- 1Key Laboratory of Zoonosis Prevention and Control of Guangdong Province, College of Veterinary Medicine, South China Agricultural University, Guangzhou, China
- 2African Swine Fever Regional Laboratory of China, Guangzhou, China
- 3Research Center for African Swine Fever Prevention and Control, South China Agricultural University, Guangzhou, China
- 4Maoming Branch, Guangdong Laboratory for Lingnan Modern Agriculture, Maoming, China
Since the first outbreak of ASFV genotype II in China in 2018, ASF has posed a significant threat to the swine industry. After the emergence of genotype I in China in 2020, the epidemic prevention and control have become more difficult. No effective commercial vaccine is currently available, and the disease is difficult to eradicate; therefore, the identification of the ASFV genotype is critical to establish biosafety control measures. In this study, a dual real-time PCR detection method based on B646L and E183L genes was developed to distinguish between ASFV genotypes I and II by specifically amplifying the genotype I E183L gene. The method is strongly specific, detects B646L and E183L genes simultaneously, and does not cross-react with PEDV, PCV, PRRSV, PRV, and CSFV. The double real-time PCR detection of ASFV genotypes I and II showed a B646L amplification curve, and only genotype I showed an E183L amplification curve, consistent with our expectations. The method has high sensitivity and the lowest copy numbers detected for recombinant plasmids B646L and E183L were 1.07 × 102 and 3.13 × 104 copies/μL, respectively. The method is reproducible, and the coefficient of variation for detecting the coefficient of variation (CV) values of the two recombinant plasmids was <2%. Seven samples were positive and 277 were negative, and the results of the two methods were consistent. The dual real-time PCR presented in this study provides a rapid detection method for the identification of ASFV genotypes I and II, which may lead to improving efficient prevention and control measures for ASF in China.
Introduction
African swine fever (ASF) is an acute, lethal, and highly contagious infectious disease caused by the African swine fever virus (ASFV) in pigs. ASF clinical features are high fever, skin cyanosis, and severe bleeding in lymph nodes and internal organs, with a mortality rate as high as 100% (1). In 1921, ASF was first reported in Kenya, East Africa, and was classified into 24 genotypes based on the sequence of the 3′-end of the B646L gene encoding the major capsid protein p72 (2, 3). Multiple ASFV genotypes were circulating in Africa until 1957, when genotype I was first discovered outside Africa, first in Portugal and successively in Europe, South America, Caribbean islands, West Africa, and East and South Africa (4). In 2007, genotype II spread from south-eastern Africa to the Caucasus region of Russia and then became prevalent in more than 30 countries and regions in Europe, Asia, and South America, causing huge economic losses to the swine industry (5, 6).
Since the discovery of ASFV genotype II in China in August 2018 (7), large-scale ASF outbreaks have occurred across the country. ASFV replicates in viral cytoplasmic factories, the presence of viral DNA within the host cell nucleus has been previously reported to be essential for productive infection. Morphological changes of promyelocytic leukemia nuclear bodies (PML-NBs) were found in ASFV-infected swine MDMs, strongly suggesting the viral modulation of cellular antiviral responses and cellular transcription. PML-NBs are involved in interferon (IFN) immune-mediated mechanisms may explain how ASFV interferes with the host immune response (8). In August 2020, our laboratory first detected and isolated ASFV genotype I in pigs. Since then, two ASFVs with genotype I possessing low virulence but high transmission has been reported in China (9). This makes the diagnosis, prevention, and control of ASF more challenging. Therefore, distinguishing between ASFV genotypes I and II is necessary before establishing any strict prevention and control strategies. As no effective vaccine against ASF is currently available in the market, the disease control relies on biosecurity prevention, rapid detection, and culling of infected animals (10). Several molecular and serological methods are currently available to identify animals infected with ASFV. Serological tests are used to determine whether animals have been exposed to ASFV, and molecular tests can detect the presence of ASFV in pigs before they develop clinical symptoms. Polymerase chain reaction (PCR) and enzyme-linked immune sorbent assay are the main routine diagnostic methods for detecting antigens or antibodies (11–14). The World Organization for Animal Health (OIE) recommends the use of validated real-time PCR methods for the diagnosis of ASF (12, 14, 15). Compared to traditional PCR, real-time PCR is fast, highly sensitive, and specific (12, 14). A variety of real-time PCR diagnostic methods for ASFV detection have been developed and validated in the market, most of which target the B646L gene (13, 14, 16–18). Studies have shown that, in addition to the B646L gene encoding the p72 protein, the tandem repeat sequence (19–21) of the central variable region in the B602L and E183L genes encoding the p54 protein are also target genes for studying the genotypic diversity of ASFV strains. Combining the B646L, E183L, and pB602L gene sequences, more data is available to support ASFV typing (19, 22, 23). It has been shown that the E183L gene sequence is a valuable candidate for genotyping in molecular epidemiological studies to identify ASFV genotype I (24). Therefore, this study established a dual real-time PCR detection method based on the ASFV B646L and E183L genes, which can identify the existing ASFV genotypes I and II through two pairs of primers and two probes. The primers and probes of the B646L gene were used to determine whether the samples were ASFV-positive, whereas the primers and probes of the E183L gene were specific to genotype I and could only detect genotype I-positive samples. Therefore, this method identifies genotypes I and II strains by specifically amplifying the E183L gene. The dual real-time PCR detection method presented in this study can provide a powerful tool for the diagnosis of ASFV genotypes I and II strains and could play a key role in the ASF prevention and control in China.
Materials and Methods
Viruses, Viral Nucleic Acids, Plasmids, and Clinical Samples
ASFV genotypes I and II nucleic acids, PEDV, PCV2, PRRSV, PRV, CSFV, recombinant plasmids pUC57-B646L, and pCAGGS-E183L were preserved by the Department of Infectious Diseases, School of Veterinary Medicine, South China Agricultural University. The 284 clinical samples were obtained from pig farms in the Guangdong Province and included 178 anticoagulated blood samples, 56 oral swabs, 27 tonsils, and 23 lymph nodes.
Primers and Probes
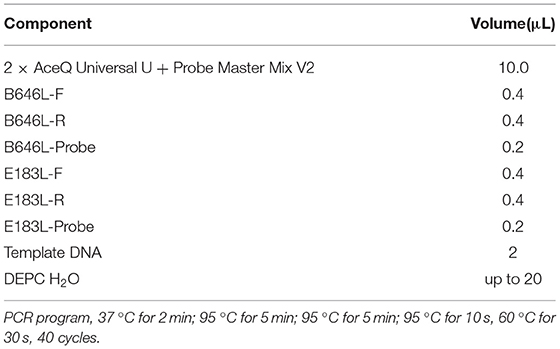
The primers and probes used for the B646L gene were recommended by China Center for Animal Disease Control (CADC) and the size of the amplified fragment was 159 bp. The primers and probes for the E183L gene were designed according to the sequences published in GenBank, using Oigo7 software, in the genotype I specific region, and the size of the amplified fragment was 73 bp. The B646L and E183L gene GenBank numbers are GZ201801.1 and NC_001659.2, respectively. The B646L gene probe 5′-end modified group was FAM, the 3′-end quencher group was MGB; E183L gene probe 5′-end modified group was HEX, 3′-end the quencher group was BHQ1, and all primers and probes were synthesized by Thermo Company (Thermo Fisher Scientific, Waltham, MA, USA). Table 1 shows the sequences of the synthesized primers and probes and the sizes of the amplified fragments.
Nucleic Acid Extraction
Tissue samples (0.1 g) were mixed with 1 mL of PBS, ground with a mortar and freeze-thaw three times. Mouth swabs were infiltrated with 1 mL of PBS, and the suspension was centrifuged at 1000 × g for 5 min at 4 °C to obtain the supernatant. Nucleic acid extraction from the blood, tissue, and mouth swabs was performed using the Axyprep Body Fluid Viral DNA/RNA Miniprep kit (Axygen, HangZhou, China). The extracted nucleic acid was obtained using the reverse transcription HiScript II 1st Strand cDNA Synthesis kit (+ gDNA wiper) (Vazyme, Nanjing, China) to obtain cDNA, which was stored at−80 °C for further use.
Dual Real-Time PCR System and Conditions
A real-time PCR 2 × AceQ Universal U kit + Probe Master Mix V2 (Vazyme, Nanjing, China) was used to process the samples, and an ExCycle-48 real-time PCR instrument (GinX, Shanghai, China) was used for double real-time PCR detection and analysis. The amplification conditions used for the reaction is shown in Table 2.
Specific Detection
Nucleic acids were extracted from PEDV, PRRSV, PRV, PCV-2, CSFV, and negative tissues and reverse-transcribed. Double real-time PCR was used to detect ASFV genotypes I and II, PEDV, PRRSV, PRV, PCV-2, CSFV, and negative pig nucleic acid. The primers and probes for double real-time PCR were specifically verified.
Sensitivity Detection and Construction of a Standard Curve
The concentrations of the recombinant plasmids pUC57-B646L and pCAGGS-E183L were determined using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and were 335.1 and 183.13 ng/μL, respectively. According to the formula to calculate the DNA copy number [dsDNA copy number (copies/μL) = (6.02 × 1023(copies/mol) × concentration (ng/μL) × 10−9)/DNA length × 660], the copy numbers for the pUC57-B646L and pCAGGS-E183L plasmids were calculated to be 1.07 × 1011 and 3.13 × 1010 copies/μL, respectively. A 10-fold serial dilution of the recombinant plasmid was used as a standard template (10−1-10−9). Three replicates of each dilution were used for real-time PCR detection, sensitivity analysis, and standard curve interpretation.
Repeated Detection
Different gradient dilutions of the recombinant plasmids pUC57-B646L and pCAGGS-E183L were used as templates for real-time PCR amplification, and each concentration was repeated three times for repeatability experiments. To verify the repeatability of the established method, the coefficient of variation (CV) was calculated according to the Ct value, CV = (standard deviation SD/mean) × 100%.
Results
Dual Real-Time PCR Specific Detection
The ASFV nucleic acids of genotypes I and II, genotypes I and II mixed samples, and negative pig tissues were determined using double fluorescence quantitative PCR. The FAM channel of the ASFV genotypes I and II and the mixed samples of both genotypes showed amplification curves (Figure 1), while the HEX channel of genotype I and the mixed samples of ASFV genotypes I and II showed amplification curves (Figure 2). This shows that, regardless of the genotype, when ASFV is present, the FAM channel for detecting the B646L gene showed an amplification curve. However, only in the presence of genotype I, the HEX channel that detects the E183L gene showed an amplification curve, which can be identified as ASFV genotype I.
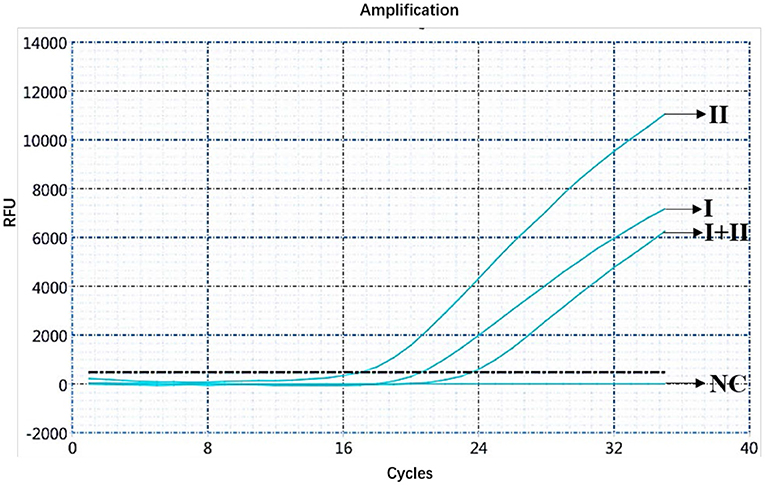
Figure 1. Amplification curve for the FAM channel determined through nucleic acid double fluorescence quantitative PCR of ASFV genotypes I and II, ASFV genotypes I and II mixed samples, and negative pig samples.
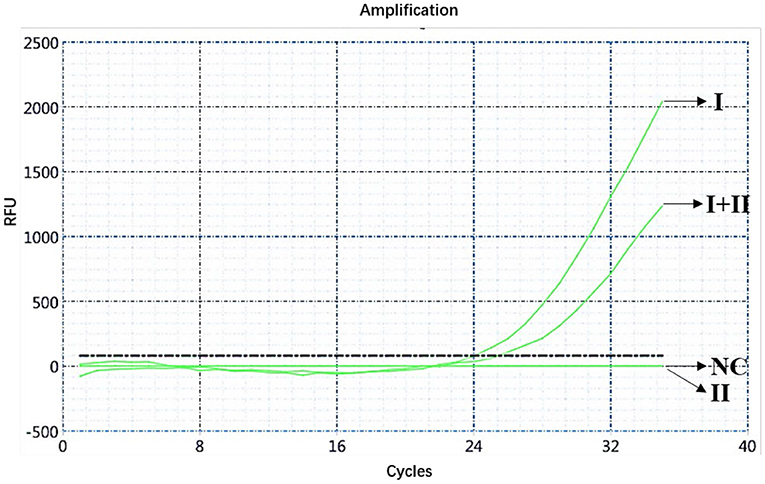
Figure 2. Amplification curve for the HEX channel determined through nucleic acid double fluorescence quantitative PCR of ASFV genotypes I and II, ASFV genotypes I and II mixed samples, and negative pig samples.
A specificity test of the dual real-time PCR detection was conducted with ASFV genotypes I and II, PEDV, PRRSV, PRV, PCV-2, CSFV, and negative pig nucleic acids as templates. PEDV, PRRSV, PRV, PCV-2, CSFV, and negative pig nucleic acid showed no amplification curve in the FAM and HEX channels, and the result was negative. The FAM channel of ASFV genotypes I and II nucleic acid samples showed amplification curves (Figure 3), and the HEX channel of the ASFV genotype I nucleic acid samples also showed amplification curves (Figure 4). The results indicate that the dual real-time PCR detection method based on B646L and E183L genes did not cross-react with the nucleic acids of other common porcine-derived viruses and could distinguish the infection of ASFV genotype I strain, which further verified that the method has good specificity.
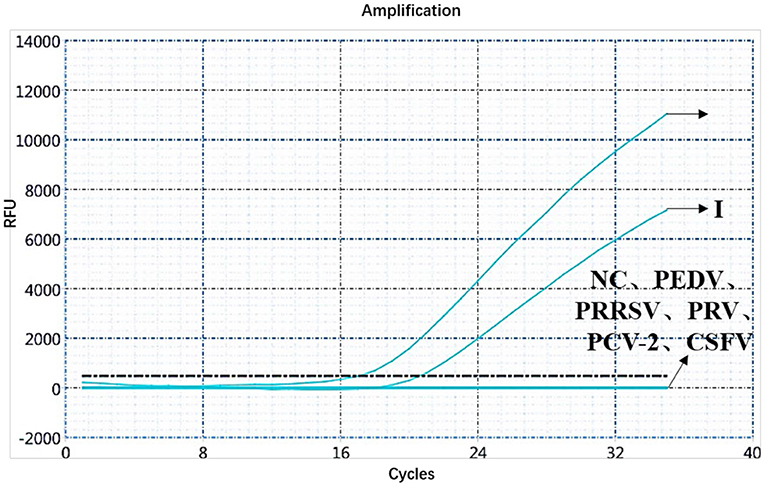
Figure 3. Amplification curve for the FAM channel of ASFV genotypes I and II, PEDV, PRRSV, PRV, PCV-2, CSFV, and negative nucleic acid pig samples determined through dual real-time PCR.
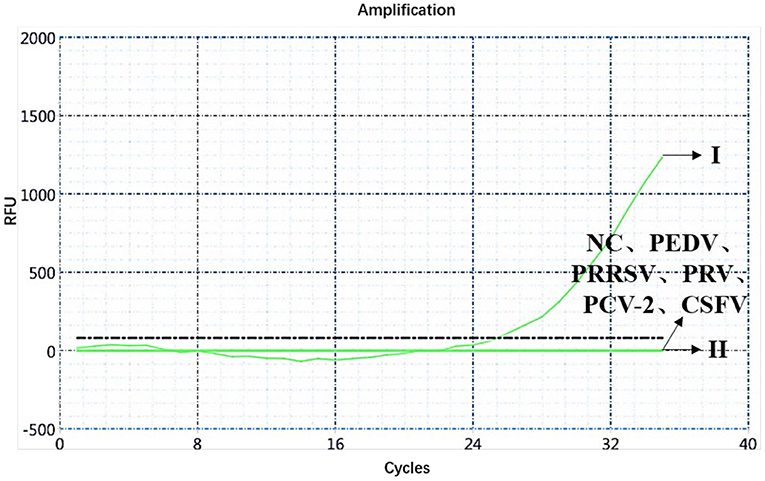
Figure 4. Amplification curve for the HEX channel of ASFV genotypes I and II, PEDV, PRRSV, PRV, PCV-2, CSFV, and negative nucleic acid pig samples determined through dual real-time PCR.
Sensitivity Determination of the Dual Real-Time PCR and Establishment of the Standard Curve
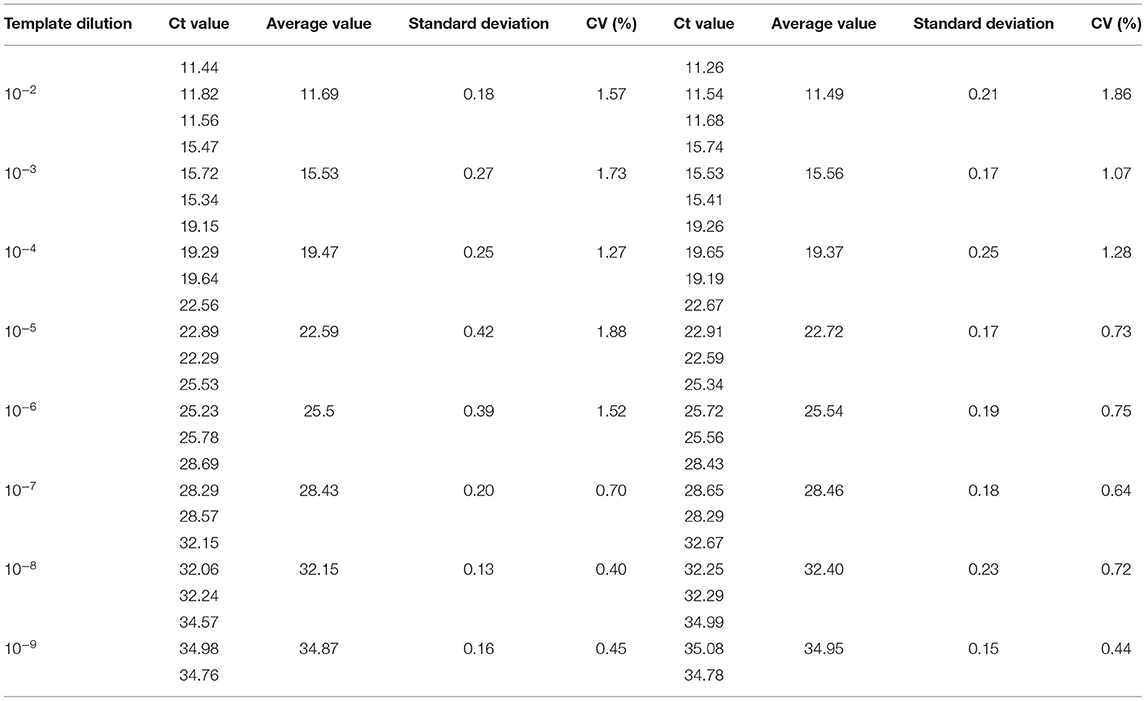
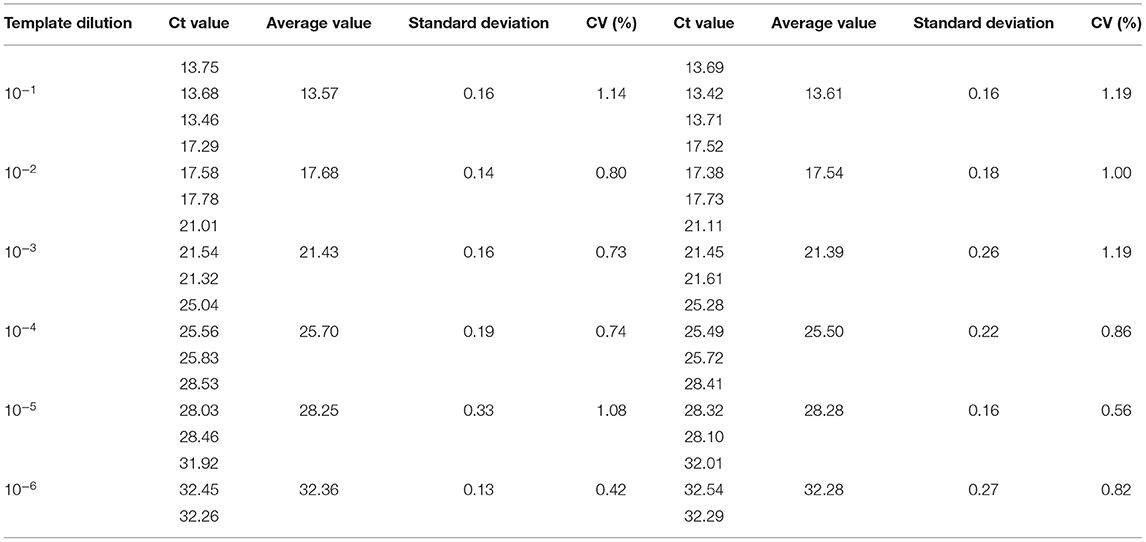
The sensitivity of the real-time PCR was determined after 10-fold gradient dilution of the recombinant plasmids pUC57-B646L and pCAGGS-E183L, and a standard curve was obtained using real-time analysis software. The results showed that the lowest copy numbers for pUC57-B646L and pCAGGS-E183L determined through real-time PCR were 1.07 × 102 and 3.13 × 104 copies/μL, respectively. This shows that the primers and probes used for this method detected both genes with high sensitivity (Figure 5). The slopes of the standard curves for the two plasmids, pUC57-B646L and pCAGGS-E183L, were −3.258 and −3.639, respectively, and the R2 values of the standard curves were 0.994 and 0.997, respectively (Figure 6), indicating that each diluted sample showed a good linear relationship.
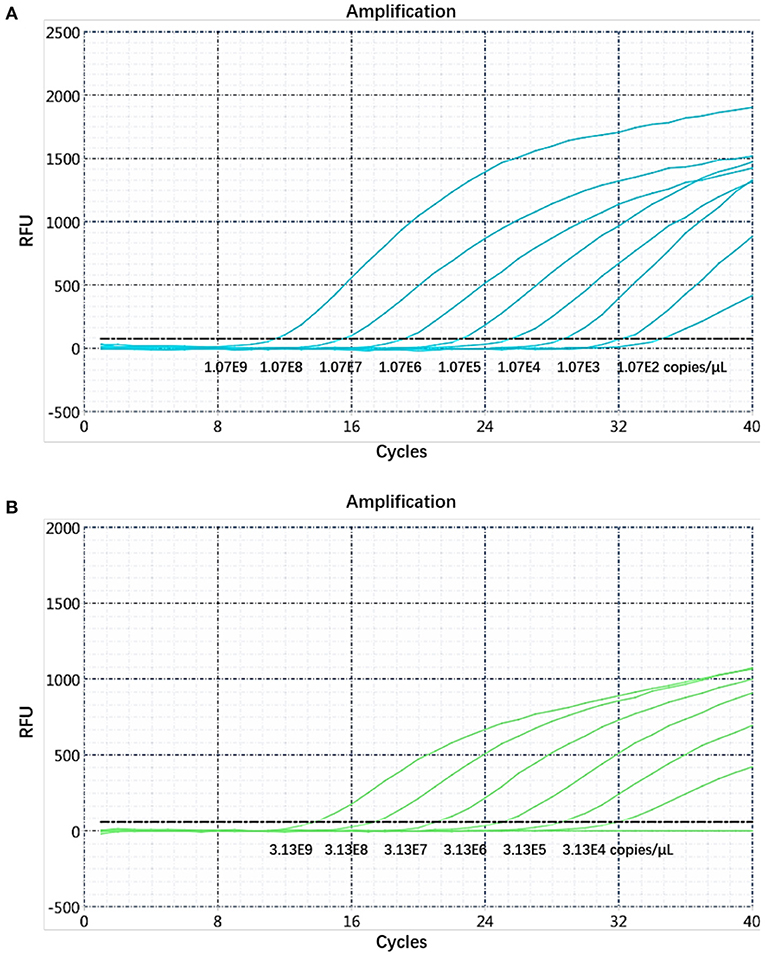
Figure 5. Determination of the dual real-time PCR sensitivity. (A) B646L gene sensitivity test. (B) E183L gene sensitivity test.
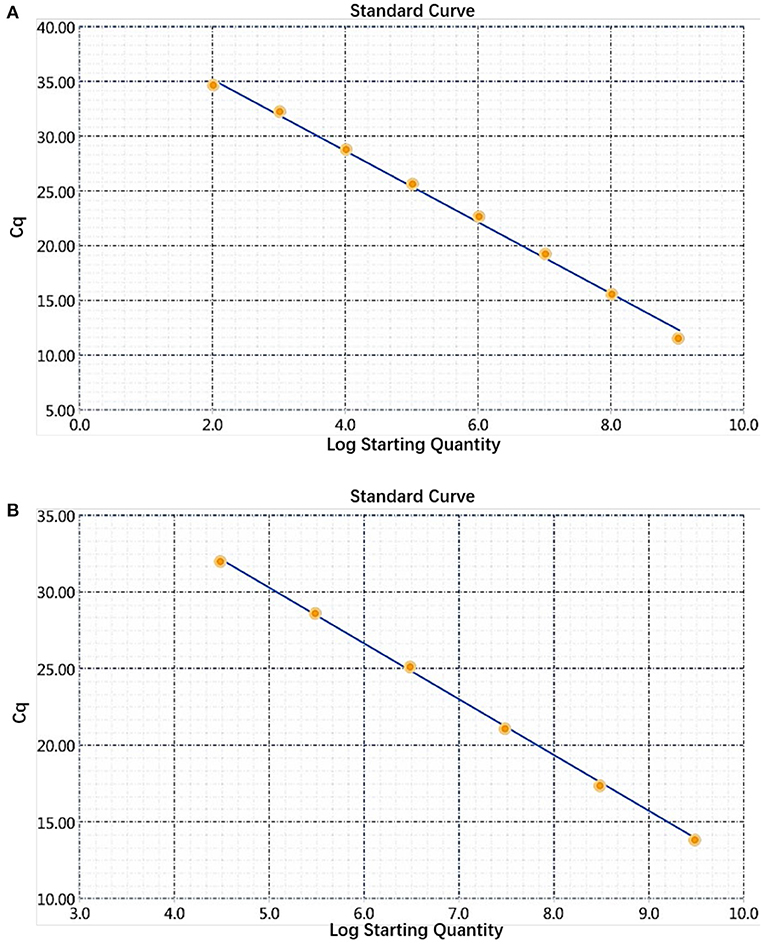
Figure 6. Establishment of a dual real-time PCR standard curve. (A) Standard curve for plasmid pUC57-B646L, y = −3.258x + 41.69, R2 = 0.994. (B) Standard curve for plasmid pCAGGS-E183L, y = −3.639x + 48.47, R2 = 0.997.
Repeatability and Reproducibility of the Dual Real-Time PCR
Repeatability and reproducibility experiments were performed using 10-fold serial dilutions of the recombinant plasmids, pUC57-B646L and pCAGGS-E183L, and calculated according to the Ct values of the results. The intra-assay CV for pUC57-B646L was 0.40–1.88% (Table 3) and the intra-assay CV for pCAGGS-E183L was 0.42–1.14% (Table 4). The inter-assay CV of pUC57-B646L was 0.441.86% (Table 3) and the inter-assay CV of pCAGGS-E183L was 0.56–1.19% (Table 4). The CV values were all <2%, indicating that the method has good repeatability.
Analysis of Clinical Samples
Clinical samples [284] from pigs from farms in the Guangdong Province were analyzed using the dual fluorescent quantitative PCR. The analysis showed 7 samples (1–7) to be positive for ASFV B646L and E183L genes, and the remaining 277 samples to be negative, which was consistent with the results of the fluorescence quantitative PCR method recommended by the OIE (Figure 7). Then, the samples were subjected to high-temperature and high-pressure treatments according to biosafety operation specifications. Among the seven ASFV-positive samples, only the FAM channel showed the amplification curve of the B646L gene, indicating that the seven positive samples were presented genotype II infection (Figure 7). The seven positive samples were collected from three ASFV-infected pigs from the Xiangzhou District, Zhuhai City, Guangdong Province. The information was published by the Ministry of Agriculture and Rural Affairs of the People's Republic of China: http://www.moa.gov.cn/gk/yjgl_1/yqfb/201812/t20181219_6165233.htm. Nucleic acids were prepared and preserved at the National African Swine Fever Regional Laboratory (Guangzhou, China).

Figure 7. Analysis of clinical samples. (A) Amplification curve for the FAM channel determined through nucleic acid dual fluorescence quantitative PCR of pig clinical samples (284). Pig clinical samples (284) were analyzed using dual real-time PCR; seven samples were ASFV-positive. (B) Amplification curve for the HEX channel determined through nucleic acid dual fluorescence quantitative PCR of pig clinical samples (284) and the remaining 277 samples were negative.
Discussion
ASF is a high-mortality and economically important viral disease of domestic and wild boars that causes acute hemorrhagic fever, with a fatality rate of up to 100% (5, 25). ASFV was first reported in Kenya 100 years ago, and today ASF poses a great threat to the swine industry worldwide; however, the clinical symptoms and case fatality rates of ASF change as the disease progresses (26). After the discovery of the acutely infective ASFV genotype II in China in 2018, due to imperfect prevention and control measures, ASFV genotype I, with lower mortality and stronger transmission ability, too was discovered in 2020. Both low virulent genotype II and new-emerging genotype I ASFVs showed lower pathogenicity but high transmissibility, which caused chronic and persistent infections in pigs. Pigs infected with these low virulent ASFV strains have milder clinical symptoms but constantly shed via oral and rectal routes at a low level, which poses a great challenge for the early diagnosis and control of the disease (9). As China is a big country of pig raising, the epidemic of ASF has brought a serious threat to the development of China's animal husbandry and the economic stability of the pig industry. Therefore, the prevention and control of ASF is the top priority of my country's animal husbandry. Epidemiological studies have shown that the entry of ASFV into non-ASF areas is mainly related to the construction of international airports and use of contaminated seaport waste to feed pigs (19). Combined with extensive commercial trade, ASF-free countries are always at risk of ASFV being introduced into their territories. As there is currently no efficient commercial vaccine for ASF, control and eradication strategies are mainly based on the early screening and implementation of strict biosecurity prevention and control measures. Therefore, laboratories must have rapid and sensitive detection procedures, and the identification of strain genotypes is critical for ASF control and eradication. It has been reported that part of the gene encoding the p72 protein and the E183L gene encoding the structural protein p54 can be used to identify the ASFV genotype. By combining the sequences of these two genes, a high-resolution method for identifying the virus genotype can be established (27). In this study, as ASFV genotypes I and II strains are prevalent in China, a novel dual fluorescence quantitative PCR detection method based on B646L and E183L genes was developed to identify these ASFV genotypes. If only the B646L gene is detected, it means that the test sample is positive for ASFV genotype II; if the B646L and E183L genes are detected, it means that the test sample is positive for genotype I. It is worth noting that only when the B646L gene is detected, the test sample can be considered ASFV positive; otherwise, it is considered negative. The specific detection of this method showed that the primers and probes used did not cross-react with other swine-derived viruses commonly found in pig farms, and the lowest copy numbers determined for B646L and E183L were 1.07 × 102 and 3.13 × 104 copies/μL, respectively. The standard curve presented a good linear relationship, and the experimental results were true and reliable. At the same time, 284 clinical swine samples were analyzed using the dual real-time PCR detection method, and seven ASFV-positive samples were found, which was consistent with the detection method recommended by the OIE. Therefore, the use of this detection method provides a powerful tool for identifying the ASFV genotypes strains in China to assess the origin and transmission trajectory of ASF outbreaks more accurately in epidemic areas and establish a solid foundation for ASF epidemiological investigation and epidemic prevention and control.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author Contributions
QG and ZZ designed the study. QG and YF were involved in the acquisition of data, analysis, figure preparation, and supervised the study. YY and HW contributed to some of the laboratory experiments and data analysis. YL and TG helped revise the manuscript. QG drafted the original paper. All authors read and approved the final manuscript.
Funding
This work was supported by the Key-Area Research and Development Program of Guangdong Province (grant number 2019B020211003), the National Natural Science Foundation of China (grant number 31941004), the Science and Technology Project of Guangdong Pig Industrial Park (grant number GDSCYY2020-024), and the China Agriculture Research System of MOF and MARA (grant number car35).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Petrov A, Schotte U, Pietschmann J, Dräger C, Beer M, Anheyer-Behmenburg H, et al. Blome, S. Alternative sampling strategies for passive classical and African swine fever surveillance in wild boar. Vet Microbiol. (2014) 174:607–8. doi: 10.1016/j.vetmic.2014.07.030
2. Gao L, Sun XD, Yang HL, Xu QG, Li J, Kang JK, et al. Epidemic situation and control measures of African Swine fever outbreaks in China 2018-2020. Transbound Emerg Dis. (2021) 68:2676–86. doi: 10.1111/tbed.13968
3. Muangkram Y, Sukmak M, Wajjwalku W. Phylogeographic analysis of African swine fever virus based on the p72 gene sequence. Genet Mol Res. (2015) 14:4566–74. doi: 10.4238/2015.May.4.15
4. Lubisi BA, Bastos ADS, Dwarka RM, Vosloo W. Molecular epidemiology of African swine fever in East Africa. Arch Virol. (2005) 150:2439–52. doi: 10.1007/s00705-005-0602-1
5. Costard S, Mur L, Lubroth J, Sanchez-Vizcaino JM, Pfeiffer DU. Epidemiology of African swine fever virus. Virus Res. (2013) 173:191–7. doi: 10.1016/j.virusres.2012.10.030
6. Malogolovkin A, Burmakina G, Titov I, Sereda A, Gogin A, Baryshnikova E, et al. Comparative analysis of African swine fever virus genotypes and serogroups. Emerg Infect Dis. (2015) 21:312–5. doi: 10.3201/eid2102.140649
7. Zhao DM, Liu RQ, Zhang XF, Li F, Wang JF, Zhang JW, et al. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg Microbes Infect. (2019) 8:438–47. doi: 10.1080/22221751.2019.1590128
8. Margarida S, Carlos M, Fernando F. Early intranuclear replication of African swine fever virus genome modifies the landscape of the host cell nucleus. Virus Res. (2015) 210:1–7. doi: 10.1016/j.virusres.2015.07.006
9. Sun EC, Huang LY, Zhang XF, Zhang JW, Shen D D, Zhang ZJ, et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg Microbes Infect. (2021) 10:2183–93. doi: 10.1080/22221751.2021.1999779
10. Oura, Edwards CAL, Batten LCA. Virological diagnosis of African swine fever-comparative study of available tests. Virus Res. (2013) 173:150–58. doi: 10.1016/j.virusres.2012.10.022
11. Abad JM, Pariente F, Hernández L, Lorenzo E. A quartz crystal microbalance assay for detection of antibodies against the recombinant African swine fever virus attachment protein p12 in swine serum. Anal Chim Acta. (1998) 368:183–9. doi: 10.1016/S0003-2670(98)00205-0
12. King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, et al. Development of a TaqMan® PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods. (2003) 107:53–61. doi: 10.1016/s0166-0934(02)00189-1
13. Tignon M, Gallardo C, Iscaro C, Hutet E, Van D, Stede Y, et al. Development and inter-laboratory validation study of an improved new real-time PCR assay with internal control for detection and laboratory diagnosis of African swine fever virus. J Virol Methods. (2011) 178:161–70. doi: 10.1016/j.jviromet.2011.09.007
14. Fernández-Pinero J, Gallardo C, Elizalde M, Robles A, Gómez C, Bishop R, et al. Molecular diagnosis of African swine fever by a new real-time PCR using universal probe library. Transbound Emerg Dis. (2003) 60:48–58. doi: 10.1111/j.1865-1682.2012.01317.x
15. AgüEro M, Fernández J, Romero L, Sánchez MC, Arias M, Sánchez-Vizcaíno JM. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J Clin Microbiol. (2003) 41:4431–4. doi: 10.1128/JCM.41.9.4431-4434.2003
16. Zsak L, Borca M V, Risatti GR, Zsak A, French RA, Lu Z, et al. Preclinical diagnosis of African swine fever in contact-exposed swine by a real-time PCR assay. J Clin Microbiol. (2005) 43:112–9. doi: 10.1128/JCM.43.1.112-119.2005
17. Wang A, Jia R, Liu Y, Zhou J, Qi Y, Chen Y, et al. Development of a novel quantitative real-time PCR assay with lyophilized powder reagent to detect African swine fever virus in blood samples of domestic pigs in China. Transbound Emerg Dis. (2020) 67:284–97. doi: 10.1111/tbed.13350
18. Wang Y, Xu L, Noll L, Stoy C, Porter E, Fu J, et al. Development of a real-time PCR assay for detection of African swine fever virus with an endogenous internal control. Transbound Emerg Dis. (2020) 67:2446–54. doi: 10.1111/tbed.13582
19. Gallardo C, Ademun AR, Nieto R, Nantima N, Arias M, Martín E, et al. Genotyping of African swine fever virus (ASFV) isolates associated with disease outbreaks in Uganda in 2007. Afr J Biotechnol. (2011) 10:3488–97. doi: 10.5897/ajb10.1439
20. Lubisi BA, Dwarka RM, Meenowa D, Jaumally R. An investigation into the first outbreak of African swine fever in the Republic of Mauritius. Transbound Emerg Dis. (2009) 56:178–88. doi: 10.1111/j.1865-1682.2009.01078.x
21. Nix RJ, Gallardo C, Hutchings G, Blanco E, Dixon L K. Molecular epidemiology of African swine fever virus studied by analysis of four variable genome regions. Arch Virol. (2006) 151:2475–94. doi: 10.1007/s00705-006-0794-z
23. Bastos AD, Penrith ML, Macome F, Pinto F, Thomson GR. Co-circulation of two genetically distinct viruses in an outbreak of African swine fever in Mozambique: no evidence for individual co-infection. Vet Microbiol. (2004) 103:169–82. doi: 10.1016/j.vetmic.2004.09.003
24. Carmina G, Dufton MM, Joseph MM, Marisa A, Evans AT, Alejandro S, et al. Enhanced discrimination of African swine fever virus isolates through nucleotide sequencing of the p54, p72, and pB602L (CVR) genes. Virus Genes. (2009) 38:85–95. doi: 10.1007/s11262-008-0293-2
25. Herman MC, Michihito S, Yona S, Gabriel G, David S, Paul F, et al. Evidence for exposure of asymptomatic domestic pigs to African swine fever virus during an inter-epidemic period in Zambia. Transbound Emerg Dis. (2020) 67:2741–52. doi: 10.1111/tbed.13630
26. Emmanuel CH, Kouame VK, Jenna EA, Leonce K, Yao MK, Hugues PG, et al. Re-emergence of genotype I of African swine fever virus in Ivory Coast. Transbound Emerg Dis. (2019) 66:882–96. doi: 10.1111/tbed.13098
Keywords: ASFV, dual real-time PCR, B646L gene, E183L gene, genotypes I and II
Citation: Gao Q, Feng Y, Yang Y, Luo Y, Gong T, Wang H, Gong L, Zhang G and Zheng Z (2022) Establishment of a Dual Real-Time PCR Assay for the Identification of African Swine Fever Virus Genotypes I and II in China. Front. Vet. Sci. 9:882824. doi: 10.3389/fvets.2022.882824
Received: 24 February 2022; Accepted: 25 April 2022;
Published: 01 June 2022.
Edited by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Fernando Costa Ferreira, University of Lisbon, PortugalKaichuang Shi, Guangxi Center for Animal Disease Control and Prevention, China
Copyright © 2022 Gao, Feng, Yang, Luo, Gong, Wang, Gong, Zhang and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guihong Zhang, Z3VpaG9uZ3poJiN4MDAwNDA7c2NhdS5lZHUuY24=; Zezhong Zheng, emV6aG9uZyYjeDAwMDQwO3NjYXUuZWR1LmNu
†These authors have contributed equally to this work
 Qi Gao1,2,3†
Qi Gao1,2,3† Heng Wang
Heng Wang Lang Gong
Lang Gong Guihong Zhang
Guihong Zhang Zezhong Zheng
Zezhong Zheng