains, each gene of 4 pathogenic M. hyopneumoniae strains ES-2, 168, 7,448, and 7,422 and each gene of 1 nonpathogenic M. hyopneumoniae strain J were aligned based on the different levels of identity (0.75, 0.8, 0.85, 0.9, 0.95, and 0.99) at the amino acid level. Venn diagram of the results showed that the number of common genes shared by 4 pathogenic M. hyopneumoniae strains and 1 nonpathogenic M. hyopneumoniae strain J were, respectively, 686 (identity = 0.75), 687 (identity = 0.80), 683 (identity = 0.85), 678 (identity = 0.90), 666 (identity = 0.95), and 521 (identity = 0.99). Excluding the common genes shared by pathogenic and nonpathogenic strains, 12 (identity = 0.75), 10 (identity = 0.80), 12 (identity = 0.85), 13 (identity = 0.90), 12 (identity = 0.95), and 25 (identity = 0.99) genes were found to be shared by all 4 pathogenic M. hyopneumoniae strains at the amino acid level, respectively (Figure 6). Further analysis of the genes which are present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strains indicated that 34 genes specific to pathogenic M. hyopneumoniae strains were detected at the amino acid level, of which 18 were hypothetical proteins, and another 16 were annotated in NCBI database (Table 1). By BlastN, 2 out of 34 genes specific to pathogenic M. hyopneumoniae strains at the amino acid level were absent in the nonpathogenic M. hyopneumoniae J strain at the nucleotide level (Table 2). ES-2_00484 gene was only identified in pathogenic M. hyopneumoniae strains 168, 168-L, 7422, and 7448. A partial sequence of the ES-2_00744 gene was only identified in pathogenic M. hyopneumoniae strains 232, 168-L, 7,422, and 7,448. Although the pathogenicity of M. hyopneumoniae KM014 is unclear, both genes were identified in the strain.
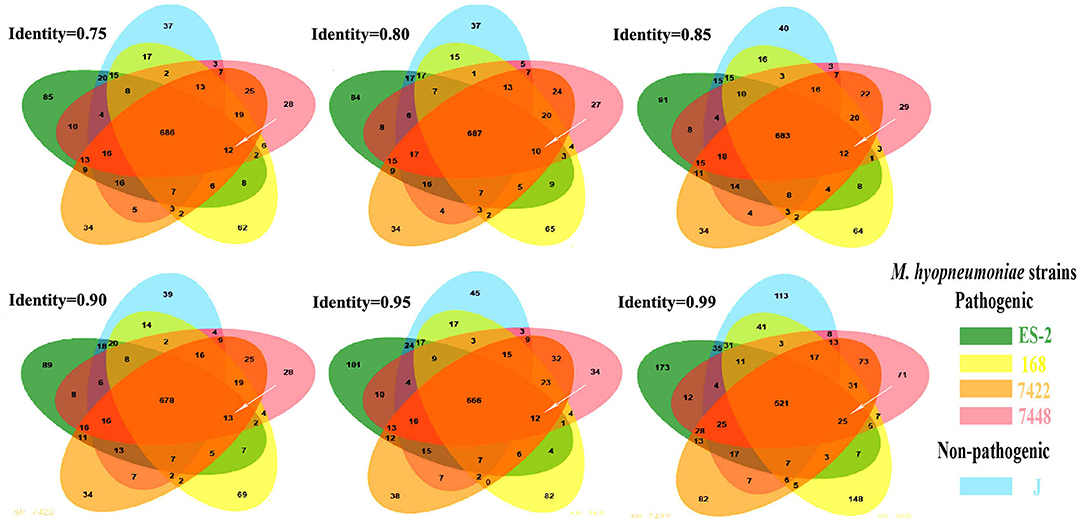
Figure 6. The genes specific to pathogenic M. hyopneumoniae strains. These Venn diagrams were developed. After the genes shared by pathogenic M. hyopneumoniae (ES-2, 168, 7422, and 7448) strains and nonpathogenic M. hyopneumoniae strain J were eliminated based on different levels of identity. 12, 10, 12, 13, 12, and 25 (white arrows) genes specific to pathogenic M. hyopneumoniae strains were, respectively, showed at the levels of identity = 0.75, 0.80, 0.85, 0.90, 0.95, and 0.99.
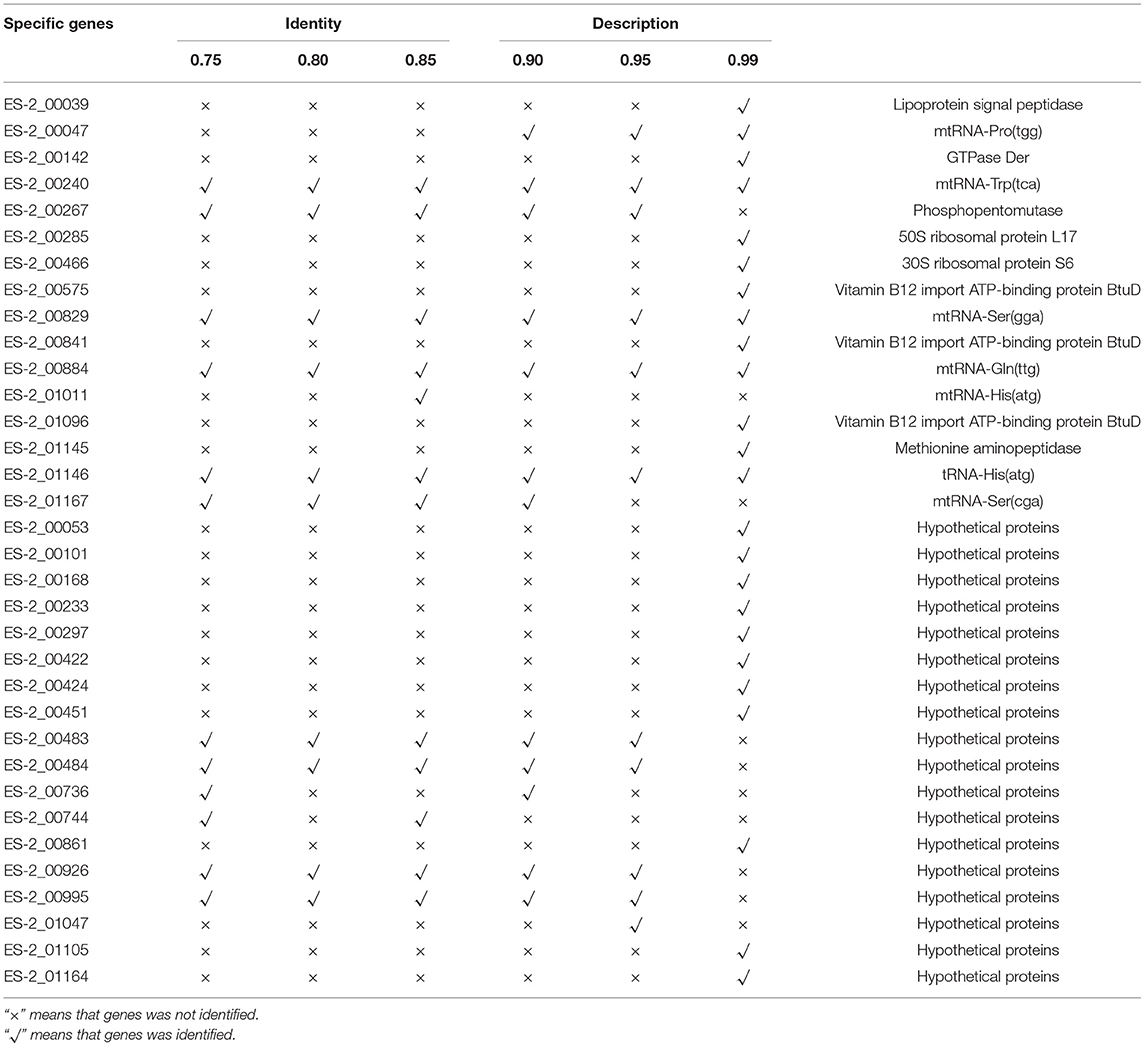
Table 1. The genes present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strains at the amino acid level.
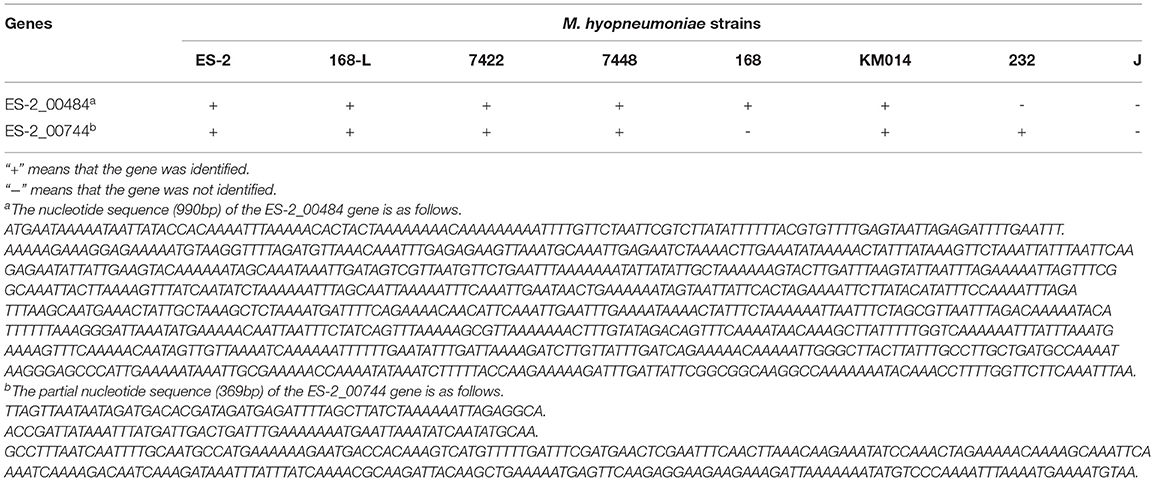
Table 2. The gene present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strain at the nucleotide level.
At present, Chinese native black pigs are suffering from severe M. hyopneumoniae infections. However, little is known about the growth characteristics and pathogenicity of M. hyopneumoniae which infects Chinese black pigs, especially Enshi black pigs. To better understand and treat M. hyopneumoniae infection in Enshi black pigs, the previously reported methods (7, 19, 37) were used to isolate M. hyopneumoniae in the lungs of Enshi black pigs. Unfortunately, due to the inability to completely remove M. hyorhinis, M. hyopneumoniae could not be isolated from pathological samples containing the fast-growing M. hyorhinis after repeating tests. To effectively remove M. hyorhinis, an improved method of multiple pulmonary-point sampling and continuous passage culturing based on previous methods reported (7, 19, 37) was used in this study.
By using the multiple pulmonary-point sampling and continuous passage culturing method, a M. hyopneumoniae strain was initially isolated from the lungs of an Enshi black pig (Figure 1). To further purify the M. hyopneumoniae strain initially isolated, the M. hyopneumoniae strain initially isolated was grown on a solid medium. A single colony was cultured in a liquid medium to obtain the pure M. hyopneumoniae strain. After three times of purification, the M. hyopneumoniae ES-2 strain was successfully isolated. Subsequently, this study determined that the M. hyopneumoniae ES-2 strain was most sensitive to kanamycin (2 μg/ml), which will provide a theoretical basis for the rapid and effective treatment of M. hyopneumoniae ES-2 strain infections.
By further study on the growth characteristics of M. hyopneumoniae ES-2 strain, we found that the modified Friis medium was most suitable for the growth of strain ES-2. The maximum viability of ES-2 strain in a modified Friis medium reached 1010 CCU/ml at approximately 2 days of incubation (Figure 3). Our SEM and TEM exhibited that ES-2 strain cells were round or oval, and the diameters of most cells were between 400 and 1,000 nm (Figure 2), which was consistent with the reports by other researchers (1) that M. hyopneumoniae cells were coccoid or coccobacillary, and their mean diameters range from 0.5 to 0.8 μm. Previous studies reported that pathogenic M. hyopneumoniae caused mycoplasma-like pneumonia lesions in the parts of the cardiac, apical, diaphragmatic, and accessory lobes, and led to the ciliostasis, clumping, and final loss of the cilia on the epithelial surface of the trachea, bronchi, and bronchioles (12). In agreement with previous reports, our findings showed that the lungs of the pigs inoculated with M. hyopneumoniae ES-2 strain exhibited the mycoplasma-like pneumonia lesions (Figure 4F), the increased mononuclear cells in peribronchiolar (Figure 4J), and the ciliostasis, clumping, and final loss of cilia by SEM (Figures 4G–I). These results revealed that ES-2 is a pathogenic M. hyopneumoniae strain. To further discover the virulence factor of M. hyopneumoniae ES-2 strain, 25 mycoplasmas-virulence-related genes previously reported were aligned with each gene of pathogenic and nonpathogenic M. hyopneumoniae. Results showed that 11 mycoplasmas-virulence-related genes were found to be present in all reported M. hyopneumoniae strains, and that only the P200 gene, which was crucial for M. pneumoniae to glide and colonize in the differentiated bronchial epithelium, was discovered to be present in ES-2 strain (Supplementary Table S1). Therefore, we speculate that the putative p200 plays an important role in the pathogenesis of the M. hyopneumoniae ES-2 strain.
In general, M. hyopneumoniae, as one of the main causes of complex porcine respiratory diseases, which works together with other pathogens (PCV-2, PRRSV, etc.), plays a crucial role in the swine respiratory disease (45, 46). Moreover, not only M. hyopneumoniae can cause Mycoplasma-like pneumonia lesions (44) but also viruses, particularly, the swine influenza virus may cause similar pneumonia lesions (13, 28). However, there was no effective method to distinguish commensal agents from potential pathogenic agents in pigs suffering from respiratory disease (47). Therefore, it is very implicated and difficult to diagnose and treat porcine respiratory disease caused by multiple pathogens infections. Considering the frequent clinical outbreaks of respiratory diseases, the development of a new method that could not only detect the presence of a pathogen but also identify whether it is pathogenic or nonpathogenic in the infected pig lungs will contribute to the accurate prevention and effective treatment of complex porcine respiratory diseases. Therefore, it is very important to discover the genes specific to pathogenic M. hyopneumoniae strains.
In this study, the phylogenetic tree revealed that pathogenic and nonpathogenic M. hyopneumoniae strains form a group (Figure 5). Therefore, pathogenic and nonpathogenic M. hyopneumoniae strains cannot be distinguished by phylogenetic analysis. To distinguish between pathogenic and nonpathogenic M. hyopneumoniae strains, this study explores the genes specific to pathogenic M. hyopneumoniae strains by comparative genomics. In previous studies, there were only several M. hyopneumoniae strains reported, and their complete genome sequences (24, 31–33) have been available in the GenBank. In these reported M. hyopneumoniae, only the J strain was nonpathogenic, which was often considered the reference strain (23, 48, 49). Although the J strain was a highly passaged strain that is incapable of colonizing pigs, the strain as a nonpathogenic M. hyopneumoniae was often compared with pathogenic M. hyopneumoniae strains in comparative genomics (23, 24). In the present study, the pathogenic M. hyopneumoniae ES-2 strain was isolated and its complete genome was sequenced by the PacBio method (Supplementary Table S1). Venn diagram developed at different levels of identity showed that 34 genes specific to pathogenic M. hyopneumoniae strains were found (Figure 6 and Table 1) at the amino acid level. Further analysis identified that 2 genes are present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strains based on amino acid and nucleotide level indicated (Table 2).
In summary, this study successfully isolated and purified a pathogenic M. hyopneumoniae ES-2 strain from the lungs of Enshi black pigs, which will help us quickly determine the susceptibility of M. hyopneumoniae ES-2 strain to drugs, and guide the clinical rapid and accurate drug selection. In addition, the phylogenetic tree revealed that there may be a selective preference in genome grouping and natural recombination among different Mycoplasma species. These results may develop the foundation for Mycoplasma grouping in the future. Further research results identified that 2 genes are present in pathogenic M. hyopneumoniae strains but absent in nonpathogenic M. hyopneumoniae strains at the amino acid and nucleotide level, which may be predicted to be used as a molecular marker to determine whether M. hyopneumoniae strains are pathogenic or nonpathogenic in clinical diagnosis. Therefore, it is meaningful and important to prevent and treat M. hyopneumoniae infection in Chinese black pigs by studying the characteristics of M. hyopneumoniae strain ES-2.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The animal study was reviewed and approved by Animal Care and Use Committee of Wuhan Polytechnic University (No: WPU202201002).
YZha, BZ, HC, and CT contributed to the conception and design of the study. YZhu organized the database. YZha, BZ, and YZhu performed the statistical analysis. YZha, BZ, and CT wrote the first draft of the manuscript. ML and XW wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
This work was supported by grants from the National Natural Science Foundation of China (32102674), the National Key R&D Program of China (2017YFD0500202), the Hubei Province Natural Science Foundation for Innovative Research Groups (2016CFA015), the earmarked fund for China Agriculture Research System (CARS-35), and the Hubei Agricultural Science Innovation Centre (2016-620- 000-001-039).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.883416/full#supplementary-material
1. Blanchard B, Vena MM, Cavalier A, Le LJ, Gouranton J, Kobisch M. Electron microscopic observation of the respiratory tract of SPF piglets inoculated with Mycoplasma hyopneumoniae. Vet Microbiol. (1992) 30:329–41. doi: 10.1016/0378-1135(92)90020-T
2. Kwon D, Choi C, Chae C. Chronologic localization of Mycoplasma hyopneumoniae in experimentally infected pigs. Vet Pathol. (2002) 39:584–87. doi: 10.1354/vp.39-5-584
3. Caron J, Ouardani M, Dea S. Diagnosis and differentiation of Mycoplasma hyopneumoniae and Mycoplasma hyorhinis infections in pigs by PCR amplification of the p36 and p46 genes. J Clin Microbiol P. (2000) 1390–6. doi: 10.1128/JCM.38.4.1390-1396.2000
4. Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of Mycoplasmas. Microbiol Mol Biol Rev. (1998) 62:1094–156. doi: 10.1128/MMBR.62.4.1094-1156.1998
5. Li S, Fang LR, Liu W, Song T, Zhao FW, Zhang RX, et al. Quantitative proteomic analyses of a pathogenic strain and its highly passaged attenuated strain of Mycoplasma hyopneumoniae. Biomed Res Int. (2019) 2019:4165735. doi: 10.1155/2019/4165735
6. Goodwin RF. The survival of Mycoplasma suipneumoniae in liquid medium, on solid medium and in pneumonic tissue. Res Vet Sci. (1972) 13:203–4. doi: 10.1016/S0034-5288(18)34079-7
7. Friis NF. Some recommendations concerning primary isolation of Mycoplasma suipneumoniae and Mycoplasma flocculare: a survey. Nordisk Veterinaer Medicin. (1975) 27:337–9.
8. Tao Y, Shu JH, Chen J, Wu YH, He YL. A concise review of vaccines against Mycoplasma hyopneumoniae. Res Vet Sci. (2019) 123:144–52. doi: 10.1016/j.rvsc.2019.01.007
9. Sorensen V, Ahrens P, Barfod K, Feenstra AA, Feld NC, Friis NF, et al. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and evaluation of four diagnostic assays. Vet Microbiol. (1997) 54:23–34. doi: 10.1016/S0378-1135(96)01266-7
10. Fano E, Pijoan C, Dee S. Dynamics and persistence of Mycoplasma hyopneumoniae infection in pigs. Can J Vet Res. (2005) 69:223–8.
11. DeBey MC, Ross RF. Ciliostasis and loss of cilia induced by Mycoplasma hyopneumoniae in porcine tracheal organ cultures. Infect Immun. (1994) 62:5312–8. doi: 10.1128/iai.62.12.5312-5318.1994
12. Mebus CA, Underdahl NR. Scanning electron microscopy of trachea and bronchi from gnotobiotic pigs inoculated with Mycoplasma hyopneumoniae. Am J Vet Res. (1977) 38:1249–54.
13. Thacker EL. Immunology of the porcine respiratory disease complex. Vet Clin N Am Food Animal Pract. (2001) 17:551–65. doi: 10.1016/S0749-0720(15)30006-2
14. Nathues H, Spergser J, Rosengarten R, Kreienbrock L, Beilage EG. Value of the clinical examination in diagnosing enzootic pneumonia in fattening pigs. Vet J. (2012) 193:443–7. doi: 10.1016/j.tvjl.2012.01.013
15. Nathues H, Woeste H, Doehring S, Fahrion AS, Doherr MG, Beilage EG. Herd specific risk factors for Mycoplasma hyopneumoniae infections in suckling pigs at the age of weaning. Acta Vet Scand 55. (2013) 30. doi: 10.1186/1751-0147-55-30
16. Bhat P, Singh ND, Leishangthem GD, Kaur A, Mahajan V, Banga HS, et al. Histopathological and immunohistochemical approaches for the diagnosis of mycoplasmosis in pig population of Punjab. Indian J Vet Pathol. (2017) 41:134–7. doi: 10.5958/0973-970X.2017.00032.3
17. Leman AD, Stein T, Straw B, Hilley HD. Pneumonia of swine; its cost and value of control. Mod Vet Pract. (1982) 63:195–8.
18. Goodwin RF, Pomeroy AP, Whittlestone P. Characterization of Mycoplasma suipneumoniae: a mycoplasma causing enzootic pneumonia of pigs. J Hyg (Cambridge). (1967) 65:85–96. doi: 10.1017/S0022172400045563
19. Cook BS, Beddow JG, Manso-Silvan L, Maglennon GA, Rycroft AN. Selective medium for culture of Mycoplasma hyopneumoniae. Vet Microbiol. (2016) 195:158–64. doi: 10.1016/j.vetmic.2016.09.022
20. Liu W, Feng Z, Fang L, Zhou Z, Li Q, Li S, et al. Complete genome sequence of Mycoplasma hyopneumoniae strain 168. J Bacteriol. (2011) 193:1016–7. doi: 10.1128/JB.01305-10
21. Minion FC, Lefkowitz EJ, Madsen ML, Cleary BJ, Swartzell SM, Mahairas GG. The genome sequence of Mycoplasma hyopneumoniae strain 232, the agent of swine mycoplasmosis. J Bacteriol. (2004) 186:7123–33. doi: 10.1128/JB.186.21.7123-7133.2004
22. Han JM, Park BS, Shin DJ, Song SY, Jeong YJ, Lee N. Complete Genome Sequence of Mycoplasma hyopneumoniae Strain KM014, a Clinical Isolate from South Korea. Genome Announc. (2017) 5:e01012–17. doi: 10.1128/genomeA.01012-17
23. Siqueira FM, Thompson CE, Virginio VG, Gonchoroski T, Reolon L, Luiz Almeida G, et al. New insights on the biology of swine respiratory tract mycoplasmas from a comparative genome analysis. BMC Genom. (2013) 14:175. doi: 10.1186/1471-2164-14-175
24. Vasconcelos AT, Ferreira HB, Bizarro CV, Bonatto SL. Swine and poultry pathogens: the complete genome sequences of two strains of Mycoplasma hyopneumoniae and a strain of Mycoplasma synoviae. J Bacteriol. (2005) 187:5568–77. doi: 10.1128/JB.187.16.5568-5577.2005
25. Thacker EL. Diagnosis of Mycoplasma hyopneumoniae. Animal Health Res Rev. (2005) 5:317–20. doi: 10.1079/AHR200491
26. Calsamiglia M, Pijoan C, Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diagn Invest. (1999) 11:246–51. doi: 10.1177/104063879901100307
27. Maes D, Verbeke W, Vicca J, Verdonck M, de Kruif A. Benefit to cost of vaccination against Mycoplasma hyopneumoniae in pig herds under Belgian market conditions from 1996 to 2000. Livestock Prod Sci. (2003) 83:85–93. doi: 10.1016/S0301-6226(03)00039-3
28. Sibila M, Pieters M, Molitor T, Maes D, Haesebrouck F, Segales J. Current perspectives on the diagnosis and epidemiology of Mycoplasma hyopneumoniae infection. Vet J. (2009) 181:221–31. doi: 10.1016/j.tvjl.2008.02.020
29. Hansen MS, Pors SE, Jensen HE, Bille-Hansen V, Bisgaard M, Flachs EM, et al. An investigation of the pathology and pathogens associated with porcine respiratory disease complex in Denmark. J Comp Pathol. (2010) 143:120–31. doi: 10.1016/j.jcpa.2010.01.012
30. Bochev I. Porcine respiratory disease complex (PRDC): a review. II Diagnostics, treatment and prevention. Bulg J Vet Med. (2008) 4:219–34.
31. Pinto PM, Klein CS, Zahaand A, Ferreira HB. Comparative proteomic analysis of pathogenic and non-pathogenic strains from the swine pathogen Mycoplasma hyopneumoniae. Proteome Sci. (2009). doi: 10.1186/1477-5956-7-45
32. Pinto PM, Chemale G, de Castro LA, Ferreira HB. Proteomic survey of the pathogenic Mycoplasma hyopneumoniae strain 7448 and identification of novel post-translationally modified and antigenic proteins. Vet Microbiol. (2007) 121:83–93. doi: 10.1016/j.vetmic.2006.11.018
33. Paes JA, Machado LDPN, Dos Anjos Leal FM, De Moraes SN, Moura H, Barr JR, et al. Comparative proteomics of two Mycoplasma hyopneumoniae strains and Mycoplasma flocculare identified potential porcine enzootic pneumonia determinants. Virulence. (2018) 9:1230–46. doi: 10.1080/21505594.2018.1499379
34. The plos one staff. Correction: Unravelling the transcriptome profile of the swine Respiratory tract mycoplasmas. PLoS ONE. (2014) 9:e116122. doi: 10.1371/journal.pone.0116122
35. Hu QH, Han XG, Zhou XJ, Ding SY, Ding C, Yu SQ. Characterization of biofilm formation by Riemerella anatipestifer. Vet Microbiol. (2010) 144:429–36. doi: 10.1016/j.vetmic.2010.02.023
36. Hammerschmidt S, Wolff S, Hocke A, Rosseau S. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun P. (2005) 4653–67. doi: 10.1128/IAI.73.8.4653-4667.2005
37. Kobisch M, Friis NF. Swine mycoplasmoses. Rev sci tech Off Int Epiz. (1996) 15:1569–605. doi: 10.20506/rst.15.4.983
38. Vicca J, Stakenborg T, Maes D, Butaye P, Peeters J, de Kruif A, et al. (2003). Evaluation of virulence of Mycoplasma hyopneumoniae field isolates. Vet Microbiol. 97, 177–190. doi: 10.1016/j.vetmic.2003.08.008
39. Chin CS, Alexander DH, Marks P, Klammer AA. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Meth. (2013) 10:563–9. doi: 10.1038/nmeth.2474
40. Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. (2007) 23:673–9. doi: 10.1093/bioinformatics/btm009
41. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MT, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. (2015) 31:3691–3. doi: 10.1093/bioinformatics/btv421
42. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
43. Kobisch M, Blanchard B, Le Potier MF. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and resistance to reinfection. Vet Res. (1993) 24:67–77.
44. Jordan JL, Chang HY, Balish MF, Holt LS. Protein P200 Is dispensable for Mycoplasma pneumoniae hemadsorption but not gliding motility or colonization of differentiated bronchial epithelium. Infection and immunity P. (2007) 518–22. doi: 10.1128/IAI.01344-06
45. Dee SA. The porcine respiratory disease complex: Are subpopulations important? Swine Health Prod. (1996) 4:147–9.
46. Opriessnig T, Giménez-Lirola LG, Halbur PG. Polymicrobial respiratory disease in pigs. Animal Health Res Rev. (2011) 12:133–48. doi: 10.1017/S1466252311000120
47. Fablet C, Marois-Créhan C, Simon G, Grasland B, Jestin A, Kobisch M, et al. Infectious agents associated with respiratory diseases in 125 farrow-to-finish pig herds: a cross-sectional study. Vet Microbiol. (2012). doi: 10.1016/j.vetmic.2011.12.015
48. Maes D, Segales J, Meyns T, Sibila M, Pieters M, Haesebrouck F. Control of Mycoplasma hyopneumoniae infections in pigs. Vet Microbiol. (2008) 126:297–309. doi: 10.1016/j.vetmic.2007.09.008
49. Villarreal I, Maes D, Meyns T, Gebruers F, Calus D, Pasmans F, et al. Infection with a low virulent Mycoplasma hyopneumoniae isolate does not protect piglets against subsequent infection with a highly virulent M. hyopneumoniae isolate. Vaccine. (2009) 27:1875–9. doi: 10.1016/j.vaccine.2008.12.005
Keywords: Mycoplasma hyopneumoniae, Chinese native black pig, swine enzootic pneumonia, ES-2 strain, characteristics
Citation: Zong B, Zhu Y, Liu M, Wang X, Chen H, Zhang Y and Tan C (2022) Characteristics of Mycoplasma hyopneumoniae Strain ES-2 Isolated From Chinese Native Black Pig Lungs. Front. Vet. Sci. 9:883416. doi: 10.3389/fvets.2022.883416
Received: 25 February 2022; Accepted: 25 May 2022;
Published: 30 June 2022.
Edited by:
Mayara Fernanda Maggioli, Oklahoma State University, United StatesReviewed by:
Shaohui Wang, Shanghai Veterinary Research Institute (CAAS), ChinaCopyright © 2022 Zong, Zhu, Liu, Wang, Chen, Zhang and Tan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yanyan Zhang, emhhbmd5YW55YW5hMzA1QDE2My5jb20=; Chen Tan, dGFuY2hlbkBtYWlsLmh6YXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.