- 1Section Veterinary Public Health, Department of Paraclinical Sciences, Faculty of Veterinary Science, University of Pretoria, Pretoria, South Africa
- 2Department of Agriculture and Animal Health, College of Agriculture and Environmental Sciences, University of South Africa, Johannesburg, South Africa
- 3Department of Medical Microbiology, University of Pretoria, Pretoria, South Africa
- 4Tshwane Academic Division, National Health Laboratory Service, Pretoria, South Africa
Background: Hospital-acquired infections (HAIs) are associated with increased mortality, morbidity, and an economic burden due to costs associated with extended hospital stays. Furthermore, most pathogens associated with HAIs in veterinary medicine are zoonotic. This study used published data to identify organisms associated with HAIs and zoonosis in veterinary medicine. Furthermore, the study also investigated the antimicrobial-susceptibility profile of these bacterial organisms.
Methods: A systematic literature review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines. Search terms and five electronic databases were used to identify studies published over 20 years (2000–2020). The risk of bias was assessed using the “Strengthening the Reporting of Observational Studies in Epidemiology-Vet” (STROBE-Vet) checklist.
Results: Out of the identified 628 papers, 27 met the inclusion criteria for this study. Most studies (63%, 17/27) included were either from small animal or companion animal clinics/hospitals, while 5% (4/27) were from large animal clinics/hospitals inclusive of bovine and equine hospitals. Hospital-acquired bacteria were reported from environmental surfaces (33%, 9/27), animal clinical cases (29.6%, 8/27), and fomites such as cell phones, clippers, stethoscopes, and computers (14.8%, 4/27). Staphylococcus spp. was the most (63%; 17/27) reported organism, followed by Escherichia coli (19%; 5/27), Enterococcus spp. (15%, 4/27), Salmonella spp. (15%; 4/27), Acinetobacter baumannii (15%, 4/27), Clostridioides difficile (4%, 1/27), and Pseudomonas aeruginosa (4%; 1/27). Multidrug-resistant (MDR) organisms were reported in 71% (12/17) of studies linked to Methicillin-resistant Staphylococcus aureus (MRSA), Methicillin-resistant Staphylococcus pseudintermedius (MRSP), Enterococcus spp., Salmonella Typhimurium, A. baumannii, and E. coli. The mecA gene was identified in both MRSA and MRSP, the blaCMY-2 gene in E. coli and Salmonella spp., and the vanA gene in E. faecium isolate. Six studies reported organisms from animals with similar clonal lineage to those reported in human isolates.
Conclusion: Organisms associated with hospital-acquired infections and zoonosis have been reported from clinical cases, environmental surfaces, and items used during patient treatment and care. Staphylococcus species is the most reported organism in cases of HAIs and some isolates shared similar clonal lineage to those reported in humans. Some organisms associated with HAIs exhibit a high level of resistance and contain genes associated with antibiotic resistance.
1. Introduction
Hospital-acquired infections (HAIs) in both veterinary and human medicine are associated with increased mortality, and morbidity, and are an economic burden due to the increased cost of extended hospital stay and treatment options (1, 2). The most reported HAIs include surgical wounds, urinary tract, and gastrointestinal infections (1–3) and are often associated with bacteria such as Enterococcus species (spp.), Escherichia coli, Staphylococcus spp., Enterobacter spp., Klebsiella spp., Acinetobacter spp., and Pseudomonas spp. (3–6).
Available evidence suggests that HAIs associated with Enterococcus spp., Escherichia coli, K. pneumoniae, and S. aureus are on the increase in veterinary medicine (7–10). There are also reports of vancomycin-resistant enterococci (VRE), multidrug-resistant (MDR) E. coli, carbapenem-resistant A. baumannii, carbapenem-resistant P. aeruginosa, carbapenem-resistant and extended-spectrum β-lactamase (ESBL) producing Enterobacteriaceae (3–6, 11–13), with limited treatment options and poor prognosis (1, 5, 6, 11, 13). It is estimated that 60% of emerging infectious diseases are likely to come from animals (14, 15). Of concern is that bacteria associated with HAIs in veterinary settings could be contributing to the emergence of these new diseases (6, 16). Since the veterinary hospital environment is a human-animal interface, it remains a potential source of zoonotic pathogens (6, 17). Therefore, veterinary healthcare workers and animal owners are at an increased risk of contracting various zoonotic infections (14, 15). This is likely to put financial stress on the human health system especially in developing countries (18). In view of this, continuous surveillance of hospital-acquired and zoonotic pathogens in veterinary medicine should be done to better quantify the risk of transmission to personnel and animal owners (19, 20).
Systematic review studies have suggested that improving surveillance systems is critical in the prevention of HAIs and in reducing the emergence of antimicrobial-resistant pathogens (6, 21). Therefore, a holistic approach is needed to investigate the types of disease agents, hosts, the antimicrobial-resistance profile of the organism, and the virulence of the organisms associated with HAIs in veterinary medicine (17).
This study describes the occurrence and antimicrobial-susceptibility profiles of bacterial organisms associated with HAIs and zoonosis in veterinary medicine. It addresses the following research questions: (1) Which bacteria associated with HAIs and zoonotic diseases have been reported in veterinary hospitals? (2) What are the antimicrobial resistance profiles of these bacteria?
2. Materials and methods
The systematic literature review was conducted using the Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) guidelines (22). Keywords and synonyms used in various databases included hospital-acquired organism or infection, nosocomial organism or infection, animal to animal infections, zoonotic infection, zoonosis, animal to human infections, veterinary hospital, and veterinary clinic.
2.1. Information source
Search terms and electronic databases used in this study are provided in Table 1. Since each database has a different search function, alternate search terms appropriate for each database were used. Boolean operators were utilized in all searches. A data search was conducted between June 2020 and December 2020. A follow-up search was performed in January 2021, however, there were no additional studies considered based on the inclusion criteria. Mendeley reference manager was used to store all studies and documents retrieved.
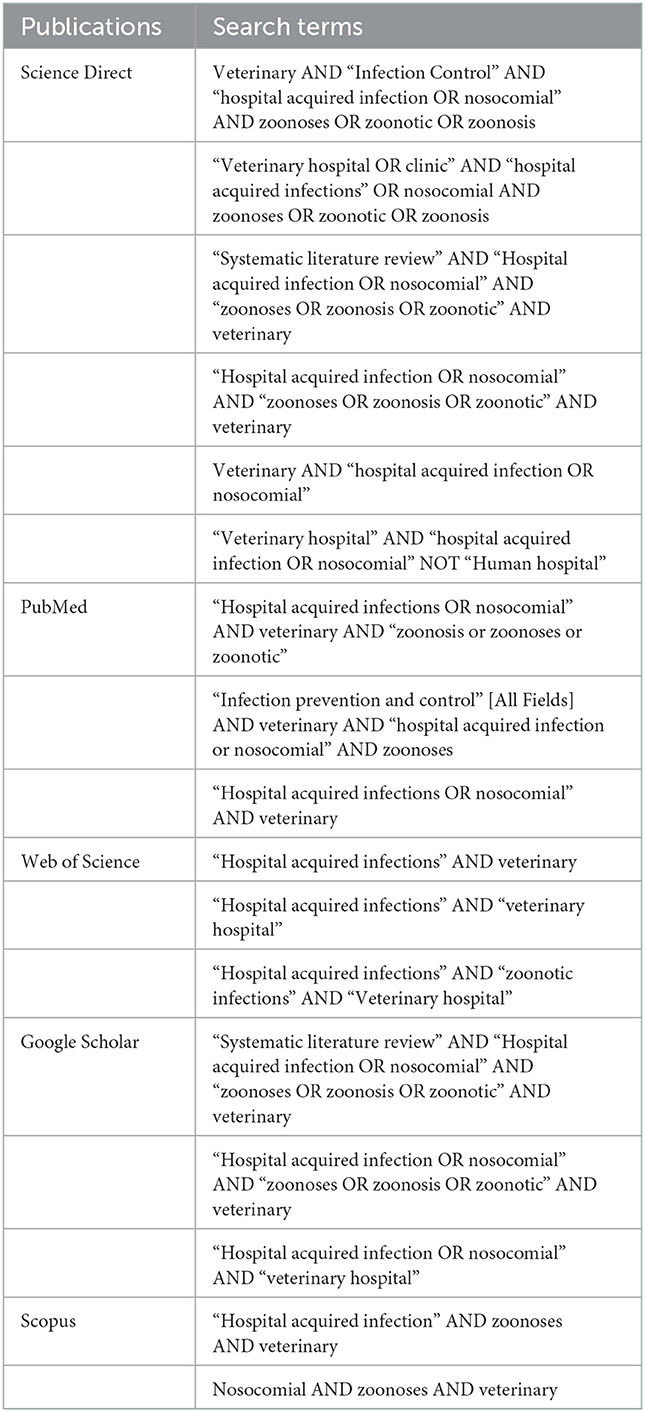
Table 1. Search terms and databases utilized to search for articles included in this review about hospital-acquired and/or zoonotic infections in veterinary facilities between 2000 and 2020.
2.2. Eligibility criteria
Only manuscripts published in peer-reviewed journals were considered for inclusion in this study. Primary research articles written in English and published between 2000 and 2020 were selected. The microbiological data included bacterial isolates from HAI cases, hospital environmental screening, fomites from veterinary hospitals, and zoonotic cases in veterinary hospitals. In addition, the antimicrobial resistance profiles of the different bacteria were also extracted. The inclusion and exclusion criteria are listed in Table 2. Two investigators (DC, DN) independently screened the titles and abstracts from the searches. Any disagreements were settled by discussion. The use of either the CLSI or EUCAST guidelines was not considered an eligibility criterion in this study since some studies report potential discrepancies between the results of the antimicrobial resistance based on CLSI and EUCAST (23), while others report comparable antibiotic susceptibility rates between CLSI and EUCAST breakpoints (24, 25).
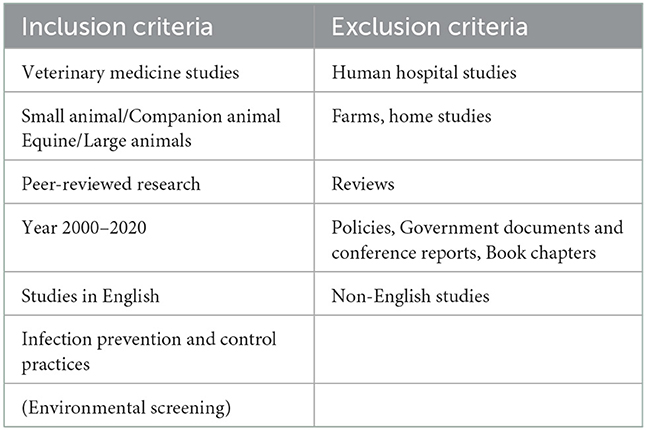
Table 2. Inclusion and exclusion criteria of articles reporting on hospital-acquired and/or zoonotic infections in veterinary facilities between 2000 and 2020.
2.3. Study selection and data items
For each study that met the selection criteria for inclusion, the following data were extracted: author, year, the theme of study (HAIs or zoonotic studies), and the antimicrobial resistance profile.
3. Results
3.1. Study selection
Initially a total of 628 studies were identified. After initial screening, 330 articles remained. Based on the eligibility screening criteria, 48 studies remained and were further critically assessed. A total of 27 studies met the inclusion criteria and were further analyzed (Figure 1).
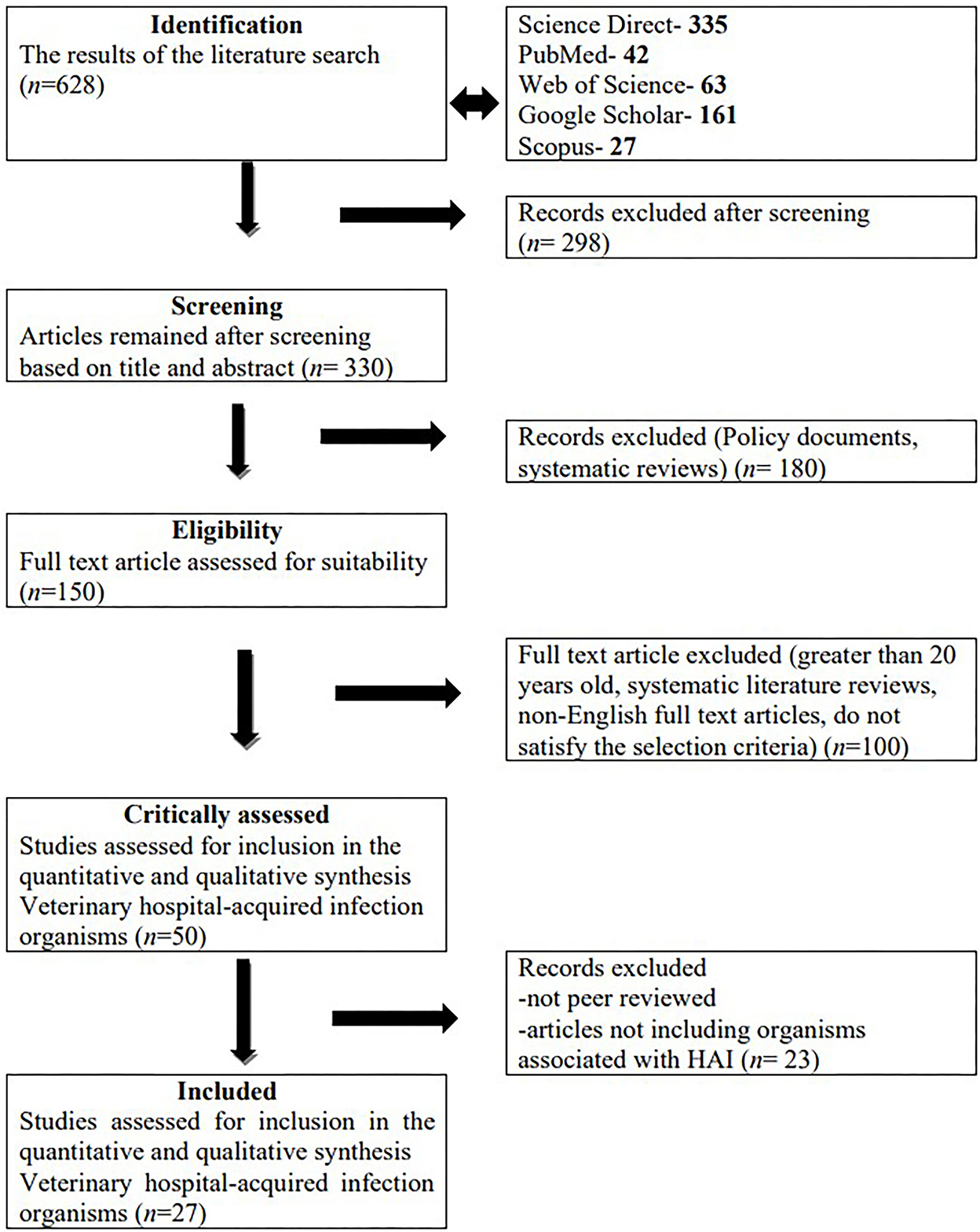
Figure 1. Summary of study selection and exclusion using the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines.
3.2. Risk of bias
Strengthening the Reporting of Observational Studies in Epidemiology (STROBE-Vet) statement is a 22-item tool that allows a systematic way of reporting on veterinary observational studies. The STROBE statement was developed to guide the reporting of observational studies related to human health. These methods have been adopted and used for standardized reporting guidelines for observational studies in veterinary medicine (21, 26). Identified studies that met the inclusion criteria were cross-sectional and cohort studies (26). Each study was assessed individually according to each of the 22 items.
Items were considered to have been reported sufficiently if the studies provided a detailed abstract and clear title (item one), background, and rationale (item two), stated the objectives (item three), presented key elements of the study design (item four), described the sample size (item 11), reported outcomes for the study (items 14 and 15), provided estimates and parameters (item 16), summarized key results regarding study objectives (item 18 and 19), interpreted results (item 20), discussed the results (item 21), and stated the funding source as well as the role of authors as described by Sergeant et al. (26).
Only two studies (7%, 2/27) reported on all STROBE-Vet items (27, 28). Based on STROBE-Vet, item 1 was partially attained by 19/27 (70%) studies as they excluded the study design and was fully attained by 8 (29%) studies. Items 6, 13,14, and 20 were fully attained by all the studies, Items 2, 4, 5, and 16 were fully attained by 26 (96%) of the studies, items 3,15,17, and were fully attained by 25 (93%) of the studies, item 7 and 18 were fully attained by 24 (89%) of the studies, items 9 and 19 were fully attained by 21 (78%) of the studies, items 11 and 21 were fully attained by 20 (74%) studies and item 10 was fully attained by 63% of the studies. Twelve (12; 44%) studies provided the funding sources, twelve (12; 44%) studies declared no conflict of interest, three studies (3; 11%) mentioned the contribution of each author, and three (3, 11%) provided ethical clearance declarations (Annexure A).
3.3. Sources of data
All the studies reviewed were observational. More than half (18; 67%) of the reported studies were cross-sectional studies, three (11%) were case-controlled studies (reported following an outbreak), and six (22%) were retrospective studies.
Twenty-four (89%) studies focused on a specific bacterium, whereas the other three studies (11%) (17, 29, 30) reported generally on the bacteria associated with HAIs. Most studies (78%) (9, 19, 20, 30–47) investigated the occurrence of HAIs in a single facility, five (19%) (3, 17, 27–29) studies investigated multiple facilities in an area, and one (4%) (48) study did not specify the area of study.
Seventeen (17/27, 63%) studies were from either the small animal or companion animal clinics/hospitals (3, 9, 17, 20, 27–33, 37, 41–44, 48). followed by both bovine (38, 42, 49, 50) (4/27, 15%) and equine medicine (19, 46, 47). Three (3/27, 11%) studies were a combination of small animals, large animals, and poultry (35, 36, 40). One (1/27, 4 %) study did not identify the type of veterinary clinic or hospital (39).
Within the hospital settings, bacteria associated with HAIs were reported from environmental surfaces (9/27; 33%) (9, 17, 20, 36, 39, 40, 42, 44, 47), animal cases (8/27; 30%) (3, 19, 28, 31, 33, 35, 43, 47), and commonly used fomites such as clothing, cell phones, clippers, stethoscopes, and computers (4/27, 15%) (29, 32, 36, 41). Only three studies (3/27, 11%) isolated bacteria from humans who have regular contact with animals (17, 32, 38).
The antimicrobial resistance profile of the different organisms was provided in eighteen (17/27, 63%) studies (3, 9, 17, 20, 28, 31, 33–37, 39, 40, 42, 43, 46, 47), while nine (9/27, 33%) studies did not report on the antimicrobial resistance patterns (19, 27, 29, 30, 32, 38, 41, 44, 48). Thirteen studies (13/27, 48%) further characterized the microorganisms using pulsed-field gel electrophoresis (PFGE) and polymerase chain reaction (PCR) assays (3, 19, 20, 28, 31, 33–35, 37, 40, 45–47).
3.4. Bacterial species associated with hospital-acquired infections
Staphylococcus spp. were the most (17/27, 63%) reported pathogens associated with HAIs, followed by Escherichia coli (5/27; 19%), Enterococcus spp. (4/27; 15%), Salmonella spp. (4/27; 15%), A. baumannii (4/27; 15%), C. difficile (1/27; 4%), and P. aeruginosa. (1/27; 4%). Enterococcus faecalis (3/4; 75%) and E. faecium (3/4; 75%) were the most reported among the Enterococcus species.
Among the Staphylococcus spp., 11 (11/17, 65%) were MRSA and six (6/17, 35%) were methicillin-resistant S. pseudintermedius (MRSP). Three out of five (3/5; 60%) studies reported MDR Escherichia coli isolates and one (1/5; 20%) study reported an extended spectrum β-lactamase (ESBL) producing E. coli. Meanwhile, vancomycin-resistant enterococci were reported in one (1/4; 25%) study. Salmonella Typhimurium was reported as the common serotype in two of the four (2/4; 50%) studies. The other two of the four (2/4; 50%) studies reported the presence of MDR Salmonella (Table 3).
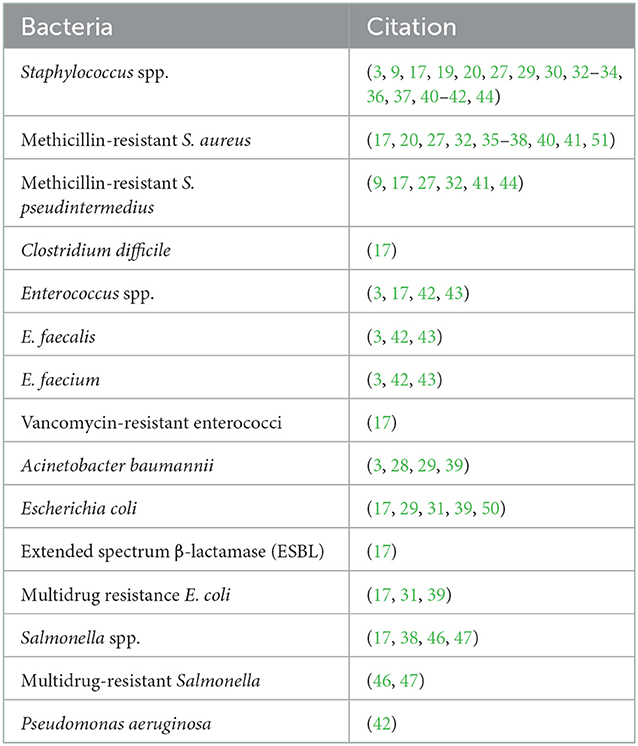
Table 3. Organism reported in hospital-acquired and/or zoonotic infections in veterinary facilities between 2000 and 2020.
3.5. Sources of organisms associated with hospital-acquired infections
The following pathogens were detected in the hospital environmental surfaces, namely MRSA (17, 20, 37, 40), MRSP (42, 44), ESBL-producing E. coli isolates (17), VRE (17), A. baumannii (39), C. difficile (17) and P. aeruginosa (42). Common pathogens identified from hospital fomites included: MRSA (19, 32, 36, 41), MRSP (9, 17, 41), Enterococcus faecalis (42), and A. baumannii (29, 32, 39).
Among patients in hospital settings, MRSA was isolated from companions (35) and equine animals (19, 34). Multidrug resistant Escherichia coli was isolated from companion and bovine animals (31, 50). Additionally, Enterococcus faecium, Enterococcus faecalis (3, 46) and A. baumannii (3) were isolated from companion animals. Salmonella species were also isolated from patients (38, 47), healthy animals (46), and the hospital environment (17, 46) (Table 4).
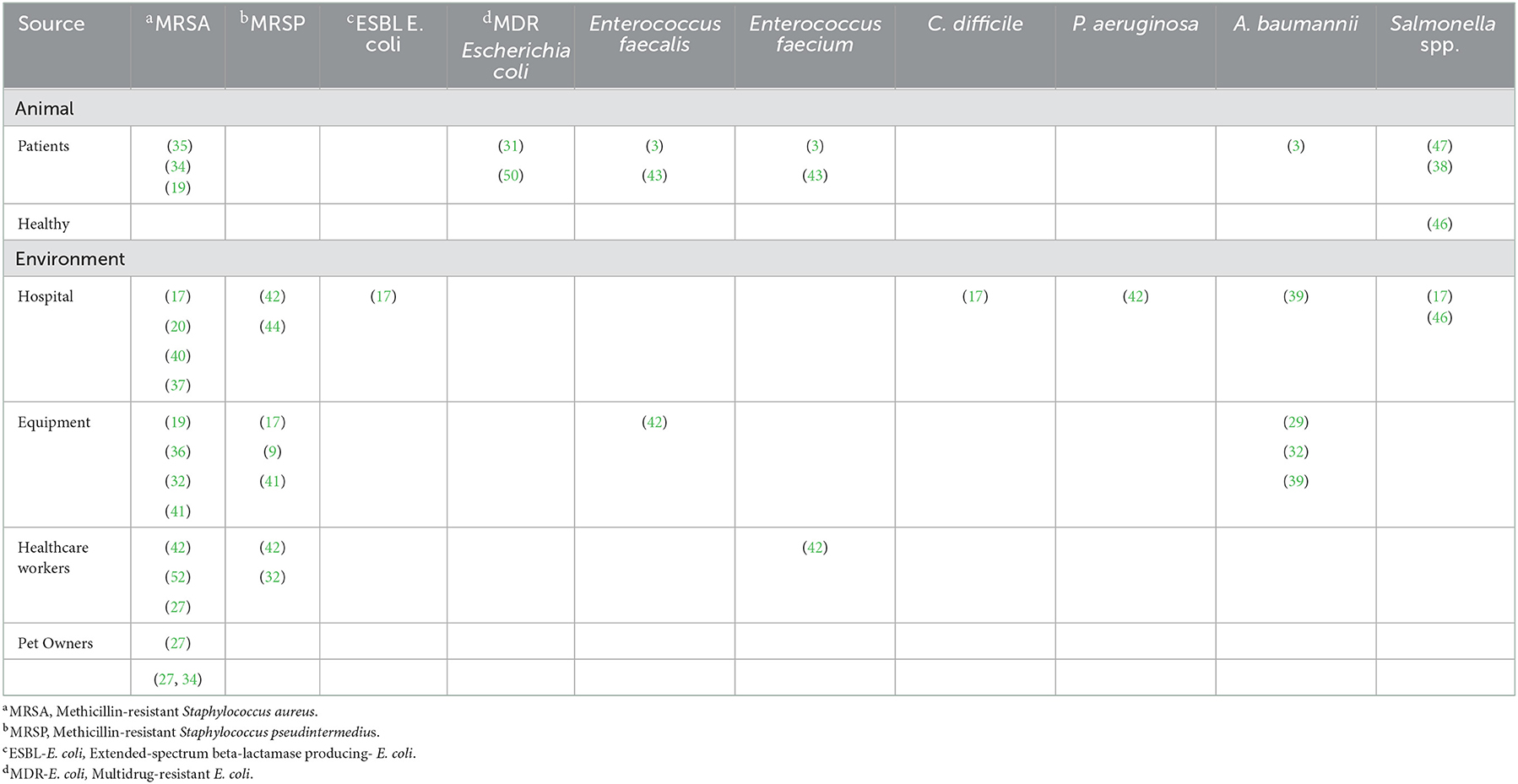
Table 4. Sources of hospital acquired organisms based on the systematic reviewed papers published from 2000 to 2020.
The healthcare workers (HCWs) harbored MRSA (27, 37, 42), MRSP (32, 42), E. faecium (42) and two studies reported MRSA among pet owners (27, 34). In addition, van Duijkeren et al. (35) and Hoet et al. (20) reported on the zoonotic potential of MRSA with van Duijkeren (35) identifying MRSA clusters in animals with a similar clonal lineage to that reported in humans (Table 6).
3.6. Antimicrobial resistance patterns of bacteria associated with hospital acquired infections
3.6.1. Phenotypic resistance
Out of the 27 studies reviewed, 17 (63%) conducted an antimicrobial susceptibility test on the isolates. Among these, 12 (71%) studies reported isolates resistant to more than one antimicrobial. Bacteria resistant to multiple drugs identified included MRSA (20, 34, 40, 45), MRSP (42), A. baumannii (28, 39), E. coli (31, 48), Salmonella Typhimurium (46, 47), E. faecalis and E. faecium (43).
Methicillin-resistant Staphylococcus aureus isolates showed resistance toward ampicillin, amoxicillin, oxacillin, clindamycin, gentamycin, ciprofloxacin, cephalexin, enrofloxacin, cefuroxime, chloramphenicol, erythromycin, and kanamycin while MRSP isolates showed resistance toward azithromycin, oxacillin, penicillin, clindamycin, gentamycin, tetracycline, and ciprofloxacin. Clostridioides difficile showed resistance toward rifampin, moxifloxacin, and chloramphenicol. Enterococcus faecalis and E. faecium showed resistance toward ampicillin, tetracycline, ciprofloxacin, enrofloxacin, erythromycin, and rifampicin (43). Enterococcus faecium was also reported to be resistant to amoxicillin and vancomycin (42).
Acinetobacter baumannii exhibited resistance to amoxicillin, tetracycline (39), ciprofloxacin (28) and imipenem (28). While E. coli showed resistance to ampicillin, cefoxitin, oxacillin, and penicillin (31, 48) and Salmonella was resistant to ampicillin, amoxicillin, cefoxitin, gentamycin, tetracycline, chloramphenicol, rifampicin, and streptomycin (47) (Table 5).
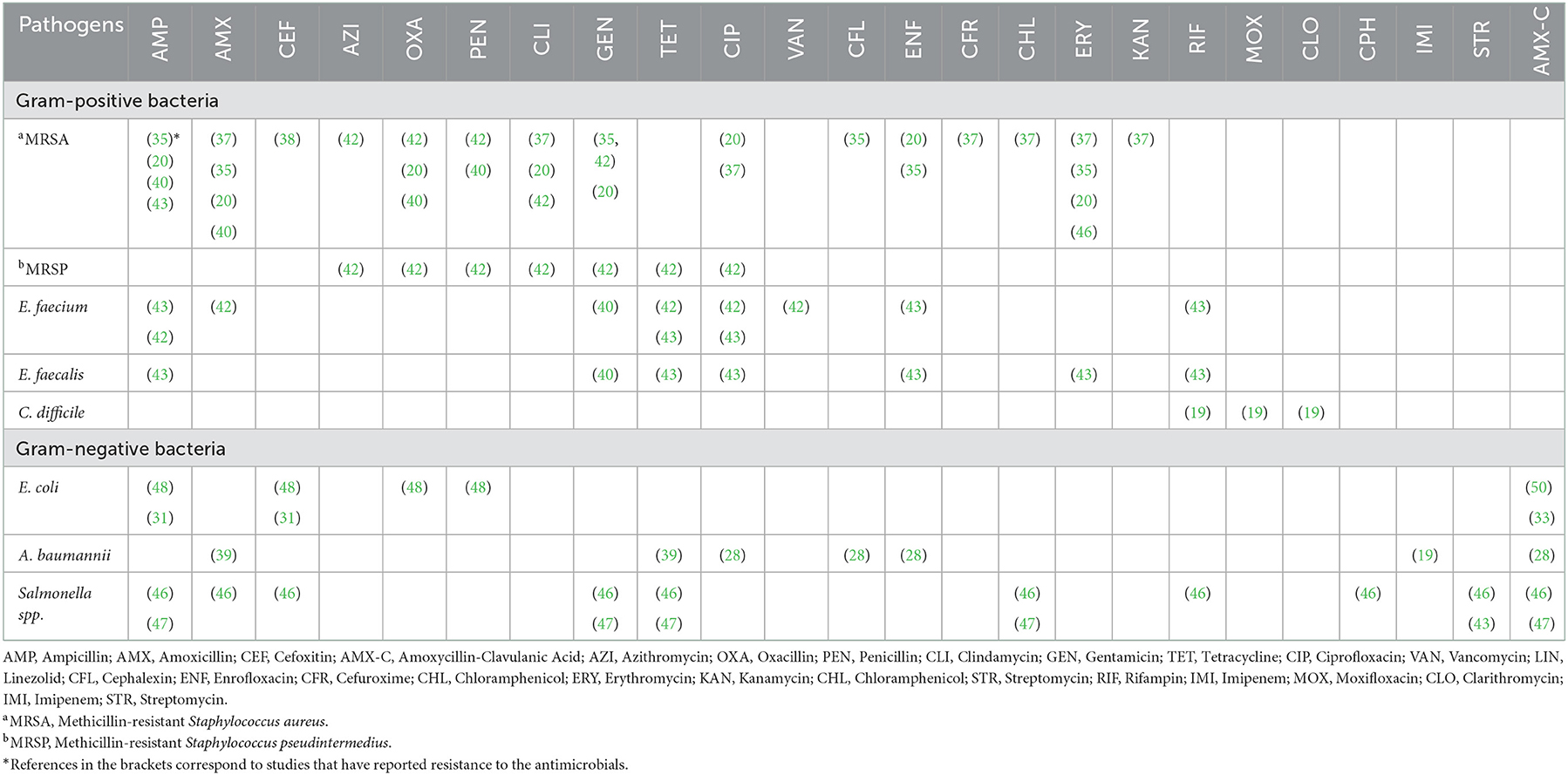
Table 5. Phenotypic antimicrobial resistance profile of hospital-acquired infection organisms based on the systematically reviewed papers published from 2000 to 2020.
3.6.2. Antimicrobial genes
Among Staphylococcus species, mecA was reported in five MRSA studies (20, 35, 36, 40, 49) and two MRSP studies (9, 42). β-lactamase gene (blaCMY − 2 gene) was reported in Salmonella spp. (47) and E. coli isolates (17, 31, 48). While the vancomycin-resistant gene (vanA gene) was reported by one E. faecium study (42). The flo gene was identified in one E. coli study (31) (Table 6).
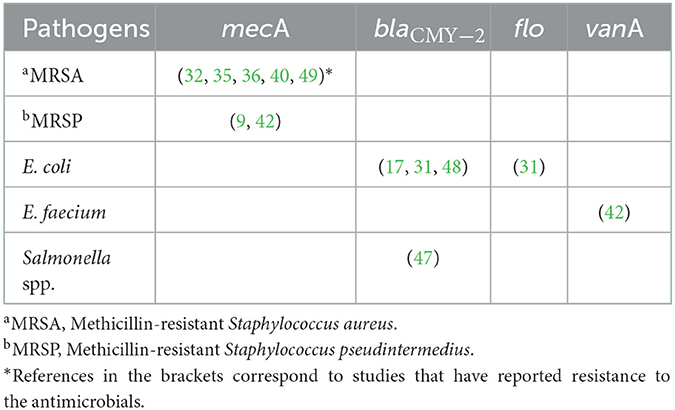
Table 6. The antimicrobial resistant genes isolated from bacteria associated with hospital-acquired infections data published between 2000 and 2020.
3.7. Zoonotic diseases
Six (22%) studies (20, 34, 35, 40, 45, 50) reported organisms associated with HAIs that are zoonotic in nature. For example, MRSA with a SCCmec type IV isolated in humans (28) has also been isolated in hospitalized horses (45) and hospitalized dogs (40). Similarly, three studies reported clonal MRSA lineage in animals similar to that previously reported in humans (34, 35, 40). The plasmid DH108/30218 from E. coli isolates which is similar to a cassette (18-ESBL 188) reported in humans (50) has been identified.
4. Discussion
Hospital-acquired infections and zoonosis are increasingly becoming a global concern (53). In addition, there is an increasing prevalence of resistance among these organisms to commonly used antimicrobials. Most studies that have investigated HAIs and their antimicrobial resistance profiles are in human medicine. In view of this, studies on the occurrence and resistant profile of organisms associated with hospital-acquired and zoonotic infections in veterinary medicine are needed. In this study, bacterial organisms associated with hospital-acquired and zoonotic infections isolated were identified. Furthermore, most of the organisms identified were multidrug-resistant or harbored resistant genes. Several sources of bacterial organisms associated with HAIs including HCWs, commonly used instruments, fomites, and contaminated hospital environments were also identified.
4.1. Hospital-acquired bacterial infections
Bacteria associated with HAIs identified MRSA, MRSP, Enterococcus spp., A. baumannii, P. aeruginosa, C. difficile, E. coli, and Salmonella spp., (3, 17, 20, 29, 30). The presence of these bacterial pathogens within veterinary settings is a public health concern and emphasizes the need for the implementation of infection prevention and control measures to eliminate these pathogens. The patient microbiota, healthcare workers, fomites, and the hospital environment were identified as possible sources of organisms associated with HAIs. Therefore, control measures being implemented should be source-specific and moment-specific during patient care (54).
4.1.1. Sources of bacterial organisms associated with hospital acquired infections
Identification of sources of organisms associated with HAIs in veterinary settings is critical to reducing the risk of transmission to patients and humans. Therefore, it is not surprising that most studies have largely focused on the hospital environment and commonly used instruments as potential reservoirs for organisms associated with HAIs (29, 32, 39, 41, 44). Furthermore, there are ongoing epidemiological studies to understand the relationship between environmental cleanliness and the risk of transmission of HAIs in veterinary settings (4).
The intensive care unit (ICU), surgical ward, in-house laboratory, and consultation rooms were the most important environmental sources of bacteria associated with HAIs in veterinary hospitals (17, 20, 30, 36, 42). Furthermore, environmental surfaces with human contact tend to have higher contamination levels compared to those without human contact (17, 20, 40, 42), suggesting that humans may play a major role in the transmission of these organisms within the hospital environment. This is further emphasized by studies that have isolated similar pathogens strains from the environment and hands of HCWs (28, 47, 53). Therefore, HCWs in veterinary hospitals must be trained on hand hygiene compliance to reduce the risk of transmission of HAI organisms.
Fomites served as sources of HAI organisms and facilitated transmission between animal patients, the hospital environment, and humans (32). Fomites such as clippers, personnel clothing (32, 39), cell phones (41), stethoscopes (39), and weighing scales (39) were reported to be contaminated with bacteria associated with HAIs. Therefore, the development and implementation of cleaning and disinfection protocols to prevent transmission is needed (2). In addition, all surgical materials, instruments, and other fomites which increase the possibility of transmission of these organisms must be sterilized before use (36).
4.1.2. Methicillin-resistant Staphylococcus aureus
Methicillin-resistant Staphylococcus aureus was among the most common organism associated with HAIs in this study (30, 37). The organisms were reported in wound infection (19, 35), invasive procedures (19, 35), skin infections (34), asymptomatic animals (45), septic arthritis, pneumonia, incisional site infection, and rhinitis (19). Studies done in veterinary medicine also reported Staphylococcus strains similar to those reported in humans were reported in this study (19, 27, 45). For example, Loeffler et al. (27) in the UK identified MRSA clones (CC22 and CC30) among humans working with or in close proximity to animals suggesting transmission between animals and humans is precise (37).
Unhygienic environmental conditions are a major source of MRSA (20, 40, 44). Therefore, implementing effective infection prevention and control (9, 30, 36, 37, 40, 52) and screening animals before hospitalization will reduce the spread of MRSA in veterinary hospitals. This is likely to reduce costs associated with increased length of hospital stay (19, 20, 27, 36, 45).
Most MRSA isolates in this study were resistant to β-lactam, 2nd generation cephalosporins, lincosamides, and aminoglycosides. While one study reported intermediate susceptibility to vancomycin among MRSA isolates (36). The presence of vancomycin resistance is concerning as it is the last resort for the treatment of MRSA in humans. Similarly, the presence of β-lactam resistance among staphylococci facilitated by the mecA gene (20, 35–37, 40, 49) is likely to contribute to resistance to other antimicrobials with a β-lactam ring (9, 40, 42, 45). Therefore, the implementation and constant review of infection control protocols are needed to help reduce the risk of the transfer of resistance genes to other organisms (9, 55–57). Without these interventions, patient care and treatment will likely be negatively impacted (36, 40).
4.1.3. Methicillin-resistant Staphylococcus pseudintermedius
Methicillin-resistant Staphylococcus pseudintermedius like MRSA has emerged as a leading cause of opportunistic infections in companion animals (32, 42). The organism has been reported in asymptomatic animals, implant-associated surgical sites (41), fomites (9, 32, 41), and in the environment within the veterinary hospital (44). Therefore, colonized, and contaminated areas remain potential sources of hospital-acquired infections (32).
Areas in the veterinary facilities have been shown to harbor MRSP. These include surfaces such as tables, chairs, floors, and surgical environments (58–60). Moreover, some MRSP organisms are able to survive cleaning and disinfection (58, 59). In view of the potential resistance to disinfectants coupled with ineffective cleaning, these areas can become a source of infection for susceptible animals. Notwithstanding, some disinfectants if used at the correct concentrations are effective against MRSP (61, 62).
Of concern is that MRSP is highly resistant to antimicrobials commonly used for the treatment of S. pseudintermedius infections (63–65). These organisms have been isolated from the environment and hands of HCWs (42), which is concerning as it limits treatment options. Similar to MRSA, MRSP can acquire the mecA gene (42). Shoen et al. (9) showed coagulase positive S. pseudintermedius commonly isolated from the skin of dogs can acquire the mecA gene from a coagulase-negative S. epidermidis commonly found in humans.
The zoonotic cases associated with MRSP are not common (32). However, an MRSA spa type 18/t338 from animal-related fomites has been reported in humans (41). The rise in the number of MRSP cases between dogs, pet owners, and veterinary staff is concerning, therefore, effective hand hygiene should be performed before and after contact with the patient, as well as after contact with potentially contaminated environmental sites within veterinary hospitals.
4.1.4. Enterococcus species
Enterococcus species are commensal of the gut flora of cats and dogs (3, 43). However, they are also opportunistic pathogens (3). In recent years, Enterococcus species have emerged as causes of HAIs in veterinary medicine associated with urinary tract infections (UTIs) (66). The transmission is mainly due to fecal contaminated fomites or environmental surfaces (29). These organisms can survive in a hospital environment for a long period. Furthermore, they can survive high temperatures and disinfectants such as chlorine and alcohol (42).
Enterococcus faecium and E. faecalis are the most predominant species reported in dogs (30, 43), hospital environments and in hands of HCWs (42). Of the two species, E. faecalis is the predominant enterococci. Multidrug-resistant enterococci have also been reported as a commensal and pathogenic organism (3, 42, 43). The presence of MDR among Enterococcus species has largely been attributed to overuse and misuse of antimicrobials (42, 43). It is also possible that some may have acquired resistance through other mechanisms including genetic transfer or mutation (43). For example, resistance to erythromycin has been associated with the methylation of the ribosomal target site of these antibiotics (42, 67). Nonetheless, the presence of MDR enterococci is likely to impact patient care in veterinary hospitals (42).
Of concern is the emergence of vancomycin-resistant E. faecium (42) which is an important antimicrobial in the treatment of enterococci infections (43, 67) and is mediated by the presence of vanA genes. These genes are important as they confer multidrug resistance and may be transmitted to other bacterial species such as Staphylococcus and create even bigger problems in the treatment of HAIs (42). Furthermore, these genes can also be transferred from animals to humans (3, 43).
4.1.5. Clostridioides difficile
Clostridioides difficile is found in the hospital environmental and it is difficult to eradicate (17). Both humans and animals are asymptomatically carriers of the organism. In humans, its presence has been attributed to the overuse of antimicrobials. However, in veterinary medicine there is limited information about the organism. Therefore, future studies should look at whether the overuse of antimicrobials could be a driver of C. difficile in veterinary settings (17). The ability of the pathogen to survive harsh environmental conditions and resistance to most disinfectants makes it a suitable indicator of the effective IPC measures (17). Therefore, it is possible that this organism can also be used as an indicator of effective infection prevention and control in veterinary hospitals.
4.1.6. Acinetobacter baumannii
Acinetobacter baumannii causes life-threatening infections in both humans and animals. This organism has been reported in UTIs, pyothorax, upper airway obstruction, bloodstream infection, and wound infections in animals (39). In infected animals, it is associated with increased morbidity and prolonged length of hospital stay (68). Acinetobacter baumannii survives on dry surfaces (29, 39, 69). Therefore, commonly used fomites, bed rails, cages, and examination tables could serve as reservoirs for A. baumannii.
The organism can survive stressful environmental conditions and remains viable on different surfaces (70). However, if used at correct concentrations, sodium hypochlorite (bleach) and 70% ethanol are effective against A. baumannii (70, 71). Lanjri et al. (71) observed that chlorhexidine digluconate was effective against A. baumannii. La Forgia et al. (72) also reported that sodium hypochlorite was effective in reducing the incidence rate of A. baumannii in hospital settings. In light of this findings, choosing the correct disinfectant is important in reducing cases of A. baumannii in hospital settings.
Most A. baumannii are multiple drug resistant with a high prevalence of resistance toward cephalexin, enrofloxacin, amoxicillin-clavulanic acid, sulphamethoxazole-trimethoprim, and tetracycline (39). Resistance to the above antimicrobials is concerning as these antimicrobials are commonly used for the treatment of bacterial infections in small animal medicine (68). In addition, the blaOXA-51 gene reported in an A. baumannii isolate from pigs has also been reported in humans (67).
4.1.7. Escherichia coli
Escherichia coli is commonly reported in UTIs and bloodstream infections (17, 31, 39, 73). The bacterium spreads from patient to patient via fecal contaminated hands of HCWs and shared equipment (31). Given, that environmental surfaces could potentially be a reservoir of E. coli, measures to minimize fecal contamination in companion animal hospitals including cleaning and disinfection of the hospital environment should be implemented. Moreover, Sanchez et al. (31) shows the transfer of E. coli isolates with similar antimicrobial resistance patterns between two different animals admitted to the same ICU.
In this study, E. coli isolates exhibited resistance toward cephalosporins and β-lactams including amoxycillin-clavulanic acid. This broad-spectrum antimicrobial resistance among E. coli is attributed to the presence of ampC like gene, blaCMY2 (17, 31), which has been identified to be of public health concern (17). Another study reported resistance among E. coli isolates to chloramphenicol mainly due to the presence of cmlA homologue flo among gram-negative bacteria (31). The presence of these genes has also been linked to the development of resistance to other commonly used antibiotics such as gentamycin, spectinomycin, and sulfadimethoxine (31, 39, 50). Considering this resistance, strict guidelines should be implemented on the prudent use of antimicrobials in veterinary medicine.
4.1.8. Salmonella species
Although most animals are asymptomatic carriers of Salmonella spp., they shed the bacterium in high quantities through their feces resulting in Salmonella outbreaks in equine veterinary hospitals (46, 47). Furthermore, infections associated with Salmonella species have also been reported in bovine with diarrhea, fever, dehydration (38) and colic in horses (47). In affected animals, the disease is characterized by high morbidity and mortality. There is a potential spread of organisms and the occurrence of zoonotic infection that may result in the closure of facilities and a loss of income for the hospital (38, 46). Therefore, personnel working in close contact with infected animals are at an increased risk of infection (46).
Managing transmission in the veterinary settings remains a challenge as Salmonella can persist in the environment for a long time. Rodents and contaminated feed could also be a source (46, 47). Therefore, biosecurity measures must be intensified in veterinary hospitals to reduce the risk of transmission. Additionally, education programs can also be developed targeting specific aspects of hygiene, movement control, and cleanliness of equipment.
Salmonella isolates were resistant to ceftiofur, gentamycin, amoxicillin, ampicillin, streptomycin, and trimethoprim/ sulfadiazine (46, 47). One study reported the presence of the cephalomycinase gene, blacmy2 (47) which has been associated with cephalosporin resistance among Salmonella species. This gene has also been reported to mediate resistance to amoxicillin, amoxicillin-clavulanic acid, cephalothin, cefoxitin, ceftiofur, and ceftriaxone (47).
5. Conclusion
Organisms associated with hospital-acquired and zoonotic diseases were reported from clinical cases, environmental surfaces, and items used in veterinary service. The hospital environment with human contact was the most reported source of organisms associated with HAIs. These results suggest that humans play a crucial role in the transmission of HAIs in veterinary hospitals.
Among the organisms reported, MRSA and MRSP were the most reported HAI organisms in veterinary facilities. Other organisms identified include E. coli, C. difficile, A. baumannii, Salmonella spp., and Enterococcus species. Some of these isolates reported in veterinary settings share similar clonal lineage to those reported in humans. Some organisms exhibit a high prevalence of antimicrobial resistance and contain genes known to be associated with antibiotic resistance.
These results suggest that strict infection prevention and control practices must be in place, monitored and modified when necessary to curb the occurrence and transmission of organisms associated with HAIs in veterinary hospitals. In addition, continuous surveillance of HAI organisms and their antimicrobial resistance patterns in veterinary hospitals should be emphasized. Further research needs to be done on C. difficile as a potential indicator of effective infection prevention and control practices in veterinary facilities.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
DS was involved in study design, data analysis, interpretation of results, and writing of manuscript as well as extensive editing of the manuscript. DQ was involved in study design, data management, analysis, and interpretation as well as reviewing of the manuscript draft. JO and MK were involved in study design, data analysis, and interpretation as well as editing of the manuscript. All authors have read and approved the final manuscript.
Acknowledgments
The authors would like to extend their appreciation to the National Research Foundation for providing the scholarship funding.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1087052/full#supplementary-material
References
1. Eggimann P, Pittet D. Infection control in the ICU. Chest. (2001) 120:2059–93. doi: 10.1378/chest.120.6.2059
2. Traverse M, Aceto H. Environmental cleaning and disinfection. Vet Clin North Am - Small Anim Pract. (2015) 45:299–330. doi: 10.1016/j.cvsm.2014.11.011
3. Boerlin P, Eugster S, Gaschen F, Straub R, Schawalder P. Transmission of opportunistic pathogens in a veterinary teaching hospital. Vet Microbiol. (2001) 82:347–59. doi: 10.1016/S0378-1135(01)00396-0
4. Dancer SJ. The role of environmental cleaning in the control of hospital-acquired infection. J Hosp Infect. (2009) 73:378–85. doi: 10.1016/j.jhin.2009.03.030
5. Stull JW, Weese JS. Hospital-associated infections in small animal practice. Vet Clin North Am Small Anim Pract. (2015) 45:217–33. doi: 10.1016/j.cvsm.2014.11.009
6. Walther B, Tedin K, Lübke-Becker A. Multidrug-resistant opportunistic pathogens challenging veterinary infection control. Vet Microbiol. (2017) 200:71–8. doi: 10.1016/j.vetmic.2016.05.017
7. Guardabassi L. Veterinary hospital-acquired infections: the challenge of MRSA and other multidrug-resistant bacterial infections in veterinary medicine. Vet J. (2012) 193:307–8. doi: 10.1016/j.tvjl.2012.04.005
8. van der Kolk JH, Endimiani A, Graubner C, Gerber V, Perreten V. Acinetobacter in veterinary medicine, with an emphasis on Acinetobacter baumannii. J Glob Antimicrob Resist. (2019) 16:59–71. doi: 10.1016/j.jgar.2018.08.011
9. Shoen HRC, Rose SJ, Ramsey SA, de Morais H, Bermudez LE. Analysis of Staphylococcus infections in a veterinary teaching hospital from 2012 to 2015. Comp Immunol Microbiol Infect Dis. (2019) 66:101332. doi: 10.1016/j.cimid.2019.101332
10. Aires-de-Sousa M. Methicillin-resistant Staphylococcus aureus among animals: current overview. Clin Microbiol Infect. (2017) 23:373–80. doi: 10.1016/j.cmi.2016.11.002
11. Ramsamy Y, Essack SY, Sartorius B, Patel M, Mlisana KP. Antibiotic resistance trends of ESKAPE pathogens in Kwazulu-Natal, South Africa: a five-year retrospective analysis. Afr J Lab Med. (2018) 7:2225–2002. doi: 10.4102/ajlm.v7i2.887
12. Stull JW, Weese JS. Infection control in veterinary small animal practice. Vet Clin North Am Small Anim Pract. (2015) 45:xi–xii. doi: 10.1016/j.cvsm.2014.12.001
13. Mulani MS, Kamble EE, Kumkar SN, Tawre MS, Pardesi KR. Emerging strategies to combat ESKAPE pathogens in the era of antimicrobial resistance: A review. Front Microbiol. (2019) 10:539. doi: 10.3389/fmicb.2019.00539
14. Mazet JAK, Clifford DL, Coppolillo PB, Deolalikar AB, Erickson JD, Kazwala RR. A “One health” approach to address emerging zoonoses: the HALI project in Tanzania. PLoS Med. (2009) 6:12. doi: 10.1371/journal.pmed.1000190
15. Day MJ, Breitschwerdt E, Cleaveland S, Karkare U, Khanna C, Kirpensteijn J, et al. Surveillance of zoonotic infectious disease transmitted by small companion animals. Emerg Infect Dis. (2012) 18:e1. doi: 10.3201/eid1812.120664
16. Taylor LH, Latham SM, Woolhouse MEJ. Risk factors for human disease emergence. Philos Trans R Soc B Biol Sci. (2001) 356:983–9. doi: 10.1098/rstb.2001.0888
17. Murphy CP, Reid-Smith RJ, Boerlin P, Weese JS, Prescott JF, Janecko N, et al. Escherichia coli and selected veterinary and zoonotic pathogens isolated from environmental sites in companion animal veterinary hospitals in southern Ontario. Can Vet J. (2010) 51:963–72.
18. Molyneux D, Hallaj Z, Keusch GT, McManus DP, Ngowi H, Cleaveland S, et al. Zoonoses and marginalised infectious diseases of poverty: where do we stand? Parasit Vectors. (2011) 4:106. doi: 10.1186/1756-3305-4-106
19. Weese JS, Rousseau J, Willey BM, Archambault M, McGeer A, Low DE. Methicillin-resistant Staphylococcus aureus in horses at a Veterinary Teaching Hospital: Frequency, characterization, and association with clinical disease. J Vet Intern Med. (2006) 20:182–6. doi: 10.1111/j.1939-1676.2006.tb02839.x
20. Hoet AE, Johnson A, Nava-Hoet RC, Bateman S, Hillier A, Dyce J, et al. Environmental methicillin-resistant staphylococcus aureus in a veterinary teaching hospital during a nonoutbreak period. Vector Borne Zoonotic Dis. (2011) 11:609–15. doi: 10.1089/vbz.2010.0181
21. Willemsen A, Cobbold R, Gibson J, Wilks K, Lawler S, Reid S. Infection control practices employed within small animal veterinary practices—a systematic review. Zoonoses Public Health. (2019) 66:439–57. doi: 10.1111/zph.12589
22. Kitchenham B. Procedures for Performing Systematic Reviews: Joint Technical Report. Keele (2004). p. 1–26. Available online at: https://www.inf.ufsc.br/~aldo.vw/kitchenham.pdf
23. Cusack TP, Ashley EA, Ling CL, Rattanavong S, Roberts T, Turner P, et al. Impact of CLSI and EUCAST breakpoint discrepancies on reporting of antimicrobial susceptibility and AMR surveillance. Clin Microbiol Infect. (2019) 25:910–1. doi: 10.1016/j.cmi.2019.03.007
24. Kassim A, Omuse G, Premji Z, Revathi G. Comparison of clinical laboratory standards institute and european committee on antimicrobial susceptibility testing guidelines for the interpretation of antibiotic susceptibility at a university teaching hospital in Nairobi, Kenya: a cross-sectional stud. Ann Clin Microbiol Antimicrob. (2016) 15:21. doi: 10.1186/s12941-016-0135-3
25. Suravaram S, Hada V, Ahmed Siddiqui I. Comparison of antimicrobial susceptibility interpretation among Enterobacteriaceae using CLSI and EUCAST breakpoints. Indian J Med Microbiol. (2021) 39:315–9. doi: 10.1016/j.ijmmb.2021.05.004
26. Sargeant JM, O'Connor AM, Dohoo IR, Erb HN, Cevallos M, Egger M, et al. Methods and processes of developing the strengthening the reporting of observational studies in epidemiology – veterinary (STROBE-Vet) statement. Zoonoses Public Health. (2016) 63:651–61. doi: 10.1111/zph.12314
27. Loeffler A, Pfeiffer DU, Lloyd DH, Smith H, Soares-Magalhaes R, Lindsay JA. Meticillin-resistant Staphylococcus aureus carriage in UK veterinary staff and owners of infected pets: new risk groups. J Hosp Infect. (2010) 74:282–8. doi: 10.1016/j.jhin.2009.09.020
28. Pailhoriès H, Belmonte O, Kempf M, Lemarié C, Cuziat J, Quinqueneau C, et al. Diversity of Acinetobacter baumannii strains isolated in humans, companion animals, and the environment in Reunion Island: an exploratory study. Int J Infect Dis. (2015) 37:64–9. doi: 10.1016/j.ijid.2015.05.012
29. Mount R, Schick AE, Lewis TP, Newton HM. Evaluation of bacterial contamination of clipper blades in small animal private practice. J Am Anim Hosp Assoc. (2016) 52:95–101. doi: 10.5326/JAAHA-MS-6355
30. Aksoy E, Boag A, Brodbelt D, Grierson J. Evaluation of surface contamination with staphylococci in a veterinary hospital using a quantitative microbiological method. J Small Anim Pract. (2010) 51:574–80. doi: 10.1111/j.1748-5827.2010.00994.x
31. Sanchez S, Stevenson MAMC, Hudson CR, Maier M, Buffington T, Dam Q, et al. Characterization of multidrug-resistant Escherichia coli isolates associated with nosocomial infections in dogs. J Clin Microbiol. (2002) 40:3586–95. doi: 10.1128/JCM.40.10.3586-3595.2002
32. Singh A, Walker M, Rousseau J, Monteith GJ, Weese JS. Methicillin-resistant staphylococcal contamination of clothing worn by personnel in a veterinary teaching hospital. Vet Surg. (2013) 42:643–8. doi: 10.1111/j.1532-950X.2013.12024.x
33. Bagcigil FA, Moodley A, Baptiste KE, Jensen VF, Guardabassi L. Occurrence, species distribution, antimicrobial resistance and clonality of methicillin- and erythromycin-resistant staphylococci in the nasal cavity of domestic animals. Vet Microbiol. (2007) 121:307–15. doi: 10.1016/j.vetmic.2006.12.007
34. Weese JS, Caldwell F, Willey BM, Kreiswirth BN, McGeer A, Rousseau J, et al. An outbreak of methicillin-resistant Staphylococcus aureus skin infections resulting from horse to human transmission in a veterinary hospital. Vet Microbiol. (2006) 114:160–4. doi: 10.1016/j.vetmic.2005.11.054
35. van Duijkeren E, Box ATA, Heck MEOC, Wannet WJB, Fluit AC. Methicillin-resistant staphylococci isolated from animals. Vet Microbiol. (2004) 103:91–7. doi: 10.1016/j.vetmic.2004.07.014
36. Stella AE, Lima TF, Moreira CN, De Paula EMN. Characterization of Staphylococcus aureus Strains Isolated from Veterinary Hospital. Int J Microbiol. (2020) 2020. doi: 10.1155/2020/2893027
37. Heller J, Armstrong SK, Girvan EK, Reid SWJ, Moodley A, Mellor DJ. Prevalence and distribution of meticillin-resistant Staphylococcus aureus within the environment and staff of a university veterinary clinic. J Small Anim Pract. (2009) 50:168–73. doi: 10.1111/j.1748-5827.2008.00695.x
38. Cummings KJ, Divers TJ, McDonough PL, Switt AM, Wiedmann M, Warnick LD. Temporal clusters of bovine Salmonella cases at a veterinary medical teaching hospital, 1996-2007. Vector-Borne Zoonotic Dis. (2010) 10:471–9. doi: 10.1089/vbz.2009.0068
39. Ng S, Saleha A, Bejo S, Dhaliwal G. Occurrence of multidrug-resistant Acinetobacter baumannii and Escherichia coli in veterinary healthcare facilities in Klang valley, Malaysia. J Vet Malaysia. (2016) 28:12–6.
40. Rojas I, Barquero-Calvo E, Van Balen JC, Rojas N, Muñoz-Vargas L, Hoet AE. High prevalence of multidrug-resistant community-acquired methicillin-resistant Staphylococcus aureus at the largest veterinary teaching hospital in Costa Rica. Vector-Borne Zoonotic Dis. (2017) 17:645–53. doi: 10.1089/vbz.2017.2145
41. Julian T, Singh A, Rousseau J, Weese JS. Methicillin-resistant staphylococcal contamination of cellular phones of personnel in a veterinary teaching hospital. BMC Res Notes. (2012) 5:193. doi: 10.1186/1756-0500-5-193
42. Oh Y-N, Baek JY, Kim SH, Kang B-J, Youn H-Y. Antimicrobial Susceptibility and Distribution of Multidrug-Resistant Organisms Isolated from Environmental Surfaces and Hands of Healthcare Workers in a Small Animal Hospital. Jpn J Vet Res. (2018) 66:193–202.
43. Ossiprandi MC, Bottarelli E, Cattabiani F, Bianchi E. Susceptibility to vancomycin and other antibiotics of 165 Enterococcus strains isolated from dogs in Italy. Comp Immunol Microbiol Infect Dis. (2008) 31:1–9. doi: 10.1016/j.cimid.2007.08.003
44. Lutz EA, Hoet AE, Pennell M, Stevenson K, Buckley TJ. Nonoutbreak-related airborne Staphylococcus spp in a veterinary hospital. Am J Infect Control. (2013) 41:648–51. doi: 10.1016/j.ajic.2012.08.011
45. Van den Eede A, Martens A, Lipinska U, Struelens M, Deplano A, Denis O, et al. High occurrence of methicillin-resistant Staphylococcus aureus ST398 in equine nasal samples. Vet Microbiol. (2009) 133:138–44. doi: 10.1016/j.vetmic.2008.06.021
46. Soza-Ossandón P, Rivera D, Tardone R, Riquelme-Neira R, García P, Hamilton-West C, et al. Widespread environmental presence of multidrug-resistant salmonella in an equine veterinary hospital that received local and international horses. Front Vet Sci. (2020) 7:346. doi: 10.3389/fvets.2020.00346
47. Ward MP, Brady TH, Couëtil LL, Liljebjelke K, Maurer JJ, Ching CW. Investigation and control of an outbreak of salmonellosis caused by multidrug-resistant Salmonella typhimurium in a population of hospitalized horses. Vet Microbiol. (2005) 107:233–40. doi: 10.1016/j.vetmic.2005.01.019
48. Sidjabat HE, Chin JJC, Chapman T, Wu K, Ulett GC, Ong CY, et al. Colonisation dynamics and virulence of two clonal groups of multidrug-resistant Escherichia coli isolated from dogs. Microbes Infect. (2009) 11:100–7. doi: 10.1016/j.micinf.2008.10.014
49. Moon JS, Lee AR, Kang HM, Lee ES, Kim MN, Paik YH, et al. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J Dairy Sci. (2007) 90:1176–85. doi: 10.3168/jds.S0022-0302(07)71604-1
50. Mulvey MR, Susky E, McCracken M, Morck DW, Read RR. Similar cefoxitin-resistance plasmids circulating in Escherichia coli from human and animal sources. Vet Microbiol. (2009) 134:279–87. doi: 10.1016/j.vetmic.2008.08.018
51. Weese JS, Dick H, Willey BM, McGeer A, Kreiswirth BN, Innis B, et al. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Vet Microbiol. (2006) 115:148–55. doi: 10.1016/j.vetmic.2006.01.004
52. Heller J, Innocent GT, Denwood M, Reid SWJ, Kelly L, Mellor DJ. Assessing the probability of acquisition of meticillin-resistant Staphylococcus aureus (MRSA) in a dog using a nested stochastic simulation model and logistic regression sensitivity analysis. Prev Vet Med. (2011) 99:211–24. doi: 10.1016/j.prevetmed.2010.10.007
53. Milton AAP. Nosocomial infections and their surveillance in veterinary hospitals. Adv Anim Vet Sci. (2015) 3:1–24. doi: 10.14737/journal.aavs/2015/3.2s.1.24
54. Sebola DC, Boucher C, Maslo C. Hand hygiene compliance in the intensive care unit of the Onderstepoort Vet Acad Hosp. (2019) 1–12. doi: 10.21203/rs.2.17130/v1
55. Gortel K, Campbell KL, Kakoma I, Whittem T, Schaeffer DJ, Weisiger RM. Methicillin resistance among staphylococci isolated from dogs. Am J Vet Res. (1999) 60:1526–30.
56. Soedarmanto I, Kanbar T, Ülbegi-Mohyla H, Hijazin M, Alber J, Lämmler C, et al. Genetic relatedness of methicillin-resistant Staphylococcus pseudintermedius (MRSP) isolated from a dog and the dog owner. Res Vet Sci. (2011) 91:e25–7. doi: 10.1016/j.rvsc.2011.01.027
57. Gagetti P, Wattam AR, Giacoboni G, De Paulis A, Bertona E, Corso A, et al. Identification and molecular epidemiology of methicillin resistant Staphylococcus pseudintermedius strains isolated from canine clinical samples in Argentina. BMC Vet Res. (2019) 15:1–12. doi: 10.1186/s12917-019-1990-x
58. Fungwithaya P, Sontigun N, Boonhoh W, Boonchuay K, Wongtawan T. Antimicrobial resistance in Staphylococcus pseudintermedius on the environmental surfaces of a recently constructed veterinary hospital in Southern Thailand. Vet World. (2022) 15:1087. doi: 10.14202/vetworld.2022.1087-1096
59. Ishihara K, Shimokubo N, Sakagami A, Ueno H, Muramatsu Y, Kadosawa T, et al. Occurrence and molecular characteristics of methicillin-resistant Staphylococcus aureus and methicillin-resistant staphylococcus pseudintermedius in an academic veterinary hospital. Appl Environ Microbiol. (2010) 76:5165–74. doi: 10.1128/AEM.02780-09
60. Perkins AV, Sellon DC, Gay JM, Lofgren ET, Moore DA, Jones LP, et al. Prevalence of methicillin-resistant Staphylococcus pseudintermedius on hand-contact and animal-contact surfaces in companion animal community hospitals. Can Vet J. (2020) 61:613.
61. Pariser M, Gard S, Gram D, Schmeitzel L. An in vitro study to determine the minimal bactericidal concentration of sodium hypochlorite (bleach) required to inhibit meticillin-resistant Staphylococcus pseudintermedius strains isolated from canine skin. Vet Dermatol. (2013) 24:632–4. doi: 10.1111/vde.12079
62. Frosini SM, Bond R, King R, Feudi C, Schwarz S, Loeffler A. Effect of topical antimicrobial therapy and household cleaning on meticillinresistant Staphylococcus pseudintermedius carriage in dogs. Vet Rec. (2022) 190:e937. doi: 10.1002/vetr.937
63. Marsilio F, Di Francesco CE, Di Martino B. Coagulase-positive and coagulase-negative staphylococci animal diseases. Pet-to-Man Travel Staphylococci A World Prog. (2018) 43–50. doi: 10.1016/B978-0-12-813547-1.00004-2
64. Gillaspy AF, Iandolo JJ. Staphylococcus. Encycl Microbiol. (2009) 293–303. doi: 10.1016/B978-012373944-5.00237-6
65. Brooks MR, Padilla-Vélez L, Khan TA, Qureshi AA, Pieper JB, Maddox CW, et al. Prophage-mediated disruption of genetic competence in Staphylococcus pseudintermedius. mSystems. (2020) 5:1–22. doi: 10.1128/mSystems.00684-19
66. Comerlato CB, de Resende MCC, Caierão J, d'Azevedo PA. Presence of virulence factors in Enterococcus faecalis and Enterococcus faecium susceptible and resistant to vancomycin. Mem Inst Oswaldo Cruz. (2013) 108:590–5. doi: 10.1590/S0074-02762013000500009
67. Argudín MA, Deplano A, Meghraoui A, Dodémont M, Heinrichs A, Denis O, et al. Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics. (2017) 6:138. doi: 10.3390/antibiotics6020012
68. Maragakis LL, Perl TM. Acinetobacter baumannii: Epidemiology, antimicrobial resistance, and treatment options. Clin Infect Dis. (2008) 46:1254–63. doi: 10.1086/529198
69. Fournier PE, Richet H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis. (2006) 42:692–9. doi: 10.1086/500202
70. Bravo Z, Orruño M, Navascues T, Ogayar E, Ramos-Vivas J, Kaberdin VR, et al. Analysis of Acinetobacter baumannii survival in liquid media and on solid matrices as well as effect of disinfectants. J Hosp Infect. (2019) 103:e42–52. doi: 10.1016/j.jhin.2019.04.009
71. Lanjri S, Uwingabiye J, Frikh M, Abdellatifi L, Kasouati J, Maleb A, et al. In vitro evaluation of the susceptibility of Acinetobacter baumannii isolates to antiseptics and disinfectants: comparison between clinical and environmental isolates. Antimicrob Resist Infect Control. (2017) 6:36. doi: 10.1186/s13756-017-0195-y
72. La Forgia C, Franke J, Hacek DM, Thomson RB, Robicsek A, Peterson LR. Management of a multidrug-resistant Acinetobacter baumannii outbreak in an intensive care unit using novel environmental disinfection: a 38-month report. Am J Infect Control. (2010) 38:259–63. doi: 10.1016/j.ajic.2009.07.012
Keywords: hospital acquired infections (HAIs), zoonosis, veterinary, antimicrobial resistance (AMR), nosocomial, antimicrobials, multi-drug resistance
Citation: Sebola DC, Oguttu JW, Kock MM and Qekwana DN (2023) Hospital-acquired and zoonotic bacteria from a veterinary hospital and their associated antimicrobial-susceptibility profiles: A systematic review. Front. Vet. Sci. 9:1087052. doi: 10.3389/fvets.2022.1087052
Received: 01 November 2022; Accepted: 19 December 2022;
Published: 09 January 2023.
Edited by:
Faham Khamesipour, Shahid Beheshti University of Medical Sciences, IranReviewed by:
Babafela Awosile, Texas Tech University, United StatesManuela Oliveira, Universidade de Lisboa, Portugal
Copyright © 2023 Sebola, Oguttu, Kock and Qekwana. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daniel N. Qekwana,  bmVuZW5lLnFla3dhbmEmI3gwMDA0MDt1cC5hYy56YQ==
bmVuZW5lLnFla3dhbmEmI3gwMDA0MDt1cC5hYy56YQ==
 Dikeledi C. Sebola
Dikeledi C. Sebola James W. Oguttu
James W. Oguttu Marleen M. Kock
Marleen M. Kock Daniel N. Qekwana
Daniel N. Qekwana