- 1Aklilu Lemma Institute of Pathobiology, Addis Ababa University, Addis Ababa, Ethiopia
- 2International Livestock Research Institute, Addis Ababa, Ethiopia
- 3Ethiopian Public Health Institute, Addis Ababa, Ethiopia
- 4Department of Veterinary Biosciences, The Ohio State University, Columbus, OH, United States
- 5Department Microbial Immunity and Infection, The Ohio State University, Columbus, OH, United States
- 6Infection Diseases Institute, The Ohio State University, Columbus, OH, United States
- 7International Livestock Research Institute, Nairobi, Kenya
- 8Natural Resources Institute, Chatham, United Kingdom
Escherichia coli O157:H7 is an emerging foodborne pathogen of public health importance. The objectives of this study were to estimate the prevalence and evaluate the antimicrobial susceptibility pattern and multidrug-resistant profile of E. coli O157:H7 isolated from raw beef sold in butcher shops in Addis Ababa, Ethiopia. A total of 384 raw beef samples were collected from randomly selected butcher shops across the 10 sub-cities of Addis Ababa. E. coli O157:H7 was isolated following ISO-16654:2001 standard, and isolates were tested for resistance to 13 antimicrobial agents using the Kirby–Bauer disk diffusion method. Out of the 384 retail raw beef samples examined, 14 (3.64%) (95% CI = 1.77–5.51%) carried E. coli O157:H7 serotype. Of the 14 E. coli O157:H7 isolates, 8 (57.14%) were found to be resistant to three or more antimicrobial categories. The frequency of resistant phenotype was more common for ampicillin (92.8%), nitrofurantoin (92.8%), and tetracycline (50%). Multidrug-resistant E. coli O157:H7 were present in raw beef sold in butcher shops in Addis Ababa. Thus, more stringent monitoring of antimicrobial use in both human and animal populations should be implemented. In addition, further studies should be conducted to understand the E. coli O157:H7 points of contamination and define appropriate risk mitigation strategies.
Introduction
Escherichia coli O157:H7 is an emerging bacterial zoonotic foodborne pathogen of global significance for which cattle is the primary reservoir (1). Cattle shed the bacteria into the environment in their faces, which are then transmitted to humans primarily through the consumption of contaminated raw or undercooked meat (2, 3). The contamination of cattle carcasses or beef can occur during processing and manipulation, such as skinning, evisceration in slaughterhouse, and distribution to butcher shops (4).
While cattle that carry E. coli O157:H7 are asymptomatic, infected humans show clinical manifestations ranging from asymptomatic (carrier state) to serious illness. The bacteria adhere to the gut wall of infected people and cause hemorrhagic colitis. Besides, the pathogen also produces toxins that can cause life-threatening complications including hemolytic uremic syndrome (HUS) and thrombotic thrombocytopenic purpura (5, 6).
Early antimicrobial treatment can prevent Shiga toxin-producing E. coli O157:H7 infection progression to the HUS (7–9). Studies have shown a significant increase in antimicrobial resistance in E. coli O157:H7 (8). This in part may be related to the overuse and misuse of antibiotics in people and food animals (10). In Ethiopia, studies have been confirmed that E. coli O157:H7 have developed different percentages of resistance against various commonly used antimicrobial drugs including ampicillin, cephalothin, streptomycin, tetracycline, trimethoprim, amikacin, amoxicillin-clavulanic acid, ciprofloxacin, nalidixic acid, streptomycin, chloramphenicol, nitrofurantoin, and erythromycin (11–18).
Ethiopian food culture includes eating raw beef “Kurt” or minced raw beef “Kitfo,” which increases people's exposure to pathogens. Despite the risk of exposure to E. coli O157:H7, limited studies on the magnitude of contamination and risk of E. coli O157:H7 and antimicrobial susceptibility has been reported, particularly from developing countries including Ethiopia (19). Such studies can provide valuable information to help in the implementation of strategies to minimize contamination levels.
Earlier studies have reported the occurrence of E. coli O157:H7 on raw beef from butcher shops in Ethiopia with results in the range of 0.8–21.9% (11, 12, 14–16). However, the previous studies tend to suffer from small samples and sampling approaches that fail to obtain a representative sample of a population of interest.
Therefore, this study was designed to estimate the prevalence and evaluate the antimicrobial susceptibility pattern and multidrug-resistant profile of E. coli O157:H7 isolated from raw beef sold in butcher shops in Addis Ababa, Ethiopia.
Materials and Methods
Study Area
The study was carried out in Addis Ababa, the capital city of Ethiopia. The city covers 540 km2 and is divided into 10 sub-cities (Figure 1). The city lies at an elevation of 2,355 m above sea level and is located at 9°1′48″N 38°44′24″E. The city has minimum, maximum, and average temperatures of 14, 21 and 17.5°C, respectively. The capital city has an estimated human population of 3.15 million.
Figure 1. Sub-cities in Addis Ababa included in the study (20).
Study Design and Sample Size Determination
A cross-sectional study was conducted from October 2018 to December 2019 to determine the prevalence and antimicrobial susceptibility pattern and multidrug-resistant profile of E. coli O157:H7 serotypes in retail raw beef samples obtained from butchery shops, in Addis Ababa, the capital city of Ethiopia.
The sample size required was calculated according to Thrusfield (21), from an expected pooled prevalence of 6.5 for the butcher shops (11, 12, 14–16) with a defined precision of 5% and a level of confidence of 95%.
where Z = z statistic for level of confidence; n = required sample size; Pexp = expected prevalence and a desired absolute precision (d) of 0.05, Z = 1.96. Therefore, the minimum sample sizes were 49 butcher shops. However, in order to increase the precision of the study, a total of 384 butcher shops were included.
Study Samples and Sampling Methods
The study samples were retail raw beef. A list of active and legally registered butcher shops within the 10 sub-cites and their distribution lines were obtained from Addis Ababa Abattoir Enterprise. A total of 384 butcher shops were selected using the simple random sampling method, and the butcher shops were visited only once A raw beef sample was purchased from each of the randomly selected butcher shops as it was sold to the consumer.
Each sample was placed in a sterile individual plastic bag. The sample was identified by its exclusive sample identification number, which was written on the plastic bag, alongside the sub-city and the date of sampling. Finally, the sample was transported to the Microbiology Laboratory of the Aklilu Lemma Institute of Pathobiology, Addis Ababa University, at cold temperature in a cool box. Upon arrival to the laboratory, the samples were stored in a refrigerator at ±4°C. The samples were processed within 6–12 h from arrival. The detection of E. coli O157:H7 was administered consistent with the protocol of ISO-16654:2001 standard (11).
Sample Preparation and Enrichment
Twenty-five grams of raw beef was weighed and cut into smaller pieces with a sterile scalpel blade on a sterile plate and put in a sterile Stomacher bag. Then, 225 ml of modified Tryptone Soya Broth (TSB) supplemented with Novobiocin (mTSB+N) (1:9) was added to the raw beef and homogenized (Stomacher 400; Seward Medical, Worthing, United Kingdom) at high speed for 2 min. The enrichment sample was then incubated aerobically at 41.5°C for 24 h.
Isolation
All enriched broths were plated on to cefixime tellurite sorbitol MacConkey agar (CT-SMAC) (Oxoid, Basingstoke, England), supplemented with 0.05 mg/L cefixime and 2.5 mg/L tellurite (Oxoid, Basingstoke, England) (CT-SMAC) (Oxoid, Basingstoke, England) and incubated at 37°C for 24 h. After the incubation period, the CT-SMAC agar plates were examined for the presence of non-sorbitol fermenter colorless colonies, and subsequently, they were sub-cultured on Rainbow agar O157 (Hayward, Berkeley Heights, NJ, USA). The plates were then incubated for 20–24 h at 37°C and observed for the presence of typical black or gray coloration on Rainbow agar O157, indicating pure colonies (22).
Biochemical Confirmation
Five typical colonies from each Rainbow agar O157 plate were sub-cultured on nutrient agar (Oxoid, Basingstoke, England) for biochemical confirmation by indole formation. The agar plates were incubated at 37°C for 18–24 h. One colony from the pure culture on nutrient agar was inoculated into a tube of tryptone/tryptophan medium (Oxoid, Basingstoke, England) and incubated at 37°C for 24 h. Then, 1 ml of Kovac's reagent (Oxoid, Basingstoke, England) was added and the tube allowed to stand at room temperature for 10 min. The formation of red color indicates a positive reaction (11).
Serological Identification of O157 and H7 Antigens
Indole-positive colonies were examined for their serological reaction with antiserum to E. coli O157:H7 using RIM E. coli O157:H7 latex test (Oxoid, Basingstoke, England). Indole-positive colonies were sub-cultured from the nutrient agar to the sorbitol MacConkey agar (Oxoid, Basingstoke, England). For every isolate to be tested, one drop of test latex was dispensed into a well of the test slide. In like manner, one drop of E. coli control latex was dispensed into a separate well of the test slide. Using a plastic stick, a portion of the non-sorbitol fermenting colony (NSFC) was removed from the sorbitol MacConkey agar (SMAC) (Oxoid, Basingstoke, England) plate and emulsified in E. coli O157 test latex on the slide and spread over the reaction area. Using a fresh plastic stick, the process was repeated with the remaining NSFC and emulsified in E. coli control, latex on the slide. The slide was rotated using circular motions for up to 1 min or until agglutination appears. For E. coli O157 positives that agglutination occurs with the E. coli O157 test latex and the control latex is negative, the isolate was streaked from sorbitol MacConkey agar (Oxoid, Basingstoke, England) to a blood agar (Oxoid, Basingstoke, England) plate and incubated at 37°C for 18–24 h. After 18–24 h incubation, the sweep of growth from the blood agar plate was emulsified in a drop of E. coli H7 test latex. Colonies giving an agglutination reaction were confirmed as E. coil O157:H7 positive.
Antimicrobial Susceptibility Testing
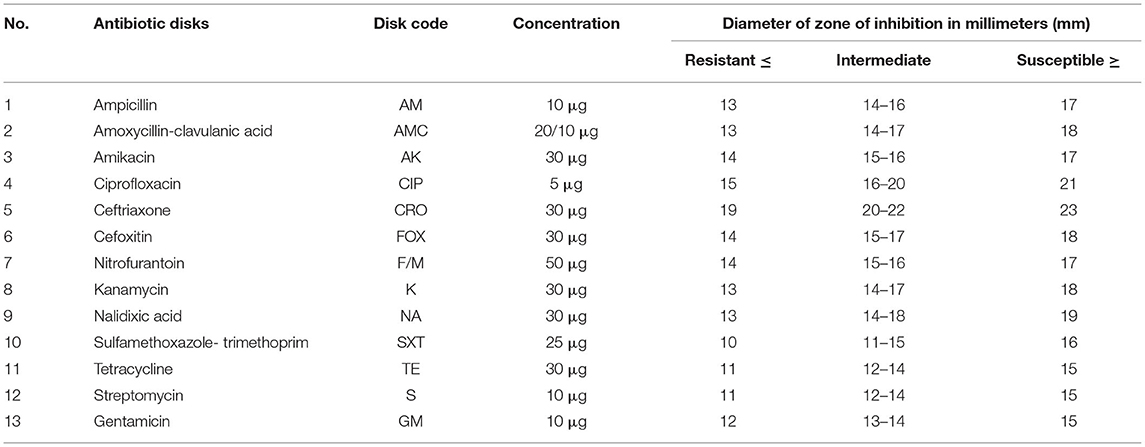
The antimicrobial susceptibility was performed, following the standard agar disk diffusion method consistent with CLSI (23) using commercial antimicrobial disks (Table 1). The antimicrobial agents were selected based on the use of antimicrobial agents in the ruminants, potential public health importance, and recommendations from the guideline of antimicrobial susceptibility testing from the Clinical and Laboratory Standards Institute (23).
Each isolated bacterial colony from pure fresh culture was transferred into a tube of 5 ml TSB (Oxoid, Basingstoke, England) and incubated at 37°C for 6 h. The turbidity of the culture broth was adjusted using sterile saline solution or added more colonies to get turbidity comparable with that of 0.5 McFarland standards. The diluted bacterial suspensions were swabbed in three directions uniformly on the surface of Mueller–Hinton agar plates using sterile cotton swabs. After the plates were dried (about 10 min), with the aid of sterile forceps, antibiotic-impregnated disks were placed to the surface of the inoculated plates. Then, the plates were incubated aerobically at 37°C for 24 h. Finally, the diameter of the inhibition zone formed around each disk was measured on a black surface using a transparent ruler by placing it over the plates. The results were classified as sensitive, intermediate, and resistant according to the CLSI (23). E. coli (ATCC 25922)-type strains were used as a positive control.
Multidrug Resistance (MDR)
Multidrug resistance (MDR) was defined as a resistance of a bacterial strain for at least one agent in three or more antimicrobial categories (24).
Ethical Consideration
The study protocol was ethically approved by the Institutional Review Board of Aklilu Lemma Institute of Pathobiology, Addis Ababa University (Minutes Ref NO: ALIPB IRB/006/2011/2018).
Data Management and Analysis
The data were entered and coded in MS Excel and then analyzed using IBM SPSS version 25.0 (25). The prevalence was determined by dividing the number of positive samples by the total number of samples examined. Descriptive statistics such as frequency and percentages were used to describe the proportion of resistant, intermediate, or susceptible strains. The difference in prevalence by sub-city was determined using the chi-square (χ2) test. A p-value < 0.05 was considered indicative of a statistically significant difference.
Results
Prevalence
Out of 384 raw beef samples examined, 14 (3.64%) (95% CI = 1.77–5.51%) were positive to E. coli O157:H7 serotypes.
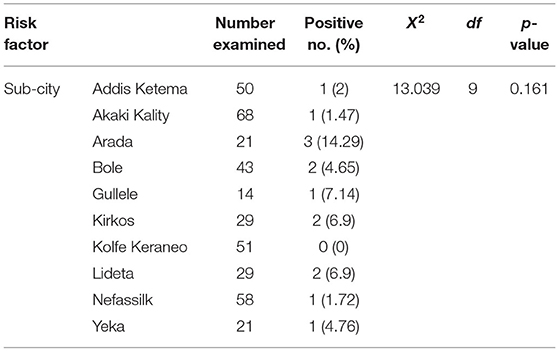
E. coli O157:H7 serotypes were detected in Addis Ketema (2%), AkakiKality (1.47%), Arada (14.29%), Bole (4.65%), Gullele (7.14%), Kirkos (69%), KolfeKeraneo (0%), Lideta (6.9%), Nefassilk (1.72%), and Yeka (4.76%). Variation in the prevalence between the butcher shops from the different sub-cities was not statistically significant (p > 0.05) (Table 2).
Antimicrobial Susceptibility Pattern
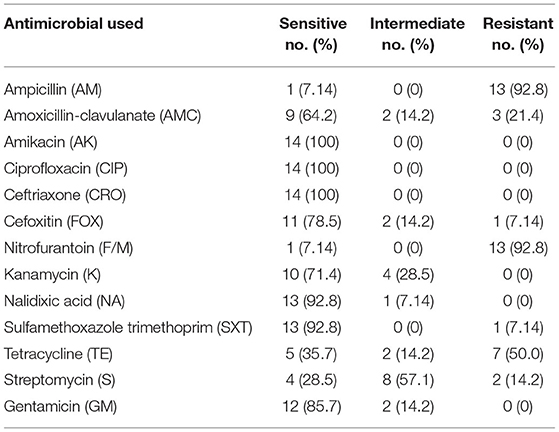
The result of the antimicrobial susceptibility test of the14 E. coli O157:H7 serotypes isolated from raw beef samples with 13 selected antimicrobial agents is shown in Table 3.
All the 14 E. coli O157:H7 serotypes' isolates from raw beef were found to be susceptible to amikacin (100%), ciprofloxacin (100%), and ceftriaxone (100%). Furthermore, the isolates showed high susceptibility to sulfamethoxazole-trimethoprim (92.8%), nalidixic acid (92.8%), gentamicin (85.7%), cefoxitin (78.5%), kanamycin (71.4%), and amoxicillin-clavulanic acid (64.2%). The results of the present study on antimicrobial sensitivity test indicated high resistance to ampicillin (92.8%), nitrofurantoin (92.8), and tetracycline (50.0%).
Multidrug Resistance Profiles
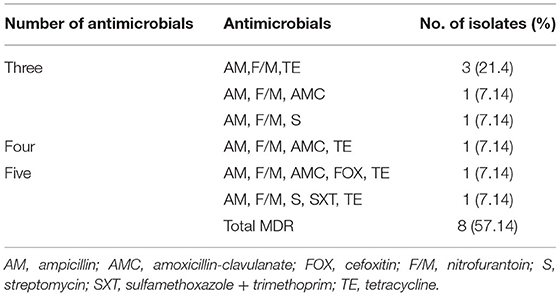
Out of the 14 E. coli O157:H7 isolates, 8 (57.14%) were found to be resistant to three or more antimicrobial categories. MDR profiles against three, four, and five antimicrobial categories were resistant to 5 (35.7%), 1 (7.1%), and 2 (14.3%), respectively. The frequency of resistant phenotype was more common for ampicillin, nitrofurantoin, and tetracycline (Table 4).
Discussion
Foodborne infections are major health concerns in developing countries including Ethiopia. The surveillance and monitoring of foodborne pathogens provide crucial information on planning, implementing, and evaluating food safety systems. Therefore, appropriate information on the contamination level and antimicrobial susceptibility of E. coli O157:H7 in retail raw beef may have implications in strengthening the surveillance system of foodborne diseases as well as is important to design prevention and control measures to decrease the risk of contamination. The prevalence of E. coli O157:H7 found in raw beef samples in the present study was 14/384 (3.64%) (95% CI = 1.77–5.51%). Similar to our findings, E. coli O157:H7 was identified in 1/25 (4%), 1/25 (4%), and 1/30 (3.3%) of raw beef samples at butcher shops in Addis Ababa, Batu, and Holetta, respectively (16). In contrast to our findings, the lower prevalence in raw beef samples was 3/150 (2%) in Hawassa and 1/125 (0.8%) in Addis Ababa and Debre Berhan (14, 15). Higher prevalence was described in butcher shops in Bishoftu 8/86 (9.3%) and 2/30 (6.7 %) and in Addis Ababa 14/64 (21.9%) (11, 12, 16). The variation of these findings might depend on different factors, e.g., abattoir, butcher conditions, sample size, and laboratory methods.
In this study, no statistically significant variation in the prevalence rate among the sub-cities butcher shops of beef samples (p > 0.05) was observed. This might be due to butcher shops sourcing their cattle carcasses from the main abattoir in the city. The small number of positives also means that a much larger sample size would be needed to identify any differences.
All of the 14 isolates of raw beef were susceptible to amikacin, ciprofloxacin, and ceftriaxone. Furthermore, the isolates showed high susceptibility to sulfamethoxazole-trimethoprim, nalidixic acid, gentamicin, cefoxitin, kanamycin, and amoxicillin-clavulanic acid. Similar findings have been reported by other researchers from Ethiopia (11, 12, 14–16, 18, 26). The E. coli O157:H7 strains isolated from raw beef had high resistance to ampicillin (92.8%), nitrofurantoin (92.8%), and tetracycline (50.0%). Similarly, studies from Ethiopia (13–16, 18, 26, 27) and Nigeria (27) revealed high resistance among E. coli O157:H7 isolates to ampicillin. However, 90% susceptibility of ampicillin was reported in Bishoftu (11). Nitrofurantoin resistance was reported in Somalia and Hawassa (13, 15). Drugs like ampicillin and nitrofurantoin have long been used for the management of various infections in Ethiopia, and high rate of resistance to these drugs might have developed as a consequence of this prolonged use (28).
Moreover, the findings of antimicrobial susceptibility test showed that 50% of E. coli O157:H7 isolates from raw beef resistance to tetracycline (14, 17). This is in agreement with previous studies from Ethiopia (11, 13, 26, 27) and Nigeria (27). This might be related to the broad use of tetracycline in the management of various infections in the livestock in Ethiopia (29).
Among the 14 E. coli O157:H7 isolates from raw beef tested, 8 (57.14%) were resistant to three or more classes of antibiotics. The occurrences of multidrug-resistant isolates (17.9–92.5%) were also reported in previous studies in Ethiopia (11–13, 17, 18). The occurrence of MDR may be associated with indiscriminate utilization of antimicrobial agents, which was not elucidated with the current study method. Furthermore, the transmission of MDR bacteria via the consumption of meat have been propounded as a potential source in Africa (30, 31).
The present study had some limitations. The use of immunomagnetic separation (IMS) with enrichment in broth culture enhances the isolation of E. coli O157 from samples with a low concentration of bacteria (32). In this study, enrichment without IMS was employed to isolate E coli O157:H7. Nevertheless, the present study revealed that multidrug-resistant E. coli O157:H7 were present in raw beef sold in butcher shops in Addis Ababa, Ethiopia. Given the low infective dose of E. coli O157:H7 [10 colony forming unit (CFU)/g] and the cultural habit of eating raw beef in the society, the current prevalence should be considered important from a public health standpoint. These findings should be communicated with government and projects working with butchers along with the information on reducing the risk. Thus, more stringent monitoring of antimicrobial use in both human and animal populations should be implemented. In addition, further studies should be conducted to understand the E. coli O157:H7 points of contamination and define appropriate risk mitigation strategies.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
AH: conceived and designed the study, conducted the study, analyzed the data, and wrote the paper. SA: conceived the study, provided guidance to its design, and reviewed the manuscript. NB: designed the study, analyzed the data, and wrote the paper. TA: conducted the study and analyzed the data. PB: reviewed the paper and wrote the paper. DG: conceived and designed study, analyzed the data, and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was conducted under the project of the International Livestock Research Institute (ILRI), funded by CGIAR Research Program on Agriculture for Nutrition and Health.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the Aklilu Lemma Institute of Pathobiology, Addis Ababa University for their cooperation.
References
1. Pal M, Mahendra R. Escherichia coli 0157: H7: an emerging bacterial zoonotic food borne pathogen of global significance. Int J Interdisc Multidisc Stud. (2016) 4:1–4.
2. Cray WC Jr, Moon HW. Experimental infection of calves and adult cattle with Escherichia Coli O157:H7. Appl Environ Microbiol. (1995) 61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995
4. Gill C. Microbiological contamination of meat during slaughter and butchering of cattle, sheep and pigs. Microbiol Meat Poult. (1998) 118–157.
5. Akomoneh EA, Esemu SN, Jerome Kfusi A, Ndip RN, Ndip LM. Prevalence and virulence gene profiles of Escherichia coli O157 from cattle slaughtered in Buea, Cameroon. PLoS ONE. (2020) 15:e0235583. doi: 10.1371/journal.pone.0235583
6. Armstrong GL, Hollingsworth J, Morris JG Jr. Emerging foodborne pathogens: Escherichia coli O157: H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol Rev. (1996) 18:29–51. doi: 10.1093/oxfordjournals.epirev.a017914
7. Amézquita-López BA, Quiñones B, Soto-Beltrán M, Lee BG, Yambao JC, Lugo-Melchor OY, et al. Antimicrobial resistance profiles of Shiga toxin-producing Escherichia coli O157 and Non-O157 recovered from domestic farm animals in rural communities in Northwestern Mexico. Antimicrob Resist Infect Control. (2016) 5:1–6. doi: 10.1186/s13756-015-0100-5
8. Mühlen S, Dersch P. Treatment strategies for infections with Shiga toxin-producing Escherichia coli. Front Cell Infect Microbiol. (2020) 10:169. doi: 10.3389/fcimb.2020.00169
9. Schroeder CM, Meng J, Zhao S, DebRoy C, Torcolini J, Zhao C, et al. Antimicrobial resistance of Escherichia coli O26, O103, O111, O128, and O145 from animals and humans. Emerg Infect Dis. (2002) 8:1409. doi: 10.3201/eid0812.0200770
10. Radostits O, Gay C, Blood D, Hinchcliff K, A A Textbook of the Diseases of Cattle Sheep Pigs Goats and Horses. Nottingham: Saunders Ltd. (2000). p. 703–739.
11. Hiko A, Asrat D, Zewde G. Occurrence of Escherichia coli O157: H7 in retail raw meat products in Ethiopia. J Infect Dev Ctries. (2008) 2:389–93. doi: 10.3855/jidc.203
12. Bekele T, Zewde G, Tefera G, Feleke A, Zerom K. Escherichia coli O157: H7 in raw meat in Addis Ababa, Ethiopia: prevalence at an abattoir and retailers and antimicrobial susceptibility. Int. J. Food Contam. (2014) 1:1–8. doi: 10.1186/s40550-014-0004-9
13. Dulo F, Feleke A, Szonyi B, Fries R, Baumann MP, Grace D. Isolation of multidrug-resistant Escherichia coli O157 from goats in the somali region of Ethiopia: a cross-sectional, abattoir-based study. PLoS ONE. (2015) 10:e0142905. doi: 10.1371/journal.pone.0142905
14. Abdissa R, Haile W, Fite AT, Beyi AF, Agga GE, Edao BM, et al. Prevalence of Escherichia coli O157: H7 in beef cattle at slaughter and beef carcasses at retail shops in Ethiopia. BMC Infect Dis. (2017) 17:1–6. doi: 10.1186/s12879-017-2372-2
15. Atnafie B, Paulos D, Abera M, Tefera G, Hailu D, Kasaye S, et al. Occurrence of Escherichia coli O157: H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol. (2017) 17:1–7. doi: 10.1186/s12866-017-0938-1
16. Beyi AF, Fite AT, Tora E, Tafese A, Genu T, Kaba T, et al. Prevalence and antimicrobial susceptibility of Escherichia coli O157 in beef at butcher shops and restaurants in central Ethiopia. BMC Microbiol. (2017) 17:1–6. doi: 10.1186/s12866-017-0964-z
17. Feleke A, Kebede D, Kiros AW. Prevalence and antibiogram of Escherichia coli O157 isolated from bovine in Jimma, Ethiopia: abattoirbased survey. Ethiop Vet J. (2017) 21:109–20. doi: 10.4314/evj.v21i2.8
18. Bedasa S Shiferaw D Abraha A Moges T Occurrence Occurrence and antimicrobial susceptibility profile of Escherichia coli O157:H7 from food of animal origin in Bishoftu town Central Ethiopia. Int. J. Food Contam. (2018). 5:1–8. doi: 10.1186/s40550-018-0064-3
19. Rahimi E, Nayebpour F. Antimicrobial resistance of Escherichia coli O 157: H7/NM isolated from feaces of ruminant animals in Iran. J Cell Anim Biol. (2012) 6:104–8. doi: 10.5897/JCAB11.082
22. Biolog. Data Collection Software Identification System User Guide, 2008. Hayward, CA: Biolog (2008).
23. CLSI. Performance standards for antimicrobial susceptibility testing. Twenty forth information supplement (M 100-524). Ser MMWR. Wayne, PA: CLSI (2014) 54:1277–80.
24. Magiorakos A-P, Srinivasan A, Carey R, Carmeli Y, Falagas M, Giske C. et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
26. Taye M, Berhanu T, Berhanu Y, Tamiru F, Terefe D. Study on carcass contaminating Escherichia coli in apparently healthy slaughtered cattle in haramaya university slaughter house with special emphasis on Escherichia coli O157:H7, Ethiopia. J Veterinar Sci Technolo. (2013) 4:132. doi: 10.4172/2157-7579.1000132
27. Ojo OE, Ajuwape AT, Otesile EB, Owoade AA, Oyekunle MA, Adetosoye AI. Potentially zoonotic shiga toxin-producing Escherichia coli serogroups in the faeces and meat of food-producing animals in Ibadan, Nigeria. Int J Food Microbiol. (2010) 142:214–21. doi: 10.1016/j.ijfoodmicro.2010.06.030
28. Eguale T, Gebreyes WA, Asrat D, Alemayehu H, Gunn JS, Engidawork E. Non-typhoidal Salmonella serotypes, antimicrobial resistance and co-infection with parasites among patients with diarrhea and other gastrointestinal complaints in Addis Ababa, Ethiopia. BMC Infect Dis. (2015) 15:1–9. doi: 10.1186/s12879-014-0722-x
29. Gemeda BA, Amenu K, Magnusson U, Dohoo I, Hallenberg GS, Alemayehu G. et al. Antimicrobial use in extensive smallholder livestock farming systems in Ethiopia: knowledge, attitudes, and practices of livestock keepers. Front Vet Sci. (2020) 7:55. doi: 10.3389/fvets.2020.00055
30. Alonso C, Zarazaga M, Sallem RB, Jouini A, Slama KB, Torres C. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol. (2017) 64:318–34. doi: 10.1111/lam.12724
31. Eibach D, Dekker D, Boahen KG, Akenten CW, Sarpong N, Campos CB. et al. Extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae in local and imported poultry meat in Ghana. Vet Microbiol. (2018) 217:7–12. doi: 10.1016/j.vetmic.2018.02.023
Keywords: Addis Ababa, antimicrobial, beef, Escherichia coli O157:H7, prevalence
Citation: Haile AF, Alonso S, Berhe N, Atoma TB, Boyaka PN and Grace D (2022) Prevalence, Antibiogram, and Multidrug-Resistant Profile of E. coli O157: H7 in Retail Raw Beef in Addis Ababa, Ethiopia. Front. Vet. Sci. 9:734896. doi: 10.3389/fvets.2022.734896
Received: 01 July 2021; Accepted: 04 January 2022;
Published: 24 February 2022.
Edited by:
Bassirou Bonfoh, Swiss Centre for Scientific Research, Côte d'IvoireReviewed by:
James Wabwire Oguttu, University of South Africa, South AfricaAgnes Kilonzo-Nthenge, Tennessee State University, United States
Copyright © 2022 Haile, Alonso, Berhe, Atoma, Boyaka and Grace. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aklilu Feleke Haile, YXRha2xpbHVAeWFob28uY29t
 Aklilu Feleke Haile
Aklilu Feleke Haile Silvia Alonso
Silvia Alonso Nega Berhe
Nega Berhe Tizeta Bekele Atoma
Tizeta Bekele Atoma Prosper N. Boyaka
Prosper N. Boyaka Delia Grace
Delia Grace