- 1Department of Veterinary Microbiology and Pathology, College of Veterinary Medicine, Washington State University, Pullman, WA, United States
- 2Department of Parasitology, Faculty of Veterinary Medicine, University of Firat, Elâzig, Turkey
- 3Parasitology and Animal Diseases Department, Veterinary Research Institute, National Research Center, Giza, Egypt
- 4Animal Disease Research Unit, Agricultural Research Service, United States Department of Agriculture, Pullman, WA, United States
Bovine babesiosis, caused by Babesia bovis, is an economically significant tick-borne disease that imposes restrictions to livestock production worldwide. Current methods to control bovine babesiosis have severe limitations and novel approaches, including transmission-blocking vaccines, are needed. Members of the widely conserved CCp family are multidomain adhesion proteins containing LCCL motifs, which are differentially expressed on gametocytes of apicomplexans, including Babesia spp. and Plasmodium spp. While Plasmodium parasites contain 6 distinct CCp genes, only three members (CCp 1-3) were previously identified in B. bovis. In this study, we describe the identification and characterization of two novel non-canonical members of the CCp gene family in B. bovis, named CCp5 and FNPA. The genes were identified in silico by TBLASTN using P. falciparum CCp family domains as queries. Unlike CCp1-3, the B. bovis CCp5 and FNPA proteins lack the LCCL canonical domain but contain other typical multidomain adhesion motifs which are present in classical CCp proteins. In addition, the B. bovis CCp5 and FNPA are in synteny with known CCp genes in related apicomplexans. Sequence analysis of these two proteins demonstrated high sequence conservation among B. bovis different isolates. Transcription, immunoblot, and immunofluorescence analyses demonstrated expression of CCp5 and FNPA in blood and in vitro induced sexual stages of B. bovis. The FNPA, in contrast to CCp5, has a predicted transmembrane domain, suggesting that it might be expressed in the surface of sexual stage parasites. Altogether, finding of this study support FNPA as a possible target of a transmission-blocking vaccine against B. bovis.
Introduction
Babesia bovis, which is transmitted by Rhipicephalus spp. ticks, is a hemoparasite responsible for bovine babesiosis, a disease that causes enormous economic losses to the cattle industry in tropical and subtropical regions worldwide. Babesia parasites have a complex lifecycle that includes the development of asexual stages in vertebrate hosts and sexual stages inside their definitive tick vectors (1–3). Sporozoites, the infectious form of B. bovis, are introduced via tick saliva into the bovine host by infected Rhipicephalus spp., where they invade and reproduce asexually as merozoites in red blood cells (RBC). Upon feeding on infected cattle, ticks may ingest B. bovis-infected RBCs and B. bovis gametogenesis is induced while in the tick midgut lumen, leading to zygote formation (4, 5). In some species of Babesia, gametocytes have been identified in host RBCs, but it is impossible to distinguish them under the light microscope (6, 7). Babesia bovis acute infection in cattle results in high fever, anorexia, inappetence, and severe intravascular hemolytic anemia. In addition, B. bovis expresses proteins that facilitate cytoadhesion of infected RBCs to capillaries, causing neurological symptoms and general organ failure, leading to rapid death of cattle, especially in immunologically naïve adult (more than one-year old) animals (3, 8, 9).
Commonly used methods to control acute bovine babesiosis combine approaches for tick management, immunization with live, attenuated, blood based Babesia vaccines, and babesicidal drugs. Despite being relatively effective, these interventions have various disadvantages and could be improved. Production of live B. bovis vaccines is expensive and laborious, requiring serial rapid passages of virulent strains in splenectomized bovines to obtain an attenuated parasite strain. In addition, these vaccines require a cold chain for deployment, are often transmissible by competent ticks, and there is a risk for the parasites to revert to virulence (1–3, 8). Thus, alternative subunit vaccines that can contribute toward effective control and eradication of bovine babesiosis are urgently needed. The current arsenal of measures against bovine babesiosis could also be greatly enhanced by the addition of transmission-blocking vaccines (TBV). Such TBV could be based on parasite sexual stage proteins, such as HAP2 (10), members of the CCp family (11) and 6cys (12–14).
CCp is highly conserved family of six multidomain adhesion proteins, containing LCCL motifs, which are differentially expressed on the surface of apicomplexan gametocytes. Previous experiments performed in the Babesia-related Plasmodium parasites demonstrated that knocking out CCp genes leads to the blocking of sexual stage development of the parasite in the mosquito vector (15–17). Three CCp genes, denominated CCp1-3, were previously identified in B. bovis and B. bigemina (11). In the current study we describe the identification of two novel genes of the B. bovis CCp family, which we named CCp5 and FNPA. The newly identified B. bovis FNPA and CCp5 proteins lack the LCCL domain characteristic of the other members of the CCp family, but do contain other functionally relevant motifs found in Plasmodium CCps (PfCCps), and their coding genes have conserved synteny with other CCp genes in related apicomplexans. Therefore, we propose CCp5 and FNPA as non-canonical members of the B. bovis CCp family. The findings of this work contribute toward a better understanding of the biology of B. bovis and support the rationale for using members of the CCp family in the development of novel TBV against B. bovis.
Materials and Methods
In silico Genes Identification by Genomic Search and Bioinformatics Analysis
The novel members of the CCp gene family were identified by performing BLAST searches (http://blast.ncbi.nlm.nih.gov/Blast.cgi) of the B. bovis genome using the amino acid sequences of CCp5/LAP3 (GenBank ID XP_001351021.1), and FNPA/LAP5 (GenBank ID XP_001348665.2) of P. falciparum 3D7 strain (18) as queries. Multiple alignments of amino acid sequences were generated using Clustal Omega Multiple Alignment (http://www.ebi.ac.uk/Tools/msa/clustalo). Phylogenetic tree prediction was generated by MEGAX program (19). Simple Molecular Architecture Research Tool “SMART” program (http://smart.embl-heidelberg.de), PROSITE program (https://prosite.expasy.org) and the Transmembrane Hidden Markov Model package 2 (TMHMM2) (http://www.cbs.dtu.dk/services/TMHMM-2.0) were used to predict domains and signal peptides in the CCp protein sequences. Synteny studies were carried out using the Piroplasma DB database (piroplasmadb.org/piro/app).
Babesia bovis in vitro Culture
The B. bovis S79-T3Bo strain was propagated in continuous microaerophilic stationary-phase culture, as previously described (10, 14) and used for gene and protein expression analyses. Genomic DNA from the B. bovis strains T2Bo (20), Mo7 (21, 22), and L17 (20) were used for gene amplification by PCR and followed by comparison of CCp5 and FNPA gene sequences.
Polymorphism and Genetic Analysis
PCR products derived from the full length CCp5 and FNPA genes from different strains of B. bovis were cloned into pCR-TOPO 2.1 vector (Invitrogen, CA, USA) and sequenced (Eurofins MWG Operon, Louisville, KY). Sequencing of PCR products was performed using the primers indicated in the Supplementary Table 1. The complete gDNA sequence for the newly identified genes were compared among four geographically distinct B. bovis strains including Texas (T2Bo) attenuated, T2Bo virulent (20), Mo7 (21, 22), and L17 (Argentina) (20). Strain-specific single nucleotide polymorphisms (SNPs) were then estimated to calculate the ratio of synonymous to non-synonymous changes (12–14). SNAP (http://hcv.lanl.gov/content/sequence/SNAP/SNAP.html) was used to investigate ω (dN/dS ratio) as follows: ω > 1 indicates positive selection, as the selection has caused some amino acid substitution; and ω <1 indicates occurrence of purifying selection and a high degree of sequence conservation (23). The Multiple Alignment using Fast Fourier Transform (MAFFT) (24) was used for DNA sequence alignment. The Molecular Evolutionary Genetics Analysis (MegaX) (19) was used to generate phylogenetic relationship among CCp family members. Nucleotide substitutions were manually calculated.
In vitro Induction of B. bovis Sexual Stages
Babesia bovis infected-RBCs were induced to form sexual stages in vitro using 100 μM xanthurenic acid (XA) (Sigma, St. Louis, MO, USA), as previously described (10, 25). Cultures were incubated up to 48 h at 27 C with 5% CO2, and collected at 0, 12, 24- and 48-h post-induction for RNA extraction. In addition, blood smears were prepared from induced and non-induced parasite cultures at different time points and stained with Giemsa stain to be further analyzed under light microscope for morphological changes.
RNA Isolation, CDNA Synthesis and Transcription Analysis for CCp Gene Family Members
Total RNA was extracted from induced and non-induced sexual stage B. bovis cultures. Parasites were collected in Trizol (Thermo Fisher Scientific, Waltham, MA, USA) and RNA extractions were performed using a phenol-based protocol. Two hundred ng of total RNAs were utilized for cDNA synthesis using the Superscript III™ cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer's protocol. Synthesized cDNA was used for PCR and quantitative PCR (qPCR) to test the transcript levels of CCp5 and FNPA in B. bovis parasite. The qPCRs were performed in a CFX96? Real-Time PCR Detection System using the SsoFast™ EvaGreen® Supermix (Bio-Rad, USA). The cycling conditions consisted of an enzyme activation step of 95°C for 30 s followed by 40 cycles of 95°C denaturation for 5 s and annealing/extension of 60°C for 5 s. Reactions were performed in duplicate in 20 μl using 200 nM of each primer and 2 μl of a 1/20 dilution of cDNA as template. The B. bovis gapdh gene (BBOV_II002540) was used as a reference gene and transcription level of CCp genes was normalized to blood stage (non-induced) parasites using the formula: relative expression (sample) = 2[Cq(control)−Cq(sample)], where time zero was used as Cq control. Supplementary Table 1 shows the primers used to evaluate the cDNA levels for CCp5 and FNPA. RT-PCR amplicons were cloned into pCR-TOPO 2.1 vector (Invitrogen, CA, USA) and sequenced (Eurofins MWG Operon, Louisville, KY). CCp5 and FNPA transcript levels were compared in blood and kinete stages using previously reported B. bovis RNAseq datasets (26).
Synthetic Peptide Design and Polyclonal Antibody Generation
Synthetic peptides ranging from 20 to 25 amino acids were produced based on the sequence of CCp5 and FNPA, as follows: Bo-CCp5 SRRHVTNASFSLFEDPSDSSNSDTS (aa16–40); and Bo-FNPA—GDINQDVVSPKDFTIPSGTD (aa 43–62). Peptides conjugated with keyhole limpet hemocyanin (KLH) were used for the immunization of rabbits (n = 2) as previously described (10, 11, 14) (Pacific Immunology Ramona, CA, USA) (protocol #11/30/20. Ref. SOP-1 for CCp5, protocol #12/28/20. Ref. SOP-1 for FNPA). The rabbit polyclonal antibodies were used in immunoblot and immunofluorescence assays to investigate the expression of the CCp5 and FNPA proteins.
Immunoblot Assays
Immunoblots were performed using a standard protocol, as previously described (11). Briefly, membranes were blocked in 5% non-fat milk and incubated separately with anti-CCp5 rabbit polyclonal antibodies (1:50), anti-FNPA rabbit polyclonal antibodies (1:50), and pre-immune polyclonal rabbit serum (1:50). The membranes were incubated with an appropriate HRP conjugated secondary antibody (goat anti-rabbit IgG peroxidase conjugate; Life Biosciences, Boston, MA, USA). The immune complexes were revealed using an enhanced chemiluminescence method (ECL™) (Amersham).
Immunofluorescence Assays (IFA)
Non-induced and induced B. bovis cultures were collected at 0, 12, 24, and 48 h and used for IFA. Cells were washed with 1X PBS at 900xg and pellets suspended with 1X PBS containing 3% bovine serum albumin (BSA). A smear was made by taking 5 μl from each cell preparation to a glass microscope slide. The slides were air dried, fixed for 5 min in cold acetone, and blocked with 1X PBS containing 10% BSA for 30 min. Then, slides were incubated with rabbit anti-CCp5 or -FNPA sera (1:50) in 1X PBS-10% BSA for 1 h. The slides were washed 3 times with 1X PBS and incubated in 10% BSA with goat anti-rabbit IgG AlexaFluor®555 (Thermo Fisher, Waltham, MA, USA), and then again washed 3 times with 1X PBS and mounted with a drop of Prolong™ Gold Anti-fade with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher, Waltham, MA, USA) and cover slip. The slides were analyzed using a Leica Sp8-X White Light Laser point scanning confocal microscope (Leica Microsystems, Wetzlar, Germany).
Results
Identification of Two New Non-canonical CCp Genes in B. bovis
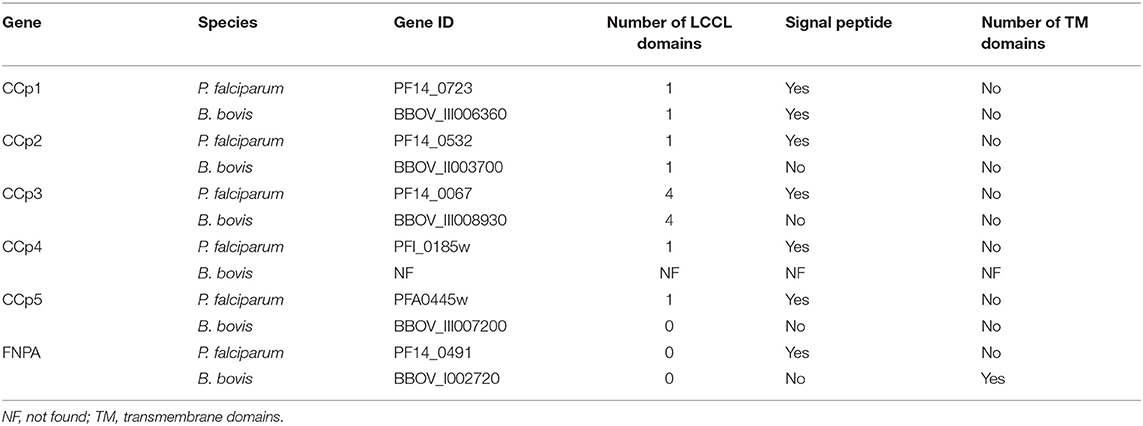
Three CCp gene family members were previously identified in B. bovis and denominated CCp1, CCp2 and CCp3 (11). However, database searches on the B. bovis genome using TBLASTN with the P. falciparum CCp family domains as queries resulted in the identification of two additional and previously unnoticed CCp-like genes, which we termed here as CCp5 and FNPA. The CCp5 and FNPA genes were originally annotated as BBOV_III007200 and BBOV_I002720 (PA14 domain containing proteins), respectively (Table 1). Single-copy B. bovis CCp5 and FNPA genes are located between positions 1,564,074 and 1,567,232 bp of chromosome 3, and positions 270,945 and 273,803 bp of chromosome 1, respectively, as schematically shown in Figure 1. While the B. bovis CCp5 gene lacks introns, the FNPA gene contains three introns (Table 2). The CCp5 and FNPA genes encode for proteins with predicted molecular weights of 117.5 and 98.9 KDa, respectively (Table 2).
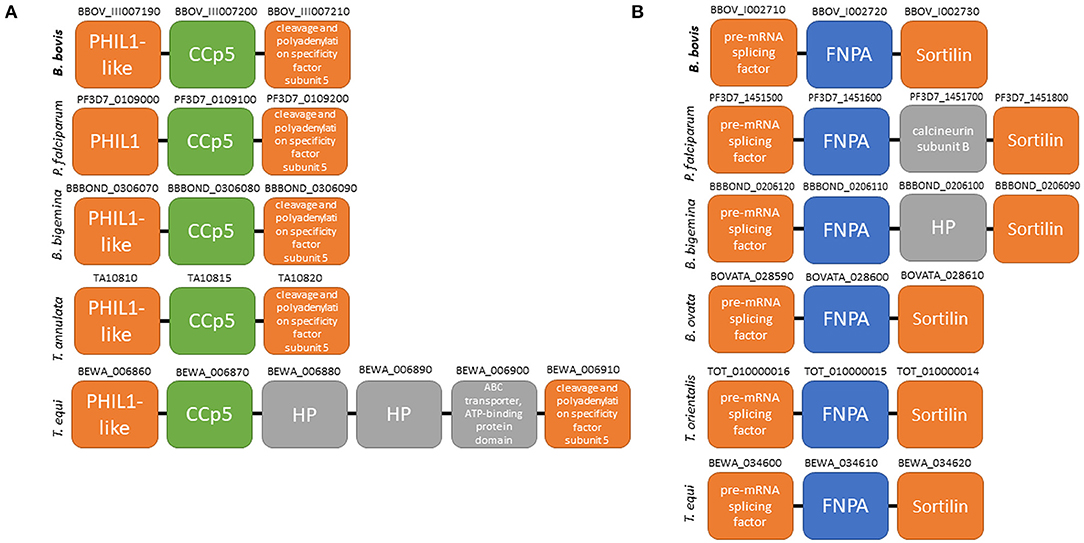
Figure 1. Schematic representation of gene localization and synteny maps of the B. bovis CCp5 (A) and FNPA (B) genes (bold) among several piroplasm species.
Synteny maps for CCp5 and FNPA genes show conservation among several distinct piroplasm species, including B. bigemina (Figure 1). However, similar homology and synteny analysis searches failed in identifying a gene homologous to CCp4 in the B. bovis genome (Supplementary Figure 1).
Analysis of domain architectures, using two different programs, confirmed the lack of LCCL domains in the CCp5 and FNPA proteins in B. bovis, However, a fibronectin type 2 domain (FN2), a collagen-binding domain, followed by a domain (PA14), similar to anthrax protective antigen, with potential roles in carbohydrate recognition were found in identical arrangements among these two proteins (Supplementary Figure 2). These domains are known to mediate calcium dependent interactions (27). A transmembrane domain was found only in B. bovis FNPA (Table 1; Figure 2), suggesting that this protein might be inserted in the cell membrane, and the possible exposure of functionally relevant protein domains on the surface of the parasite. A phylogenetic analysis performed with the five known CCp members of B. bovis together with similar CCp proteins of other related piroplasm species showed formation of five different clades corresponding to CCp1-3, FNPA, and CCp5 (Figure 3). Interestingly, a similar domain architecture search performed on the FNPA and CCp5 proteins of B. bigemina and Theileria also failed in detecting recognizable LCCL domains. Therefore, database searches, together with phylogenetic and synteny analysis, support the notion presented hereby that B. bovis FNPA and CCp5 are true homologs of the FNPA and CCp5 proteins of other related apicomplexan organisms. Also, results suggest that canonical LCCL domains might not be present in the CCp5 and FNPA proteins in Babesia and Theileria organisms.
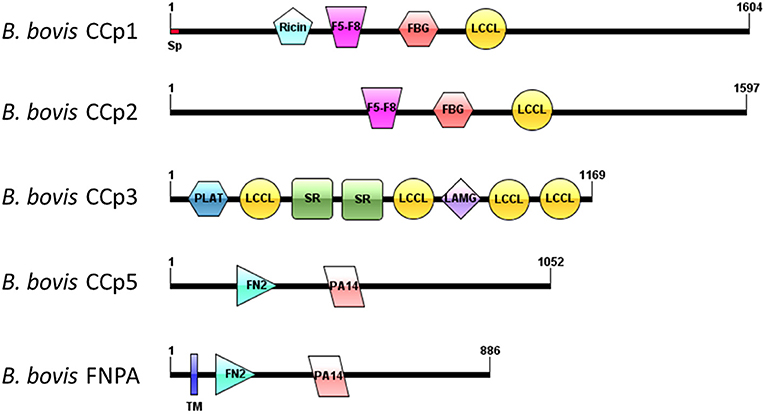
Figure 2. Schematic domain architectures representation of all CCp family members of B. bovis, including the novel non-canonical members CCp5 and FNPA. Diagrams were constructed using IBS program (28), version 1.0. Domain abbreviations are as follows: Sp, signal peptide; Ricin, Ricin domain, F5-F8, F5-F8 type C domain, FBG, Fibrinogen-related domains; LCCL, Limulus coagulation factor domain; PLAT, Polycystin-1, Lipoxygenase, Alpha-Toxin; SR, Scavenger receptor domain; LAMG, Laminin G domain; FN2, Fibronectine type 2 domain; PA14, Anthrax PA domain; TM, transmembrane domain.

Figure 3. Phylogenetic analyses of piroplasm CCp family protein sequences by maximum likelihood method based on the Whelan and Goldman + Freq. model (29). Sequences were obtained from the GenBank database. The newly identified B. bovis CCp5 and FNPA genes are shown in bold. The CCp4 gene sequence of P. falciparum was used as outgroup.
Cp5 and FNPA Genes Are Highly Conserved Among Geographically Distinct B. bovis Strains
The complete gDNA sequence for CCp5 and FNPA genes was compared among four different B. bovis strains including T2Bo attenuated, T2Bo virulent, Mo7, and L17 (Argentina). Sequence comparisons showed that CCp5 and FNPA genes are highly conserved among these four B. bovis strains (96 to 100% identity) (Supplementary Figure 3). Moreover, the calculated synonymous and non-synonymous S/N ratios with the parameter, ω (ω = dN/dS), as an indicator of potential selection pressures are shown in Table 3. In all cases, ω parameters <1 was obtained, providing no support for positive selection for the CCp5 and FNPA genes. Altogether, these observations suggest that these two genes are not likely exposed to immune selection in the vertebrate host. The full DNA sequences of the CCp5 and FNPA genes derived from these four distinct strains, and their alignment are shown in Supplementary Figures 4A,B, respectively. In addition, the CCp5 and FNPA genes are also well-conserved among different Babesia spp. and other piroplasm species (Supplementary Figure 5).
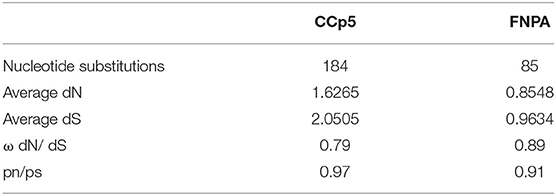
Table 3. Polymorphism and average SNPs of the CCp5 and FNPA genes among different distinct B. bovis strains.
CCp5 and FNPA Genes Are Expressed in Blood and Sexually Induced Stages of B. bovis
The levels of CCp5 and FNPA transcripts were investigated by RT-PCR analysis performed on RNA extracted from cultured non-induced and in vitro sexual-stage induced B. bovis parasites. The RAP-1 gene was also similarly amplified by nested PCR as control for cDNA quality of all stages (30), and the sexual stage marker 6cysA gene (14) was amplified for sexual stage development control (Figure 4A). The qPCR transcription analysis demonstrated differential expression of CCp5 and FNPA genes in blood and sexual stages (Figure 4B). The transcription levels of CCp5 were similar between sexually induced and blood stage parasites at 12 and 24 h post-sexual stage induction, but it significantly increased in sexually induced parasites at 48 h post induction. However, the levels of expression of the FNPA gene significantly increased at 12, 24 and 48 h post-sexual stage induction, compared to the non-induced blood stage parasites. The peak for FNPA gene expression was observed at 48 h after sexual stage induction by temperature.
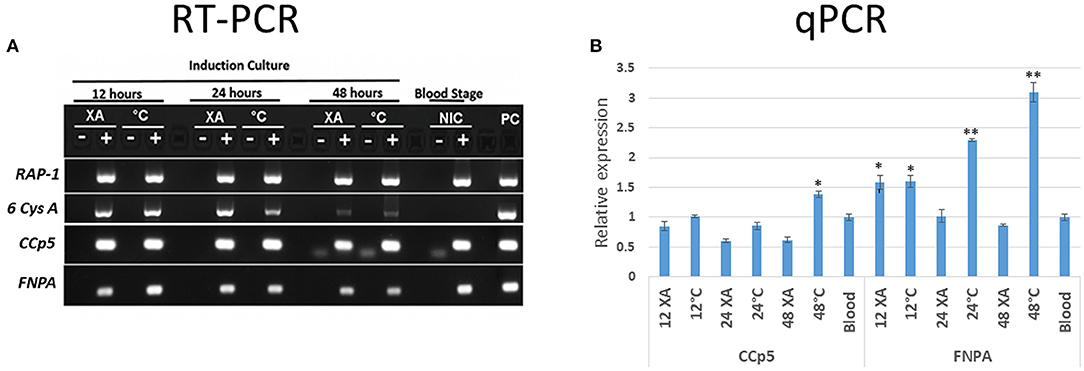
Figure 4. (A) The RT-PCR analysis for the detection of RAP-1, 6-Cys A, CCp5 and FNPA transcripts was performed using non-induced (blood stage-NIC) and induced (12, 24 and 48 h) in vitro cultures (XA, xanthurenic acid induction; C, temperature induction; PC, positive control. (B) Relative expression of B. bovis CCp5 and FNPA by non-induced (blood stage) and induced stages (12, 24, and 48 h) in vitro cultures. *p = 0.0.012–0.0028; **p < 0.0001 (p value compared with blood stage).
In addition, the B. bovis RNAseq datasets were used to compare the transcript levels of CCp5 and FNPA at the blood and kinete stages (Figure 5). Overall, these comparisons showed that the blood stage levels of CCp5 and FNPA transcript are significantly higher than the kinete stage levels (Figure 5).
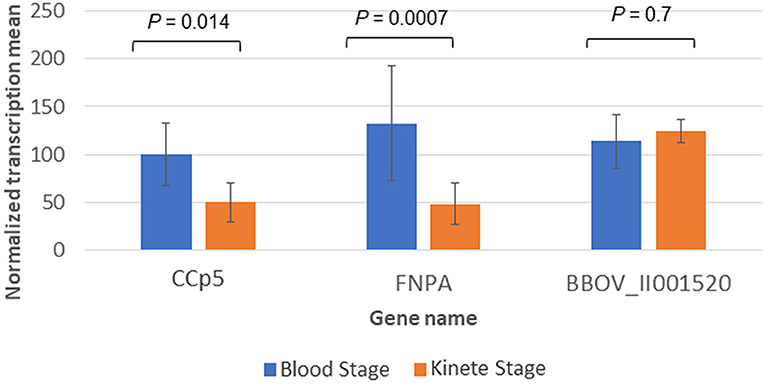
Figure 5. Differential expression of CCp5 and FNPA between B. bovis blood and kinete stages, respectively in the RNA-Seq datasets. The 26S proteasome non-ATPase gene (BBOV_II001520) was used as a control gene for blood and kinete stages.
Expression of CCp5 and FNPA proteins were also examined in in vitro non-induced and sexual stage induced (48 h-temperature induction) parasites by immunoblot analysis using rabbit polyclonal anti-CCp5 and FNPA peptides (Figure 6). Results showed that anti-CCp5 and anti-FNPA peptide antibodies weakly reacted with blood stage proteins of expected molecular weight of 117 and 97 KDa, respectively (Figures 6A,B). In contrast, reactivity of the monoclonal antibody BABB35 against MSA-1 was strongly evident in both, blood and sexual stage lysates (Figure 6C). Therefore, consistent with the transcription analysis, CCp5 and FNPA protein expression were also confirmed in blood and sexual stage parasites.
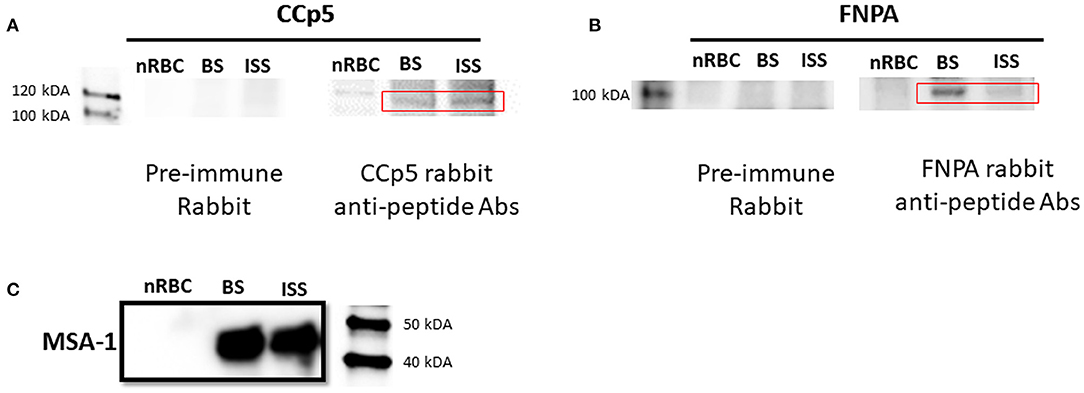
Figure 6. Immunoblot analysis using rabbit polyclonal antibodies against CCp5 peptide (A) and FNPA peptide (B) performed on blood stage (BS) and induced sexual stage (ISS) B. bovis parasites. The monoclonal antibody BABB35, reactive with MSA-1, was used as a control (C).
Immunofluorescence assays using rabbit polyclonal anti-CCp5 and FNPA peptides also revealed expression of CCp5 and FNPA proteins in in vitro non-induced (blood stage-BS) and induced (12–24–48 h-XA induction) parasites. Fluorescence signals were more intense for sexually in vitro induced forms of the parasite compared to blood stage or non-induced forms (Figure 7). Pre-immune rabbit sera did not show any IFA reactivity against blood stage or in vitro induced sexual stages (Supplementary Figure 6).
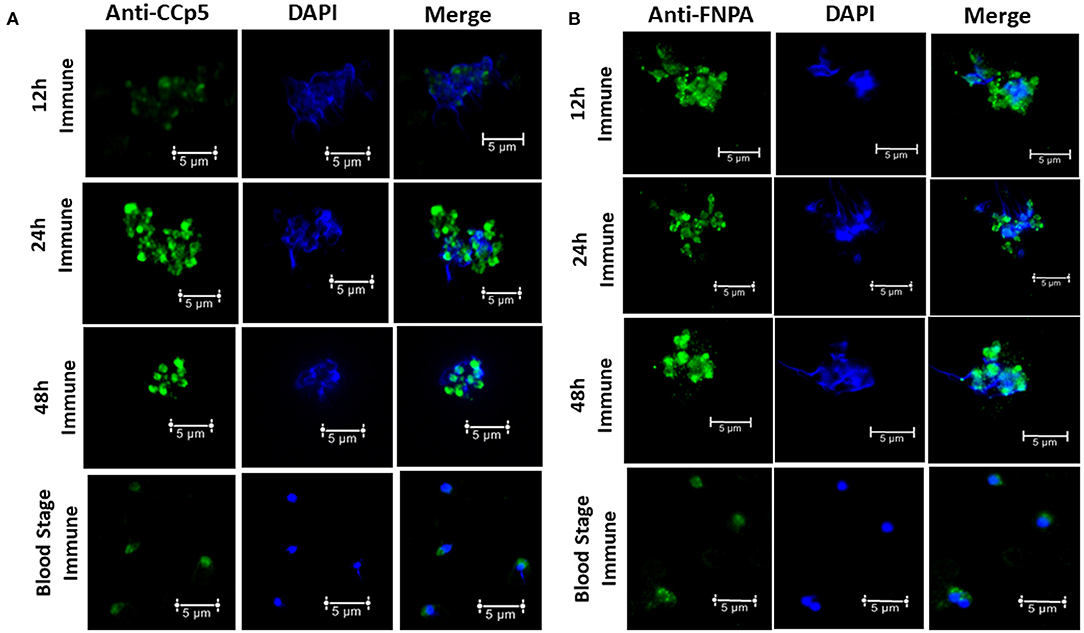
Figure 7. Immunofluorescence assays demonstrating the expression of CCp5 (A) and FNPA (B) in blood stage and in vitro induced sexual stages (12–24–48 h) of B. bovis.
Discussion
The CCp protein family, which has an important role in sporozoite development and infectivity, consists of six members with a modular structure composed of multiple protein, lipid and carbohydrate binding domains, including LCCL domains in Plasmodium parasites. However, while the Pf CCp1-Pf CCp5 proteins contain at least one LCCL domain, Pf FNPA, which lacks such domain, has also been included as a member of the Pf CCp protein family due to its structural similarity to Pf CCp5 (15–17). The FNPA acronym is derived from the domains FN2 and PA14 present in this proteins (15). Although six LCCL protein family members have been identified in P. falciparum, only genes encoding homologs of CCp1-3 proteins have been reported in B. bovis and B. bigemina so far (11). In this study, we describe the presence of two additional single gene-copy CCp-related genes (CCp5 and FNPA) in the genome of B. bovis and B. bigemina, as previously described in Plasmodium spp. (16, 31). While B. bovis FNPA is structurally similar to Pf FNPA, the B. bovis, B. bigemina and T. equi CCp5 proteins contain no canonical LCCL domains, unlike Pf CCp5. However, the B. bovis CCp5 clusters with other Babesia sp. and Plasmodium parasites in the phylogenetic analysis presented in this study. This finding, together with conserved synteny, supports the denomination of this protein as B. bovis CCp5, despite the lack of the canonical LCCL domain.
Interestingly, no genes encoding for a protein equivalent to Pf CCp4 were found in the genome of Babesia parasites, either by homology searches or by synteny analysis, suggesting that this protein might play a significant role during the life cycle of Plasmodium but not for Babesia parasites. This may correlate with the observation that the CCp gene family encodes for proteins with parasite functions operating during sexual and other stages expressed in their arthropod vectors. Thus, the lack of CCp4 genes might be related to the many differences that occur among the Babesia and Plasmodium life cycles, including the fact that Babesia parasites are transovarially transmitted, and the different nature of their arthropod hosts vectors in comparison with Plasmodium spp. (ticks vs. mosquitos). Alternatively, it is also possible that other CCp genes present in Babesia spp. play similar or overlapping functional roles as the Plasmodium CCp4.
CCp orthologs are also present in the rodent malaria species Plasmodium berghei, where they were termed LAP proteins. Several studies have been carried out to determine the function of this protein family in P. berghei (15, 32). Genetic crossover studies have shown that PbCCp/LAP proteins are female-specific markers (33). In addition, recent studies on GFP fusions of PbCCp3, PbCCp1 and PbCCp5 have shown protein expression in macrogametocytes and accumulation of these proteins in crystalloid organelles that form in the ookinete and persist until the early oocyst stage (34, 35). Crystalloids are transient specialized structures of malaria parasites which are present in mosquito-specific ookinetes and young oocyst stages of the parasite. They appear to play an important role in protein trafficking and sporozoite transmission and could be exploited as new targets for control of malaria transmission (36). Knockout of LAP1 (CCp3) or LAP3 (CCp5) was shown to completely abolish crystalloid formation in P. berghei (37). While sporozoites are formed in Babesia parasites, crystalloids and oocysts have not been so far reported, and whether CCp proteins may play similar roles in Babesia spp. remains unknown, and more research is needed.
The CCp5 and FNPA nucleotide and amino acid sequences were found to be highly conserved among otherwise distinct B. bovis isolates, and even among other related apicomplexans. High levels of sequence conservation among geographical strains is a very important criteria for selecting effective vaccine candidate antigens. Sequence comparisons of the CCp5 and FNPA genes were also used to investigate whether immune selection pressure in the bovine host affects this gene family. The calculated ω (dN/dS) ratios for CCp5 and FNPA are <1, suggesting that these gene family members are unlikely exposed to host immune system selective forces.
In this study, we demonstrated transcription of the CCp5 and FNPA genes in both blood and in vitro sexual stages of B. bovis. We also showed that the expression of CCp5 and FNPA genes is up regulated in B. bovis sexual stages (Figure 4B). In addition, according to the RNAseq data, the CCp5 and FNPA transcript levels in the kinete stage were found to be downregulated compared with blood stages. These results demonstrate that the expression of CCp5 and FNPA genes needs to be tightly regulated and exert different transcriptional patterns at developmental stages of B. bovis, which may reflect distinct functional requirements. Furthermore, relatively low abundance of CCp5 and FNPA proteins were observed in both blood and sexual stages by immunoblot analysis. Consistently, Pf CCp5 and Pf FNPA have been reported to be less abundant in gametocytes than the other four Pf CCp proteins (31). In addition, Pf CCp5 also showed transcript expression in both asexual and sexual stage Plasmodium parasites (16). In this study, we demonstrated expression of CCp5 and FNPA proteins in blood and sexual stages by immunoblot and fixed IFA. Although a more intense fluorescent signal was observed in in vitro induced sexual stages than in blood stage parasites, we are limited to interpret this observation as a strong evidence of increased protein expression because sexual stage parasites tend to aggregate (38), which could result in more intense IFA signals and mislead the interpretation of the results.
Ideally, TBV candidate antigens should be widely conserved and expressed on the surface of the parasites, where they can be possible targets of antibodies. Bioinformatics analysis suggests that the CCp5 and FNPA proteins lack detectable signal peptides, but FNPA, unlike CCp5, has a transmembrane domain, suggesting that it might also be exposed on the surface of sexual stage parasites. Taking this data together, FNPA fulfills, at least in part, the criteria to be considered as a transmission blocking vaccine candidate.
Overall, the data in this study demonstrate the presence of CCp5 and FNPA genes in B. bovis as non-canonical members of the CCp family, which might play important roles for the development of the parasite stages in ticks and vertebrates. Future studies need to be focused on knock out/knock in approaches of these genes in B. bovis for functional analyses.
Conclusion
In this study, bioinformatics, pattern of expression, and localization analysis of CCp5 and FNPA genes belonging to the B. bovis CCp family were performed. Findings from this study demonstrated expression of CCp5 and FNPA genes and proteins in both blood and sexual stages of B. bovis, but their expression is increased in in vitro induced sexual stage parasites Furthermore, expression analysis, serology data, and the presence of a TM domain suggest that FNPA may be surface exposed during tick stages of the parasites, and thus, a possible candidate target for TBV.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author/s.
Author Contributions
CS, RB, and SO: conceptualization. SO, JL, and HA: investigation. SO, CS, HA, JL, and RB: formal analysis, writing, review, and editing. SO, HA, and CS: visualization. CS: supervision. CS and SO: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Scientific and Technological Council of Turkey (TUBITAK) 2219 Grant Program, the International Development Research Center (IDRC) [Livestock Vaccine Innovation Fund (Grant 108525), funded by the Canadian Government and the Bill and Melinda Gates Foundation], the USDA CRIS Project 2090- 32000-039-00-D, and the USDA National Institute of Food and Agriculture (NIFA) (Award Number: 2020-67015-31809, Proposal Number: 2019-05375, Accession Number: 1022541).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Paul Lacy for the excellent technical and administrative support. We also express our gratitude to Lan He, Jinna Navas, Manuel Rojas, Hala Hussein, Rubikah Vimonish, Janaina Capelli-Peixoto, Wendell Johnson and Massaro Ueti for their assistance. We thank Vignesh Ambothi Rathinasamy and Brian Cooke for fruitful discussions.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.833183/full#supplementary-material
References
1. Uilenberg G. Babesia - A historical overview. Vet Parasitol. (2006) 138:3–10. doi: 10.1016/j.vetpar.2006.01.035
2. Schnittger L, Rodriguez AE, Florin-Christensen M, Morrison DA. Babesia: a world emerging. Infect Genet Evol. (2012) 12:1788–809. doi: 10.1016/j.meegid.2012.07.004
3. Suarez CE, Noh S. Emerging perspectives in the research of bovine babesiosis and anaplasmosis. Vet Parasitol. (2011) 180:109–25. doi: 10.1016/j.vetpar.2011.05.032
4. Jalovecka M, Hajdusek O, Sojka D, Kopacek P, Malandrin L. The complexity of piroplasms life cycles. Front Cell Infect Microbiol. (2018) 8:248. doi: 10.3389/fcimb.2018.00248
5. Jalovecka M, Sojka D, Ascencio M, Schnittger L. Babesia life cycle - when phylogeny meets biology. Trends Parasitol. (2019) 35:356–68. doi: 10.1016/j.pt.2019.01.007
6. Lobo CA, Rodriguez M, Cursino-Santos JR. Babesia and red cell invasion. Curr Opin Hematol. (2012) 19:170–5. doi: 10.1097/MOH.0b013e328352245a
7. Rudzinska MA SA, Riek RF, Lewengrub SJ, Piesman J. Intraerythrocytic gametocytes of Babesia microti and their maturation in ticks. Can J Zool. (1979) 57:424–34. doi: 10.1139/z79-050
8. Bock R, Jackson L, De Vos A, Jorgensen W. Babesiosis of cattle. Parasitology. (2004) 129:S247–S69. doi: 10.1017/S0031182004005190
9. Ozubek S, Bastos RG, Alzan HF, Inci A, Aktas M, Suarez CE. Bovine babesiosis in turkey: impact, current gaps, and opportunities for intervention. Pathogens. (2020) 9:1041. doi: 10.3390/pathogens9121041
10. Hussein HE, Bastos RG, Schneider DA, Johnson WC, Adham FK, Davis WC, et al. The Babesia bovis hap2 gene is not required for blood stage replication, but expressed upon in vitro sexual stage induction. Plos Neglect Trop D. (2017) 11:e0005965. doi: 10.1371/journal.pntd.0005965
11. Bastos RG, Suarez CE, Laughery JM, Johnson WC, Ueti MW, Knowles DP. Differential expression of three members of the multidomain adhesion CCp family in Babesia bigemina, Babesia bovis and Theileria equi. PLoS One. (2013) 8:e0005965. doi: 10.1371/journal.pone.0067765
12. Alzan HF, Bastos RG, Ueti MW, Laughery JM, Rathinasamy VA, Cooke BM, et al. Assessment of Babesia bovis 6cys A and 6cys B as components of transmission blocking vaccines for babesiosis. Parasite Vector. (2021) 14:210. doi: 10.1186/s13071-021-04712-7
13. Alzan HF, Cooke BM, Suarez CE. Transgenic Babesia bovis lacking 6-Cys sexual-stage genes as the foundation for non-transmissible live vaccines against bovine babesiosis. Ticks Tick-Borne Dis. (2019) 10:722–8. doi: 10.1016/j.ttbdis.2019.01.006
14. Alzan HF, Lau AOT, Knowles DP, Herndon DR, Ueti MW, Scoles GA, et al. Expression of 6-cys gene superfamily defines Babesia bovis sexual stage development within Rhipicephalus microplus. PLoS One. (2016) 11:e0163791. doi: 10.1371/journal.pone.0163791
15. Kuehn A, Simon N, Pradel G. Family members stick together: multi-protein complexes of malaria parasites. Med Microbiol Immun. (2010) 199:209–26. doi: 10.1007/s00430-010-0157-y
16. Pradel G, Hayton K, Aravind L, Iyer LM, Abrahamsen MS, Bonawitz A, et al. A multidomain adhesion protein family expressed in Plasmodium falciparum is essential for transmission to the mosquito. J Exp Med. (2004) 199:1533–44. doi: 10.1084/jem.20031274
17. Pradel G, Wagner C, Mejia C, Templeton TJ. Plasmodium falciparum: co-dependent expression and co-localization of the Pf CCp multi-adhesion domain proteins. Exp Parasitol. (2006) 112:263–8. doi: 10.1016/j.exppara.2005.11.010
18. Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. (2002) 419:498–511. doi: 10.1038/nature01097
19. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
20. Lau AOT, Kalyanaraman A, Echaide I, Palmer GH, Bock R, Pedroni MJ, et al. Attenuation of virulence in an apicomplexan hemoparasite results in reduced genome diversity at the population level. BMC Genomics. (2011) 12:410. doi: 10.1186/1471-2164-12-410
21. Hines SA, McElwain TF, Buening GM, Palmer GH. Molecular characterization of Babesia bovis merozoite surface proteins bearing epitopes immunodominant in protected cattle. Mol Biochem Parasitol. (1989) 37:1–9. doi: 10.1016/0166-6851(89)90096-0
22. Rodriguez SD, Buening GM, Green TJ, Carson CA. Cloning of Babesia bovis by in vitro cultivation. Infect Immun. (1983) 42:15–8. doi: 10.1128/iai.42.1.15-18.1983
23. Massingham T, Goldman N. Detecting amino acid sites under positive selection and purifying selection. Genetics. (2005) 169:1753–62. doi: 10.1534/genetics.104.032144
24. Katoh K, Standley DM. MAFFT: multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. (2013) 30:772–80. doi: 10.1093/molbev/mst010
25. Mosqueda J, Falcon A, Alvarez JA, Ramos JA, Oropeza-Hernandez LF, Figueroa JV. Babesia bigemina sexual stages are induced in vitro and are specifically recognized by antibodies in the midgut of infected Boophilus microplus ticks. Int J Parasitol. (2004) 34:1229–36. doi: 10.1016/j.ijpara.2004.07.003
26. Ueti MW, Johnson WC, Kappmeyer LS, Herndon DR, Mousel MR, Reif KE, et al. Comparative analysis of gene expression between Babesia bovis blood stages and kinetes allowed by improved genome annotation. Int J Parasitol. (2021) 51:123–36. doi: 10.1016/j.ijpara.2020.08.006
27. Rigden DJ, Mello LV, Galperin MY. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem Sci. (2004) 29:335–9. doi: 10.1016/j.tibs.2004.05.002
28. Liu WZ, Xie YB, Ma JY, Luo XT, Nie P, Zuo ZX, et al. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. (2015) 31:3359–61. doi: 10.1093/bioinformatics/btv362
29. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. (2001) 18:691–9. doi: 10.1093/oxfordjournals.molbev.a003851
30. Figueroa JV, Chieves LP, Johnson GS, Buening GM. Multiplex polymerase chain-reaction based assay for the detection of Babesia bigemina, Babesia bovis and Anaplasma marginale DNA in bovine blood. Vet Parasitol. (1993) 50:69–81. doi: 10.1016/0304-4017(93)90008-B
31. Scholz SM, Simon N, Lavazec C, Dude MA, Templeton TJ, Pradel G. Pf CCp proteins of Plasmodium falciparum: gametocyte-specific expression and role in complement-mediated inhibition of exflagellation. Int J Parasitol. (2008) 38:327–40. doi: 10.1016/j.ijpara.2007.08.009
32. Trueman HE, Raine JD, Florens L, Dessens JT, Mendoza J, Johnson J, et al. Functional characterization of an LCCL-lectin domain containing protein family in Plasmodium berghei. J Parasitol. (2004) 90:1062–71. doi: 10.1645/GE-3368
33. Raine JD, Ecker A, Mendoza J, Tewari R, Stanway RR, Sinden RE. Female inheritance of malarial lap genes is essential for mosquito transmission. PLoS Pathog. (2007) 3:e30. doi: 10.1371/journal.ppat.0030030
34. Carter V, Shimizu S, Arai M, Dessens JT. PbSR is synthesized in macrogametocytes and involved in formation of the malaria crystalloids. Mol Microbiol. (2008) 68:1560–9. doi: 10.1111/j.1365-2958.2008.06254.x
35. Saeed S, Carter V, Tremp AZ, Dessens JT. Plasmodium berghei crystalloids contain multiple LCCL proteins. Mol Biochem Parasit. (2010) 170:49–53. doi: 10.1016/j.molbiopara.2009.11.008
36. Dessens JT, Tremp AZ, Saeed S. Crystalloids: fascinating parasite organelles essential for malaria transmission. Trends Parasitol. (2021) 37:581–4. doi: 10.1016/j.pt.2021.04.002
37. Saeed S, Tremp AZ, Dessens JT. Biogenesis of the crystalloid organelle in Plasmodium involves microtubule-dependent vesicle transport and assembly. Int J Parasitol. (2015) 45:537–47. doi: 10.1016/j.ijpara.2015.03.002
38. Hussein HE, Johnson WC, Taus NS, Capelli-Peixoto J, Suarez CE, Mousel MR, et al. Differential expression of calcium-dependent protein kinase 4, tubulin tyrosine ligase, and methyltransferase by xanthurenic acid-induced Babesia bovis sexual stages. Parasite Vector. (2021) 14:395. doi: 10.1186/s13071-021-04902-3
Keywords: Babesia bovis, bovine babesiosis, sexual stages, transmission blocking vaccine, CCp protein family
Citation: Ozubek S, Alzan HF, Bastos RG, Laughery JM and Suarez CE (2022) Identification of CCp5 and FNPA as Novel Non-canonical Members of the CCp Protein Family in Babesia bovis. Front. Vet. Sci. 9:833183. doi: 10.3389/fvets.2022.833183
Received: 10 December 2021; Accepted: 10 January 2022;
Published: 15 February 2022.
Edited by:
Hany M. Ibrahim, Menoufia University, EgyptReviewed by:
Mahmoud Rezk Ali AbouLaila, Damanhour University, EgyptMünir Aktaş, Firat University, Turkey
Copyright © 2022 Ozubek, Alzan, Bastos, Laughery and Suarez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sezayi Ozubek, c2V6YXlpLm96dWJlayYjeDAwMDQwO3dzdS5lZHU=; c296dWJlayYjeDAwMDQwO2ZpcmF0LmVkdS50cg==; Carlos E. Suarez, Y2FybG9zLnN1YXJleiYjeDAwMDQwO3VzZGEuZ292
 Sezayi Ozubek
Sezayi Ozubek Heba F. Alzan1,3
Heba F. Alzan1,3 Reginaldo G. Bastos
Reginaldo G. Bastos Carlos E. Suarez
Carlos E. Suarez