- 1Division of Animal and Nutritional Science, West Virginia University, Morgantown, WV, United States
- 2Department of Medicine, Surgery, and Biomedical Sciences, Joan C. Edwards School of Medicine, Marshall University, Huntington, WV, United States
- 3Agricultural Research Station, Fort Valley State University, Fort Valley, GA, United States
We applied whole blood transcriptome analysis and gene set enrichment analysis to identify pathways associated with divergent selection for low or high RFI in beef cattle. A group of 56 crossbred beef steers (average BW = 261 ± 18.5 kg) were adapted to a high-forage total mixed ration in a confinement dry lot equipped with GrowSafe intake nodes for period of 49 d to determine their residual feed intake (RFI). After RFI determination, whole blood samples were collected from beef steers with the lowest RFI (most efficient; low-RFI; n = 8) and highest RFI (least efficient; high-RFI; n = 8). Prior to RNA extraction, whole blood samples collected were composited for each steer. Sequencing was performed on an Illumina NextSeq2000 equipped with a P3 flow. Gene set enrichment analysis (GSEA) was used to analyze differentially expressed gene sets and pathways between the two groups of steers. Results of GSEA revealed pathways associated with metabolism of proteins, cellular responses to external stimuli, stress, and heat stress were differentially inhibited (false discovery rate (FDR) < 0.05) in high-RFI compared to low-RFI beef cattle, while pathways associated with binding and uptake of ligands by scavenger receptors, scavenging of heme from plasma, and erythrocytes release/take up oxygen were differentially enriched (FDR < 0.05) in high-RFI, relative to low-RFI beef cattle. Taken together, our results revealed that beef steers divergently selected for low or high RFI revealed differential expressions of genes related to protein metabolism and stress responsiveness.
Introduction
Residual feed intake (RFI), a measure of feed efficiency, continues to be of great economic importance due to increasing cost of animal feeds (1). Residual Feed Intake is the difference between an animal's actual feed intake and its predicted feed intake for a given level of maintenance and body weight gain (1). Feed efficient animals consume less than expected and have a low (negative) RFI, while inefficient animals consume more than expected and have a high (positive) RFI. Thus, beef cattle selected for low RFI have decreased feed costs because they consume less dry matter when compared with high-RFI beef cattle while maintaining similar growth performance.
Due to the great economic importance of RFI, the biological mechanisms underlying variation in this trait have always been of great interest; however, these mechanisms have not been fully understood. Difference in RFI has been suggested to be an indication of differences in metabolism rather than differences in growth performance because the trait is phenotypically independent of growth performance (1). Several studies have applied whole transcriptome analysis of several tissues such as liver and ruminal epithelium to further understand the biological mechanisms regulating feed efficiency traits including RFI in beef cattle (2–4). For instance, Kong et al. (3) analyzed the rumen epithelial transcriptome from low-RFI and high-RFI beef steers and observed increased tissue morphogenesis and greater expression of mitochondrial genes in low-RFI compared to high-RFI steers. Mukiibi et al. (4) analyzed liver tissue transcriptome profile and observed differential expressions of genes involved in nutrient metabolisms and cellular development in beef steers divergent for low and high RFI. However, these studies involve invasive sample collection procedures. Despite the convenience of collection and relatively non-invasive accessibility of blood in ruminants, very few attempts have been made to apply whole-blood transcriptome to understand the biological mechanisms associated with RFI in animals. Genes expressed in peripheral blood cells have been demonstrated to reflect physiological changes in different body tissues and can highlight biological processes related to overall metabolisms. Therefore, the objective of this study was to analyze the whole-blood transcriptome data of beef steers via gene-set enrichment analysis to identify key pathways associated with divergent selection for low or high RFI in beef cattle.
Materials and Methods
Animals and Sample Collection
A total of 56 crossbred growing beef steers with average BW of 261.3 ± 18.5 kg were fed a high-forage total mixed ration (TMR; primarily consisting of triticale silage; rye grass silage; and a ration balancing Supplementary Table 1) in five confinement dry lot pens (15 by 47 m2), each served by three GrowSafe 8000 (GrowSafe Systems Ltd., Airdrie, Alberta, Canada) feeding nodes to measure individual feed intake and two In-Pen Weighing Positions (IPW Positions, Vytelle LLC) to measure daily BW for a total of 49 d after 15-d adjustment period to the feeding facilities. The use of IPW Positions has enabled the measurement of individual animal BW multiple times in a day (5). The IPW Positions measure the partial BW by weighing the front end of an animal (6). The IPW positions were positioned at a water trough in each pen such that an animal must place its front feet on the scale in order to drink (7). The partial BW of the animals was measured every second the animals stayed on the scale while drinking. More details on the accuracy, use and applicability of IPW positions have been described in previous studies (5, 6). In this study, approximately 617 ± 92 daily BW data points (after filtering outliers) per animal were generated and were regressed on time to calculate beginning BW, mid-test BW, and average daily gain (ADG) of each animal. Animal ADG and metabolic mid-test BW (mid-test BW0.75) were regressed against individual average daily intake, and RFI was calculated as the residual or the difference between the predicted value of the regression and the actual measured value based on the following equation: Y = β0 + β1X1 + β2X2 + ε, where Y is the observed DMI (kg/d), β0 is the regression intercept, β1 and β2 are the partial regression coefficients, X1 is the mid-test metabolic BW (kg), X2 is the ADG (kg/d), and ε indicates the RFI [kg/d; (8)]. After the determination of RFI values for all animals, the most-efficient with the lowest RFI (low-RFI; n = 8) and the least-efficient with the highest RFI (high-RFI; n = 8) beef steers were selected, kept separate from others, and kept on the same diet for additional 21 d (designated in this study as d 50–70). On d 56, 63, and 70, 10 mL of blood samples were collected before morning feeding into tubes containing sodium heparin. Immediately after collection, subsamples (500 μL each) were transferred into RNA-protect tubes (Cat. No. 76554; Qiagen) containing a reagent that lyses blood cells and stabilizes intracellular RNA and stored at −80°C until later analysis.
RNA Extraction, Library Preparation, and Sequencing
Prior to RNA extraction, whole blood samples collected on d 56, 63, and 70 were composited for each steer. Total RNA was extracted from the composited samples using RNeasy Protect Animal Blood kit (Cat. No. 73224; Qiagen) following the manufacturer's instructions. RNA concentration was measured using a NanoDrop One C spectrophotometer with an A260:A280 ratio from 1.8 to 2.0 (Thermo Fisher Scientific, Waltham, MA, USA). All RNA samples had RNA integrity numbers >8.0. Dual indexed RNA Libraries were prepared from 100 to 250 ng of total RNA per sample using the KAPA RNA HyperPrep Kit with RiboErase (Human, Mouse, Rat) Globin Reduction method in the WVU Genomics Core according to the kit manufacturer's instructions. Library quality was assessed by electrophoretic analysis on the Agilent 4,200 TapeStation system with High Sensitivity D1000 screentape. RNA libraries were sequenced in a dual indexed 2 × 50 paired-end run on an Illumina NextSeq2000 equipped with a P3 flow.
Data and Statistical Analysis
For the RNA-seq data, reads were trimmed using Trimmomatic v 0.39 to remove low-quality base calls and adapter sequences (9), and then aligned to the Bovine reference genome ARS-UCD1.2 (10) using HISAT2 v 2.2.1 (11). Resulting files were sorted and indexed, and PCR and optical duplicate reads were marked using SamTools v1.12 (12). The numbers of reads mapping to each gene for each sample were counted using the R/Bioconductor package GenomicAlignments v 1.26.0 (13). Log2 fold change values were computed using DESeq2 version 1.30.1 (14). We used gene set enrichment analysis (GSEA), a pathway enrichment method that utilizes predefined gene sets from the reactome pathways (15), to analyze differentially expressed gene sets using the R/Bioconductor package fgsea v 1.16.0. The GSEA was performed to determine the key pathways that were enriched or inhibited by considering the expression levels of sets of biologically related genes (16). Genes identified by DESeq2 as expressing over a minimal threshold were ranked by Log2 fold change and analyzed by the GSEA algorithm (17). The altered pathways were filtered based on FDR ≤ 0.05 and were arranged in the order of their normalized enrichment scores.
Results
The average RFI values of low- and high-RFI steers were−1.93 and 2.01, respectively. An average of 36 million reads per sample was generated (Supplementary Table 2). Results of GSEA revealed gene sets (pathways) associated with metabolism of proteins, cellular responses to external stimuli, stress, heat stress, and regulation of HSF-1-mediated heat shock response were differentially inhibited (FDR = 0.01) in high-RFI compared to low-RFI beef cattle (Table 1, Supplementary Figure 1). The gene set associated with metabolism of proteins consists of 248 genes, and 85 of which were leading edge genes (significantly enriched genes). Both cellular response to external stimuli and cellular response to stress shared the same nineteen leading edge genes including HSPA1A, HSPH1, BAG2, DNAJA1, DNAJB1, H3C13, H2BC7, H4C2, ELOC, JUN, and HSPA4. Five of the leading edge genes (HSPA1A, HSPH1, BAG2, HSPA4, and DNAJB1) associated with cellular response to external stimuli and stress were also leading edge genes in gene sets associated with response to heat stress and regulation of HSF-1-mediated heat shock response (Table 1).
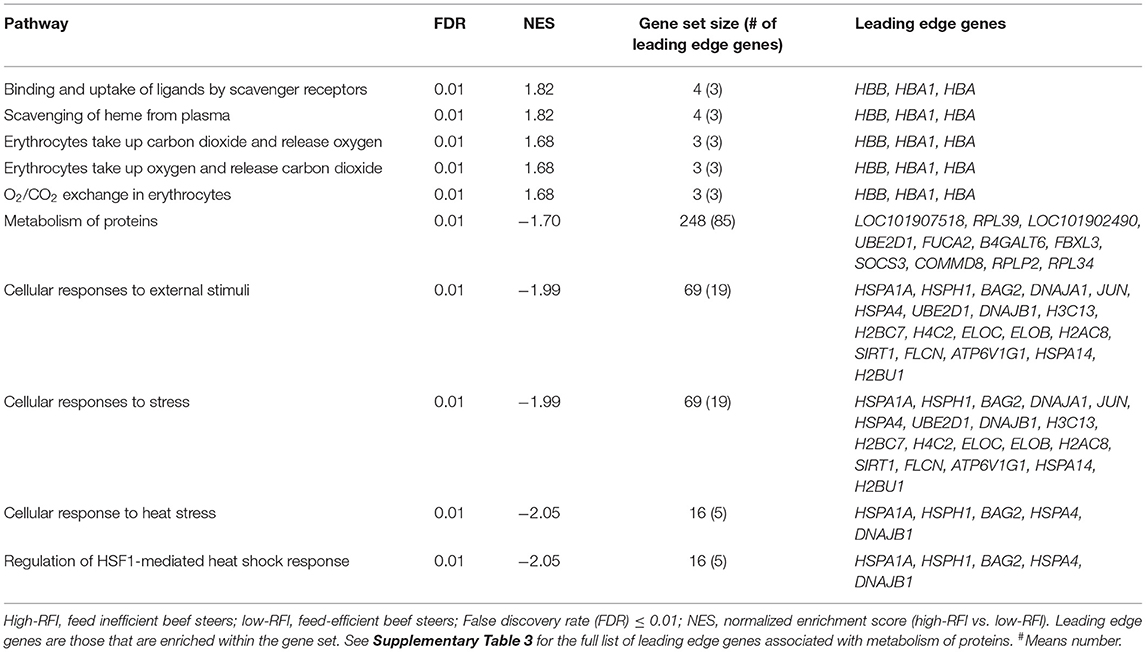
Table 1. Altered pathways identified by Gene Set Enrichment Analysis in high-RFI compared to low-RFI beef steers.
Gene sets associated with binding and uptake of ligands by scavenger receptors, scavenging of heme from plasma, erythrocytes take up/release carbon dioxide and release/take up oxygen share the same leading edge genes (HBB, HBA1, and HBA) and were all differentially enriched (FDR < 0.05) in high-RFI, relative to low-RFI beef cattle (Table 1, Supplementary Figure 2).
Discussion
Understanding the biological mechanisms regulating feed efficiency using easily accessible and non-invasive sample such as blood is essential to the future of livestock production systems in terms of profitability and animal welfare concern.
In this study, protein metabolism is the most enriched metabolic pathway based on the number of leading edge genes (such as LOC101907518, RPL39, LOC101902490, UBE2D1, FUCA2) in the gene set. In addition to the function of amino acids as the building blocks of proteins, amino acids regulate key metabolism essential for growth, performance, reproduction, and immunity (18). Research studies have shown that protein (amino acids) metabolism is essential for optimizing efficiency of nutrient absorption and metabolism to enhance immunity against diseases and stress, growth performance, and milk production of animals (18). Several published articles have identified protein metabolism as one of the most important metabolic processes associated with RFI in animals (19, 20). In our previous study, we identified plasma amino acid metabolites as the most significant metabolic signatures associated with RFI in beef cattle (20). Elolimy et al. (21) reported differences in signaling mechanisms controlling protein turnover in ruminal epithelium of beef cattle divergent for low- or high-RFI. In a similar study, Kong et al. (3) reported increased expression of genes involved in protein and cell turnover in the ruminal epithelium of low-RFI beef cattle, compared with high-RFI beef cattle. 4 performed RNA-seq analysis of liver tissue in beef cattle divergent for low and high RFI and observed downregulation of genes involved in amino acid degradation and urea synthesis in low-RFI beef cattle. In fact, some studies have reported significant association of blood metabolites involved in urea cycle with RFI in beef cattle (22, 23). Our results and those of others that utilized tissues with relatively more invasive collection methods suggest that amino acid metabolism plays a considerable role in regulating RFI of beef cattle and its enrichment in low-RFI beef steers probably explains their similar growth performance with high-RFI beef steers despite lower DMI.
Amino acids play a functional role in regulating stress response, including oxidative stress, in animals (24). Stress response has significant implication on health and production efficiency of animals (25). In fact, difference in stress responsiveness has been suggested to contribute to variation in feed efficiency of beef cattle (19, 26). In this study, we observed downregulation of gene set including HSPA1A, HSPH1, BAG2, and DNAJA1 associated with cellular responses to external stimuli, stress, heat stress, and regulation of HSF-1-mediated heat shock response in high-RFI beef steers, which suggests that these steers are more susceptible to stress. When an animal can no longer cope with a stressor, level of blood cortisol increases via activation of hypothalamic–pituitary–adrenal axis (HPA) axis which causes a fight or flight response that increases energy expenditure. Thus, stress response in animals is often determined by blood cortisol level and activity of the HPA axis (27). In a study that determined the response of beef heifers to an exogenous adrenocorticotropic hormone (ACTH) challenge, there was a positive association of plasma cortisol level with RFI status and low-RFI had a lower cortisol response than high-RFI heifers indicating that low-RFI heifers coped better with the stress challenge (28). Richardson et al. (19) and Gomes et al. (29) reported lower blood levels of cortisol in low-RFI beef cattle when compared to high-RFI beef cattle. A similar result was observed in crossbred rams following ACTH challenge (26). Taken together, downregulation of genes associated with cellular responses to external stimuli, stress, heat stress, and regulation of HSF-1-mediated heat shock response in high-RFI beef steers suggests that low-RFI steers have better adaptive mechanisms to cope with environmental stressors, thereby, reducing energy expenditure and increasing energy availability for improved growth performance and better feed efficiency.
In this study, we observed enrichment of gene sets (HBB, HBA1, and HBA) associated with erythrocytes take up/release carbon dioxide, release/take up oxygen, scavenging of heme from plasma, and binding and uptake of ligands by scavenger receptors in high-RFI, relative to low-RFI beef cattle. Erythrocytes contain hemoglobins which carry oxygen to the body and are continuously exposed to high oxygen content which pre-disposes them to oxidative stress damage (30, 31). Heme scavenger proteins, such as hemopexin and alpha-1-microglobulin, scavenge extracellular heme, a physiological ligand (32), synthesized from hemoglobin degradation via the activity of heme-oxygenase, an enzyme that is inducible by stressors such as oxygen free radicals (33, 34). Previous investigations have shown that cellular expression of alpha-1-microglobulin is enriched during increased oxidative stress and heme exposure (30, 34). In ruminants, oxidative stress has been implicated in many pathophysiological conditions that are relevant for growth performance, reproduction, and health (35). In fact, several studies have shown that oxidative damage of cell organelles and biomolecules is a source of energy drain and negatively affects several cellular processes including lipid and protein metabolism (36, 37). The major source of intracellular reactive oxygen species production is the mitochondria (38) and previous studies have reported higher mitochondrial ROS production in less feed-efficient compared to high feed efficient animals (36, 39, 40). In addition, Casal et al. (41) reported increased hepatic abundance of protein carbonyls and thiobarbituric acid reactive species, which are products of protein and lipid oxidative damage, and reduced protein expression of antioxidant enzymes, including mitochondrial manganese superoxide dismutase, in high-RFI when compared with low-RFI beef steers. Similarly, Tizioto et al. (42) observed upregulation of oxidative stress-induced transcription factors in muscle of high-RFI beef steers. Therefore, it is reasonable to speculate that enrichment of genes associated with erythrocytes take up/release carbon dioxide, scavenging of heme from plasma, and binding and uptake of ligands by scavenger receptors in high-RFI compared to low-RFI beef steers suggests that they may be more prone to oxidative stress, thereby resulting in reduced efficiency of energy use for metabolic processes such as growth.
It is important to note that though whole blood transcriptome data might encompass gene activities of several body tissues and organs including liver, kidney, muscles, and rumen, the contribution of each tissue to the whole blood transcriptome is not known and should be determined in future studies. In addition, biological validation of the RNA-Seq data on selected genes by RT-qPCR is also needed to confirm the results of this study.
Conclusion
Results of GSEA of whole blood transcriptome data in beef steers divergently selected for low or high RFI revealed differential expression of genes related to protein metabolism, erythrocytes take up/release carbon dioxide and release/take up oxygen, and stress responsiveness. These results are similar to those of several studies that utilized other tissues including liver, muscle, and ruminal epithelium. Thus, this study demonstrates the suitability of whole blood transcriptome data for understanding the biological mechanisms regulating RFI in animals. Due to the small number of animals used in this study and the effect of different diets and breeds on RFI ranking, further validation using a larger cohort of beef cattle fed different diets is needed to confirm these findings.
Data Availability Statement
All raw and processed sequencing data were submitted to the Gene Expression Omnibus (GEO) at the National Center for Biotechnology Information (NCBI) and can be accessed via accession number GSE198068.
Ethics Statement
The animal study was reviewed and approved by the Institutional Animal Care and Use Committees of West Virginia University (protocol number 1608003693).
Author Contributions
IO and APC designed the experiment. GT and MI conducted the experiment. JD analyzed the RNA-Seq data. IO, GT, and APC drafted the manuscript. All authors reviewed and approved the final manuscript.
Funding
This work was funded by West Virginia University Experimental Station (scientific article number 3430) in support of U.S. Department of Agriculture hatch multi-state regional project W-3010.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.848027/full#supplementary-material
References
1. Koch RM, Swiger LA, Chambers D, Gregory KE. Efficiency of food use in beef cattle. J Anim Sci. (1963) 22:486–94. doi: 10.2527/jas1963.222486x
2. Alexandre PA, Kogelman LJ, Santana MH, Passarelli D, Pulz LH, Fantinato-Neto P, et al. Liver transcriptomic networks reveal main biological processes associated with feed efficiency in beef cattle. BMC Genomics. (2015) 16:s12864–015. doi: 10.1186/s12864-015-2292-8
3. Kong RS, Liang G, Chen Y, Stothard P, Guan I. Transcriptome profiling of the rumen epithelium of beef cattle differing in residual feed intake. BMC Genomics. (2016) 17:592. doi: 10.1186/s12864-016-2935-4
4. Mukiibi R, Vinsky M, Keogh KA, Fitzsimmons C, Stothard P, Waters SM, et al. Transcriptome analyses reveal reduced hepatic lipid synthesis and accumulation in more feed efficient beef cattle. Sci Rep. (2018) 8:7303. doi: 10.1038/s41598-018-25605-3
5. MacNeil MD, Berry DP, Clark SA, Crowley JJ, Scholtz MM. Evaluation of partial body weight for predicting body weight and average daily gain in growing beef cattle. Transl Anim Sci. (2021) 5:1–12. doi: 10.1093/tas/txab126
6. Wells RS, Interrante SM, Sakkuma SS, Walker RS, Butler TJ. Accuracy of the VYTELLE SENSE in-pen weighing positions. Applied Anim Sci. (2021) 37:626–34. doi: 10.15232/aas.2021-02183
7. Benfield D, Garossino K, Sainz RD, Kerley MS, Huisma C. Conversion of high-frequency partial body weights to total body weight in feedlot cattle. J Anim Sci. (2017) 95:241–2. doi: 10.2527/asasann.2017.495
8. Durunna ON, Mujibi FDN, Goonewardene L, Okine EK, Wang Z, Moore SS. Feed efficiency differences and re-ranking in beef steers fed grower and finisher diets. J Anim Sci. (2011) 89:158–67. doi: 10.2527/jas.2009-2514
9. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
10. Rosen BD, Bickhart DM, Schnabel RD, Koren S, Elsik CG, Tseng E, et al. De novo assembly of the cattle reference genome with single-molecule sequencing. Gigascience. (2020) 9:giaa021. doi: 10.1093/gigascience/giaa021
11. Kim D, Langmead B, Salzberg SL. HISAT: a fast-spliced aligner with low memory requirements. Nat Methods. (2015) 12:357–60. doi: 10.1038/nmeth.3317
12. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence alignment/map format and SAMtools. Bioinformatics. (2009) 25:2078–89. doi: 10.1093/bioinformatics/btp352
13. Lawrence M, Huber W, Pages H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. (2013) 9:e1003118. doi: 10.1371/journal.pcbi.1003118
14. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550–8. doi: 10.1186/s13059-014-0550-8
15. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. (2020) 48:498–503. doi: 10.1093/nar/gkz1031
16. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g: Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc. (2019) 14:482–517. doi: 10.1038/s41596-018-0103-9
17. Luo W, Friedman MS, Shedden K, Hankenson K, Woolf PJ. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. (2009) 10:161. doi: 10.1186/1471-2105-10-161
18. Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. (2009) 37:1–17. doi: 10.1007/s00726-009-0269-0
19. Richardson E, Herd R. Biological basis for variation in residual feed intake in beef cattle. 2 Synthesis of results following divergent selection. Anim Prod Sci. (2004) 44:431–40. doi: 10.1071/EA02221
20. Taiwo G, Idowu M, Collins S, Sidney T, Wilson M, Pech-Cervantes A, et al. Chemical group-based metabolome analysis identifies candidate plasma biomarkers associated with residual feed intake in beef steers. Front Anim Sci. (2022) 2:783314. doi: 10.3389/fanim.2021.783314
21. Elolimy AA, Abdel-Hamied E, Hu L, McCann JC, Shike DW, Loor JJ. Residual feed intake in beef cattle is associated with differences in protein turnover and nutrient transporters in ruminal epithelium. J Anim Sci. (2019) 97:2181–7. doi: 10.1093/jas/skz080
22. Jorge-Smeding E, Renand G, Centeno D, Pétéra M, Durand S, Polakof S, et al. Metabolomics reveals changes in urea cycle associated to residual feed intake in growing heifers. In: Energy and Protein Metabolism and Nutrition. Wageningen: Wageningen Academic (2019). p. 231–2. doi: 10.3920/978-90-8686-891-9_50
23. Goldansaz SA, Markus S, Berjanskii M, Rout M, Guo AC, Wang Z, et al. Candidate serum metabolite biomarkers of residual feed intake and carcass merit in sheep. J Anim Sci. (2020) 98:10. doi: 10.1093/jas/skaa298
24. Coleman DN, Lopreiato V, Alharthi A, Loor JJ. Amino acids and the regulation of oxidative stress and immune function in dairy cattle. J Anim Sci. (2020) 98:S175–93. doi: 10.1093/jas/skaa138
25. Lyles JL, Calvo-Lorenzo MS, Bill E. Kunkle Interdisciplinary Beef Symposium: Practical developments in managing animal welfare in beef cattle: What does the future hold? J. Anim Sci. (2014) 92:5334–44. doi: 10.2527/jas.2014-8149
26. Knott S, Cummins L, Dunshea F, Leury B. Rams with poor feed efficiency are highly responsive to an exogenous adrenocorticotropin hormone (ACTH) challenge. Domest Anim Endocrinol. (2008) 34:261–8. doi: 10.1016/j.domaniend.2007.07.002
27. Ralph C, Tilbrook A. The hypothalamo-pituitary-adrenal (HPA) axis in sheep is attenuated during lactation in response to psychosocial and predator stress. Domest Anim Endocrinol. (2016) 55:66–73. doi: 10.1016/j.domaniend.2015.11.003
28. Kelly AK, Lawrence P, Earley B, Kenny DA, McGee M. Stress and immunological response of heifers divergently ranked for residual feed intake following an adrenocorticotropic hormone challenge. J Anim Sci Biotechnol. (2017) 8:65. doi: 10.1186/s40104-017-0197-x
29. Gomes RC, Sainz RD, Leme PR. Protein metabolism, feed energy partitioning, behavior patterns and plasma cortisol in Nellore steers with high and low residual feed intake. Revista Brasileira de Zootecnia. (2013) 42:44–50. doi: 10.1590/S1516-35982013000100007
30. Olsson MG, Allhorn M, Olofsson T, Akerstrom B. Up-regulation of alpha1-microglobulin by hemoglobin and reactive oxygen species in hepatoma and blood cell lines. Free Radic Biol Med. (2007) 42:842–51. doi: 10.1016/j.freeradbiomed.2006.12.017
31. Maurya PK, Kumar P, Chandra P. Biomarkers of oxidative stress in erythrocytes as a function of human age. World J Methodol. (2015) 5:216–22. doi: 10.5662/wjm.v5.i4.216
32. Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. (2007) 14:1207–13. doi: 10.1038/nsmb1344
33. Nielsen MJ, Moller HJ, Moestrup SK. Hemoglobin and heme scavenger receptors. Antioxid Redox Signal. (2010) 12:261–73. doi: 10.1089/ars.2009.2792
34. Kalapotharakos G, Murtoniemi K, Åkerström B, Hämäläinen E, Kajantie E, Räikkönen K, et al. Plasma heme scavengers alpha-1-microglobulin and hemopexin as biomarkers in high-risk pregnancies. Front Physiol. (2019) 10:300. doi: 10.3389/fphys.2019.00300
35. Miller JK, Brzezinska-Slebodzinska E, Madsen FC. Oxidative stress, antioxidants, and animal function. J Dairy Sci. (1993) 76:2812–23. doi: 10.3168/jds.S0022-0302(93)77620-1
36. Bottje WG, Carstens G. Association of mitochondrial function and feed efficiency in poultry and livestock species. J Anim Sci. (2009) 87:48–63. doi: 10.2527/jas.2008-1379
37. Radi R. Oxygen radicals, nitric oxide, and peroxynitrite: redox pathways in molecular medicine. Proc Natl Acad Sci USA. (2018). 115:5839–48. doi: 10.1073/pnas.1804932115
38. Boveris A, Chance B. The mitochondrial generation of hydrogen peroxide. general properties and effect of hyperbaric oxygen. Biochem J. (1973) 134:707–16. doi: 10.1042/bj1340707
39. Iqbal M, Pumford N, Lassiter K, Tang ZX, Wing T, Cooper M, et al. Compromised liver mitochondrial function and complex activity in low feed efficient broilers within a single genetic line associated with higher oxidative stress and differential protein expression. Poult Sci. (2005) 84:933–41. doi: 10.1093/ps/84.6.933
40. Grubbs JK, Fritchen AN, Huff-Lonergan E, Dekkers JC, Gabler NK, Lonergan SM. Divergent genetic selection for residual feed intake impacts mitochondria reactive oxygen species production in pigs. J Anim Sci. (2013) 91:2133–40. doi: 10.2527/jas.2012-5894
41. Casal A, Garcia-Roche M, Navajas EA, Cassina A, Carriquiry M. Differential hepatic oxidative status in steers with divergent residual feed intake phenotype. Animal. (2020) 14:78–85. doi: 10.1017/S1751731119001332
Keywords: protein metabolism, cellular response, feed efficiency, oxidative stress, heat stress
Citation: Taiwo GA, Idowu M, Denvir J, Cervantes AP and Ogunade IM (2022) Identification of Key Pathways Associated With Residual Feed Intake of Beef Cattle Based on Whole Blood Transcriptome Data Analyzed Using Gene Set Enrichment Analysis. Front. Vet. Sci. 9:848027. doi: 10.3389/fvets.2022.848027
Received: 03 January 2022; Accepted: 08 March 2022;
Published: 18 April 2022.
Edited by:
Rajib Deb, National Research Centre on Pig (ICAR), IndiaReviewed by:
Jon Schoonmaker, Purdue University, United StatesSara Pegolo, University of Padua, Italy
John B. Hall, University of Idaho, United States
Copyright © 2022 Taiwo, Idowu, Denvir, Cervantes and Ogunade. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ibukun M. Ogunade, ibukun.ogunade@mail.wvu.edu
 Godstime A. Taiwo1
Godstime A. Taiwo1 Modoluwamu Idowu
Modoluwamu Idowu James Denvir
James Denvir Ibukun M. Ogunade
Ibukun M. Ogunade