- Department of Clinical Sciences, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
For both human and veterinary patients, non-infectious intestinal disease is a major cause of morbidity and mortality. To improve treatment of intestinal disease, large animal models are increasingly recognized as critical tools to translate the basic science discoveries made in rodent models into clinical application. Large animal intestinal models, particularly porcine, more closely resemble human anatomy, physiology, and disease pathogenesis; these features make them critical to the pre-clinical study of intestinal disease treatments. Previously, large animal model use has been somewhat precluded by the lack of genetically altered large animals to mechanistically investigate non-infectious intestinal diseases such as colorectal cancer, cystic fibrosis, and ischemia-reperfusion injury. However, recent advances and increased availability of gene editing technologies has led to both novel use of large animal models in clinically relevant intestinal disease research and improved testing of potential therapeutics for these diseases.
Introduction
Gastrointestinal disease accounts for over 3.0 million hospitalizations and over '135.9 billion in health care expenditures per year (1). To lessen this incredible burden to patients and our healthcare system, animal models play a critical role in discovery of intestinal disease pathogenesis and therapeutic innovation. For successful clinical translation, it is critical that animal models are properly validated. The criteria used to validate an animal model include certifying similarity in biology and clinical presentation between model and human disease (face validity), confirming that clinical interventions produce similar effects (predictive ability), and demonstrating that the target under investigation has a similar role in the model compared to human clinical disease (target validity) (2). Of intestinal disease animal models, rodents are historically preferred for use due to their low cost and maintenance, rapid reproduction, and readily available rodent-specific reagents. However, it is now widely recognized that rodents do not fully mimic human disease, physiology, immunology, or drug metabolism, thus limiting their use as pre-clinical models for disease treatment (3–7). For example, despite promising pre-clinical murine anti-cancer therapeutic studies, success rate of these therapeutics in human clinical trial is only around 5% (4, 8). Furthermore, the small size of rodents makes it difficult to model and advance surgical and endoscopic techniques. The differences between rodents and humans have left gaps in both basic science research and pre-clinical model development for intestinal diseases.
To better represent both human physiology and disease, and aid in the discovery of new treatments, porcine models are gaining popularity. With similar genome, size and architecture of the intestine, omnivorous diet, microbiome, immunology, and physiology to humans, pigs are increasingly the preferred model of enteric diseases (Table 1) (3, 6, 13, 17–23). The large size of the pig allows for multiple, longitudinal sampling from the same individual. Their large litter size of around 12 piglets allows for ease of gender and sibling matching. These attributes reduce both experimental variation as well as overall animal use. Additionally, for toxicology and drug discovery testing, pigs have oral and parenteral dosing rates similar to humans as well as similar responses to a variety of drug classes (24).
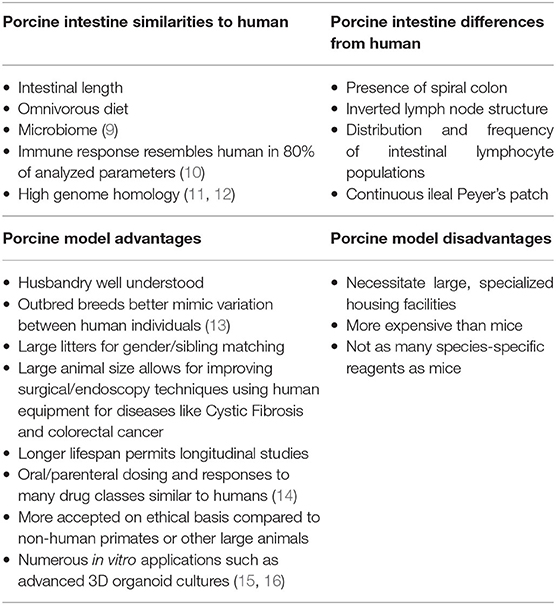
Table 1. Summary of comparisons between human and porcine intestinal physiology and anatomy as well as advantages and disadvantages of porcine models.
Grossly, both human and porcine adult intestine are at a ratio of 0.1 length per kilogram of body weight (25). The anatomy of the small intestine is similar between pig and human, though the large intestine varies slightly in the pig due to a larger cecum, lack of appendix, and the presence of the spiral colon (Figure 1) (3). Microscopically, for both species, the small and large intestine are comprised of a single layer of epithelial cells, interspersed with intra-epithelial lymphocytes, to serve as a barrier between luminal contents and systemic circulation. The single layer epithelium covers villus projections (present only in small intestine) and extends down into the crypts of Lieberkuhn. Located at the crypt-base are the intestinal epithelial stem cells (ISCs), which are responsible for renewing the epithelial cell populations on a continuous 3–5 day cycle (17, 27–29). In the human, the ISCs are interspersed with Paneth cells, a specialized secretory cell type that both supports ISC function and releases antimicrobial factors into the intestinal lumen (30, 31). While a similar cell population expressing the same biomarkers and intracellular structures has been identified in the pig, the porcine Paneth cell has yet to be fully defined (15, 32).
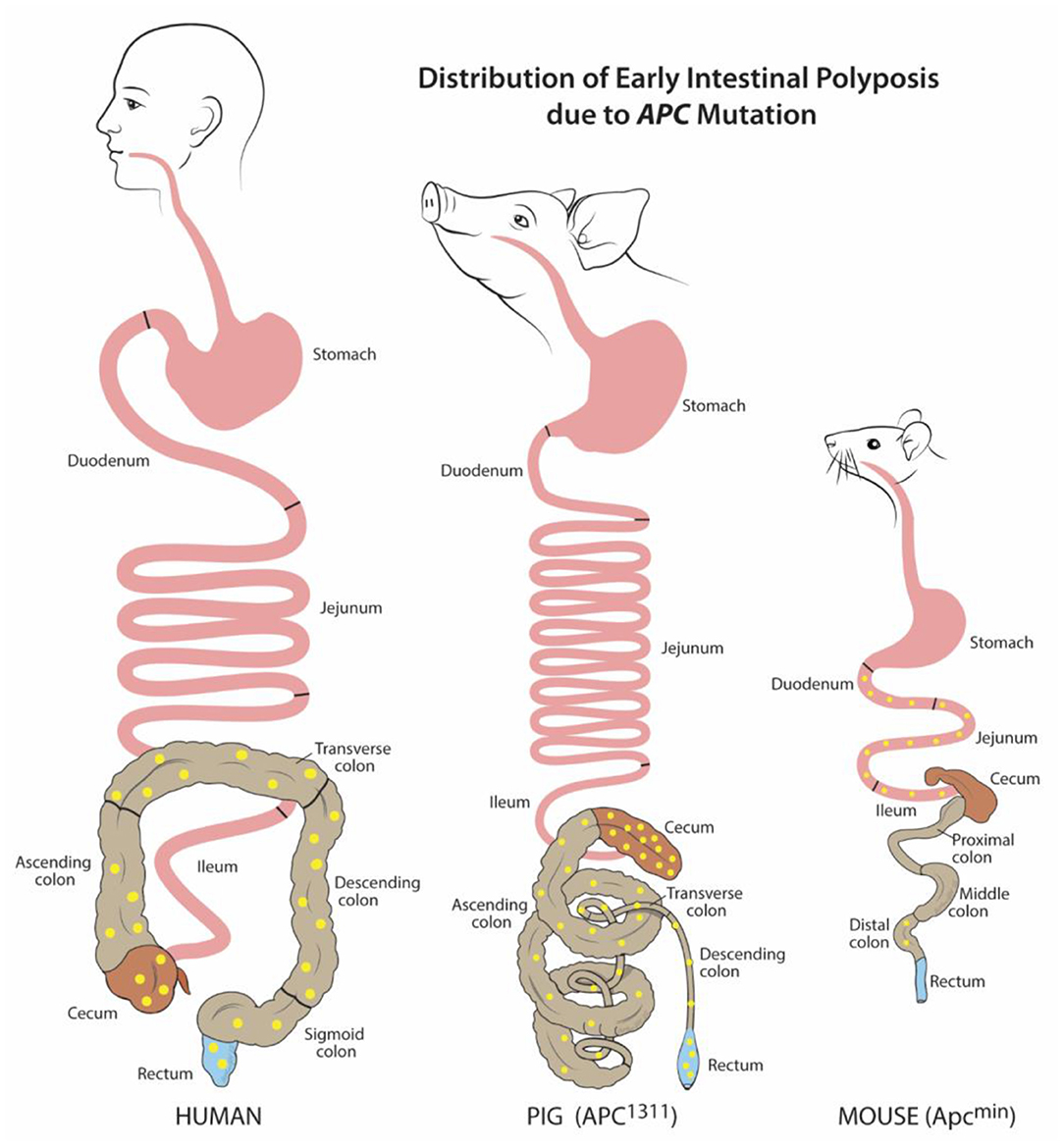
Figure 1. Distribution of early intestinal polyposis due to APC mutation. Human and porcine (APC1311) early adenoma/polyp distribution is similar within the colon and rectum, while murine (Apcmin) is mostly localized to the small intestine. Adenoma/polyps shown in yellow. Figure adapted from Gonzalez et al. and Flisikowska et al. (21, 26).
Beneath the epithelial barrier is the lamina propria compartment. While the lamina propria is made up of a mix of structural elements including blood and lymph vessels, connective tissue, and mesenchymal cells, immune cells constitute a major population, including dendritic cells and lymphocytes. These immune cells are responsible for discriminating between harmless luminal antigens and potential enteropathogens (33–35). Some differences exist between the immunologic organization between human and pig intestine such as the distribution and frequency of lamina propria and intraepithelial lymphocyte populations, the inverted structure of porcine peripheral and gut-associated lymph nodes, and the aggregated lymphoid follicles (Peyer's patches) which form one long continuous band in the porcine ileum (22, 29, 35). However, despite these differences, pigs are still recognized as useful models in several enteric immunologic studies including infectious disease, oral vaccination, small bowel transplantation, food hypersensitivity, and immune development (13, 22, 36–39).
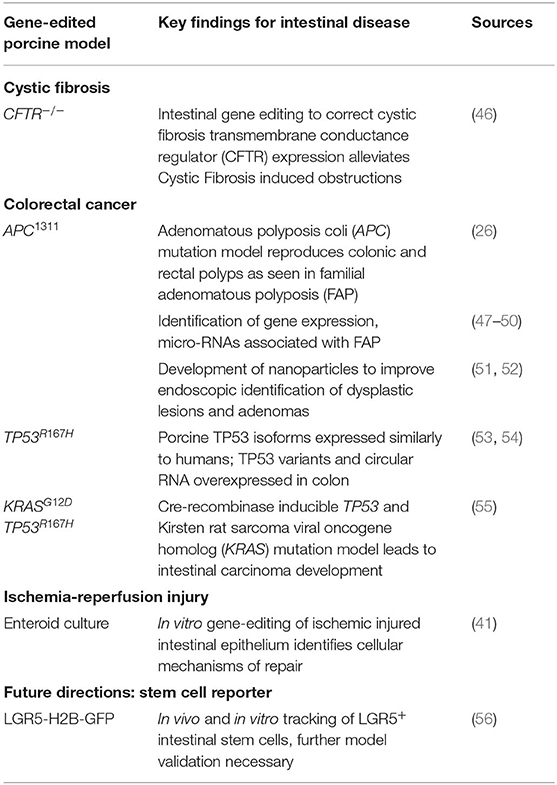
With these similarities in both physiology and architecture between human and porcine intestine (Table 1), porcine models of various intestinal injuries such as ischemia-reperfusion injury, intestinal transplantation, short gut syndrome, and necrotizing enterocolitis have progressed the field of gastroenterology (37, 40–44). Furthermore, in vitro advancements continue to broaden the utility of porcine enteric disease models. These advancements include an increase in porcine specific reagents and the use of primary intestinal epithelial cell culture in 2-D monolayers, 3-D organoid culture, and co-culture with microbes and lamina-propria derived cells to better understand intestinal barrier function (15, 16, 19, 20, 40, 41, 45). However, for more mechanistic studies and to better understand human genetic diseases in the wide array of intestinal maladies, advancements in porcine gene-edited models are needed. Fortunately, enhanced strategies to edit the porcine genome and develop transgenic models, as well as approaches to genetically modify porcine-derived intestinal organoids, have increased the availability of pre-clinical modeling for Cystic Fibrosis (CF), colorectal cancer (CRC), and ischemia-reperfusion injury (Table 2). This review summarizes the current use of pre-clinical, gene edited porcine intestinal disease and injury models and evaluates future additional needs to ultimately improve treatment of intestinal disease.
Gene Therapy in Porcine Cystic Fibrosis Models to Alleviate Intestinal Obstruction
Cystic Fibrosis (CF) is a life-threatening disease due to various mutations in the CF transmembrane conductance regulator (CFTR) gene (57). This critical gene encodes for an anion channel widely expressed in epithelium including lung, pancreas, kidney, and intestine; its loss of function inhibits chloride and bicarbonate transport across cell membranes. This leads to thick mucoid secretions with low pH, subsequent pathogen colonization, and dysregulated inflammation (57, 58). While cause of death in patients afflicted with CF is primarily due to respiratory failure, intestinal disease also contributes significantly to patient morbidity (59). Up to 20% of infants with CF suffer from meconium ileus at birth, followed by distal intestinal obstructive syndrome due to intestinal atresia, diverticulosis, and microcolon (46, 57). While genetically modified murine models of CF assisted in basic understanding of CFTR functions, these mice fail to fully recreate human clinical disease (59). These limitations in the mouse models impeded further discovery of disease pathogenesis and potential therapeutics.
In 2008, in an effort to develop an animal model that better represents CF clinical disease, Rogers et al. utilized recombinant adeno-associated virus (rAAV) vectors to create the first CFTR-null piglets (CFTR−/−). These piglets also demonstrated immediate clinical signs of intestinal obstruction similar to that in human infants with CF (60). These clinical signs include intestinal atresia, microcolon, and diverticulosis (57). Other gene editing strategies have since been used to generate CFTR−/− pigs including bacterial artificial chromosome vectors (61) as well as rAAVs to introduce a point mutation within the CFTR gene, CFTR-ΔF508 mutation (59, 62). This specific CFTR mutation is the most common CF-causing mutation in human patients – it accounts for about 70% of CF alleles (62). Murine models with this same induced point mutation fail to develop airway disease, pushing the need for a porcine model of this common mutation. While CFTR-ΔF508 porcine models display the same features of human disease in the lung and intestine, the rate of meconium ileus is 100% in the newborn pigs, in contrast to a rate of 20% in human infants (46). As in humans, meconium ileus requires immediate medical and/or surgical correction, which inhibited the use of the porcine model due to cost and complexity. These factors pushed researchers to utilize additional gene editing techniques to alleviate meconium ileus in the porcine model. Stoltz et al. were able to correct this phenotype by inducing CFTR expression under the control of intestinal fatty acid-binding protein (iFABP) in CFTR−/− pig fibroblasts (46). These findings indicated that correcting the expression of CFTR by gene editing in the intestine is sufficient to prevent intestinal obstruction. Further work in the porcine CF models is necessary to identify exactly how much CFTR function is required for proper intestinal function. With these findings, novel gene therapy approaches can be developed such as somatic tissue gene editing to restore endogenous CFTR function. It is critical that this research is done in a porcine model, similarly sized to humans, so that delivery of therapeutics can be modeled using human equipment such as endoscopy. The information gained from a translational porcine model of CF will lead to the development of innovative gene therapy treatment approaches to alleviate CF-induced intestinal obstruction.
Improved Modeling and Detection of Colorectal Cancer in Gene Edited Pigs
As of 2020 in the US, colorectal cancer (CRC) is the second leading cause of cancer related deaths for men and women combined (63). For many patients, including those with precursor familial adenomatous polyposis condition (FAP), CRC begins with germline or somatic mutations in the tumor-suppressor, adenomatous polyposis coli (APC) gene. This key driver mutation initiates polyp formation in the colonic epithelium. Subsequent compounding epigenetic changes and genetic mutations progresses tumorigenesis through the adenoma-carcinoma sequence, often culminating in metastatic cancer (64). For reasons unknown, CRC incidence has recently risen for young and middle-aged adults. Furthermore, many CRC tumor subtypes exist which remain without treatments (63). Without a doubt, there is a critical need for animal models to better understand CRC pathogenesis and develop targeted therapies. Attempts to recreate the polyp-adenoma-carcinoma pathogenesis sequence by mutating Apc (Apc+/min) in the mouse usually only leads to non-invasive, non-metastatic neoplasia. Furthermore, this neoplasia only typically occurs in the murine small intestine instead of the colon (Figure 1) (26, 65–67). Moreover, the small size of mice and critical differences in drug metabolism make these mice impractical for development of human CRC drug therapies, progress in endoscopic imaging techniques, or improving surgical interventions.
To overcome these barriers and create an improved, human-scale CRC model, the first gene-targeted APC mutated pig line was developed by inserting a translational stop signal at codon 1311 (APC1311). This mutation is orthologous to the germline mutations in patients with familial adenomatous polyposis condition (FAP) (26). In these genetically modified pigs, colonic, and rectal polyps and adenomas develop such as those found in human FAP and CRC patients (Figure 1) (68). Since their development, these pigs have contributed greatly to the discovery of epigenetic modifications, dysplastic polyp premalignant progression, and the function of genes other than APC that contribute to the varying severity and progression of disease in patients with FAP (47–49, 69). One pivotal study by Stachowiak et al., using APC1311 pigs, was the first to reveal that microRNAs are associated with premalignant transformation of colon polyps and can serve as potential useful biomarkers of disease development (50). Tan et al. attempted to replicate the APC mutation porcine model by transcription activator-like effector nucleases (TALENs) introduction of a stop signal at codon 902; however, these pigs have yet to develop CRC phenotype (70).
In addition to the APC1311 line, other mutated tumor suppressor gene porcine models are now available to study tumorigenesis. One such group carries a latent Cre-activated tumor protein p53 gene (TP53) mutated allele (TP53R167H) (53). This mutation is orthologous to the oncogenic human mutant TP53 allele that plays a role in numerous human cancers including CRC. Previously, studies using the APC1311 pigs reported that in severe cases of polyposis, there is an increase in expression of polymorphic TP53 (49). A more recent study using the TP53R167H pig demonstrated that these pigs express TP53 isoforms in a more similar manner to humans, further underscoring the benefits of porcine cancer models compared to murine. The TP53R167H porcine model showed that TP53 variants and circular RNA are overexpressed in the colon, indicating likely oncogenic function (54). These findings highlight the important role of porcine oncogenic models to improve our understanding of the genetic and epigenetic changes that contribute to CRC pathogenesis. Further developments on the TP53R167H model include the addition of inducible Kirsten rat sarcoma viral oncogene homolog (KRAS) mutation (55, 71). Mutated KRAS is present in over 25% of human tumors, including CRC, and is one of the more commonly activated oncogenes (72). Schook et al. showed that these Cre recombinase inducible KRASG12D TP53R167H transgenic pigs developed rapid and reproducible mesenchymal tumors. For more specific study of intestinal cancer, Callesen et al. refined the combined KRAS and TP53 mutation model by directing the control of the inducible recombination events under an intestinal epithelial specific gene promoter, which led to development of duodenal carcinoma. Further model establishment studies using the KRASG12D TP53R167H pigs are warranted to establish carcinoma development in the lower intestine in order to better understand CRC carcinogenesis in a translational large animal model.
While these gene-targeted porcine CRC models have served an important role for cellular mechanistic studies, they can also afford clinicians and researchers opportunities to improve endoscopy skills for minimally invasive surgery, conduct longitudinal sampling and monitoring during treatment, and enhance detection systems for pre-malignancies in a human-sized animal model. In general, porcine models have been popular for testing and advancing endoscopic techniques, particularly for colonoscopy (73–75). Early diagnostic detection of colon dysplasia and adenomas typically relies on white-light endoscopy (76). However, due to the subtle appearance of adenomas in situ, early-stage CRC lesions are often missed, especially in patients with abnormal colonic mucosa due to inflammatory bowel disease (51, 52). To improve real-time detection of colorectal adenomas, one group developed biodegradable near-infrared fluorescent silica nanoparticles (FSNs) (52). These FSNs, administered intravascularly, permeate into cancerous tissue and ‘mark’ a lesion because tumor and dysplasia-associated blood and lymph vessels are typically leaky (52, 77). Additionally, development of these biodegradable FSNs ensured no long-term sequestration within the body, as is typical of traditional nanoparticles (52). In that study, nanoparticle application was tested by administering FSNs to APC1311 pigs intravenously. Twenty-four hours later, colons were surveyed using near-infrared fluorescence-assisted white light endoscopy and adenomas were successfully highlighted by the FSNs (52). Since this study, additional work has successfully tested other probes in the APC1311 pigs to serve as CRC polyp markers (51). This pre-clinical application testing made possible by the APC1311 pigs was critical to develop new techniques to accurately identify clinically significant colorectal dysplasia in human patients.
Gene Editing in Porcine In vitro Models of Ischemia-Reperfusion Injury
Ischemic injury occurs when there is reduction or complete loss of blood flow to an organ. In the intestine, ischemic events can occur due to numerous pathologic events including thrombi, emboli, shock, cardiac insult, mechanical obstruction such as a hernia or intussusception, or necrotizing enterocolitis (21). In all of these disease states, the decrease in blood flow to the intestine diminishes the oxygen supply necessary for normal cellular metabolism. Cell damage and apoptosis quickly follows, particularly within the intestinal epithelium that is responsible for maintaining a critical barrier between harmful luminal microbes and systemic vasculature. Microbial translocation across a compromised epithelial barrier can develop into systemic inflammatory response syndrome, intestinal necrosis, and remote organ failure. Ultimately, this disease progression results in over 50% patient mortality (78). To lessen the high mortality rate, intestinal ischemia animal models are critical to better understand the pathophysiology of ischemic injury, identify factors driving epithelial repair, and develop potential therapeutics (40–42, 79).
The process of ischemia-induced epithelial cell loss, as described in numerous animal models, begins at the villus tip and progressively extends down to the crypt-base intestinal epithelial stem cell (ISC) compartment with increasing durations of ischemia (40, 80–83). In the ISC compartment, two populations of ISCs exist: active, proliferating ISCs that are sensitive to injury (aISCs) and quiescent, reserve ISCs that are injury resistant (rISCs). These two populations were first described using murine models (84, 85). However, small rodent models are unable to accurately represent severe human ischemic injury, likely due to differences in intestinal microvascular anatomy and overall small intestinal size (17). Thus, the impact of ischemic injury on these two ISC populations was largely undescribed until the introduction of a porcine surgical model of mesenteric vascular occlusion (16, 17). With similar sized intestine and more similar microvascular anatomy as humans, pigs make for a better model of intestinal ischemia (17, 86). Using the surgical porcine model of mesenteric vascular occlusion, researchers identified that severe ischemic injury differentially impacts the two known ISC populations: aISCs undergo apoptosis while rISCs are preserved and are likely responsible for epithelial recovery after injury (40). In this model, rISCs were identified in vivo by expression of the known ISC biomarker homeodomain only protein X (HOPX) (85). When ischemic-injured tissue, initially enriched in HOPX+ rISCs, was recovered in vivo for up to 3 days post injury, increased signs of crypt-base epithelial regeneration corresponded to a decrease in HOPX expression (41). Until this point, HOPX, a known tumor-suppressor gene in other cell types, had served as merely a biomarker of rISCs (87). To clarify the potential role of HOPX as a controller of ISC proliferation after severe ischemia, genetic modification of the porcine ischemic injury model was necessary.
Whole animal, genetically-modified porcine models have yet to be used in intestinal ischemia-reperfusion studies. However, recent advances in genetic modification of porcine intestinal crypt culture are a promising first step for more mechanistic studies. Culture techniques for porcine enteroids, 3D organoids derived from intestinal stem cells that recapitulate the intestinal epithelium, have been well described (15, 16, 28). Using lentivirus, Khalil et al. were the first group to genetically modify uninjured porcine intestinal crypts to create GFP expressing enteroids (88). To better understand epithelial recovery after ischemic injury and the specific role of HOPX function in epithelial crypt cells, Stewart et al. utilized adenovirus mediated transduction of short hairpin RNA to silence HOPX within ischemic injured crypt epithelium and showed that HOPX serves to suppress cellular proliferation in resultant enteroids (41). This novel advancement in porcine enteroids encourages future experiments of in vitro gene editing to improve our understanding of repair mechanisms in clinically relevant intestinal ischemia.
Future Directions: Development of Transgenic Porcine Intestinal Stem Cell Reporter Models
To improve studies of the cellular mechanisms involved in various types of intestinal injury and cancer, transgenic reporter porcine models are warranted for in vivo and in vitro cell tracking of intestinal cell populations. Intestinal epithelial stem cell reporter pigs are of particular interest as ISCs play critical roles in both the generation of colorectal cancer and in epithelial barrier regeneration during homeostasis and disease (89–91). With the development of high efficiency genome editing tools, one group generated a novel porcine cell reporter model via CRISPR-Cas9 insertion of fused histone 2B (H2B) to green fluorescent protein (GFP) under the ACTB locus (92). With successful ubiquitous nuclear expression of GFP demonstrated within these pigs, the same group went on to insert the H2B-GFP sequence under control of leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5), a known biomarker expressed by ISCs (LGR5-H2B-GFP) (56, 84). Histologic sections of colon demonstrated nuclear GFP in the crypt base cell populations in this novel model. However, to truly utilize this translational model and isolate ISCs for study of intestinal disease, further work is necessary to conclude that these GFP expressing cells are in fact LGR5+ ISCs. With this information, researchers would then have access to the first ever large animal ISC reporter, making way for improved translational studies of colorectal cancer development and intestinal barrier function and repair.
Future Directions: Transgenic Porcine Models of Inflammatory Bowel Disease
Inflammatory Bowel Disease (IBD) is a multifactorial disease that is typically categorized as Crohn's Disease (CD) or Ulcerative Colitis (UC). These syndromes are characterized by inflammation of the intestinal mucosa, influx of immune cells, and dysregulated cytokine production. Subsequently, patients suffer from episodes of abdominal pain, diarrhea, bloody stools, and weight loss (93). Historically, gene edited rodent models have been used to determine underlying etiologies and test therapeutic targets (94). Murine models of IBD include knock-outs of cytokines such as IL-10, TGF-β, IL-2, and IL-23. Loss of these cytokines disrupts regulation of inflammation in the intestine, leading to intestinal lesions similar to those seen in IBD (95–98). Mice have also been engineered to over express signals such as IL-7 or STAT4 to upregulate immune cell activity and induce IBD (99, 100). However, given the immunological differences known between man and mouse (5, 7), alternative models that better emulate human immune physiology are needed to test surgical interventions and pharmaceutical therapeutics. A host of factors have been identified to contribute to IBD including gut microbiota, environmental factors, and abnormal innate and adaptive immune responses (93). Pigs, with similar intestinal microbiota, immunology, and anatomy to humans, are the clear choice for IBD models (3, 6, 13, 18–23). Chemical-induction of IBD by dextran sulfate sodium (DSS) or trinitrobenzenesulfonic acid (TNBS) has been shown to reproduce intestinal lesions in the pig similar to those found in UC and CD, respectively (101–105). These models have been used to test advanced endoscopic techniques to correct strictures and supplemental amino acid therapy. To study IBD on a more mechanistic level within a translationally relevant large animal model, transgenic induced IBD porcine models, parallel to the IBD murine models, are needed to mimic the specific immune cell dysregulations seen with IBD.
Conclusions
The need for large animal models, particularly porcine, to improve pre-clinical intestinal disease translational research is well known. As described in this review, innovative applications of gene-edited porcine models of Cystic Fibrosis, colorectal cancer, and ischemia-reperfusion injury have progressed both the mechanistic understanding of disease pathophysiology as well as led to novel therapeutic treatment development. With continued improvement of gene-editing systems such as CRISPR/Cas, additional porcine models to track intestinal stem cells or simulate disorders such as Inflammatory Bowel Disease can be made available to further progress translational intestinal disease research.
Author Contributions
CS drafted and edited the manuscript. LG edited and revised the manuscript. CS and LG approved the final version of the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Peery AF, Crockett SD, Murphy CC, Lund JL, Dellon ES, Williams JL, et al. Burden and cost of gastrointestinal, liver, and pancreatic diseases in the United States: update 2018. Gastroenterology. (2019) 156:254–72.e11. doi: 10.1053/j.gastro.2018.08.063
2. Denayer T, Stöhr T, Van Roy M. Animal models in translational medicine: Validation and prediction. New Horiz. Transl Med. (2014) 2:5–11. doi: 10.1016/j.nhtm.2014.08.001
3. Kararli TT. Comparison of the gastrointestinal anatomy, physiology, and biochemistry of humans and commonly used laboratory animals. Biopharm Drug Dispos. (1995) 16:351–80. doi: 10.1002/bdd.2510160502
4. Mak IW, Evaniew N, Ghert M. Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res. (2014) 6:114–8.
5. Mestas J, Hughes CCW. Of mice and not men: differences between mouse and human immunology. J Immunol. (2004) 172:2731. doi: 10.4049/jimmunol.172.5.2731
6. Lunney Joan K, Van Goor A, Walker Kristen E, Hailstock T, Franklin J, Dai C. Importance of the pig as a human biomedical model. Sci Transl Med. (2021) 13:eabd5758. doi: 10.1126/scitranslmed.abd5758
7. Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci USA. (2013) 110:3507–12. doi: 10.1073/pnas.1222878110
8. Hutchinson L, Kirk R. High drug attrition rates—where are we going wrong? Nat Rev Clin Oncol. (2011) 8:189–90. doi: 10.1038/nrclinonc.2011.34
9. Heinritz SN, Mosenthin R, Weiss E. Use of pigs as a potential model for research into dietary modulation of the human gut microbiota. Nutr Res Rev. (2013) 26:191–209. doi: 10.1017/S0954422413000152
10. Dawson HD, Reece JJ, Urban JF Jr. A comparative analysis of the porcine, murine, and human immune systems. Vet Immunol Immunopathol. (2009) 128:309. doi: 10.1016/j.vetimm.2008.10.211
11. Hart EA, Caccamo M, Harrow JL, Humphray SJ, Gilbert JGR, Trevanion S, et al. Lessons learned from the initial sequencing of the pig genome: comparative analysis of an 8 Mb region of pig chromosome 17. Genome Biol. (2007) 8:R168. doi: 10.1186/gb-2007-8-8-r168
12. Malkas LH, Herbert BS, Waleed A, Dobrolecki LE, Liu Y, Agarwal B, et al. A cancer-associated PCNA expressed in breast cancer has implications as a potential biomarker. Proc Natl Acad Sci. (2006) 103:19472–7. doi: 10.1073/pnas.0604614103
13. Meurens F, Summerfield A, Nauwynck H, Saif L, Gerdts V. The pig: a model for human infectious diseases. Trends Microbiol. (2012) 20:50–7. doi: 10.1016/j.tim.2011.11.002
14. Schelstraete W, Clerck LD, Govaert E, Millecam J, Devreese M, Deforce D, et al. Characterization of porcine hepatic and intestinal drug metabolizing cyp450: comparison with human orthologues from a quantitative, activity and selectivity perspective. Sci Rep. (2019) 9:9233. doi: 10.1038/s41598-019-45212-0
15. van der Hee B, Loonen LMP, Taverne N, Taverne-Thiele JJ, Smidt H, Wells JM. Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res. (2018) 28:165–71. doi: 10.1016/j.scr.2018.02.013
16. Stewart AS, Freund JM, Blikslager AT, Gonzalez LM. Intestinal stem cell isolation and culture in a porcine model of segmental small intestinal ischemia. J Vis Exp. (2018) 135:57647. doi: 10.3791/57647
17. Nejdfors P, Ekelund M, Jeppsson B, Westrom BR. Mucosal in vitro permeability in the intestinal tract of the pig, the rat, and man: species- and region-related differences. Scand J Gastroenterol. (2000) 35:501–7. doi: 10.1080/003655200750023769
18. Käser T. Swine as biomedical animal model for T-cell research—Success and potential for transmittable and non-transmittable human diseases. Mol Immunol. (2021) 135:95–115. doi: 10.1016/j.molimm.2021.04.004
19. Dawson HD, Lunney JK. Porcine cluster of differentiation (CD) markers 2018 update. Res Vet Sci. (2018) 118:199–246. doi: 10.1016/j.rvsc.2018.02.007
20. Dawson HD, Sang Y, Lunney JK. Porcine cytokines, chemokines and growth factors: 2019 update. Res Vet Sci. (2020) 131:266–300. doi: 10.1016/j.rvsc.2020.04.022
21. Gonzalez LM, Moeser AJ, Blikslager AT. Porcine models of digestive disease: the future of large animal translational research. Transl Res. (2015) 166:12–27. doi: 10.1016/j.trsl.2015.01.004
22. Mair KH, Sedlak C, Käser T, Pasternak A, Levast B, Gerner W, et al. The porcine innate immune system: an update. Dev Comp Immunol. (2014) 45:321–43. doi: 10.1016/j.dci.2014.03.022
23. Nguyen TLA, Vieira-Silva S, Liston A, Raes J. How informative is the mouse for human gut microbiota research? Dis Model Mech. (2015) 8:1–16. doi: 10.1242/dmm.017400
24. Walters EM, Prather RS. Advancing swine models for human health and diseases. Mo Med. (2013) 110:212–5.
25. Patterson JK, Lei XG, Miller DD. The pig as an experimental model for elucidating the mechanisms governing dietary influence on mineral absorption. Exp Biol Med. (2008) 233:651–64. doi: 10.3181/0709-MR-262
26. Flisikowska T, Merkl C, Landmann M, Eser S, Rezaei N, Cui X, et al. A porcine model of familial adenomatous polyposis. Gastroenterology. (2012) 143:1173–5.e7. doi: 10.1053/j.gastro.2012.07.110
27. Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. Bioessays. (2002) 24:91–8. doi: 10.1002/bies.10028
28. Gonzalez LM, Williamson I, Piedrahita JA, Blikslager AT, Magness ST. Cell lineage identification and stem cell culture in a porcine model for the study of intestinal epithelial regeneration. PLoS ONE. (2013) 8:e66465. doi: 10.1371/journal.pone.0066465
29. Vega-López MA, Arenas-Contreras G, Bailey M, González-Pozos S, Stokes CR, Ortega MG, et al. Development of intraepithelial cells in the porcine small intestine. Dev Immunol. (2001) 8:147–58. doi: 10.1155/2001/25301
30. Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, et al. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. (2011) 469:415–8. doi: 10.1038/nature09637
31. Wehkamp J, Chu H, Shen B, Feathers RW, Kays RJ, Lee SK, et al. Paneth cell antimicrobial peptides: topographical distribution and quantification in human gastrointestinal tissues. FEBS Lett. (2006) 580:5344–50. doi: 10.1016/j.febslet.2006.08.083
32. Dekaney CM, Bazer FW, Jaeger LA. Mucosal morphogenesis and cytodifferentiation in fetal porcine small intestine. Anat Rec. (1997) 249:517–23.
33. Hunyady B, Mezey E, Palkovits M. Gastrointestinal immunology: cell types in the lamina propria–a morphological review. Acta Physiol Hung. (2000) 87:305–28. Available online at: http://europepmc.org/article/MED/11732886
34. Powell Anne E, Wang Y, Li Y, Poulin Emily J, Means Anna L, Washington Mary K, et al. The Pan-ErbB negative regulator lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. (2012) 149:146–58. doi: 10.1016/j.cell.2012.02.042
35. Burkey TE, Skjolaas KA, Minton JE, Board-Invited Review. Porcine mucosal immunity of the gastrointestinal tract1. J Anim Sci. (2009) 87:1493–501. doi: 10.2527/jas.2008-1330
36. Helm RM, Furuta GT, Stanley JS, Ye J, Cockrell G, Connaughton C, et al. A neonatal swine model for peanut allergy. J Allergy Clin Immunol. (2002) 109:136–42. doi: 10.1067/mai.2002.120551
37. Gruessner RWG, Nakhleh RE, Benedetti E, Pirenne J, Belani KG, Beebe D, et al. Combined liver—total bowel transplantation has no immunologic advantage over total bowel transplantation alone: a prospective study in a porcine model. Arch Surg. (1997) 132:1077–85. doi: 10.1001/archsurg.1997.01430340031004
38. Azevedo MP, Vlasova AN, Saif LJ. Human rotavirus virus-like particle vaccines evaluated in a neonatal gnotobiotic pig model of human rotavirus disease. Expert Rev Vaccines. (2013) 12:169–81. doi: 10.1586/erv.13.3
39. Stokes CR, Bailey M, Wilson AD. Immunology of the porcine gastrointestinal tract. Vet Immunol Immunopathol. (1994) 43:143–50. doi: 10.1016/0165-2427(94)90130-9
40. Gonzalez LM, Stewart AS, Freund J, Kucera CR, Dekaney CM, Magness ST, et al. Preservation of reserve intestinal epithelial stem cells following severe ischemic injury. Am J Physiol-Gastrointest Liver Physiol. (2019) 316:G482–G94. doi: 10.1152/ajpgi.00262.2018
41. Stewart AS, Schaaf CR, Luff JA, Freund JM, Becker TC, Tufts SR, et al. HOPX+ injury-resistant intestinal stem cells drive epithelial recovery after severe intestinal ischemia. Am J Physiol-Gastrointest Liver Physiol. (2021) 321:G588–602. doi: 10.1152/ajpgi.00165.2021
42. Yandza T, Tauc M, Saint-Paul MC, Ouaissi M, Gugenheim J, Hebuterne X. The pig as a preclinical model for intestinal ischemia-reperfusion and transplantation studies. J Surg Res. (2012) 178:807–19. doi: 10.1016/j.jss.2012.07.025
43. Llanos JC, Bakonyi Neto A, Lerco MM, Clark RMO. Polachini do Valle A, Sousa MMF. Induction of short gut syndrome and transplantation in a porcine model. Transplant Proc. (2006) 38:1855–6. doi: 10.1016/j.transproceed.2006.06.085
44. Sangild PT, Siggers RH, Schmidt M, Elnif J, Bjornvad CR, Thymann T, et al. Diet- and colonization-dependent intestinal dysfunction predisposes to necrotizing enterocolitis in preterm pigs. Gastroenterology. (2006) 130:1776–92. doi: 10.1053/j.gastro.2006.02.026
45. Cui T, Theuns S, Desmarets LMB, Xie J, De Gryse GMA, Yang B, et al. Establishment of porcine enterocyte/myofibroblast co-cultures for the growth of porcine rota- and coronaviruses. Sci Rep. (2018) 8:15195. doi: 10.1038/s41598-018-33305-1
46. Stoltz DA, Rokhlina T, Ernst SE, Pezzulo AA, Ostedgaard LS, Karp PH, et al. Intestinal CFTR expression alleviates meconium ileus in cystic fibrosis pigs. J Clin Invest. (2013) 123:2685–93. doi: 10.1172/JCI68867
47. Sikorska A, Stachowiak M, Flisikowska T, Stachecka J, Flisikowski K, Switonski M. Polymorphisms of CSF1R and WISP1 genes are associated with severity of familial adenomatous polyposis in APC(1311) pigs. Gene. (2020) 759:144988. doi: 10.1016/j.gene.2020.144988
48. Perkowska A, Flisikowska T, Perleberg C, Flisikowski K, Stachowiak M, Nowacka-Woszuk J, et al. The expression of TAP1 candidate gene, but not its polymorphism and methylation, is associated with colonic polyp formation in a porcine model of human familial adenomatous polyposis. Anim Biotechnol. (2020) 31:306–13. doi: 10.1080/10495398.2019.1590377
49. Sikorska A, Flisikowska T, Stachowiak M, Kind A, Schnieke A, Flisikowski K, et al. Elevated expression of p53 in early colon polyps in a pig model of human familial adenomatous polyposis. J Appl Genet. (2018) 59:485–91. doi: 10.1007/s13353-018-0461-6
50. Stachowiak M, Flisikowska T, Bauersachs S, Perleberg C, Pausch H, Switonski M, et al. Altered microRNA profiles during early colon adenoma progression in a porcine model of familial adenomatous polyposis. Oncotarget. (2017) 8:96154–60. doi: 10.18632/oncotarget.21774
51. Yim JJ, Harmsen S, Flisikowski K, Flisikowska T, Namkoong H, Garland M, et al. A protease-activated, near-infrared fluorescent probe for early endoscopic detection of premalignant gastrointestinal lesions. Proc Nat Acad Sci. (2021) 118:e2008072118. doi: 10.1073/pnas.2008072118
52. Rogalla S, Flisikowski K, Gorpas D, Mayer AT, Flisikowska T, Mandella MJ, et al. Biodegradable fluorescent nanoparticles for endoscopic detection of colorectal carcinogenesis. Adv Funct Mater. (2019) 29:1904992. doi: 10.1002/adfm.201904992
53. Leuchs S, Saalfrank A, Merkl C, Flisikowska T, Edlinger M, Durkovic M, et al. Inactivation and inducible oncogenic mutation of p53 in gene targeted pigs. PLoS ONE. (2012) 7:e43323. doi: 10.1371/journal.pone.0043323
54. Niu G, Hellmuth I, Flisikowska T, Pausch H, Rieblinger B, Carrapeiro A, et al. Porcine model elucidates function of p53 isoform in carcinogenesis and reveals novel circTP53 RNA. Oncogene. (2021) 40:1896–908. doi: 10.1038/s41388-021-01686-9
55. Callesen MM, Árnadóttir SS, Lyskjær I, Ørntoft M-BW, Høyer S, Dagnæs-Hansen F, et al. A genetically inducible porcine model of intestinal cancer. Mol Oncol. (2017) 11:1616–29. doi: 10.1002/1878-0261.12136
56. Polkoff KM, Chung J, Simpson SG, Gleason K, Piedrahita JA. In vitro validation of transgene expression in gene-edited pigs using CRISPR transcriptional activators. CRISPR J. (2020) 3:409–18. doi: 10.1089/crispr.2020.0037
57. Yan Z, Stewart ZA, Sinn PL, Olsen JC, Hu J, McCray PB, et al. Ferret and pig models of cystic fibrosis: prospects and promise for gene therapy. Hum Gene Ther Clin Dev. (2014) 26:38–49. doi: 10.1089/humc.2014.154
58. Zhou ZP, Yang LL, Cao H, Chen ZR, Zhang Y, Wen X-Y, et al. In vitro validation of a CRISPR-mediated CFTR correction strategy for preclinical translation in pigs. Hum Gene Ther. (2019) 30:1101–16. doi: 10.1089/hum.2019.074
59. Rogers CS, Hao Y, Rokhlina T, Samuel M, Stoltz DA Li Y, et al. Production of CFTR-null and CFTR-ΔF508 heterozygous pigs by adeno-associated virus–mediated gene targeting and somatic cell nuclear transfer. J Clin Invest. (2008) 118:1571–7. doi: 10.1172/JCI34773
60. Meyerholz DK, Stoltz DA, Pezzulo AA, Welsh MJ. Pathology of gastrointestinal organs in a porcine model of cystic fibrosis. Am J Pathol. (2010) 176:1377–89. doi: 10.2353/ajpath.2010.090849
61. Klymiuk N, Mundhenk L, Kraehe K, Wuensch A, Plog S, Emrich D, et al. Sequential targeting of CFTR by BAC vectors generates a novel pig model of cystic fibrosis. J Mol Med. (2012) 90:597–608. doi: 10.1007/s00109-011-0839-y
62. Ostedgaard Lynda S, Meyerholz David K, Chen J-H, Pezzulo Alejandro A, Karp Philip H, Rokhlina T, et al. The ΔF508 mutation causes CFTR misprocessing and cystic fibrosis–like disease in pigs. Sci Transl Med. (2011) 3:74ra24. doi: 10.1126/scitranslmed.3001868
63. Siegel RL, Miller KD, Goding Sauer A, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin. (2020) 70:145–64. doi: 10.3322/caac.21601
64. Huang D, Sun W, Zhou Y, Li P, Chen F, Chen H, et al. Mutations of key driver genes in colorectal cancer progression and metastasis. Cancer Metastasis Rev. (2018) 37:173–87. doi: 10.1007/s10555-017-9726-5
65. Boivin GP, Washington K, Yang K, Ward JM, Pretlow TP, Russell R, et al. Pathology of mouse models of intestinal cancer: consensus report and recommendations. Gastroenterology. (2003) 124:762–77. doi: 10.1053/gast.2003.50094
66. Stastna M, Janeckova L, Hrckulak D, Kriz V, Korinek V. Human colorectal cancer from the perspective of mouse models. Genes. (2019) 10:788. doi: 10.3390/genes10100788
67. Su L-K, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. (1992) 256:668–70. doi: 10.1126/science.1350108
68. Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. (2012) 487:330–7. doi: 10.1038/nature11252
69. Flisikowska T, Stachowiak M, Xu H, Wagner A, Hernandez-Caceres A, Wurmser C, et al. Porcine familial adenomatous polyposis model enables systematic analysis of early events in adenoma progression. Sci Rep. (2017) 7:6613. doi: 10.1038/s41598-017-06741-8
70. Tan W, Carlson Daniel F, Lancto Cheryl A, Garbe John R, Webster Dennis A, Hackett Perry B, et al. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Nat Acad Sci. (2013) 110:16526–31. doi: 10.1073/pnas.1310478110
71. Schook LB, Collares TV, Hu W, Liang Y, Rodrigues FM, Rund LA, et al. A Genetic Porcine Model of Cancer. PLoS ONE. (2015) 10:e0128864. doi: 10.1371/journal.pone.0128864
72. Beganoyic S. Clinical significance of the kras mutation. Bosn J Basic Med Sci. (2009) 9:S17–20. doi: 10.17305/bjbms.2009.2749
73. Obstein KL, Valdastri P. Advanced endoscopic technologies for colorectal cancer screening. World J Gastroenterol. (2013) 19:431–9. doi: 10.3748/wjg.v19.i4.431
74. Choi WJ, Moon J-H, Min JS, Song YK, Lee SA, Ahn JW, et al. Real-time detection system for tumor localization during minimally invasive surgery for gastric and colon cancer removal: in vivo feasibility study in a swine model. J Surg Oncol. (2018) 117:699–706. doi: 10.1002/jso.24922
75. Ciuti G, Skonieczna-Zydecka K, Marlicz W, Iacovacci V, Liu H, Stoyanov D, et al. Frontiers of robotic colonoscopy: a comprehensive review of robotic colonoscopes and technologies. J Clin Med. (2020) 9:1648. doi: 10.3390/jcm9061648
76. Winawer S, Fletcher R, Rex D, Bond J, Burt R, Ferrucci J, et al. Colorectal cancer screening and surveillance: clinical guidelines and rationale-update based on new evidence. Gastroenterology. (2003) 124:544–60. doi: 10.1053/gast.2003.50044
77. Jiang T, Jin K, Liu X, Pang Z. 8-Nanoparticles for tumor targeting. In: Jana S, Maiti S, Jana S, editors. Biopolymer-Based Composites. Cambridge, MA: Woodhead Publishing (2017). p. 221–67.
78. Reginelli A, Genovese EA, Cappabianca S, Iacobellis F, Berritto D, Fonio P, et al. Intestinal ischemia: US-CT findings correlations. Crit Ultrasound J. (2013) 5:S7. doi: 10.1186/2036-7902-5-S1-S7
79. Nolan LS, Wynn JL, Good M. Exploring clinically-relevant experimental models of neonatal shock and necrotizing enterocolitis. Shock. (2020) 53:596–604. doi: 10.1097/SHK.0000000000001507
80. Chiu C-J, McArdle AH, Brown R, Scott HJ, Gurd FN. Intestinal Mucosal Lesion in Low-Flow States: I. A morphological, hemodynamic, and metabolic reappraisal. Arch Surg. (1970) 101:478–83. doi: 10.1001/archsurg.1970.01340280030009
81. Park PO, Haglund U, Bulkley GB, Fält K. The sequence of development of intestinal tissue injury after strangulation ischemia and reperfusion. Surgery. (1990) 107:574–80.
82. Parks DA, Granger DN. Ischemia-induced vascular changes: role of xanthine oxidase and hydroxyl radicals. Am J Physiol Gastrointest Liver Physiol. (1983) 245:G285–G9. doi: 10.1152/ajpgi.1983.245.2.G285
83. Robinson JW, Mirkovitch V, Winistörfer B, Saegesser F. Response of the intestinal mucosa to ischaemia. Gut. (1981) 22:512–27. doi: 10.1136/gut.22.6.512
84. Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. (2007) 449:1003–7. doi: 10.1038/nature06196
85. Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. (2011) 334:1420. doi: 10.1126/science.1213214
86. Bellamy JE, Latshaw WK, Nielsen NO. The vascular architecture of the porcine small intestine. Can J Comp Med. (1973) 37:56–62.
87. Katoh H, Yamashita K, Waraya M, Margalit O, Ooki A, Tamaki H, et al. Epigenetic silencing of HOPX promotes cancer progression in colorectal cancer. Neoplasia. (2012) 14:55971. doi: 10.1593/neo.12330
88. Khalil HA, Lei NY, Brinkley G, Scott A, Wang J, Kar UK, et al. A novel culture system for adult porcine intestinal crypts. Cell Tissue Res. (2016) 365:123–34. doi: 10.1007/s00441-016-2367-0
89. Potten CS, Hume WJ, Reid P, Cairns J. The segregation of DNA in epithelial stem cells. Cell. (1978) 15:899–906. doi: 10.1016/0092-8674(78)90274-X
90. Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. (2009) 457:608–11. doi: 10.1038/nature07602
91. Baker A-M, Graham TA, Elia G, Wright NA, Rodriguez-Justo M. Characterization of LGR5 stem cells in colorectal adenomas and carcinomas. Sci Rep. (2015) 5:8654. doi: 10.1038/srep08654
92. Sper RB, Koh S, Zhang X, Simpson S, Collins B, Sommer J, et al. Generation of a stable transgenic swine model expressing a porcine histone 2B-eGFP fusion protein for cell tracking and chromosome dynamics studies. PloS ONE. (2017) 12:e0169242–e. doi: 10.1371/journal.pone.0169242
93. Fiocchi C. Inflammatory bowel disease: etiology and pathogenesis. Gastroenterology. (1998) 115:182–205. doi: 10.1016/S0016-5085(98)70381-6
94. Prattis S, Jurjus A. Spontaneous and transgenic rodent models of inflammatory bowel disease. Lab Anim Res. (2015) 31:47–68. doi: 10.5625/lar.2015.31.2.47
95. Lees CW, Barrett JC, Parkes M, Satsangi J. New IBD genetics: common pathways with other diseases. Gut. (2011) 60:1739. doi: 10.1136/gut.2009.199679
96. Baumgart DC, Olivier W-A, Reya T, Peritt D, Rombeau JL, Carding SR. Mechanisms of intestinal epithelial cell injury and colitis in interleukin 2 (IL2)-deficient mice. Cell Immunol. (1998) 187:52–66. doi: 10.1006/cimm.1998.1307
97. Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. (1992) 359:693–9. doi: 10.1038/359693a0
98. Berg DJ, Davidson N, Kühn R, Müller W, Menon S, Holland G, et al. Enterocolitis and colon cancer in interleukin-10-deficient mice are associated with aberrant cytokine production and CD4(+) TH1-like responses. J Clin Invest. (1996) 98:1010–20. doi: 10.1172/JCI118861
99. Simpson SJ, Shah S, Comiskey M, de Jong YP, Wang B, Mizoguchi E, et al. T Cell–mediated pathology in two models of experimental colitis depends predominantly on the interleukin 12/signal transducer and activator of transcription (Stat)-4 Pathway, but is not conditional on interferon γ expression by T cells. J Exp Med. (1998) 187:1225–34. doi: 10.1084/jem.187.8.1225
100. Watanabe M, Ueno Y, Yajima T, Okamoto S, Hayashi T, Yamazaki M, et al. Interleukin 7 transgenic mice develop chronic colitis with decreased interleukin 7 protein accumulation in the colonic mucosa. J Exp Med. (1998) 187:389–402. doi: 10.1084/jem.187.3.389
101. Lukas M, Kolar M, Ryska O, Juhas S, Juhasova J, Kalvach J, et al. Novel porcine model of Crohn's disease anastomotic stricture suitable for evaluation and training of advanced endoscopic techniques. Gastrointest Endosc. (2021) 93:250–6. doi: 10.1016/j.gie.2020.05.063
102. Ziv Y, Nevler A, Willenz E, Doron O, Zbar A, Shperber A, et al. A novel porcine model for chemically inducible Crohn's-like reaction. Isr Med Assoc J. (2015) 17:19–23. Available online at: http://europepmc.org/abstract/MED/25739171
103. Kim CJ, Kovacs-Nolan J, Yang C, Archbold T, Fan MZ, Mine Y. L-cysteine supplementation attenuates local inflammation and restores gut homeostasis in a porcine model of colitis. Biochim Biophys Acta. (2009) 1790:1161–9. doi: 10.1016/j.bbagen.2009.05.018
104. Kim CJ, Kovacs-Nolan JA, Yang C, Archbold T, Fan MZ, Mine Y. l-Tryptophan exhibits therapeutic function in a porcine model of dextran sodium sulfate (DSS)-induced colitis. J Nutr Biochem. (2010) 21:468–75. doi: 10.1016/j.jnutbio.2009.01.019
Keywords: cystic fibrosis, colorectal cancer, ischemia-reperfusion injury, genetically altered models, intestinal disease, translational porcine model, stem cell reporter model
Citation: Schaaf CR and Gonzalez LM (2022) Use of Translational, Genetically Modified Porcine Models to Ultimately Improve Intestinal Disease Treatment. Front. Vet. Sci. 9:878952. doi: 10.3389/fvets.2022.878952
Received: 18 February 2022; Accepted: 27 April 2022;
Published: 20 May 2022.
Edited by:
Stefano Guido, University of Edinburgh, United KingdomReviewed by:
Christopher K. Tuggle, Iowa State University, United StatesFrançois J. M. A. Meurens, Agroalimentaire et de l'alimentation de Nantes-Atlantique (Oniris), France
Copyright © 2022 Schaaf and Gonzalez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liara M. Gonzalez, bG1nb256YTRAbmNzdS5lZHU=
 Cecilia R. Schaaf
Cecilia R. Schaaf Liara M. Gonzalez
Liara M. Gonzalez