- 1School of Agriculture, Ningxia University, Yinchuan, China
- 2Key Laboratory of Ruminant Molecular Cell Breeding, Ningxia Hui Autonomous Region, Yinchuan, China
Healthy mammary gland is essential for milk performance in dairy cows. MicroRNAs (miRNAs) are the key molecules to regulate the steady state of mammary gland in dairy cows. This study investigated the potential role of miR-29c in bovine mammary epithelial cells (bMECs). RNA sequencing (RNA-seq) was used to measure the transcriptome profile of bovine mammary epithelial cells line (MAC-T) transfected with miR-29c inhibitor or negative control (NC) inhibitor, and then differentially expressed genes (DEGs) were screened. The results showed that a total of 42 up-regulated and 27 down-regulated genes were found in the miR-29c inhibitor group compared with the NC inhibitor group. The functional enrichment of the above DEGs indicates that miR-29c is a potential regulator of oxidative stress and inflammatory response in bMECs through multiple genes, such as forkhead box O1 (FOXO1), tumor necrosis factor-alpha (TNF-α), and major histocompatibility complex, class II, DQ alpha 5 (BoLA-DQA5) in the various biological process and signaling pathways of stress-activated mitogen-activated protein kinase (MAPK) cascade, Epstein-Barr virus infection, inflammatory bowel disease, etc. The results imply that miR-29c plays an important role in a steady state of bMECs or cow mammary gland and may be a potential therapeutic target for mastitis in dairy cows.
Introduction
The bovine mammary epithelial cells (bMECs) are the most important cell group of mammary tissues, which act in milk synthesis and innate immunity (1). When the cow mammary gland is exposed to exogenous pathogenic bacteria, bMECs are stimulated to produce reactive oxygen species (ROS), nitric oxide (NO), and pro-inflammatory cytokines, thereby initiating oxidative stress and inflammatory response (2, 3). In the process of dairy farming, the high incidence of mastitis affects the health and economic benefits of dairy cows (4).
MicroRNAs (miRNAs) are small non-coding RNA with a length of about 22 nt that can inhibit the expression levels of genes by targeting the 3' untranslated region (3' UTR) of the messenger RNAs (mRNAs), thereby regulating the biological processes of humans and other animals (5, 6). Previous studies observed that miRNAs could regulate the immunity, proliferation and apoptosis of bMECs. miR-21-3p promotes triglyceride synthesis of bMECs through direct and targeted regulation of elongation-of-very-long-chain (ELOVL) fatty acid elongase 5 (ELOVL5) expression (7). miR-24-3p targets multiple endocrine neoplasia type 1 (MEN1)/menin in bovine mammary epithelial cells line (MAC-T) and regulates cell proliferation and milk protein synthesis in the form of negative feedback (8). miR-146a targets the toll-like receptor 4 (TLR4)/tumor necrosis factor receptor (TNFR)-associated factor 6 (TRAF6)/nuclear factor kappa B (NF-κB) pathway and regulates lipopolysaccharide (LPS)-induced inflammatory response in bMECs by a negative feedback mechanism (9). miR-29 family regulates the synthesis and secretion of milk components, cell proliferation, and apoptosis by targeting DNA-methyltransferase 3A/3B (DNMT-3A/-3B) in bMECs (10). In mice and humans, the miR-29 family is a series of small RNA molecules closely related to inflammation or apoptosis (11–13), and miR-29c can target leukemia inhibitory factor (LIF) in primary intestinal epithelial cells (ICEs) and is involved in the regulation of proliferation, apoptosis, and immune function, thereby regulating Ulcerative colitis (UC) (14). Previous study found that the expression of miR-29c was down-regulated in mammary tissues samples of cows with Escherichia coli-induced mastitis (15) and cows with clinical mastitis (16). However, there is no report on the involvement of miR-29c in bMECs immune response regulation.
In this study, RNA sequencing (RNA-seq), Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were used to analyze the effects of miR-29c inhibition on the transcriptome profile of bMECs to explore the role of miR-29c in bMECs.
Materials and Methods
Cell Culture and Identification
The previously stored MAC-T cells (17) in liquid nitrogen were resuscitated and cultured with 10 mL Dulbecco's Modified Eagle Medium/Nutrient Mixture F-12 (DMEM/F12) medium (Hyclone, UT, USA) containing 10% fetal bovine serum (System Biosciences, Mountain View, CA, USA) in a 5% CO2 humidified incubator at 37°C and then passaged every 2 days.
MAC-T cells were identified by immunofluorescence analysis with epithelial marker cytokeratin 18 (18). The cells were inoculated in a six-well plate (NEST, Wuxi, Jiangsu, China). When the cell confluence was 80%, the medium was discarded. The cells were rinsed three times with PBS, and fixed at room temperature for 20 min with pre-cooled 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA), then added 0.5% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA), and incubated at room temperature for 5 min. After blocking for 1 h at room temperature in 2% BSA. the cells were incubated with primary antibody for cytokeratin 18 (1:500, Santa Cruz, Dallas, TX, USA) overnight at 4°C. The cells was incubated with Cy3-labeled goat anti-mouse IgG (1:200, Beyotime, Shanghai, China) for 1 h at room temperature, then incubated with DAPI (Beyotime, Shanghai, China) for 10 min at room temperature, followed by seal the coverslip and imaging under fluorescence inverted microscope (Olympus, Shinjuku-ku, Tokyo, Japan).
Cell Transfection
The miR-29c inhibitor and NC (negative control) inhibitor were designed and synthesized by Guangzhou RiboBio Co., Ltd. (Guangzhou, China). MAC-T cells were inoculated into a six-well cell culture plate (NEST, Wuxi, Jiangsu, China), a fresh medium was added before transfection when the cell confluence was 60~70%. X-treme GENE™ HP DNA Transfection Reagent (Roche, Basel, Switzerland) was used for instantaneous transfection of miR-29c inhibitor (100 nM) or NC inhibitor (100 nM) according to the manufacturer's instructions. The transfection efficiency was observed under fluorescence inverted microscope (Olympus, Shinjuku-ku, Tokyo, Japan) and detected by quantitative real-time polymerase chain reaction (qRT-PCR) of miR-29c in the transfected cells. The MAC-T cells were harvested at 48 h after transfection for RNA extraction.
Construction of Transcriptome Libraries and Sequencing
Total RNA of 6 MAC-T cells samples, including 3 replicates of miR-29c inhibitor transfected cells and 3 replicates of NC inhibitor transfected cells, were extracted by TRIzol reagent (Takara, Beijing, China) for constructing transcriptome libraries. The integrity of the RNA samples were detected by agarose gel electrophoresis, and the purity was detected by Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA). The optical density (OD260/OD280) of all RNA samples ranged from 1.8 to 2.0, indicating that the RNA samples were qualified. Then, 1 μg total RNA was used to construct transcriptome library by ABclonal mRNA-seq Lib Prep Kit (ABclonal, Wuhan, Hubei, China), and mRNA was enriched with magnetic beads with oligo (dT). mRNA was broken into short fragments by fragmentation buffer and used as a template. First-strand of complementary DNA (cDNA) was synthesized by random hexamers, and then the second-strand of cDNA was synthesized by adding buffer, deoxynucleotide triphosphate (dNTPs), DNA polymerase I, and RNase H. Then, AMPure XP beads were used to purify double-stranded cDNA. Purified double-stranded cDNA ends were repaired. A-tails were added, and sequencing adapters were connected. Then, AMPure XP beads were used to select segment sizes. Polymerase chain reaction (PCR) amplification was performed, and AMPure XP beads were used to purify the PCR products to get the final library. After the library's construction, the insert size and effective concentration of the library were detected to ensure the quality of the library. After the quality inspection, the 6 libraries were sequenced on Illumina Novaseq 6000 (Illumina, Inc., San Diego, CA, USA) high-throughput platform in shanghai applied protein technology co. ltd. (Shanghai, China).
Quality Control and Mapping of Sequencing Data
After high-throughput sequencing, raw data were stored in FASTQ file format. Clean reads were obtained after the quality control of raw reads by Perl, such as removal of adapters sequence, filtering out low-quality reads and filtering out reads with N ratio >5%. Clean reads were compared to mapped reads in the Bos taurus reference genome UMD3.1 (http://oct2018.archive.ensembl.org/Bos_taurus/Info/Index) using HiSAT2 software (http://daehwankimlab.github.io/hisat2/).
Differential Expression Analysis of Genes
The expression level of each gene in each sample was calculated using FeatureCounts software (http://subread.sourceforge.net/). The gene expression level was expressed as FPKM (number of bases per million compared segments to transcript per thousand) (19). DESeq2 (http://bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to compare the genes expression levels of miR-29c inhibitor and NC-inhibitor groups, with P < 0.05 & | log | > 1 as the standard significance of the difference between the groups.
Reverse Transcription and qRT-PCR
The reverse transcription reactions of miR-29c and mRNAs were carried out by stem-loop structure primer (20) (Supplementary Table S1) and the mixture of Oligo (dT) primers and random 6 mers, respectively. All reverse transcription reactions were performed using 1 μg total RNA as the template and using PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Biomedical Technology Co., Ltd., Beijing, China) according to the manufacturer's instructions.
qRT-PCR was used to detect the transfection efficiency of miR-29c and validate the differentially expressed genes (DEGs) obtained by RNA-seq. qRT-PCR was performed using 2× M5 HiPer SYBR Premix EsTaq plus kit (Mei5 Biotechnology Co., Ltd., Beijing, China) on a CFX96 assay system (Bio-Rad Laboratories, Inc., CA, USA) in a 20 μL reaction system, including 10 μL 2 × M5 HiPer SYBR Premix Es Taq (with Til RNaseH), 0.8 μL 10 μmol/L of each of forward and reverse primers, 2.0 μL cDNA (100 ng/μL) and 6.4 μL RNase-free ddH2O. The amplification conditions were as follows: pre-denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and annealing/extension at 60°C for 30 s. The primers designed with Primer Premier 5 (PREMIER Biosoft International, Palo Alto, CA, USA) are presented in Supplementary Table S1.
The relative expression levels of miR-29c and DEGs were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and ribosomal protein S18 (RPS18) (21), and the data were calculated using the 2−ΔΔCT method (22). A significant difference was performed using SPSS 25.0 Student's t-tests to determine the statistical significance of the two groups, and P < 0.05 indicated significant difference, and P < 0.01 indicated extremely significant difference. All data were expressed as mean ± standard error of the mean (SEM).
Functional Annotation of DEGs
The R package of clusterProfiler (version: 4.0.5) (23) was used for GO and KEGG enrichment analysis of DEGs, and terms with a P < 0.05 were considered significantly enriched.
Results
Detection of miR-29c Silencing Effect in MAC-T Cells
The identification of MAC-T cells was performed by detecting the expression of epithelial cell-specific cytokeratin 18. The results showed that cytokeratin 18 was expressed in all cells (Supplementary Figure S1), suggested that the MAC-T cells used in this study were reliable.
After MAC-T cells were transfected with a Cy3-labeled NC inhibitor and incubated for 24 h, red fluorescence in MAC-T cells were clearly observed (Figure 1A). The qRT-PCR results showed that the expression of miR-29c was significantly down-regulated in miR-29c inhibitor group compared with the NC inhibitor group (P < 0.01, Figure 1B). The above results indicated that the inhibitor successfully inhibited the expression of miR-29c in MAC-T cells.
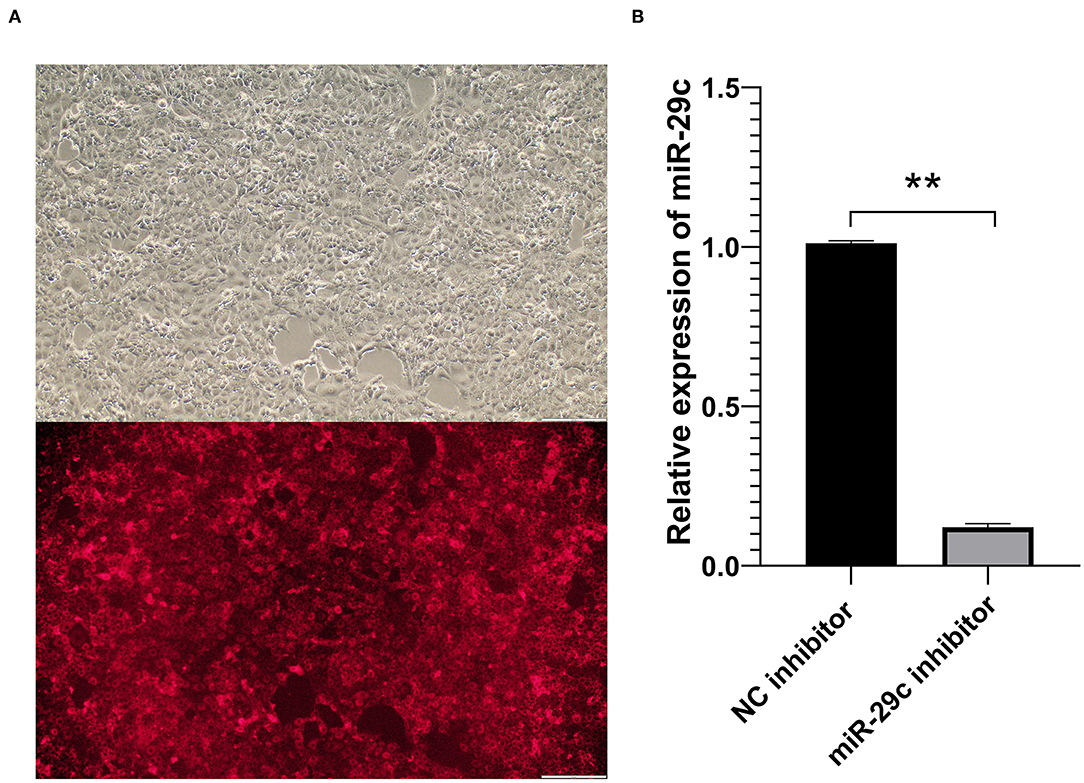
Figure 1. Transfection efficiency detection of bovine miR-29c in MAC-T cells. (A) Transfection efficiency of NC inhibitor in MAC-T cells observed by fluorescence microscopy. (B) The transfection efficiency of bovine miR-29c inhibitor in MAC-T cells were measured by qRT-PCR. Data are presented as mean ± SEM. **P < 0.01.
RNA-Seq Summary Statistics
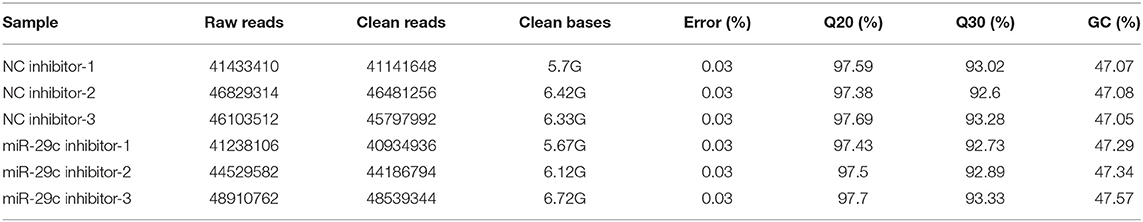
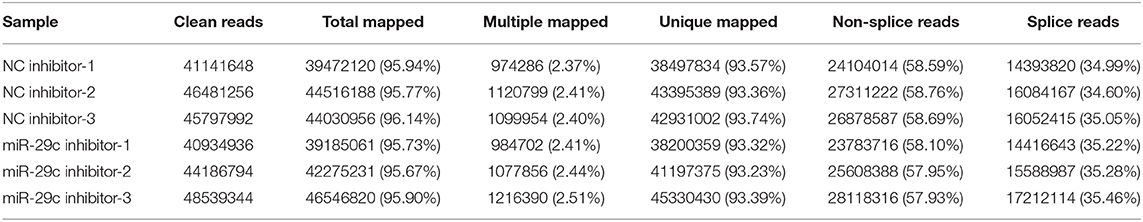
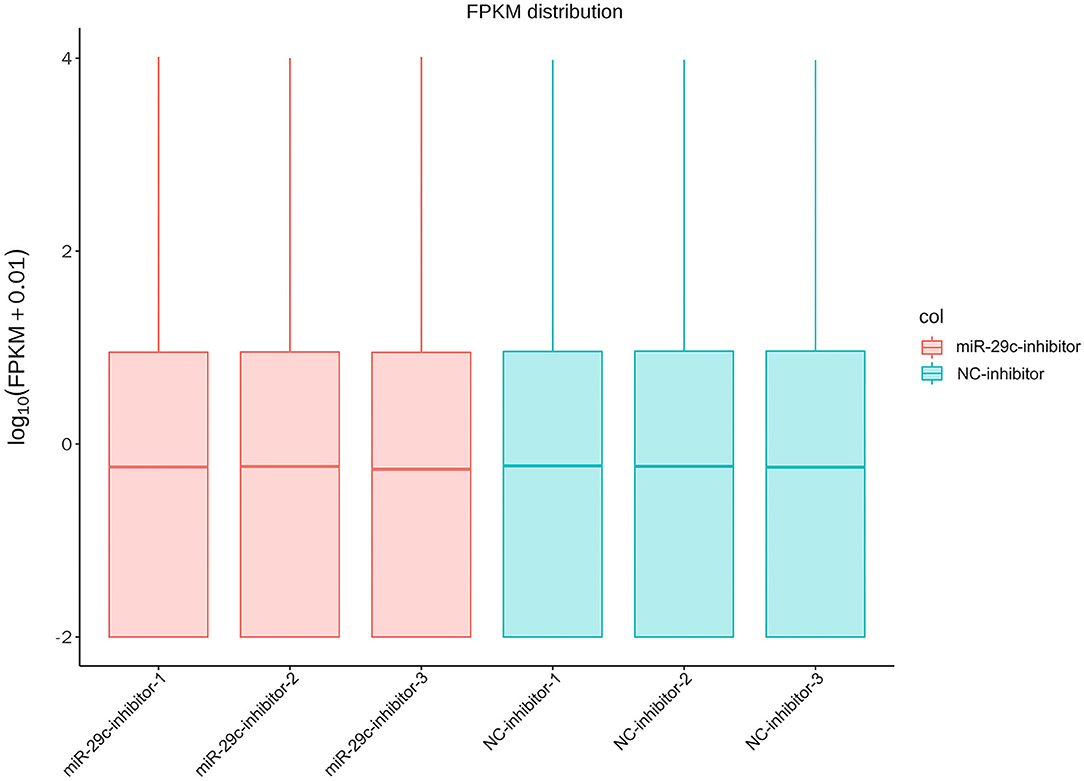
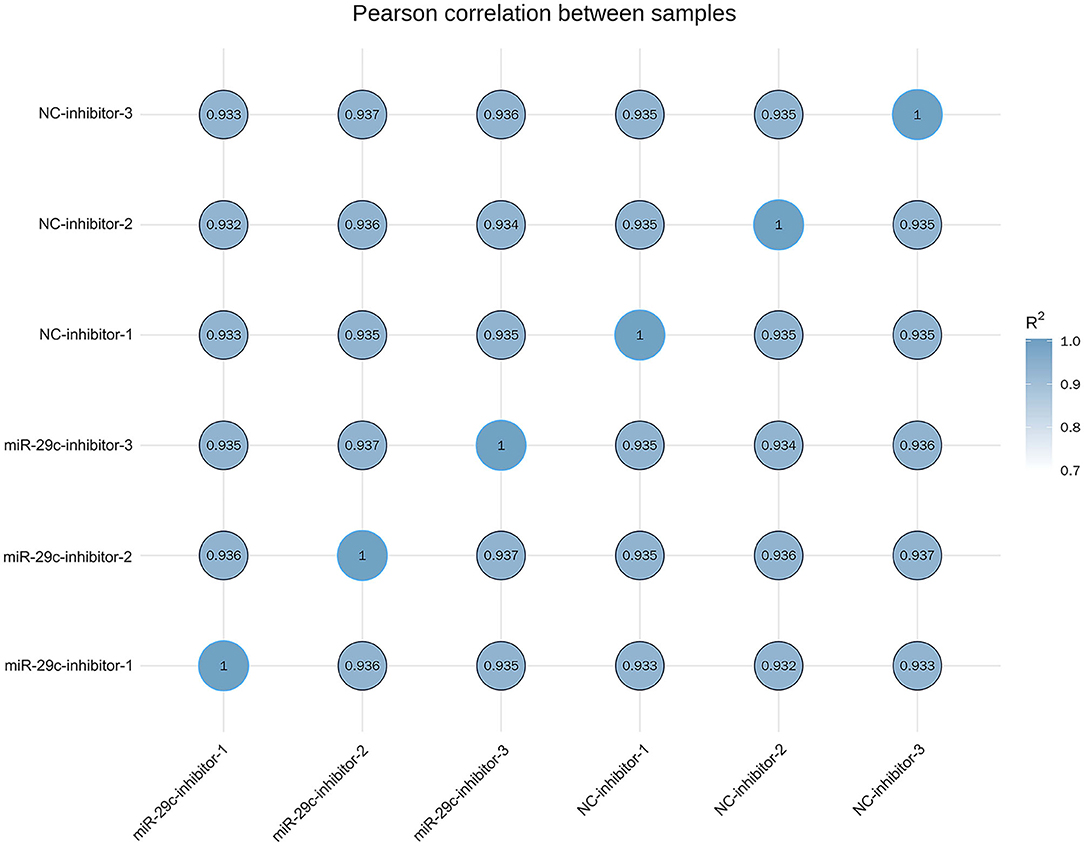
Total RNA was extracted from each sample of NC inhibitor and miR-29c inhibitor groups. Transcriptome libraries were established, and RNA-seq was performed. A total of 267 million clean reads were obtained by filtering the original data, and at least 5.67 G of the clean base was obtained from each sample, which could be further used for subsequent analysis. The Q30 of all samples were >92.6%, and the GC content ranged from 47.05 to 47.57% (Table 1). Clean reads were mapped to the bovine reference genome, and the total mapping rate reached 95.67–96.14%, among which 93.23–93.74% of clean reads had unique mapped positions on the reference sequence (Table 2). The boxplot of expression abundance (FPKM) of genes in each sample showed the overall gene expression abundance of different samples (Figure 2). Correlation analysis between samples showed that R2-values were >0.8 for each group of biological replicates (Figure 3). The above results indicated that our sequencing data met the requirements for the analysis of DEGs.
Differential Expression Analysis of Genes
With P < 0.05 and |log|>1, a total of 42 genes were significantly up-regulated and 27 genes were significantly down-regulated in the miR-29c inhibitor group compared with the NC inhibitor group (Figures 4A,B, Supplementary Table S2). Three immune-related DEGs, namely forkhead box O1 (FOXO1), tumor necrosis factor-alpha (TNF-α), and major histocompatibility complex, class II, DQ alpha 5 (BoLA-DQA5) (24–26), were significantly down-regulated in the miR-29c inhibitor group (the values of log of FOXO1, TNF-α, and BoLA-DQA5 were −1.0581, −4.1784, and −1.394, respectively). The above results suggested that miR-29c might regulate the immune response in MAC-T by regulating these genes.
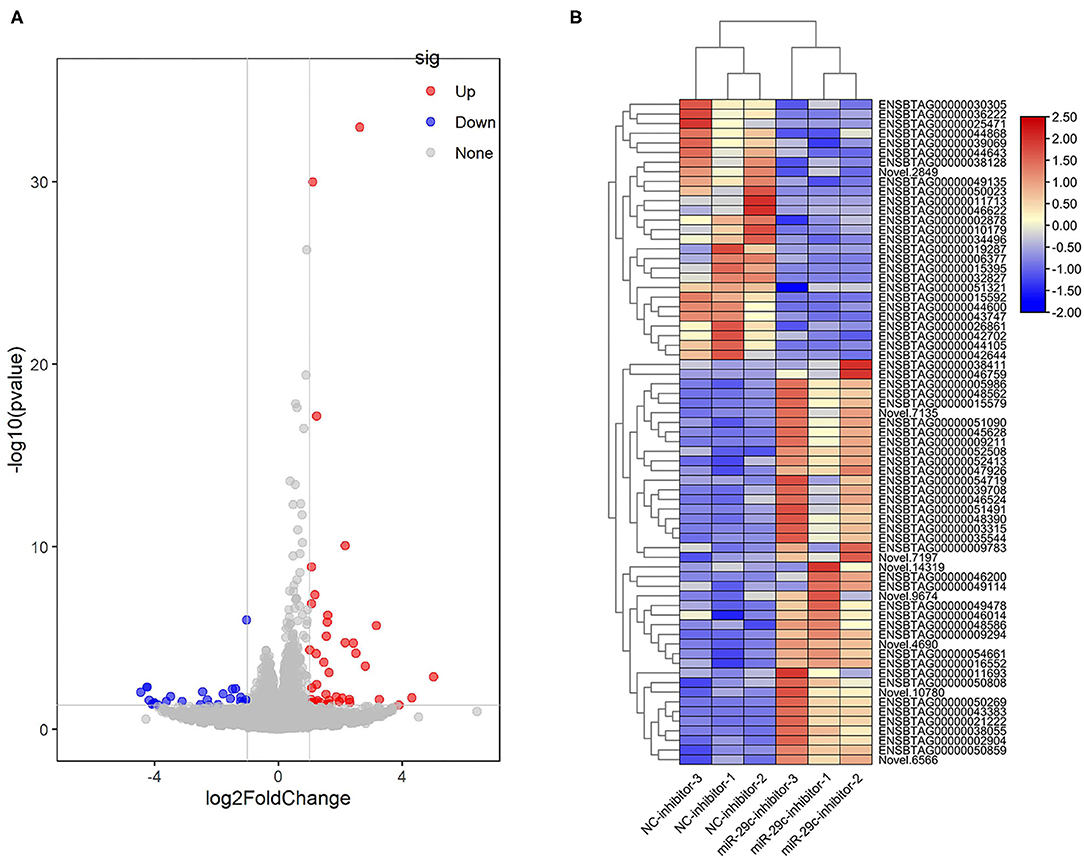
Figure 4. Analysis of DEGs in miR-29c inhibitor and NC inhibitor groups. (A) Volcano map of genes expression between the miR-29c inhibitor and NC inhibitor groups. The red points represent up-regulated genes, and the blue points represent down-regulated genes with statistical significance. (B) Cluster heat map of DEGs between the miR-29c inhibitor and NC inhibitor groups.
Validation of RNA-Seq Results by qRT-PCR
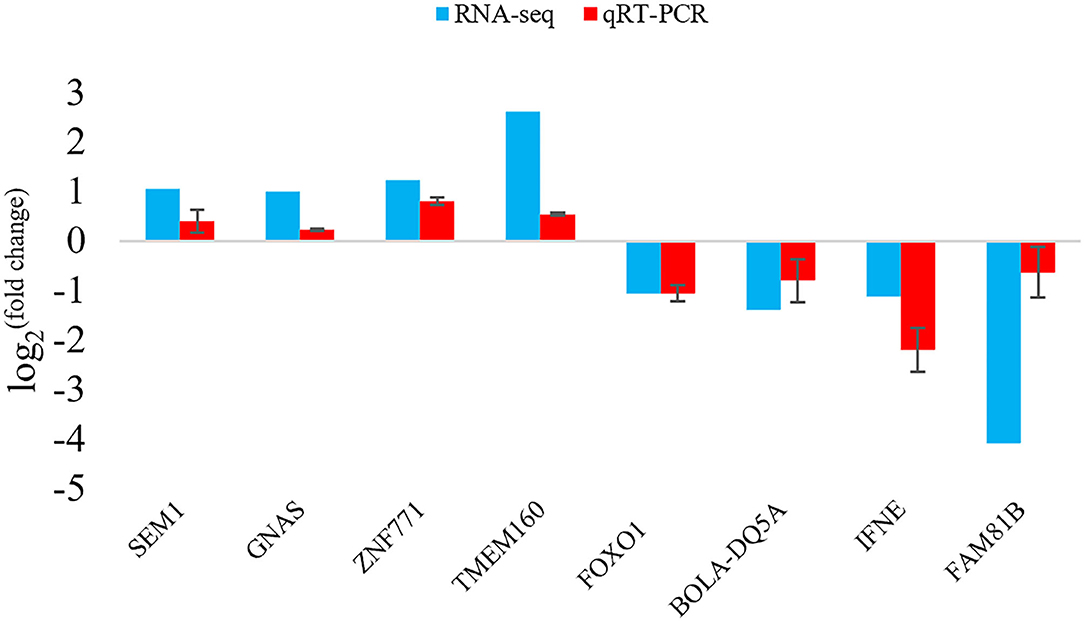
To verify the results of the deep sequencing and the DEGs obtained by RNA-seq analysis, a total of 8 DEGs were randomly selected for qRT-PCR. Comparing the qRT-PCR results with the RNA-seq results, the expression trends of 8 DEGs obtained by two methods were similar (Figure 5), indicating that the results of RNA-seq and the screening of DEGs were reliable.
GO and KEGG Analysis of DEGs
Total DEGs were annotated to cellular components (CC), molecular function (MF), and biological process (BP) through GO enrichment analysis. Figure 6, Supplementary Table S3 shows the top 10 entries in each group. The results showed that DEGs were significantly enriched in CC entries, such as anaphase-promoting complex, nuclear ubiquitin ligase complex, and proteasome accessory complex. MF annotation indicated that the DEGs might play a role in SNARE binding, protein phosphatase binding and protease binding. BP annotation showed that the DEGs might be involved in regulating stress-activated mitogen-activated protein kinase (MAPK) cascade, stress-activated protein kinase signaling cascade, and cellular response to ROS. KEGG enrichment was used to analyze the possible key signaling pathways involved in DEGs (Figure 7, Supplementary Table S4). DEGs were significantly enriched in immune-related signaling pathways, such as asthma, allograft rejection, Epstein-Barr virus infection, and inflammatory bowel disease. The analysis showed that most of these results were related to FOXO1, TNF-α, and BOLA-DQA5, which were significantly down-regulated in the miR-29c inhibitor group and were previously tested to be involved in the regulation of oxidative stress and immune response. The above results suggested that miR-29c might be involved in the regulation of oxidative stress and the immune response of MAC-T through the above-mentioned genes.
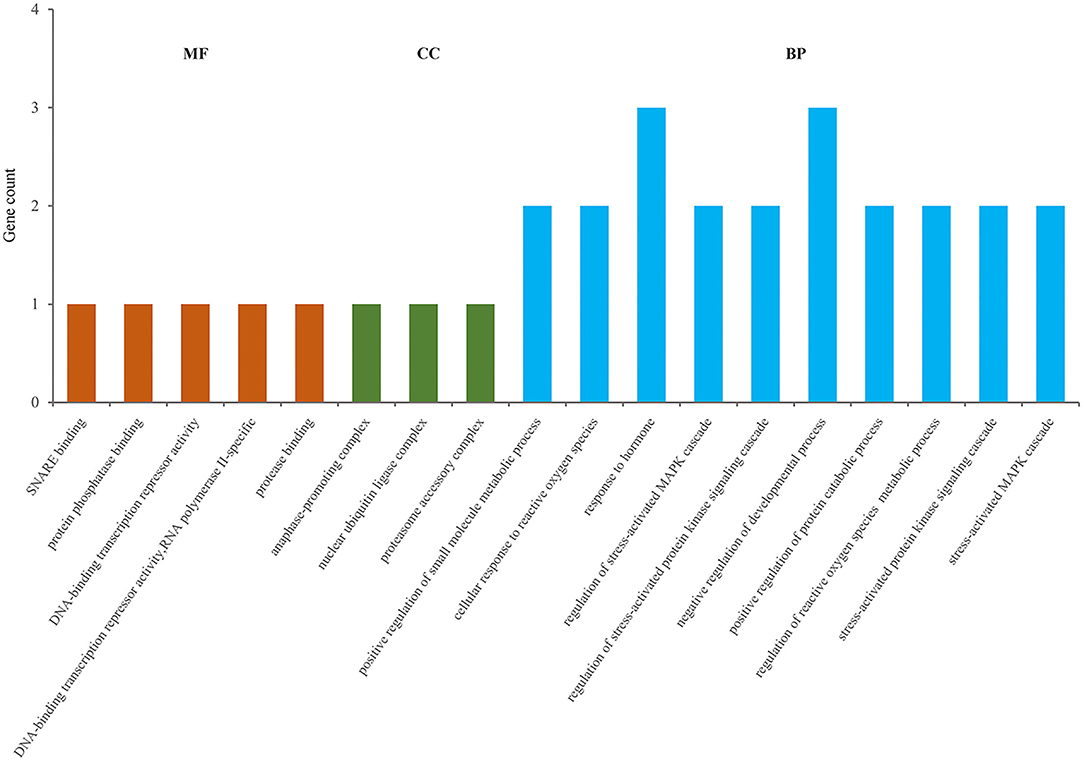
Figure 6. GO annotation of DEGs, and the significance level of enrichment was set at P < 0.05 (top10).
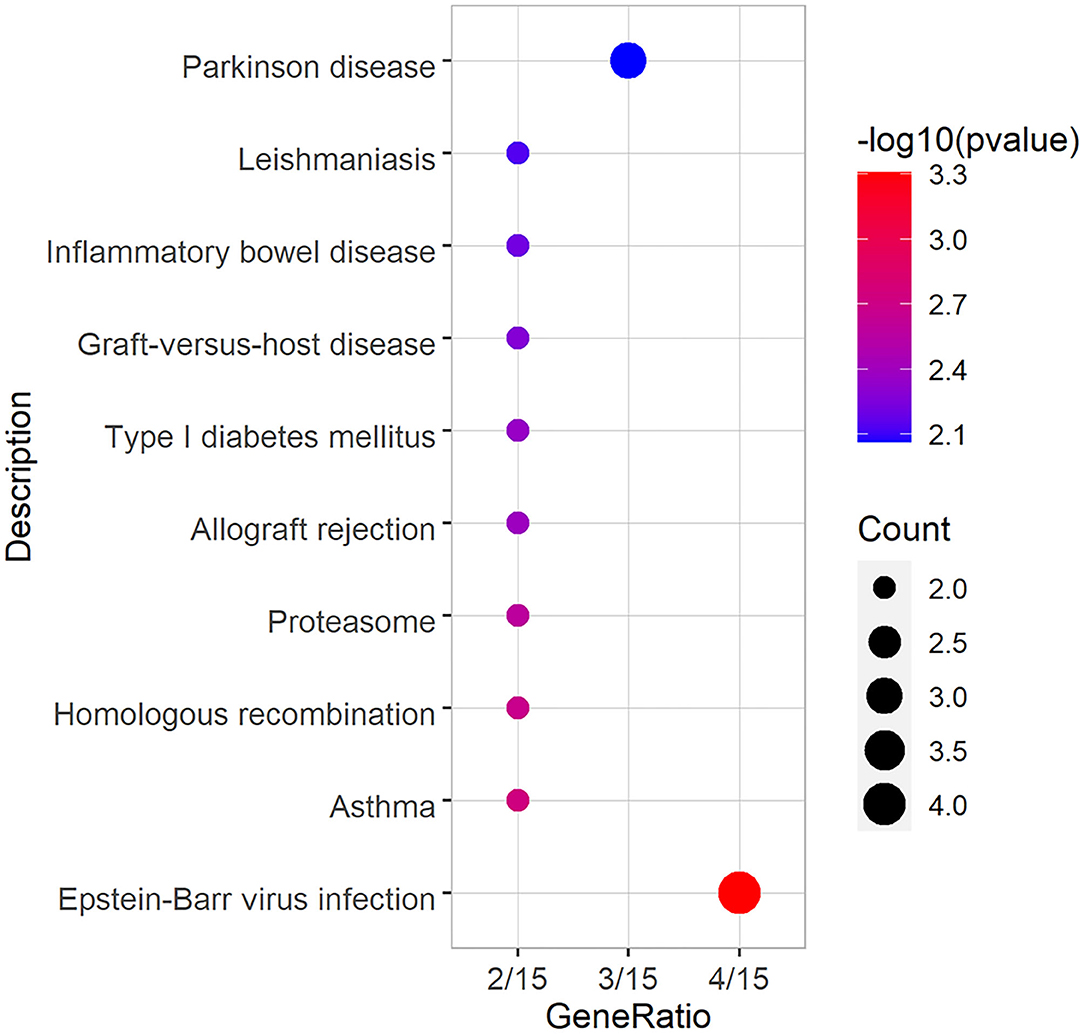
Figure 7. KEGG annotation of DEGs, and the significance level of enrichment was set at P < 0.05 (top10).
Discussion
When endogenous or exogenous ROS and stimulation factors, such as LPS, stimulate bMECs, the oxidative stress and inflammatory response generated by bMECs reduce vitality and affect the health of the mammary gland, resulting in lactation dysfunction, which is the main cause of cow mastitis (27–30). The occurrence of mastitis leads to extensive changes in transcriptome profiles, including miRNA. Some important differentially expressed miRNAs (DEmiRNAs) have been screened out in bovine mammary tissues infected with pathogenic bacteria or in LPS-induced bMECs, such as miR-145 (31), miR-146a (9), and miR-223 (32, 33), which affect the inflammatory response of bMECs by regulating the activation of immune-related signaling pathways, such as TLRs signaling pathway, NF-κB signaling pathway and Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling pathway (9, 34–36). In addition, the study have shown that miR-141 and miR-200a are closely related to the regulation of oxidative stress in bMECs (37).Thus, miR-29c, as a significant DEmiRNA related to cow mastitis, should determine its potential role in bMECs. In this study, we interfered with the expression of miR-29c in MAC-T cells using a specific inhibitor and detected its transcriptome changes relative to the NC inhibitor group by RNA-seq. A total of 69 DEGs were detected, and the result of qRT-PCR verified that result of RNA-seq were reliable. These DEGs are involved in immune-related biological processes and signaling pathways, such as stress-activated MAPK cascade, Epstein-Barr virus infection and inflammatory bowel disease signaling pathways. It is suggested that the inhibition of miR-29c can regulate the immune and stress responses of bMECs.
The MAPK cascade mainly consists of three kinases: MAPK kinase kinase, MAPK kinase, and MAPK, which are activated and phosphorylated downstream in turn and participate in regulating a variety of life processes, including cellular immunity, proliferation, and apoptosis (38–41). Similarly, oxidative stress is an important part of the cellular immune response (42, 43). Inhibition of the MAPK signaling pathway and the expression of genes related to oxidative stress can effectively reduce the inflammatory injury of bMECs induced by LPS (44, 45). Our results found that FOXO1 and TNF-α were significantly enriched in the above biological processes in bMECs. FOXO1 is an important regulator of the cellular oxidative stress response (46), it was reported that FOXO1 was significantly up-regulated in H2O2-induced H9C2 cells, while inhibition of FOXO1 could up-regulate superoxide dismutase (SOD) levels and down-regulate malondialdehyde (MDA) and lactate dehydrogenase (LDH) levels, thus reducing oxidative stress and cell apoptosis induced by H2O2 (47). Furthermore, inhibition expression of FOXO1 in the RAW264.7 cell line resulted in the down-regulation of pro-inflammatory genes, such as interleukin 1-beta (IL-1β), interleukin 6 (IL-6), and monocyte chemoattractant protein-1 (MCP-1) (48). Previous reports found that knockdown of miR-29c in breast cancer cells inhibited FOXO1 expression but promoted the proliferation, migration, and invasion of breast cancer cells (49). TNF-α is an important regulator of the immune response in mammals under physiological or pathological conditions and can regulate the signaling pathways related to immune cell proliferation and apoptosis (50, 51). The expression of the TNF-α gene is positively correlated with the severity of dairy cow mastitis. Studies have found that miR-142-5p regulates LPS-induced bMECs inflammation by targeting Bcl-2-associated athanogene (BAG5), and it is positively correlated with TNF-α, IL-1β, IL-6, and interleukin 8 (IL-8) (52). Therefore, FOXO1 and TNF-α were significantly down-regulated in the miR-29c inhibitor group, suggesting that down-regulation of miR-29c may reduce inflammation and oxidative stress, which was of great significance to the maintenance of bMECs steady state.
As a cell surface protein, soluble major histocompatibility complex (MHC) class II molecules (e.g., BoLA-DQA family) play an important role in maintaining immune homeostasis with the intracellular transport process (53). In this study, BoLA-DQA5 was significantly enriched in inflammatory and immune-related signal pathways, such as Epstein-Barr virus infection and inflammatory bowel disease. Hou et al. (54) reported that cow mastitis might be regulated by BoLA-DQA2 splice variants. In addition, studies have shown that the copy number variation of BoLA-DQA5 may be related to various infection-related phenotypes in Holstein cows (55). And in the inflammatory diseases of cows, the expression of the BoLA-DQA5 gene was significantly down-regulated in subclinical endometritis samples of cows on the 7th day of the estrus cycle compared with that of healthy cows, which may be involved in regulating antigen processing and presentation pathway (56). However, the potential effect of BoLA-DQA5 on bMECs immune response needs further study.
It is widely known that miRNAs don't encode proteins, but they can directly or indirectly regulate the expressions of many genes (57). In previous reports, miR-29c was proved to target specific protein-1 (SP1) (11) and nuclear factor of activated T cells 5 (NFAT5) (58) to inhibit the inflammatory response in Parkinson's disease. However, miR-29c promotes the inflammatory response by targeting LIF in UC (14). Interestingly, the pathway for the above diseases were also enriched in our KEGG annotated results. In addition, a study was shown that inhibition of expression of miR-29c can help to reduce inflammation caused by sepsis (59). The above studies have shown that the role of miR-29c is different in different inflammation, which further highlights the importance of studying its role in bMECs. In this study, we identified the effect of miR-29c inhibition on the gene expression of MAC-T cells from a global perspective using RNA-seq technology, and the results indicated that miR-29c may regulate inflammatory responses and oxidative stress in bMECs. Meanwhile, considered the similarities of genes among different species, the target gene of miR-29c in other species still needs to be verified in dairy cow. Consequently, we predicted the potential target genes of miR-29c using the TargetScan database (http://www.targetscan.org/vert_80/) (Supplementary Table S5), and compared them with DEGs. What's surprising to us, though, is the fact that the predicted target genes did not include in the list of the up-regulated genes (log > 1) in the miR-29c inhibitor group, the reason of which is that, the predicted target genes of miR-29c might not be identified under the criteria of DEGs screening adopted in this study. As we know, the roles of miRNA are extensive, sensitive and rapid, and the inhibition of miRNAs on target genes may be more significant at the protein level, rather than mRNA level (60, 61). In addition, the expressions of target genes of miRNAs may be affected by negative feedback and cascade regulation (62). In short, this study provides valuable evidence for the potential role of miR-29c in regulating inflammation and oxidative stress in bMECs, but its specific molecular mechanism, including target gene identification, remains to be further studied.
Conclusion
In conclusion, RNA-seq revealed that the inhibition of miR-29c in MAC-T cells could lead to the up-regulation of 42 genes and the down-regulation of 27 genes. The functional enrichment of the DEGs indicated that miR-29c might be a potential regulator of oxidative stress and inflammatory response in bMECs through multiple genes, such as FOXO1, TNF-α, and BoLA-DQA5, which enriched in stress-activated MAPK cascade, Epstein-Barr virus infection and inflammatory bowel disease signaling pathways. The above results imply that miR-29c plays an important role in the steady state of bMECs or cow mammary gland and may be a potential therapeutic target for mastitis in dairy cows.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material, and GEO DATABASE (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE195508).
Author Contributions
JY, X-PW, and Z-ML designed the experiments. JY and Q-CH completed the experiments. JY, X-PW, Z-ML, and J-PW analyzed the data. JY drafted the paper. J-PW, Q-QR, X-PW, Z-ML, YM, and D-WW revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Natural Science Foundation of China (No. 31960652), the Key Research and Development Project (Talent Introduction Project) of Ningxia Hui Autonomous Region (No. 2019BEB04002), and the Introducing Talent Research Project of Ningxia University (No. 030900001926).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors want to express their sincere appreciation to MogoEdit professional English editing company (Mogo Internet Technology Co., Ltd., Shaanxi, China) for English language editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.865415/full#supplementary-material
Supplementary Figure S1. Identification of MAC-T cells by immunofluorescence with cytokeratin 18.
Supplementary Table S1. The list of all primer sequences for qRT-PCR.
Supplementary Table S2. The list of DEGs in miR-29c inhibitor group vs. NC inhibitor group.
Supplementary Table S3. The list of GO enrichment analysis results for DEGs.
Supplementary Table S4. The list of KEGG enrichment analysis results for DEGs.
Supplementary Table S5. The list of the predicted target genes for miR-29c.
References
1. Wei ZK, Wang JJ, Wang YA, Wang CQ, Liu X, Han Z, et al. Effects of neutrophil extracellular traps on bovine mammary epithelial cells in vitro. Front Immunol. (2019) 10:1003. doi: 10.3389/fimmu.2019.01003
2. Geng ZJ, Shan XF, Lian S, Wang JF, Wu R. LPS-induced SOCS3 antagonizes the JAK2-STAT5 pathway and inhibits beta-casein synthesis in bovine mammary epithelial cells. Life Sci. (2021) 278:119547. doi: 10.1016/j.lfs.2021.119547
3. Zhao YY, Tang JN, Yang DR, Tang C, Chen J. Staphylococcal enterotoxin M induced inflammation and impairment of bovine mammary epithelial cells. J Dairy Sci. (2020) 103:8350–9. doi: 10.3168/jds.2019-17444
4. Abed AH, Menshawy A, Zeinhom M, Hossain D, Khalifa E, Wareth G, et al. Subclinical mastitis in selected bovine dairy herds in north upper egypt: assessment of prevalence, causative bacterial pathogens, antimicrobial resistance and virulence-associated genes. Microorganisms. (2021) 9:1175. doi: 10.3390/microorganisms9061175
5. Hammond SM. An overview of microRNAs. Adv Drug Deliver Rev. (2015) 87:3–14. doi: 10.1016/j.addr.2015.05.001
6. Correia De Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs' action through miRNA editing. Int J Mol Sci. (2019) 20:6249. doi: 10.3390/ijms20246249
7. Li XH, Jiang P, Yu HB, Yang YW, Xia LX, Yang RJ, et al. MiR-21-3p targets Elovl5 and regulates triglyceride production in mammary epithelial cells of cow. Dna Cell Biol. (2019) 38:352–7. doi: 10.1089/dna.2018.4409
8. Cao QQ, Li HH, Liu X, Yan ZG, Zhao M, Xu ZJ, et al. MiR-24-3p regulates cell proliferation and milk protein synthesis of mammary epithelial cells through menin in dairy cows. J Cell Physiol. (2019) 234:1522–33. doi: 10.1002/jcp.27017
9. Wang XP, Luoreng ZM, Zan LS, Li F, Li N. Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene. J Dairy Sci. (2017) 100:7648–58. doi: 10.3168/jds.2017-12630
10. Bian YJ, Lei Y, Wang CM, Wang J, Wang LN, Liu LL, et al. Epigenetic regulation of miR-29s affects the lactation activity of dairy cow mammary epithelial cells. J Cell Physiol. (2015) 230:2152–63. doi: 10.1002/jcp.24944
11. Wang RL, Yang Y, Wang H, He Y, Li C. MiR-29c protects against inflammation and apoptosis in Parkinson's disease model in vivo and in vitro by targeting SP1. Clin Exp Pharmacol Physiol. (2020) 47:372–82. doi: 10.1111/1440-1681.13212
12. Le LT, Swingler TE, Crowe N, Vincent TL, Barter MJ, Donell ST, et al. The microRNA-29 family in cartilage homeostasis and osteoarthritis. J Mol Med. (2016) 94:583–96. doi: 10.1007/s00109-015-1374-z
13. Xu W, Li ZK, Zhu XD, Xu RJ, Xu YJ. miR-29 family inhibits resistance to methotrexate and promotes cell apoptosis by targeting COL3A1 and MCL1 in osteosarcoma. Med Sci Monit. (2018) 24:8812–21. doi: 10.12659/MSM.911972
14. Guo J, Zhang RY, Zhao YQ, Wang JP. MiRNA-29c-3p promotes intestinal inflammation via targeting leukemia inhibitory factor in ulcerative colitis. J Inflamm Res. (2021) 14:2031–43. doi: 10.2147/JIR.S302832
15. LuoReng ZM, Wang XP, Zan LS. Comparison of microRNA profiles between bovine mammary glands infected with Staphylococcus aureus and Escherichia coli. Int J Biol Sci. (2018) 14:87–99. doi: 10.7150/ijbs.22498
16. Chen L, Liu XL, Li ZX, Wang HL, Liu Y, He H, et al. Expression differences of miRNAs and genes on NF-κB pathway between the healthy and the mastitis Chinese holstein cows. Gene. (2014) 545:117–25. doi: 10.1016/j.gene.2014.04.071
17. Huynh HT, Robitaille G, Turner JD. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp Cell Res. (1991) 197:191–9. doi: 10.1016/0014-4827(91)90422-Q
18. Zhao K, Liu HY, Zhou MM, Liu JX. Establishment and characterization of a lactating bovine mammary epithelial cell model for the study of milk synthesis. Cell Biol Int. (2010) 34:717–21. doi: 10.1042/CBI20100023
19. Wang JP, Hu QC, Yang J, Luoreng ZM, Wang XP, Ma Y, et al. Differential expression profiles of lncRNA following lps-induced inflammation in bovine mammary epithelial cells. Front Vet Sci. (2021) 8:758488. doi: 10.3389/fvets.2021.758488
20. Kramer MF. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol. (2011) Chapter 15: Unit 15.10. doi: 10.1002/0471142727.mb1510s95
21. Bougarn S, Cunha P, Gilbert FB, Meurens F, Rainard P. Technical note: validation of candidate reference genes for normalization of quantitative PCR in bovine mammary epithelial cells responding to inflammatory stimuli. J Dairy Sci. (2011) 94:2425–30. doi: 10.3168/jds.2010-3859
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
23. Wu TZ, Hu EQ, Xu SB, Chen MJ, Guo PF, Dai ZH, et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. (2021) 2:100141. doi: 10.1016/j.xinn.2021.100141
24. Chung S, Lee TJ, Reader BF, Kim JY, Lee YG, Park GY, et al. FoxO1 regulates allergic asthmatic inflammation through regulating polarization of the macrophage inflammatory phenotype. Oncotarget. (2016) 7:17532–46. doi: 10.18632/oncotarget.8162
25. Alam MS, Otsuka S, Wong N, Abbasi A, Gaida MM, Fan Y, et al. TNF plays a crucial role in inflammation by signaling via T cell TNFR2. Proc Natl Acad Sci USA. (2021) 118:e2109972118. doi: 10.1073/pnas.2109972118
26. Gelhaus A, Forster B, Wippern C, Horstmann RD. Evidence for an additional cattle DQA locus, BoLA-DQA5. Immunogenetics. (1999) 49:321–7. doi: 10.1007/s002510050499
27. Sun LL, Wang F, Wu ZH, Ma L, Baumrucker C, Bu DP. Comparison of selenium source in preventing oxidative stress in bovine mammary epithelial cells. Animals. (2020) 10:842. doi: 10.3390/ani10050842
28. Liu Y, Zhou M, Xu SY, Khan MA, Shi YX, Qu WJ, et al. Mycoplasma bovis-generated reactive oxygen species and induced apoptosis in bovine mammary epithelial cell cultures. J Dairy Sci. (2020) 103:10429–45. doi: 10.3168/jds.2020-18599
29. Dai H, Coleman DN, Hu L, Martinez-Cortes I, Wang M, Parys C, et al. Methionine and arginine supplementation alter inflammatory and oxidative stress responses during lipopolysaccharide challenge in bovine mammary epithelial cells in vitro. J Dairy Sci. (2020) 103:676–89. doi: 10.3168/jds.2019-16631
30. Liu MJ, Zhang C, Xu XL, Zhao X, Han ZY, Liu DD, et al. Ferulic acid inhibits LPS-induced apoptosis in bovine mammary epithelial cells by regulating the NF-kappaB and Nrf2 signalling pathways to restore mitochondrial dynamics and ROS generation. Vet Res. (2021) 52:104. doi: 10.1186/s13567-021-00973-3
31. Chen Z, Xu X, Tan TL, Chen DJ, Liang HJ, Sun KD, et al. MicroRNA-145 regulates immune cytokines via targeting FSCN1 in Staphylococcus aureus-induced mastitis in dairy cows. Reprod Domest Anim. (2019) 54:882–91. doi: 10.1111/rda.13438
32. Pu JH, Li R, Zhang CL, Chen D, Liao XX, Zhu YH, et al. Expression profiles of miRNAs from bovine mammary glands in response to Streptococcus agalactiae-induced mastitis. J Dairy Res. (2017) 84:300–8. doi: 10.1017/S0022029917000437
33. Jiao P, Wang XP, Luoreng ZM, Yang J, Jia L, Ma Y, et al. miR-223: an effective regulator of immune cell differentiation and inflammation. Int J Biol Sci. (2021) 17:2308–22. doi: 10.7150/ijbs.59876
34. Wang XP, Luoreng ZM, Zan LS, Raza SH, Li F, Li N, et al. Expression patterns of miR-146a and miR-146b in mastitis infected dairy cattle. Mol Cell Probes. (2016) 30:342–4. doi: 10.1016/j.mcp.2016.08.004
35. Li LM, Huang JM, Zhang XJ, Ju ZH, Qi C, Zhang Y, et al. One SNP in the 3′-UTR of HMGB1 gene affects the binding of target bta-miR-223 and is involved in mastitis in dairy cattle. Immunogenetics. (2012) 64:817–24. doi: 10.1007/s00251-012-0641-1
36. Zhang ZB, Guo YF, Li CY, Qiu CW, Guo MY. Selenium influences mmu-miR-155 to inhibit inflammation in Staphylococcus aureus-induced mastitis in mice. Food Funct. (2019) 10:6543–55. doi: 10.1039/C9FO01488H
37. Salama AAK, Duque M, Wang L, Shahzad K, Olivera M, Loor JJ. Enhanced supply of methionine or arginine alters mechanistic target of rapamycin signaling proteins, messenger RNA, and microRNA abundance in heat-stressed bovine mammary epithelial cells in vitro. J Dairy Sci. (2019) 102:2469–80. doi: 10.3168/jds.2018-15219
38. Liang YJ, Yang WX. Kinesins in MAPK cascade: how kinesin motors are involved in the MAPK pathway?. Gene. (2019) 684:1–9. doi: 10.1016/j.gene.2018.10.042
39. Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. (2002) 12:9–18. doi: 10.1038/sj.cr.7290105
40. Yue JC, Lopez JM. Understanding MAPK signaling pathways in apoptosis. Int J Mol Sci. (2020) 21:2346. doi: 10.3390/ijms21072346
41. Arthur JSA, Ley SC. Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol. (2013) 13:679–92. doi: 10.1038/nri3495
42. Al-Shehri SS. Reactive oxygen and nitrogen species and innate immune response. Biochimie. (2021) 181:52–64. doi: 10.1016/j.biochi.2020.11.022
43. Chen KL, Li L, Li CM, Wang YR, Yang FX, Kuang MQ, et al. SIRT7 regulates lipopolysaccharide-induced inflammatory injury by suppressing the NF-κB signaling pathway. Oxid Med Cell Longev. (2019) 2019:1–15. doi: 10.1155/2019/3187972
44. Lai JL, Liu YH, Liu C, Qi MP, Liu RN, Zhu XF, et al. Indirubin inhibits LPS-induced inflammation via TLR4 abrogation mediated by the NF-kB and MAPK signaling pathways. Inflammation. (2017) 40:1–12. doi: 10.1007/s10753-016-0447-7
45. Liu KJ, Ding T, Fang L, Cui LY, Li J, Meng X, et al. Organic selenium ameliorates Staphylococcus aureus-induced mastitis in rats by inhibiting the activation of NF-kappaB and MAPK signaling pathways. Front Vet Sci. (2020) 7:443. doi: 10.3389/fvets.2020.00443
46. Klotz LO, Sanchez-Ramos C, Prieto-Arroyo I, Urbanek P, Steinbrenner H, Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. (2015) 6:51–72. doi: 10.1016/j.redox.2015.06.019
47. Qiu ZJ, Wang L, Mao HY, Xu F, Sun B, Lian XB, et al. MiR-370 inhibits the oxidative stress and apoptosis of cardiac myocytes induced by hydrogen peroxide by targeting FOXO1. Exp Ther Med. (2019) 18:3025–31. doi: 10.3892/etm.2019.7908
48. Chen L, Gao BY, Zhang YD, Lu HY, Li XB, Pan LF, et al. PAR2 promotes M1 macrophage polarization and inflammation via FOXO1 pathway. J Cell Biochem. (2019) 120:9799–809. doi: 10.1002/jcb.28260
49. Li W, Yi J, Zheng XJ, Liu SW, Fu WQ, Ren LW, et al. MiR-29c plays a suppressive role in breast cancer by targeting the TIMP3/STAT1/FOXO1 pathway. Clin Epigenetics. (2018) 10:64. doi: 10.1186/s13148-018-0495-y
50. Varfolomeev E, Vucic D. Intracellular regulation of TNF activity in health and disease. Cytokine. (2018) 101:26–32. doi: 10.1016/j.cyto.2016.08.035
51. Hackel A, Aksamit A, Bruderek K, Lang S, Brandau S. TNF-alpha and IL-1beta sensitize human MSC for IFN-gamma signaling and enhance neutrophil recruitment. Eur J Immunol. (2021) 51:319–30. doi: 10.1002/eji.201948336
52. Lu JY, Gu BB, Lu W, Liu J, Lu J. MiR-142-5p regulates lipopolysaccharide-induced bovine epithelial cell proliferation and apoptosis via targeting BAG5. Exp Ther Med. (2021) 22:1425. doi: 10.3892/etm.2021.10860
53. Bakela K, Athanassakis I. Soluble major histocompatibility complex molecules in immune regulation: highlighting class II antigens. Immunology. (2018) 153:315–24. doi: 10.1111/imm.12868
54. Hou QL, Huang JM, Ju ZH, Li QL, Li LM, Wang CF, et al. Identification of splice variants, targeted microRNAs and functional single nucleotide polymorphisms of the BOLA-DQA2 gene in dairy cattle. Dna Cell Biol. (2012) 31:739–44. doi: 10.1089/dna.2011.1402
55. Fukunaga K, Yamashita Y, Yagisawa T. Copy number variations in BOLA-DQA2, BOLA-DQB, and BOLA-DQA5 show the genomic architecture and haplotype frequency of major histocompatibility complex class II genes in Holstein cows. Hla. (2020) 96:601 doi: 10.1111/tan.14086
56. Hoelker M, Salilew-Wondim D, Drillich M, Christine GB, Ghanem N, Goetze L, et al. Transcriptional response of the bovine endometrium and embryo to endometrial polymorphonuclear neutrophil infiltration as an indicator of subclinical inflammation of the uterine environment. Reprod Fertil Dev. (2012) 24:778–93. doi: 10.1071/RD11171
57. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. (2004) 116:281–97. doi: 10.1016/S0092-8674(04)00045-5
58. Wang RL, Li Q, He Y, Yang Y, Ma QY, Li C. miR-29c-3p inhibits microglial NLRP3 inflammasome activation by targeting NFAT5 in Parkinson's disease. Genes Cells. (2020) 25:364–74. doi: 10.1111/gtc.12764
59. Zhang BY, Yu L, Sheng Y. Clinical value and role of microRNA-29c-3p in sepsis-induced inflammation and cardiac dysfunction. Eur J Med Res. (2021) 26:90. doi: 10.1186/s40001-021-00566-y
60. Behm-Ansmant I, Rehwinkel J, Izaurralde E. MicroRNAs silence gene expression by repressing protein expression and/or by promoting mRNA decay. Cold Spring Harb Symp Quant Biol. (2006) 71:523–30. doi: 10.1101/sqb.2006.71.013
61. Stavast CJ, Erkeland SJ. The non-canonical aspects of microRNAs: many roads to gene regulation. Cells-Basel. (2019) 8:1465. doi: 10.3390/cells8111465
Keywords: dairy cow, mastitis, immune, oxidative stress, RNA-seq
Citation: Yang J, Hu Q-C, Wang J-P, Ren Q-Q, Wang X-P, Luoreng Z-M, Wei D-W and Ma Y (2022) RNA-Seq Reveals the Role of miR-29c in Regulating Inflammation and Oxidative Stress of Bovine Mammary Epithelial Cells. Front. Vet. Sci. 9:865415. doi: 10.3389/fvets.2022.865415
Received: 29 January 2022; Accepted: 22 February 2022;
Published: 01 April 2022.
Edited by:
Aline Silva Mello Cesar, University of São Paulo, BrazilReviewed by:
Mao Yongjiang, Yangzhou University, ChinaXianyong Lan, Northwest A&F University, China
Copyright © 2022 Yang, Hu, Wang, Ren, Wang, Luoreng, Wei and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xing-Ping Wang, d3hwQG54dS5lZHUuY24=; Zhuo-Ma Luoreng, bHVvcmVuemh1b21hQG54dS5lZHUuY24=
 Jian Yang
Jian Yang Qi-Chao Hu
Qi-Chao Hu Jin-Peng Wang
Jin-Peng Wang Qian-Qian Ren
Qian-Qian Ren Xing-Ping Wang
Xing-Ping Wang Zhuo-Ma Luoreng
Zhuo-Ma Luoreng Da-Wei Wei
Da-Wei Wei Yun Ma
Yun Ma