- 1College of Animal Science and National Engineering Research Center for Breeding Swine Industry, South China Agricultural University, Guangzhou, China
- 2Guangdong Wens Breeding Swine Technology Co., Ltd., Yunfu, China
- 3Guangdong Provincial Laboratory of Lingnan Modern Agricultural Science and Technology, Guangzhou, China
Runs of homozygosity (ROH) are widely used to investigate genetic diversity, demographic history, and positive selection signatures of livestock. Commercial breeds provide excellent materials to reveal the landscape of ROH shaped during the intense selection process. Here, we used the GeneSeek Porcine 50K single-nucleotide polymorphism (SNP) Chip data of 3,770 American Duroc (AD) and 2,096 Canadian Duroc (CD) pigs to analyze the genome-wide ROH. First, we showed that AD had a moderate genetic differentiation with CD pigs, and AD had more abundant genetic diversity and significantly lower level of inbreeding than CD pigs. In addition, sows had larger levels of homozygosity than boars in AD pigs. These differences may be caused by differences in the selective intensity. Next, ROH hotspots revealed that many candidate genes are putatively under selection for growth, sperm, and muscle development in two lines. Population-specific ROHs inferred that AD pigs may have a special selection for female reproduction, while CD pigs may have a special selection for immunity. Moreover, in the overlapping ROH hotspots of two Duroc populations, we observed a missense mutation (rs81216249) located in the growth and fat deposition-related supergene (ARSB-DMGDH-BHMT) region. The derived allele of this variant originated from European pigs and was nearly fixed in Duroc pigs. Further selective sweep and association analyses indicated that this supergene was subjected to strong selection and probably contributed to the improvement of body weight and length in Duroc pigs. These findings will enhance our understanding of ROH patterns in different Duroc lines and provide promising trait-related genes and a functional-altering marker that can be used for genetic improvement of pigs.
Introduction
Runs of homozygosity (ROH) are defined as a contiguous genome segment of the identical haplotype inherited from a common ancestor without recombination (1). ROH fragments are widely distributed in human and livestock genomes, and the patterns of ROH length and frequency distribution in the genome are mainly attributed to demographic history and selection (1, 2). The short ROH fragment indicates ancestral inbreeding, while the longer ROH fragment reflects recent close inbreeding (3). Hence, ROH are considered as an advanced method for assessing the degree of inbreeding (FROH) in individuals and populations, providing support for the true level of homozygosity (4). ROH patterns are not randomly distributed throughout the genome. Selection may strongly affect the distribution of ROH, and regions of genomic loci under selection tend to generate a high frequency of ROH (hotspots) (5). An increasing number of studies have confirmed that ROH hotspots are due to positive selection for economically important traits in cattle (6, 7), pigs (8–10), chickens (11, 12), goats (13, 14), and sheep (15, 16). Therefore, the identification and characterization of ROH in a population can provide new insights for uncovering the demographic history and the genetic basis of germplasm characteristics.
Duroc pigs were first developed in North America in the 1860's and have spread all over the world as one of the best-known lean pig breeds (17). Duroc pigs have the characteristics of fast growth rate, high resistance to adversity, and good carcass performance. However, due to the unspectacular female reproductive performance, Duroc pigs are currently used as terminal sires in breeding programs (18). In the past 100 years, Duroc pigs have been widely imported into many countries and bred into different lines with different features according to the preferences of breeders. From the perspective of selection, intense selection reduced the diversity of haplotype, and different selective sweeps contributed to the formation of germplasm characteristics in Duroc pigs (19). For example, numerous studies (20–27) have used the methods of selection signatures to reveal a lot of candidate genes related to growth, immunity, meat, and carcass quality traits of Duroc pigs. Currently, the ROH pattern has been used to detect genetic diversity and genomic regions putatively under strong selection in Duroc pigs. For instance, Schiavo et al. (10) used the 60 K single-nucleotide polymorphism (SNP) data of 573 Italian Duroc pigs to reveal the distribution of ROH and FROH. They also used the 80 K SNP data of 48 Italian Duroc pigs to detect an ROH island including genes that have been shown to affect body size (28). Grossi et al. (29) evaluated the FROH using the 60 K SNP data of 1,066 Canadian Duroc pigs. Gorssen et al. (30) found that the incidence of SNPs in a number of ROH hotspots of the Duroc genome was higher than 80%, and genes associated with coat color, blood physiology, and body size traits were putatively under intensive selection. Nevertheless, few studies have compared ROH patterns between different Duroc pig lines to reveal the potential differences in selection or breeding processes between lines. The aim of this study was to identify the distribution of ROHs in the genomes of 3,770 American Duroc (AD) and 2,096 Canadian Duroc (CD) pigs. Then, ROH hotspots were detected to reveal the different selection directions and the potential causal mutation related to the body size traits of the two Duroc lines.
Methods
Ethics Statement
All animal experiments used in this study were in accordance with the guidelines of the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised June 2004) and approved by the Animal Care and Use Committee of South China Agricultural University, Guangzhou, China (SCAU#2013-10).
Animals, Genotyping, and Quality Control
In this study, ear tissue samples of 3,770 AD pigs (2,280 males and 1,490 females) and 2,096 CD pigs (1,017 males and 1,079 females) were collected from two core breeding farms of Wens Foodstuffs Group Co., Ltd. (Guangdong, China) between 2013 and 2017. During the fattening period of 30–100 kg body weight, all pigs in the two groups maintained uniform feeding conditions, fine fodder, and consistent management to minimize the influence of non-genetic factors (31). The genomic DNA was extracted from ear samples following the standard phenol/chloroform method. Genotyping was performed using the GeneSeek Porcine 50 K SNP Chip, which contains 50,703 genomic SNP markers. Quality control was conducted using PLINK v1.90 software (32) under the following criteria: (1) call rates of SNPs and individuals higher than 90%; (2) the Hardy–Weinberg equilibrium higher than 10−6; (3) all unmapped SNPs and those on sex chromosomes were discarded; and (4) the minor allele frequency (MAF) was not set since pruning low MAF may ignore a large number of homozygous regions (10). After quality control, a set of all 5,866 individuals and 45,424 SNPs was retained for subsequent analyses.
Analysis of Genetic Diversity
In this section, MAF was set as 0.01, leaving 5,866 individuals and 39,416 SNPs for genetic diversity analysis. PLINK v1.90 software (32) was used to calculate the expected heterozygosity (He) and observed heterozygosity (Ho). Effective population size (Ne) was estimated by SNeP software (33) with linkage disequilibrium (LD) method. The formula as follows: Ne(t) = , where Ne(t) is the Ne t generations ago, Ct is the recombination rate t generations ago inferred by the method of Sved and Feldman (34), is the LD expectancy adjusted for sampling bias, f(Ct) is a modified function of the recombination rate based on the genetic distances with default value of 1 Mb = 1 cM, and α is a constant. Finally, nucleotide diversity (π) and fixation index (FST) within and between AD and CD pigs were calculated by VCFtools (35) software. As individual π and FST values may be subjected by genotyping and missing errors in chip data, consistent with previous literature (36, 37), we estimated these statistics using a 500-kb sliding window.
Detection and Classification of Runs of Homozygosity
ROHs were detected for each individual using PLINK v1.90 software (32) by sliding window method on the genome. The following parameters were used to define ROHs (8–10): (1) the minimum length of ROHs was 1 Mb; (2) a sliding window of 50 SNPs across the genome; (3) each window allowed one heterozygous genotype and five missing SNPs to avoid false negatives caused by occasional genotyping errors and missing genotypes; (4) the maximum gap between continuous SNPs was 1 Mb; (5) the required minimum SNP density was set to 100 kb; and (6) each ROH contained at least 63 and 68 consecutive SNPs in AD and CD pigs, respectively, which were computed by the following formula (38):
Where α is the percentage of false positive ROHs, which was set 0.05 in this study, ns is the number of SNPs per individual, ni is the number of individuals, and is the proportion of heterozygosity across all SNPs. In this study, the detected ROHs were divided into three categories for further analyses (39): 1–5, 5–10, and >10 Mb. Finally, we calculated and compared the numbers of each ROH length category and the ratio of ROH on each autosome of the two Duroc lines.
Estimation of Inbreeding Coefficient
Two methods of genomic inbreeding coefficient for each individual were calculated using PLINK v1.90 software (32): (1) SNP-based inbreeding coefficient (FHOM) was estimated with the command set to “–het.” (2) ROH-based inbreeding coefficient (FROH) was assessed for each individual according to McQuillan et al. (40) as follows: FROH = LROH / Lauto, where LROH is the total size of ROHs in the genome of each individual and Lauto is the total size of 18 autosomes of pigs covered by SNPs, which was 2.45 Gb (41). We estimated the FROH of total sizes, 1–5, 5–10, and >10 Mb. Pearson correlation analyses were calculated among different FROH classifications and FHOM in two Duroc populations.
Detection of ROH Hotspots and Coldspot
The percentage of SNP occurrences in ROHs was calculated to characterize the genomic regions of ROH hotspots. In previous studies (7, 8, 11, 15, 42), the threshold of ROH hotspots was usually set as the top 1% and 5% of the SNP occurrences. For a better comparison, similar to a previous study (30), we defined the ROH hotspots with a frequency of SNP occurrences exceeding 80% (top 1.96% in AD pigs and top 1.46% in CD pigs) as significant regions putatively under selection. In addition, we retrieved the 50 K SNP data of 11 European wild boars (EWB) from a previous study (43). Following the quality control mentioned above (MAF > 0.01), a total of 34,196 SNPs remained. Then, the values of π, Tajima's D, and FST among AD, CD, and EWB pigs were calculated using VCFtools software (35) within 500-kb sliding bins to validate the candidate regions under selection. In addition to ROH hotspot, regions without ROH in any of the animals were considered ROH coldspots (44). These regions might be produced by high recombination rates and are likely enriched for variants with severe adverse effects on fitness in homozygotes (45).
Candidate Gene, Pathway, and Functional Analyses
In this study, the average distance between SNPs in the quality control data used to detect ROH was 53.9 kb, and the minimum density of SNPs in ROH was 100 kb. In addition, a strong LD typically extends up to about 100 kb in the pig genome (46). Therefore, candidate genes were annotated via the Ensembl database (Sscrofa 11.1, http://www.ensemble.org/) at 100-kb regions (upstream 50 kb and downstream 50 kb) flanking the SNPs of ROH hotspots. The Gene Ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed for all candidate genes by Metascape database (https://metascape.org/). Meanwhile, in order to reveal the relevance of selected regions to Duroc quantitative traits, ROH hotspots were mapped and compared with the pig quantitative trait loci (QTL) database (https://www.animalgenome.org/cgi-bin/QTLdb/SS/index, release 45). We downloaded the pig QTL file and discarded QTLs with uncertain positions and length more than 1 Mb (26), resulting in 21,952 informative QTLs for analysis.
Identification of Putative Functional SNPs and Their Relationship With Production Traits
The candidate SNPs first predicted the effects on protein function using the Sorting Intolerant From Tolerant (SIFT) score via the Variant Effect Predictor (VEP) program in Ensembl database (47). SNPs with SIFT scores of ≤ 0.05 and >0.05 were predicted to be deleterious and tolerant (48), respectively. The ARSB-DMGDH-BHMT region [chromosome (SSC) 2:87,614,952–87, 935,253, nine SNPs, including a putative deleterious mutation (rs81216249)] was retrieved from the publicly available 60 K SNP data of 1,522 pigs from 107 global populations (49). The dataset includes Asian domestic pigs, Asian wild boars, Western commercial pigs (including Duroc, Large White, Landrace, Berkshire, Hampshire, and Pietrain pigs), European domestic pigs, European wild boars, African feral pigs, American feral pigs, Oceania feral pigs, and outgroup populations. Then, the haplotypes of nine SNPs were constructed using fastPHASE software (50) in global pig populations. In order to uncover the evolutionary history of the missense mutation (rs81216249) and its potential effect on the phenotype in Duroc pigs, we calculated the allele frequency of this variant in global pig breeds. Next, we downloaded the publicly available genotyping (80 K SNPs) and phenotypic (body weight and body length) data of 365 Sujiang sows (a synthetic breed derived from Chinese Jiangquhai, Fengjing, and Western Duroc pigs) from a previous study (51). The association analysis between this missense mutation and body weight and body length was implemented using “–linear” command in PLINK (32) with age and batch as covariates. The linear model is as follows: yijkl = μ+SNPj+COV1k+COV2l+eijkl, where yijkl is the body weight or body length phenotype of the i-th individual, μ is the mean term for the body weight and body length, respectively, SNPj is the fixed effect with three levels (GG, GA, and AA coding as 0, 1, and 2), COV1k is the fixed effect for age, COV2l is the fixed effect of batch, and eijkl is the random residual.
Results
Estimation of Genetic Diversity and Inbreeding Coefficient
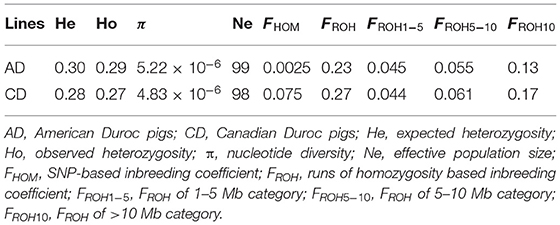
Four metrics were calculated to evaluate the genetic diversities of two Duroc lines: Ho, He, Ne, and π. As seen in Table 1, the Ho (0.29), He (0.30), and π (5.22 × 10−6) values of AD pigs were greater than those of CD (Ho = 0.27, He = 0.28, and π = 4.83 × 10−6) pigs. Then, two inbreeding coefficients based on genomic data of each individual were computed. In our previous study (27), the FROH calculated using consecutive pattern showed that the FROH of AD pigs was significantly lower than that of CD pigs. At the present study, we used the sliding window method to estimate FROH and obtained the same result, and the average FROH (0.23) of AD was significantly (p < 2.20 × 10−16) less than that of CD (FROH = 0.27) pigs (Figure 1A). In addition, the average FHOM (0.0025) of AD was also significantly (p < 2.20 × 10−16) lower than that of CD (FHOM = 0.075) pigs (Figure 1D). Similar to previous studies (8, 19), we also found a highly significant positive correlation between FHOM and FROH in AD (r = 0.89, p < 2.20 × 10−16) and CD (r = 0.84, p < 2.20 × 10−16) pigs (Supplementary Figure 1). Considering that Duroc pigs are mainly used as terminal male parents to cross with other commercial pigs to produce hybrid pigs, the selection intensity of boars may be higher than sows, which may result in higher levels of FHOM and FROH in boars. Unexpectedly, in AD pigs, the values of FHOM and FROH were significantly (p = 4.78 × 10−7 in FHOM and p = 2.78 × 10−5 in FROH) higher in females than in males (Figures 1B,E). In comparison, there was no difference in FHOM and FROH between males and females of CD pigs (Figures 1C,F). The FROH values were divided into three classes, and the FROH> 10 displayed the highest correlation coefficients with FROH in both AD (r = 0.91, p < 2.20 × 10−16) and CD (r = 0.91, p < 2.20 × 10−16) pigs (Supplementary Figure 1). According to the formula LROH = 100 / (2g × cM), where g is the generation ago, and 1 cM is approximately equal to 1 Mb (52). The ROHs were mainly accumulated within the last five generations in AD and CD pigs. The Ne was estimated from the LD method, and the results showed that AD (Ne = 99) was slightly larger than CD (Ne = 98) pigs (Table 1) in the last 13 generations. According to the classification by Hartl (53), the value of FST ranging from 0 to 0.05 indicates small genetic differentiation, 0.05 to 0.15 indicates moderate genetic differentiation, and > 0.15 indicates obvious genetic differentiation. The FST value between AD and CD pigs was 0.093, indicating a moderate degree of genetic divergence between two Duroc lines.
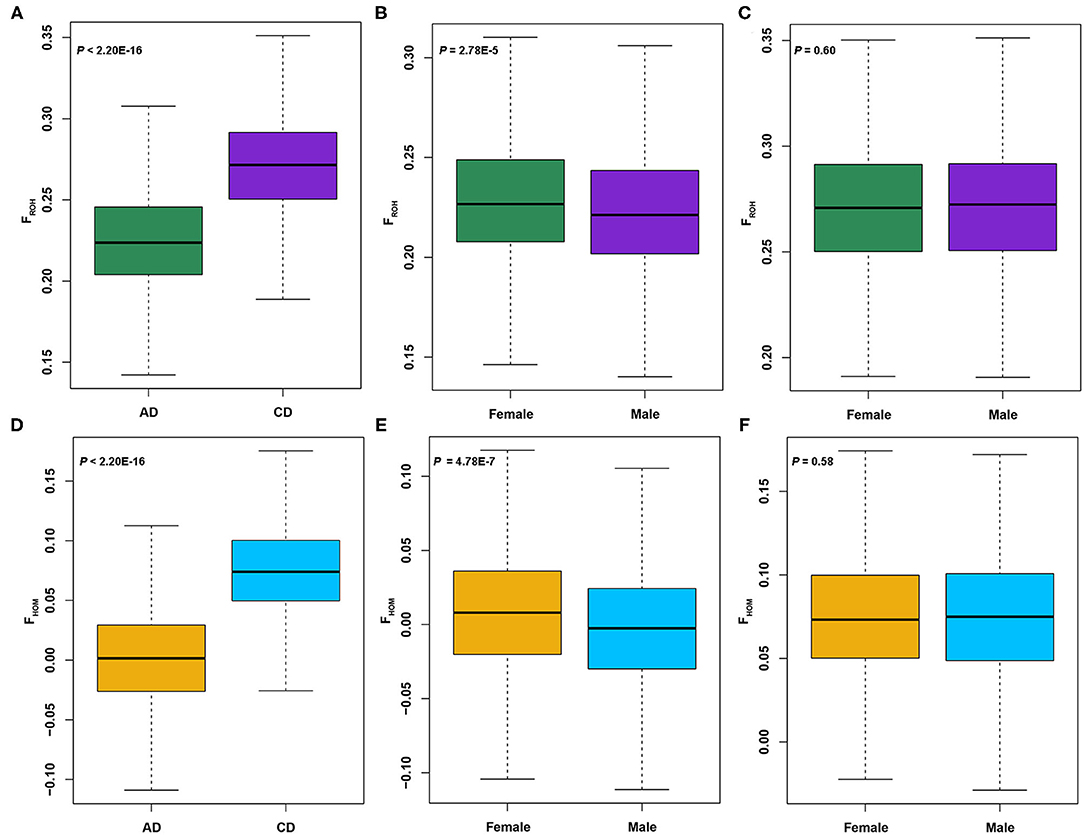
Figure 1. Analysis of the inbreeding coefficients of American Duroc (AD) and Canadian Duroc (CD) pigs based on single-nucleotide polymorphism (SNP) and runs of homozygosity (ROH). (A) FROH of two Duroc lines. (B,C) FROH in females and males of AD and CD pigs, respectively. (D) FHOM of two Duroc lines. (E,F) FROH in females and males of AD and CD pigs, respectively.
Distribution of Runs of Homozygosity
The genome-wide ROHs were assessed on 18 autosomes of all tested individuals. A sum of 256,530 and 152,877 ROHs were detected in AD and CD pigs, respectively (Supplementary Table 1). The average numbers of ROH were 68.05 and 72.94 in AD and CD pigs, respectively. The distribution of ROHs on chromosomes was uneven for the three length categories. The numbers of short fragment of ROH (1–5 Mb) took up the largest part of the total ROHs (49.04% in AD pigs and 43.81% in CD pigs). The smallest chromosome coverage of ROH by dividing the total ROH length was found on SSC10, and the largest was on SSC1 in both AD and CD pigs (Figures 2A,B). The number of ROH was significantly positively correlated with the length of each chromosome in AD (r = 0.93, p = 2.50 × 10−8) and CD (r = 0.91, p < 1.17 × 10−7) pigs. Although the number of ROH1−5Mb was the largest, the proportion of ROH in this category to the total ROH length was not the largest (19.90% in AD pigs and 16.33% in CD pigs). In comparison, ROH fragments larger than 10 Mb had the smallest number (22.36% in AD pigs and 26.67% in CD pigs) but the largest proportion of the total ROH length (55.54% in AD pigs and 61.11% in CD pigs). The total ROH length of each AD pig ranged from 3.20 to 1,240.97 Mb, while each CD pig ranged from 392.64 to 1,085.34 Mb (Figure 2C and Table 2). AD (r = 0.53, p < 2.20 × 10−16) and CD (r = 0.39, p < 2.20 × 10−16) pigs had significant correlations between the number and total length of ROHs per animal (Figure 2C). The coverage of ROH on each autosome was also estimated to compare the distribution of ROH in autosomes. In general, CD pigs had a high genome coverage of ROH in most autosomes (except for SSC4, 6, 8, and 10) than AD pigs (Figure 2D).
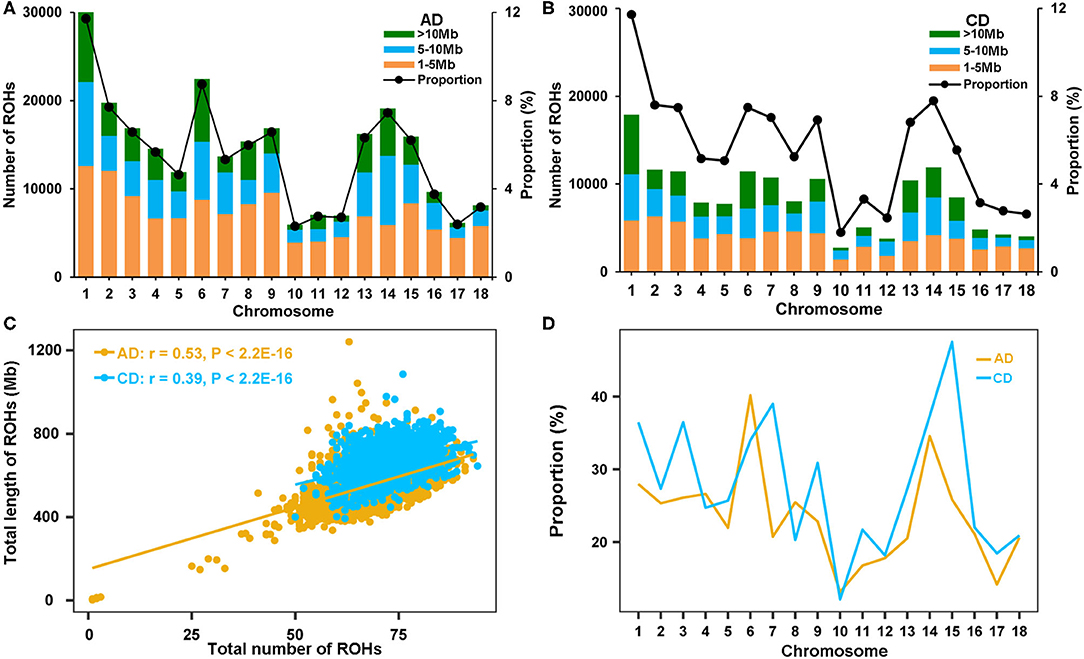
Figure 2. Summary results of ROH in two Duroc lines. (A,B) The number distribution of the three ROH length categories (bars) and average percentage of ROH to the total ROH length (lines) on each autosome of AD and CD pigs, respectively. (C) The total number of ROH and the total length of ROH, per animal, for AD and CD pigs. (D) The average percentage of ROH to chromosome length on each autosome of AD and CD pigs.
QTLs, Candidate Genes, and Functional Processes in ROH Hotspots and Coldspot
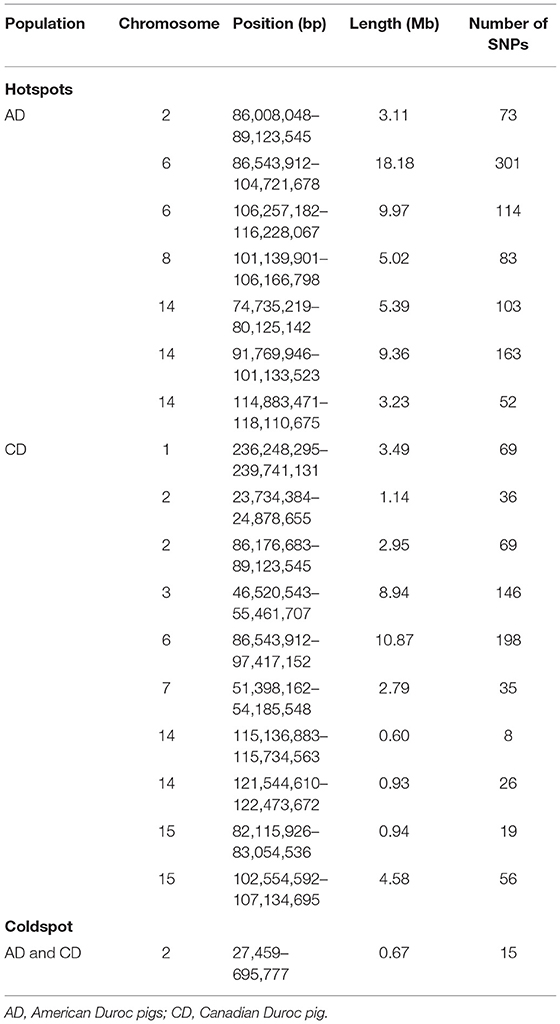
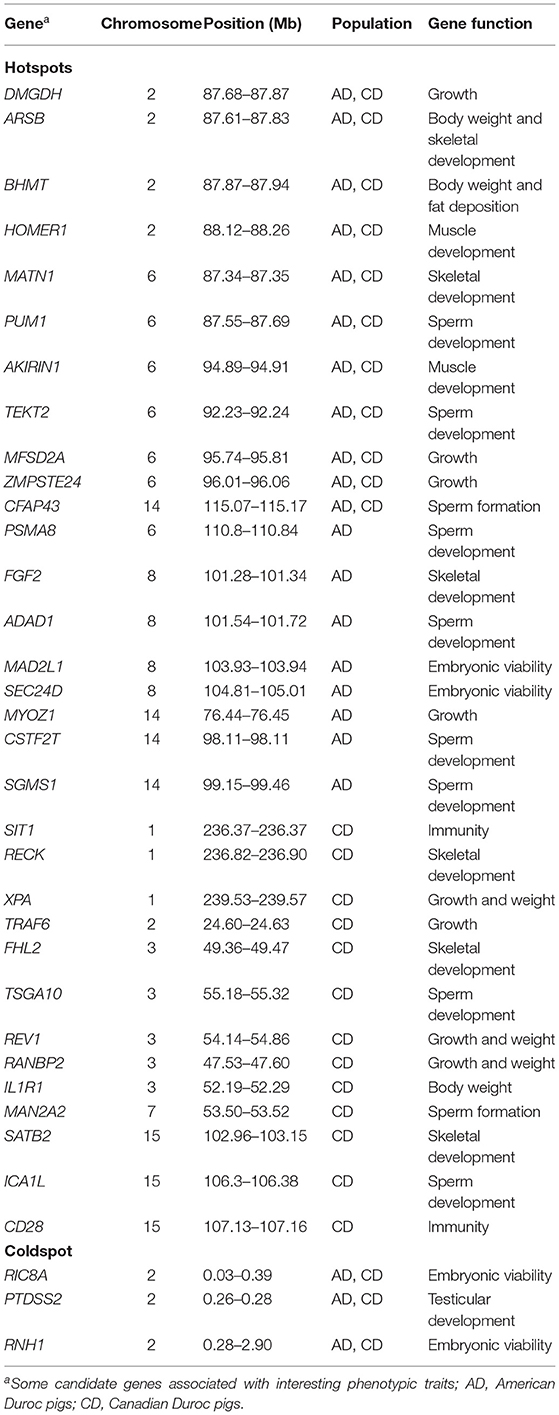
SNPs with an occurrence frequency higher than 80% in ROH were considered as candidate loci putatively under selection. In AD pigs, seven ROH hotspots were detected on four chromosomes, including 891 SNPs with a total length of 18.18 Mb. The highest number (n = 301) and occurrence frequency (92.79%) of SNPs were found on SSC6 (Figure 3A and Table 2). A total of 259 candidate genes resided in 100-kb regions surrounding the 891 SNPs (Supplementary Figure 2). For CD pigs, 10 ROH hotspots were detected, containing 663 candidate SNPs. A total of 271 candidate genes were annotated in 100-kb regions surrounding these 663 SNPs (Supplementary Figure 2). Among those ROH hotspots, the highest number (n = 198) and occurrence frequency (100.00%) of SNPs were also identified on SSC6 (Figure 3B and Table 2). A total of three overlapping ROH hotspots (on SSC2, 6, and 14, including 275 SNPs) were identified in AD and CD pigs (Figure 3 and Supplementary Figure 2). Eighty-six QTLs (41 terms) were identified in the overlapping ROH hotspots of AD and CD pigs (Supplementary Table 2). These QTLs were mainly enriched in health (“mean corpuscular volume and platelet count”), exterior (“splay leg”), meat and carcass (“capric acid content”), and production (“days to 110 kg” and “feed conversion ratio”) traits. A total of 120 overlapping genes were mainly enriched in GO terms of metabolic, muscle development, reproduction, immunity, and behavior (Figure 3D and Supplementary Table 3). Among these genes, three genes were related to sperm development (PUM1, CFAP43, and TEKT2), six genes (DMGDH, ARSB, BHMT, MATN1, MFSD2A, and ZMPSTE24) were involved in growth, and two genes (HOMER1 and AKIRIN1) were relevant to muscle development (Table 3). Meanwhile, ROH hotspots generally showed the low level of π values, Tajima's D < 0, high values of FST between Duroc and EWB pigs, and low values of FST between two Duroc lines, such as SSC2 (86,176,683-89,123,545) and SSC6 (86,543,912-97,417,152) (Figure 3C and Supplementary Figure 3).
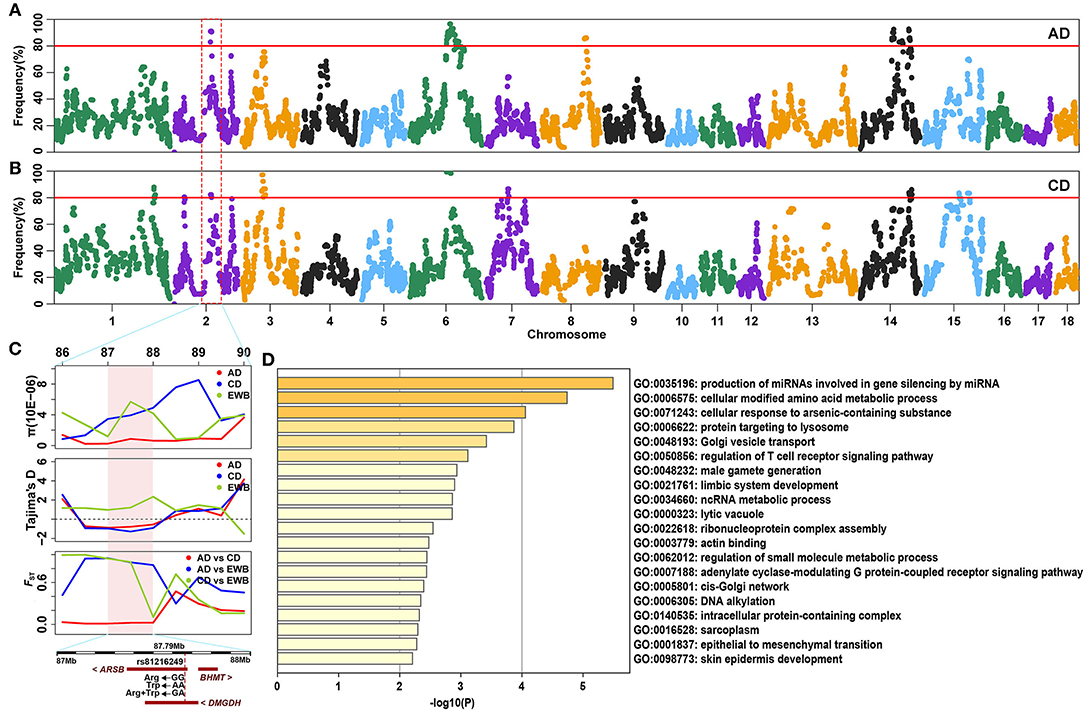
Figure 3. Detection of ROH hotspots in two Duroc lines. (A,B) Manhattan plot of the occurrence (%) of each SNP in ROHs of AD and CD pigs, respectively. The red lines correspond to the significance threshold (80%). (C) The identical ROH region (SSC2: 87.00–88.00 Mb) was detected by π, Tajima's D, and FST methods in AD and CD pigs. The significant region is colored by a pink background. (D) Top 20 significant Gene Ontology (GO) terms of overlapping genes identified by ROH.
In addition to the overlapping ROH hotspots, we also detected several population-specific ROH regions in two Duroc lines (Table 3). For AD pigs, a total of 108 (57 terms) AD-specific QTLs were retrieved for the QTLs of ROH hotspots (Supplementary Table 2). These QTLs were mainly enriched in exterior (“thoracolumbar vertebra number” and “iris pigmentation”), meat and carcass (“backfat between 3rd and 4th last ribs”), and reproduction (“age at puberty” and “number of stillborn”) traits. A total of 138 AD-specific genes were used to perform the GO and KEGG analyses. The results showed that candidate genes were mainly enriched in metabolic, muscle development, immunity, and organ development (Supplementary Table 3). Among these candidate genes, two genes (ADAD1 and PSMA8) were related to sperm development, two genes (MAD2L1 and SEC24D) were involved in embryonic development, and two genes (FGF2 and MYOZ1) were associated with growth (Table 3). For CD pigs, 479 (43 terms) CD-specific QTLs were identified (Supplementary Table 2), and these QTLs were mostly enriched in health (“CD8-positive leukocyte percentage and basophil percentage”) and exterior (“coping behavior”) traits. The GO and KEGG analyses revealed that 151 CD-specific candidate genes were mainly enriched in immunity, stress response, and metabolism (Supplementary Table 3). Among these candidate genes, three genes (TSGA10, MAN2A2, and ICA1L) were associated with sperm development, eight genes (RECK, XPA, TRAF6, FHL2, REV1, RANBP2, IL1R1, and SATB2) were relevant to growth and/or skeletal development, and two genes (SIT1 and CD28) were related to immunity. In addition, we also found low values of π and Tajima's D and high values of FST between AD/CD and EWB in population-specific ROH regions, for example, SSC8 (103,000,000-106,166,798) in AD pigs and SSC1 (236,248,295-237,000,000) and SSC3 (49,360,000-55,461,707) in CD pigs (Supplementary Figure 3).
Interestingly, an overlapping region with 15 SNPs was observed on SSC2 (0.027–0.70 Mb) of the two Duroc lines, which did not occur in any ROH (ROH coldspot). Three reproduction-related genes (RIC8A, PTDSS2, and RNH1) were annotated to this 0.67 Mb region (Table 3). Previous study showed that ROH coldspots were considered to have loci with the capability of avoiding purely lethal or cryptic mutation-critical functions (45). Therefore, this coldspot region may play an important role in the viability of Duroc pigs.
Functional Annotation of Variants
To better understand the genetic basis of ROH hotspots, we performed variant annotation on candidate loci. A total of 21 and 30 non-synonymous variants were observed in the ROH hotspots of AD and CD pigs (Supplementary Table 4), respectively. Notably, only one identical missense variant (rs81216249, g. 87792989G > A) was predicted as a functional-altering mutation (SIFT = 0) in AD and CD pigs. Five outgroup populations (Babyrousa babyrussa, Phacochoerus africanus, Sus barbatus, Sus celebensis, and Sus verrucosus) (49) were used to infer that G was an ancestral allele and A was a derived allele. This variant was highly conserved across multiple vertebrate species (Figure 4C). The frequency of derived allele (DAF) of this variant displayed a large difference between Duroc and other global pig breeds (Figures 4A,B and Supplementary Table 5). The DAF was near fixation in AD (96%), CD (90%), and other (80–95%) Duroc populations with unknown origin (49). The FST-based neighbor-joining tree (54) revealed that these Duroc populations had different origins from AD and CD pigs (Supplementary Figure 4). This indicated that a high DAF of this mutation may present in Duroc pig breed. Intriguingly, the derived allele was at high prevalence in Western commercial and European domestic pigs but nearly absent in Chinese indigenous pigs [except for three hybrid breeds, Sutai, Licha, and Neijiang pigs, which were reported to be admixed with Duroc pigs (49)] (Figure 4A and Supplementary Table 5). This variant is located in the overlapping region of three genes (ARSB-DMGDH-BHMT, SSC2:87614952-87935253) and as a missense mutation of DMGDH. The values of π, Tajima's D, and FST revealed that the region of three genes was under positive selection (Figure 3C). The haplotype heat map of this region showed that most Duroc pigs had the same haplotype, revealing that this region was under heavy selection (Supplementary Figure 5). The cluster of supergenes (ARSB-DMGDH-BHMT) has an important role in growth and fat deposition traits (55). The above results indicated that this missense mutation may play an important role in the production traits of Duroc pigs. Finally, a correlation analysis was used to detect the effect of this functional-altering variant on the phenotypes of Sujiang pigs. The results showed that rs81216249 was significantly associated with body weight (p = 0.016) and body length (p = 0.0002). The phenotypic value of pigs with the AA genotype was higher than that of pigs with the GG and GA genotypes (Figures 4D,E).
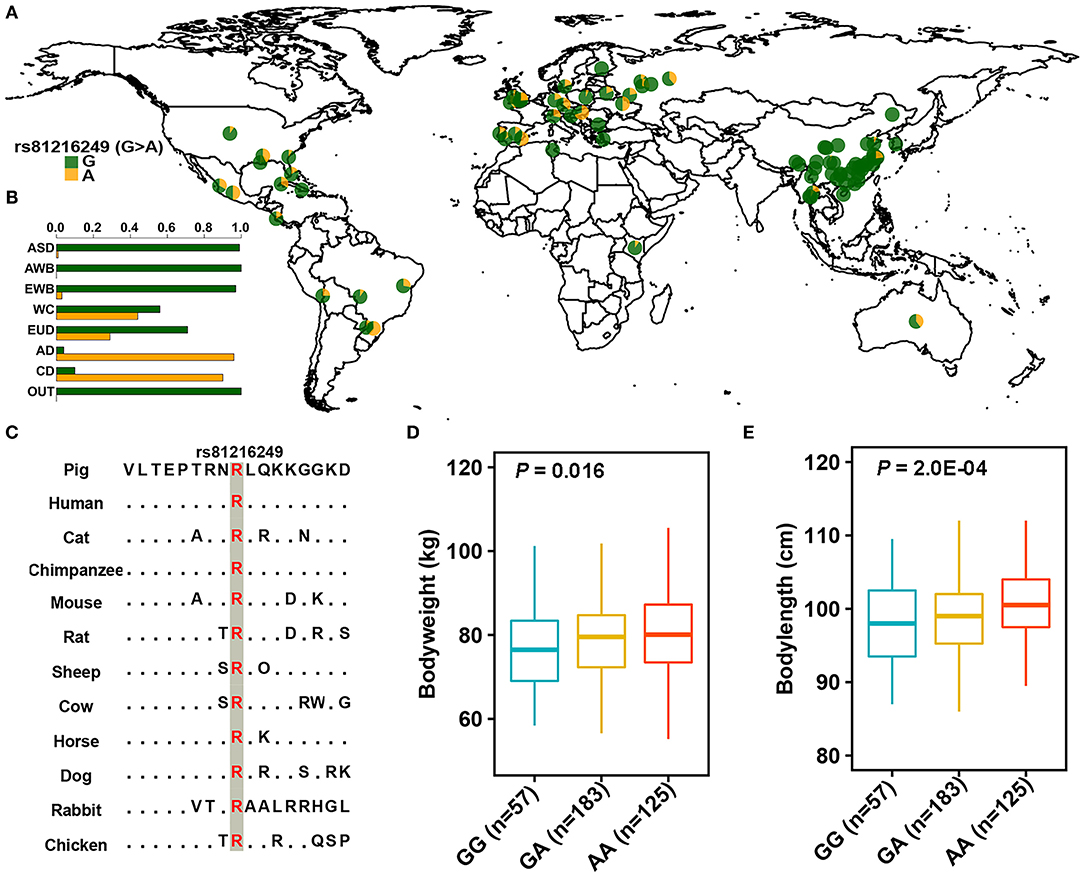
Figure 4. The distribution of rs81216249. (A,B) The allele frequency distribution of rs81216249 in global pig breeds. G and A denote ancestral and derived genotypes and are marked by green and yellow, respectively. ASD, AWB, EWB, EUD, WC, AD, CD, and OUT represent Asian domestic pigs, Asian wild boars, European wild boars, European domestic pigs, Western commercial pigs, American Duroc pigs, Canadian Duroc pigs, and outgroup populations, respectively. (C) Multispecies alignment of the protein sequences around the variant. (D,E) Association analysis of rs81216249 with body weight and body length in Sujiang pigs.
Discussion
Artificial Selection Possibly Caused the Differences in Genetic Diversity and Inbreeding Levels of the Two Duroc Lines
In our study, combining the results of six genetic diversity and inbreeding indices (Ho, He, π, Ne, FHOM, and FROH), AD pigs had an abundant genetic diversity and a low level of inbreeding than CD pigs. Similar to previous studies (8, 56), the values of FROH were larger than FHOM in two Duroc populations. This may be caused by the inability of the FHOM to differentiate between IBD and IBS alleles (57). The moderate genetic differentiation (FST = 0.093) indicated that two Duroc lines possibly experienced different demographic, inbreeding, and selection histories. FROH>10 made a major contribution to FROH of the two Duroc lines, revealing the recent reduction in genetic diversity and increase in homozygosity in two Duroc lines. Due to the differences in computational methods and parameters, tested populations, sample sizes, and SNP arrays, the Duroc populations in this study displayed different ROH numbers and lengths compared with previous studies of ROH in Duroc pigs (10, 28–30). Two Duroc populations had low degrees of FHOM, likely due to the two Duroc populations raised in the same company (Wen's Foodstuff Group Co., Ltd.), which had the better inbreeding control and breeding programs. Our previous study (58) reported that LD decays faster with distance in AD than in CD pigs, implying that CD has a greater selective strength than AD pigs. A previous study (45) showed that, compared with medium and small ROH, recent strong directional selection may have a greater impact on long ROH, because it tends to produce long haplotypes. Based on the above results, we believe that the existence of genetic differences between the two Duroc lines may be more partly due to recent selection. Moreover, compared to CD pigs, the FHOM and FROH values of females were larger than males in AD pigs. A reasonable explanation is that the traits associated with female reproduction may be selected to improve the poor fertility of AD pigs.
Overlapping ROH Hotspots Uncovered the Same Breed Characteristics of the Two Duroc Lines
ROH patterns were mainly shaped by population bottlenecks, inbreeding, genetic drift, and intensive natural and artificial selection (3). Peripolli et al. (2) suggested that a lot of ROHs were shared among livestock individuals, which may be due to selection rather than just demographic history. Duroc pig is a representative commercial pig breed that has recently been strongly selected for economic traits. Sonesson et al. (59) reported that genomic selection may also lead to the risk of long homozygous segments appearing around QTL regions related to any given trait in the populations. Therefore, the investigation of ROH islands can provide information about selection signatures that are putatively derived from various selection events, directions, and adaptations to different production systems (60). We expected that overlapping ROH regions and genes may have undergone directional selection in Duroc pigs, which contributes to their breed characteristics such as excellent growth rate, carcass traits, male reproductive capacity, and meat quality. The results of QTLs and GO and KEGG analyses were in line with this expectation, and QTLs and functional processes were more enriched in health, production, and behavior. We also observed three sperm-related genes, six growth-related genes, and two muscle development-related genes putatively under selection that are functionally related to these breed features. For instance, the PUM1 gene mediates activation and apoptosis of spermatogonia and acts as a post-transcriptional regulator of spermatogenesis in the testis (61). TEKT2 is associated with male sterility and abnormal sperm morphology and function in mice (62). Mice lacking CFAP43 show multiple morphological abnormalities of the flagella and impaired sperm motility (63). MATN1 is expressed in cartilage structures such as the trachea, nasal septum, auricle, and epiphysis (64). MFSD2A knockout mice exhibit a smaller and leaner body size (65). Mice lacking the ZMPSTE24 gene show premature signs of aging such as reduced body weight and subcutaneous fat, spinal prolapse, hair loss, and premature death (66). HOMER1 (67) and AKIRIN1 (68) are related to muscle development. Some candidate genes have been reported in previous selective sweep studies, such as HOMER1 (23, 24) and ARSB (25). These promising genes associated with economic traits may contribute to the genetic breeding process of pigs.
In addition, we focused on neighboring genes (ARSB-DMGDH-BHMT) located on the overlapping ROH hotspots (SSC2:87614952-87935253). ARSB is related to abnormal morphology of the head, nose, and tail vertebra; fat/triglyceride levels; and reduced body size at birth and adulthood in mice (69). ARSB is also the causal gene of human mucopolysaccharidosis type VI (Maroteaux–Lamy syndrome), which is related to facial deformities and short stature (70). DMGDH plays an important role in regulating the insulin-like growth factor/growth hormone (IGF-1/GH) pathway, which is an important regulator of vertebrate growth (71). BHMT is associated with body weight, fat deposition, and energy metabolism (55, 72). Three genes are genetically linked in most vertebrates, and these functionally relevant genes, in some cases called “supergene,” may play an influential role in the physiological processes that affect growth and fat deposition (55). The selective sweep analyses of π, Tajima's D, and FST confirmed that the supergene experienced strongly positive selection in Duroc pigs. Therefore, we believe that these three genes are important genes associated with the production traits of Duroc pigs, and it is worthwhile to further explore the genetic mechanism of their impact on the phenotypes.
Population-Specific ROH Hotspots Revealed Potential Differences in Breeding Objectives of the Two Duroc Lines
Compared with overlapping ROH regions, we detected the population-specific ROH hotspots to elucidate the potential differences in the breeding goals of the two Duroc populations. Population-specific ROH regions generally displayed distinct patterns of π, Tajima's D, and FST, which may reveal unique artificial selection in the two pig lines. However, some population-specific QTLs were associated with the same traits, such as “average daily gain,” “intramuscular fat content,” and “coping behavior.” The first two traits are very important to the pork industry and have been targeted for positive selection in Duroc breeding. Coping behavior is considered as the reaction behavior of pig to aversive environment (73). In recent years, the intensive feeding model has been greatly developed in the pig industry, which may lead to the selection of “coping behavior” (26). The results of GO and KEGG analyses were consist with QTLs, in which two sets of population-specific candidate genes were enriched in some functional processes with similar functions, such as immune response, metabolism, and development. Simultaneously, we also detected many candidate genes related to superior breed characteristics in two population-specific ROH hotspots, such as sperm development (PSMA8, ADAD1, CSTF2T, and SGMS1 in AD pigs and TSGA10, MAN2A2, and ICA1L in CD pigs) and growth (FGF2 and MYOZ1 in AD pigs and RECK, XPA, TRAF6, FHL2, REV1, RANBP2, IL1R1, and SATB2 in CD pigs) traits. The results revealed that CD pigs had more candidate genes related to growth than AD pigs, which may be caused by their stronger selection as mentioned above. These same QTLs, similar functional processes, and candidate genes revealed the possible polygenic basis of these economic traits in the two populations.
Besides the potentially polygenic basis, population-specific ROHs can also uncover some different characteristics in two populations. For example, QTLs associated with exterior and production were enriched in AD pigs, such as “thoracolumbar vertebra number” and “backfat between 3rd and 4th last ribs.” In contrast, most QTLs related to health were enriched in CD pigs, such as “CD8-positive leukocyte percentage” and “CD8-negative leukocyte percentage.” Candidate genes of AD pigs were more significantly enriched in metabolic process, and CD pigs were more significantly enriched in the immune system process, which was consistent with our previous study on the selection signatures of these two lines (27). We also observed two genes (MAD2L1 and SEC24D) associated with embryonic development in AD pigs and two immune-related genes (SIT1 and CD28) in CD pigs. The MAD2L1 gene is related to embryonic viability (74). Mice lacking SEC24D show early embryonic lethality (75). SIR1 knockout mice show increased susceptibility to autoimmune encephalitis (76), and CD28 affects T-cell proliferation in mice (77). Based on the above results, we infer that AD pigs may have a specific selection for female fertility, which was in line with the result that females harbored larger values of FROH and FHOM than males in AD pigs. A previous study (78) also showed that AD pigs had a lower number of stillborn and a higher rate of born alive than CD pigs in the first two parities. In comparison, CD pigs may have a specific selection for immunity, possibly because they need to possess excellent adaptability to the intensive feeding environment during the stronger selection. In addition, the number of healthy births and the rate of healthy birth of CD pigs were higher than those of AD pigs (78), which may be related to the better immunity of CD pigs. However, populations with different time dimensions (born in 2016–2017) and small sample sizes (n = 413–604) may affect the phenotypic comparison of the two lines. Considering the complexity of the genetic mechanisms of reproductive and immune traits, more direct reproductive and immune data are needed to validate and improve our results in the future.
Functional-Altering Variant Likely Played an Important Role in the Breeding Process of Duroc Pigs
The Duroc pigs were mainly developed in North America but originated in Europe (17). Considering the independent domestication of Eurasian pig breeds and the loci of Eurasian wild boar populations and Chinese native pigs were all or nearly all ancestral alleles, while European domestic and commercial pigs had derived alleles, we hypothesize that the missense mutation (rs81216249) originated from European domestic pig breeds and appeared earlier than the formation of Duroc pig breeds. The DAF of Western commercial pigs was significantly higher than that of other European domestic pigs (p < 0.01), indicating that DAF was increased in the intensive selection processes of commercial pig breeds. This missense mutation showed very strong conservation in vertebrates and was predicted to have a deleterious effect on protein function (SIFT = 0). Previous literature demonstrated that harmful mutations are expected to be maintained at low frequency due to the efficacy of purifying selection (79). This functional-altering mutation displayed a high frequency or near fixation of derived allele in Western commercial pigs, especially Duroc pigs, revealing that this mutation may be a beneficial mutation and could improve the production traits of pigs. This speculation was further verified by the SNP–phenotypic association analysis in Sujiang pigs, a synthetic breed of Chinese and Duroc pigs, in which derived allele significantly improved the body weight and length of pigs. The derived allele also existed in other Eurasian hybrid breeds, such as Sutai and Licha pigs (49), indicating that this mutation could be used for marker-assisted hybrid breeding in Chinese native pigs. In addition, many Chinese indigenous pig breeds admixed with exotic pigs due to indiscriminate crossbreeding between Chinese and European breeds, which may lead to the loss of the original characteristics and disruption of the locally adapted gene complexes (80). This variant can also be used to detect and monitor whether Chinese local pig breeds are mixed with Western commercial pig breeds (especially Duroc pigs) and provide an effective tool for the purebred preservation of Chinese indigenous pigs. Notably, we observed only one functional-altering variant in this supergene region due to the limitation of SNP density. Considering that this missense mutation is one of the potential functional-altering mutations in this supergene and that the tested population for SNP–phenotypic association analysis was not Duroc pigs, further in-depth analyses such as resequencing, RNA-sequencing, phenotypic association studies, and functional experiments are needed to definitively determine the role of candidate genes and functional-altering mutations.
Conclusion
In this study, we estimated the ROH patterns in two Duroc pig lines. The results showed that CD pigs had a low level of genetic diversity and a high inbreeding degree than AD pigs, which are possibly due to stronger selection. ROH hotspots revealed that a lot of shared genes putatively under selection were related to growth, sperm, and muscle development in two Duroc lines. Population-specific ROH hotspots indicated that AD may have a specific selection on female reproduction, while CD pigs may have a specific selection on immunity. Moreover, a functional-altering mutation was observed on the overlapping ROH hotspots of two Duroc populations, and the derived allele could significantly improve the body weight and length of pigs. Altogether, our results not only benefit the inbreeding management of two Duroc lines but also provide a series of promising genes that may affect economic traits in pigs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository and accession number can be found at: https://doi.org/10.6084/m9.figshare.8019551.v1.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Use Committee of South China Agricultural University, Guangzhou, China.
Author Contributions
ZW and JY proposed the idea of study. XW and GL performed the analysis and wrote the manuscript. JY directed the analyses and revised the manuscript. DR, ZZ, RD, JQ, SW, YJ, JH, TG, LH, EZ, ZL, and GC collected samples and performed the experiments. ZW and JY contributed the materials. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Guangdong Province (2018B030313011), the Local Innovative and Research Teams Project of Guangdong Province (2019BT02N630), and the National High-quality Lean-type Pig Breeding United Research Program of China.
Conflict of Interest
RD, GC, and ZW were employed by Guangdong Wens Breeding Swine Technology Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the staff at the pig core breeding farms of Wens Foodstuff Group Co., Ltd., (Guangdong, China) for the help in sample collection and also the researchers who kindly made their data publicly available.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.832633/full#supplementary-material
References
1. Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. (2006) 15:789–95. doi: 10.1093/hmg/ddi493
2. Peripolli E, Munari DP, Silva MVGB, Lima ALF, Irgang R, Baldi F. Runs of homozygosity: current knowledge and applications in livestock. Anim Genet. (2017) 48:255–71. doi: 10.1111/age.12526
3. Ceballos FC, Joshi PK, Clark DW, Ramsay M, Wilson JF. Runs of homozygosity: windows into population history and trait architecture. Nat Rev Genet. (2018) 19:220–34. doi: 10.1038/nrg.2017.109
4. Keller MC, Visscher PM, Goddard ME. Quantification of inbreeding due to distant ancestors and its detection using dense single nucleotide polymorphism data. Genetics. (2011) 189:237–49. doi: 10.1534/genetics.111.130922
5. Zhang QQ, Guldbrandtsen B, Bosse M, Lund MS, Sahana G. Runs of homozygosity and distribution of functional variants in the cattle genome. BMC Genomics. (2015) 16:542. doi: 10.1186/s12864-015-1715-x
6. Zhao GY, Liu YQ, Niu QH, Zheng X, Zhang TL, Wang ZZ, et al. Runs of homozygosity analysis reveals consensus homozygous regions affecting production traits in Chinese simmental beef cattle. BMC Genomics. (2021) 22:678. doi: 10.1186/s12864-021-07992-6
7. Moscarelli A, Sardina MT, Cassandro M, Ciani E, Pilla F, Senczuk G, et al. Genome-wide assessment of diversity and differentiation between original and modern brown cattle populations. Anim Genet. (2021) 52:21–31. doi: 10.1111/age.13019
8. Shi LY, Wang LG, Liu JX, Deng TY, Yan H, Zhang LC, et al. Estimation of inbreeding and identification of regions under heavy selection based on runs of homozygosity in a large white pig population. J Anim Sci Biotechno. (2020) 11:46. doi: 10.1186/s40104-020-00447-0
9. Wang XP, Zhang H, Huang M, Tang JH, Yang LJ Yu ZQ, et al. Whole-genome SNP markers reveal conservation status, signatures of selection, and introgression in Chinese Laiwu pigs. Evol Appl. (2021) 14:383–98. doi: 10.1111/eva.13124
10. Schiavo G, Bovo S, Bertolini F, Tinarelli S, Dall Olio S, Nanni Costa L, et al. Comparative evaluation of genomic inbreeding parameters in seven commercial and autochthonous pig breeds. Animal. (2020) 14:910–20. doi: 10.1017/S175173111900332X
11. Zhang JX, Nie CS, Li XH, Ning ZH, Chen Y, Jia YX, et al. Genome-wide population genetic analysis of commercial, indigenous, game, and wild chickens using 600K SNP microarray data. Front Genet. (2020) 11:543294. doi: 10.3389/fgene.2020.543294
12. Talebi R, Szmatoła T, Mészáros G, Qanbari S. Runs of homozygosity in modern chicken revealed by sequence data. G3. (2020) 10:4615–23. doi: 10.1534/g3.120.401860
13. Dadousis C, Cecchi F, Ablondi M, Fabbri MC, Stella A, Bozzi R. Keep Garfagnina alive. An integrated study on patterns of homozygosity, genomic inbreeding, admixture and breed traceability of the Italian Garfagnina goat breed. PLoS ONE. (2021) 16:e232436. doi: 10.1371/journal.pone.0232436
14. Islam R, Li YF, Liu XX, Berihulay H, Abied A, Gebreselassie G, et al. Genome-wide runs of homozygosity, effective population size, and detection of positive selection signatures in six Chinese goat breeds. Genes. (2019) 10:938. doi: 10.3390/genes10110938
15. Nosrati M, Nanaei AH, Javanmard A, Esmailizadeh A. The pattern of runs of homozygosity and genomic inbreeding in world-wide sheep populations. Genomics. (2021) 113:1407–15. doi: 10.1016/j.ygeno.2021.03.005
16. Dzomba EF, Chimonyo M, Pierneef R, Muchadeyi FC. Runs of homozygosity analysis of South African sheep breeds from various production systems investigated using OvineSNP50k data. BMC Genomics. (2021) 22:7. doi: 10.1186/s12864-020-07314-2
17. Wang LY, Wang AG, Wang LX, Li K, Yang GS, He LG, et al. Animal Genetic Resources in China Pigs. Beijing: Chinese Agriculture Press (2011).
18. Suzuki K, Shibata T, Kadowaki H, Abe H, Toyoshima T. Meat quality comparison of berkshire, duroc and crossbred pigs sired by berkshire and duroc. Meat Sci. (2003) 64:35–42. doi: 10.1016/S0309-1740(02)00134-1
19. Zhang Z, Zhang QQ, Xiao Q, Sun H, Gao HD, Yang YM, et al. Distribution of runs of homozygosity in Chinese and Western pig breeds evaluated by reduced-representation sequencing data. Anim Genet. (2018) 49:579–91. doi: 10.1111/age.12730
20. Edea Z, Hong JK, Jung JH, Kim DW, Kim YM, Kim ES, et al. Detecting selection signatures between duroc and duroc synthetic pig populations using high-density SNP chip. Anim Genet. (2017) 48:473–7. doi: 10.1111/age.12559
21. Wang K, Wu PX, Chen DJ, Zhou J, Yang XD, Jiang AA, et al. Detecting the selection signatures in Chinese Duroc, Landrace, Yorkshire, Liangshan, and Qingyu pigs. Funct Integr Genomic. (2021) 21:655–64. doi: 10.1007/s10142-021-00809-5
22. Tang Z, Fu Y, Xu J, Zhu M, Li X, Yu M, et al. Discovery of selection-driven genetic differences of Duroc, Landrace, and Yorkshire pig breeds by EigenGWAS and Fst analyses. Anim Genet. (2020) 51:531–40. doi: 10.1111/age.12946
23. Yu J, Zhao PJ, Zheng XR, Zhou L, Wang CD, Liu JF. Genome-wide detection of selection signatures in duroc revealed candidate genes relating to growth and meat quality. G3. (2020) 10:3765–73. doi: 10.1534/g3.120.401628
24. Ma YL, Zhang SX, Zhang KL, Fang CC, Xie SS, Du XY, et al. Genomic analysis to identify signatures of artificial selection and loci associated with important economic traits in duroc pigs. G3. (2018) 8:3617–25. doi: 10.1534/g3.118.200665
25. Kim ES, Ros-Freixedes R, Pena RN, Baas TJ, Estany J, Rothschild MF. Identification of signatures of selection for intramuscular fat and backfat thickness in two Duroc populations. J Anim Sci. (2015) 93:3292–302. doi: 10.2527/jas.2015-8879
26. Zhang SX, Zhang KL, Peng X, Zhan HW, Lu JH, Xie SS, et al. Selective sweep analysis reveals extensive parallel selection traits between large white and Duroc pigs. Evol Appl. (2020) 13:2807–20. doi: 10.1111/eva.13085
27. Li DS, Huang M, Zhuang ZW, Ding RR, Gu T, Hong LJ, et al. Genomic analyses revealed the genetic difference and potential selection genes of growth traits in two duroc lines. Front Vet Sci. (2021) 8:725367. doi: 10.3389/fvets.2021.725367
28. Schiavo G, Bovo S, Muñoz M, Ribani A, Alves E, Araújo JP, et al. Runs of homozygosity provide a genome landscape picture of inbreeding and genetic history of European autochthonous and commercial pig breeds. Anim Genet. (2021) 52:155–70. doi: 10.1111/age.13045
29. Grossi DA, Jafarikia M, Brito LF, Buzanskas ME, Sargolzaei M, Schenkel FS. Genetic diversity, extent of linkage disequilibrium and persistence of gametic phase in Canadian pigs. BMC Genet. (2017) 18:6. doi: 10.1186/s12863-017-0473-y
30. Gorssen W, Meyermans R, Janssens S, Buys N. A publicly available repository of ROH islands reveals signatures of selection in different livestock and pet species. Genet Sel Evol. (2021) 53:2. doi: 10.1186/s12711-020-00599-7
31. Zhuang ZW, Li SY, Ding RR, Yang M, Zheng EQ, Yang HQ, et al. Meta-analysis of genome-wide association studies for loin muscle area and loin muscle depth in two Duroc pig populations. PLoS ONE. (2019) 14:e218263. doi: 10.1371/journal.pone.0218263
32. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. (2007) 81:559–75. doi: 10.1086/519795
33. Barbato M, Orozco-terWengel P, Tapio M, Bruford MW. SNeP: a tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet. (2015) 6:109. doi: 10.3389/fgene.2015.00109
34. Sved JA, Feldman MW. Correlation and probability methods for one and two loci. Theor Popul Biol. (1973) 4:129–32. doi: 10.1016/0040-5809(73)90008-7
35. Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, et al. The variant call format and VCFtools. Bioinformatics. (2011) 27:2156–8. doi: 10.1093/bioinformatics/btr330
36. Chen M, Wang J, Wang Y, Wu Y, Fu J, Liu J. Genome-wide detection of selection signatures in Chinese indigenous Laiwu pigs revealed candidate genes regulating fat deposition in muscle. BMC Genet. (2018) 19:31. doi: 10.1186/s12863-018-0622-y
37. Kominakis A, Tarsani E, Hager-Theodorides AL, Mastranestasis I, Gkelia D, Hadjigeorgiou I. Genetic differentiation of mainland-island sheep of Greece: implications for identifying candidate genes for long-term local adaptation. PLoS ONE. (2021) 16:e257461. doi: 10.1371/journal.pone.0257461
38. Lencz T, Lambert C, DeRosse P, Burdick KE, Morgan TV, Kane JM, et al. Runs of homozygosity reveal highly penetrant recessive loci in schizophrenia. Proc Natl Acad Sci U S A. (2007) 104:19942–7. doi: 10.1073/pnas.0710021104
39. Mastrangelo S, Tolone M, Di Gerlando R, Fontanesi L, Sardina MT, Portolano B. Genomic inbreeding estimation in small populations: evaluation of runs of homozygosity in three local dairy cattle breeds. Animal. (2016) 10:746–54. doi: 10.1017/S1751731115002943
40. McQuillan R, Leutenegger A, Abdel-Rahman R, Franklin CS, Pericic M, Barac-Lauc L, et al. Runs of homozygosity in European populations. Am J Hum Genet. (2008) 83:359–72. doi: 10.1016/j.ajhg.2008.08.007
41. Gorssen W, Meyermans R, Buys N, Janssens S. SNP genotypes reveal breed substructure, selection signatures and highly inbred regions in Piétrain pigs. Anim Genet. (2020) 51:32–42. doi: 10.1111/age.12888
42. Wu F, Sun H, Lu SX, Gou X, Yan DW, Xu Z, et al. Genetic diversity and selection signatures within diannan small-ear pigs revealed by next-generation sequencing. Front Genet. (2020) 11:733. doi: 10.3389/fgene.2020.00733
43. Huang M, Zhang H, Wu ZP, Wang XP, Li DS, Liu SJ, et al. Whole-genome resequencing reveals genetic structure and introgression in pudong white pigs. Animal. (2021) 15:100354. doi: 10.1016/j.animal.2021.100354
44. Xu Z, Sun H, Zhang Z, Zhao QB, Olasege BS, Li QM, et al. Assessment of autozygosity derived from runs of homozygosity in jinhua pigs disclosed by sequencing data. Front Genet. (2019) 10:274. doi: 10.3389/fgene.2019.00274
45. Pemberton TJ, Absher D, Feldman MW, Myers RM, Rosenberg NA, Li JZ. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. (2012) 91:275–92. doi: 10.1016/j.ajhg.2012.06.014
46. Ai HS, Huang LS, Ren J. Genetic diversity, linkage disequilibrium and selection signatures in chinese and Western pigs revealed by genome-wide SNP markers. PLoS ONE. (2013) 8:e56001. doi: 10.1371/journal.pone.0056001
47. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. (2016) 17:122. doi: 10.1186/s13059-016-0974-4
48. Ng PC. SIFT predicting amino acid changes that affect protein function. Nucleic Acids Res. (2003) 31:3812–4. doi: 10.1093/nar/gkg509
49. Yang B, Cui LL, Perez-Enciso M, Traspov A, Crooijmans RPMA, Zinovieva N, et al. Genome-wide SNP data unveils the globalization of domesticated pigs. Genet Sel Evol. (2017) 52:30. doi: 10.1186/s12711-020-00549-3
50. Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. (2006) 78:629–44. doi: 10.1086/502802
51. Xu P, Ni L, Tao Y, Ma Z, Hu T, Zhao X, et al. Genome-wide association study for growth and fatness traits in Chinese Sujiang pigs. Anim Genet. (2020) 51:314–8. doi: 10.1111/age.12899
52. Howrigan DP, Simonson MA, Keller MC. Detecting autozygosity through runs of homozygosity: a comparison of three autozygosity detection algorithms. BMC Genomics. (2011) 12:460. doi: 10.1186/1471-2164-12-460
53. Daniel L, Hartl AGC. Principles of Population Genetics. Sunderland: Sinauer and Associates (2007).
54. Wang X, Wang C, Huang M, Tang J, Fan Y, Li Y, et al. Genetic diversity, population structure and phylogenetic relationships of three indigenous pig breeds from Jiangxi Province, China, in a worldwide panel of pigs. Anim Genet. (2018) 49:275–83. doi: 10.1111/age.12687
55. Card DC, Adams RH, Schield DR, Perry BW, Corbin AB, Pasquesi GIM, et al. Genomic basis of convergent island phenotypes in boa constrictors. Genome Biol Evol. (2019) 11:3123–43. doi: 10.1093/gbe/evz226
56. Freitas PHF, Wang Y, Yan P, Oliveira HR, Schenkel FS, Zhang Y, et al. Genetic diversity and signatures of selection for thermal stress in cattle and other two bos species adapted to divergent climatic conditions. Front Genet. (2021) 12:604823. doi: 10.3389/fgene.2021.604823
57. Wang J. Marker-based estimates of relatedness and inbreeding coefficients: an assessment of current methods. J Evolution Biol. (2014) 27:518–30. doi: 10.1111/jeb.12315
58. Zhuang ZW, Ding RR, Peng LL, Wu J, Ye Y, Zhou SP, et al. Genome-wide association analyses identify known and novel loci for teat number in duroc pigs using single-locus and multi-locus models. BMC Genomics. (2020) 21:344. doi: 10.1186/s12864-020-6742-6
59. Sonesson AK, Woolliams JA, Meuwissen THE. Maximising genetic gain whilst controlling rates of genomic inbreeding using genomic optimum contribution selection [Conference presentation]. In; Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, Leipzig, Germany(2010).
60. Schiavo G, Bovo S, Bertolini F. Dall'Olio S, Nanni Costa L, Tinarelli S, et al. Runs of homozygosity islands in Italian cosmopolitan and autochthonous pig breeds identify selection signatures in the porcine genome. Livest Sci. (2020) 240:104219. doi: 10.1016/j.livsci.2020.104219
61. Chen D, Zheng W, Lin AP, Uyhazi K, Zhao HY, Lin HF. Pumilio 1 suppresses multiple activators of p53 to safeguard spermatogenesis. Curr Biol. (2012) 22:420–5. doi: 10.1016/j.cub.2012.01.039
62. Tanaka H, Iguchi N, Toyama Y, Kitamura K, Takahashi T, Kaseda K, et al. Mice deficient in the axonemal protein Tektin-t exhibit male infertility and immotile-cilium syndrome due to impaired inner arm dynein function. Mol Cell Biol. (2004) 24:7958–64. doi: 10.1128/MCB.24.18.7958-7964.2004
63. Tang S, Wang X, Li W, Yang X, Li Z, Liu W, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. (2017) 100:854–64. doi: 10.1016/j.ajhg.2017.04.012
64. Aszódi A, Bateman JF, Hirsch E, Baranyi M, Hunziker EB, Hauser N, et al. Normal skeletal development of mice lacking matrilin 1: redundant function of matrilins in cartilage? Mol Cell Biol. (1999) 19:7841–5. doi: 10.1128/MCB.19.11.7841
65. Berger JH, Charron MJ, Silver DL. Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PLoS ONE. (2012) 7:e50629. doi: 10.1371/journal.pone.0050629
66. Marino G, Ugalde AP, Fernandez AF, Osorio FG, Fueyo A, Freije JMP, et al. Insulin-like growth factor 1 treatment extends longevity in a mouse model of human premature aging by restoring somatotroph axis function. Proc Natl Acad Sci. (2010) 107:16268–73. doi: 10.1073/pnas.1002696107
67. Hao XJ, Plastow G, Zhang CY, Xu ST, Hu ZQ, Yang TF, et al. Genome-wide association study identifies candidate genes for piglet splay leg syndrome in different populations. BMC Genet. (2017) 18:64. doi: 10.1186/s12863-017-0532-4
68. Rao VV, Sangiah U, Mary KA, Akira S, Mohanty A. Role of Akirin1 in the regulation of skeletal muscle fiber-type switch. J Cell Biochem. (2019) 120:11284–304. doi: 10.1002/jcb.28406
69. Smith CL, Blake JA, Kadin JA, Richardson JE, Bult CJ. Mouse genome database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. (2018) 46:D836–42. doi: 10.1093/nar/gkx1006
70. Azevedo A, Schwartz IV, Kalakun L, Brustolin S, Burin MG, Beheregaray A, et al. Clinical and biochemical study of 28 patients with mucopolysaccharidosis type VI. Clin Genet. (2004) 66:208–13. doi: 10.1111/j.1399-0004.2004.00277.x
71. Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. (1993) 75:73–82. doi: 10.1016/S0092-8674(05)80085-6
72. Millard HR, Musani SK, Dibaba DT, Talegawkar SA, Taylor HA, Tucker KL, et al. Dietary choline and betaine; associations with subclinical markers of cardiovascular disease risk and incidence of CVD, coronary heart disease and stroke: the jackson heart study. Eur J Nutr. (2018) 57:51–60. doi: 10.1007/s00394-016-1296-8
73. Wechsler B. Coping and coping strategies: a behavioural view. Appl Anim Behav Sci. (1995) 43:123–34. doi: 10.1016/0168-1591(95)00557-9
74. Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. (2000) 101:635–45. doi: 10.1016/S0092-8674(00)80875-2
75. Baines AC, Adams EJ, Zhang B, Ginsburg D. Disruption of the Sec24d gene results in early embryonic lethality in the mouse. PLoS ONE. (2013) 8:e61114. doi: 10.1371/journal.pone.0061114
76. Simeoni L, Posevitz V, Kolsch U, Meinert I, Bruyns E, Pfeffer K, et al. The transmembrane adapter protein SIT regulates thymic development and peripheral T-cell functions. Mol Cell Biol. (2005) 25:7557–68. doi: 10.1128/MCB.25.17.7557-7568.2005
77. Mandelbrot DA, Oosterwegel MA, Shimizu K, Yamada A, Freeman GJ, Mitchell RN, et al. B7-dependent T-cell costimulation in mice lacking CD28 and CTLA4. J Clin Invest. (2001) 107:881–7. doi: 10.1172/JCI11710
78. Ding R, Qiu Y, Zhuang Z, Ruan D, Wu J, Zhou S, et al. Genome-wide association studies reveals polygenic genetic architecture of litter traits in duroc pigs. Theriogenology. (2021) 173:269–78. doi: 10.1016/j.theriogenology.2021.08.012
79. Bortoluzzi C, Bosse M, Derks M, Crooijmans RP, Groenen MA, Megens HJ. The type of bottleneck matters: insights into the deleterious variation landscape of small managed populations. Evol Appl. (2019) 13:330–41. doi: 10.1111/eva.12872
Keywords: Duroc pigs, runs of homozygosity, candidate genes, missense mutation, association analysis
Citation: Wang X, Li G, Ruan D, Zhuang Z, Ding R, Quan J, Wang S, Jiang Y, Huang J, Gu T, Hong L, Zheng E, Li Z, Cai G, Wu Z and Yang J (2022) Runs of Homozygosity Uncover Potential Functional-Altering Mutation Associated With Body Weight and Length in Two Duroc Pig Lines. Front. Vet. Sci. 9:832633. doi: 10.3389/fvets.2022.832633
Received: 10 December 2021; Accepted: 24 January 2022;
Published: 08 March 2022.
Edited by:
Xiangdong Ding, China Agricultural University, ChinaReviewed by:
Xiaolei Liu, Huazhong Agricultural University, ChinaZhipeng Wang, Northeast Agricultural University, China
Qishan Wang, Zhejiang University, China
Pinghua Li, Nanjing Agricultural University, China
Copyright © 2022 Wang, Li, Ruan, Zhuang, Ding, Quan, Wang, Jiang, Huang, Gu, Hong, Zheng, Li, Cai, Wu and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenfang Wu, d3pmZW1haWxAMTYzLmNvbQ==; Jie Yang, amlleWFuZzIwMTJAaG90bWFpbC5jb20=
†These authors have contributed equally to this work
 Xiaopeng Wang
Xiaopeng Wang Guixin Li
Guixin Li Donglin Ruan
Donglin Ruan Zhanwei Zhuang1
Zhanwei Zhuang1 Jianping Quan
Jianping Quan Ting Gu
Ting Gu Linjun Hong
Linjun Hong Zicong Li
Zicong Li Gengyuan Cai
Gengyuan Cai Zhenfang Wu
Zhenfang Wu Jie Yang
Jie Yang