- 1Department of Veterinary Microbiology, West Bengal University of Animal and Fishery Sciences, Kolkata, West Bengal, India
- 2Animal Science Division, ICAR-Central Island Agricultural Research Institute, Port Blair, Andaman and Nicobar Islands, India
- 3Department of Veterinary Public Health, West Bengal University of Animal and Fishery Sciences, Kolkata, West Bengal, India
- 4Department of Veterinary Microbiology, Central Agricultural University, Aizawl, Mizoram, India
- 5ICAR-Indian Veterinary Research Institute, Eastern Regional Station, Kolkata, West Bengal, India
- 6Department of Food Hygiene and Environmental Health, University of Helsinki, Helsinki, Finland
Objectives: The present study was conducted to detect the occurrence of β-lactamase and biofilm-producing Escherichia coli, Salmonella, and Klebsiella in broilers and native fowl reared in the Andaman and Nicobar Islands, India. The study also included molecular docking experiments to confirm the nature of the catalytic domains found in the β-lactamase variants obtained and to reveal the clonal relationship of the isolates with human clinical strains from the database.
Materials and methods: A total of 199 cloacal swabs were collected from five poultry breeds/varieties (broiler, Vanraja, Desi, Nicobari, and layer) in three districts of the Andaman and Nicobar Islands. E. coli, Salmonella enterica, and Klebsiella pneumoniae were isolated by standard techniques and confirmed by PCR. Phenotypical β-lactamase producers were identified by a double-disc test. The genes (blaCTX, blaSHV, blaTEM, and blaAmpC) were screened, and selected sequences of β-lactamase variants were submitted to DDBJ. Homology modeling, model validation, and active site identification of different β-lactamase variants were done by the SWISS-MODEL. Molecular docking was performed to identify the catalytic domains of the β-lactamase variants. The selected β-lactamase sequences were compared with the Indian ESBL sequences from human clinical strains in NCBI-GenBank.
Results: In total, 425 Enterobacteriaceae strains were isolated from the collected samples. Klebsiella pneumoniae (42.58%) was found to be the most prevalent, followed by Salmonella enterica (30.82%) and E. coli (26.58%). The phenotypical antibiogram of all 425 isolates showed the highest resistance against oxytetracycline (61–76%) and the lowest against gentamicin (15–20%). Phenotypical production of β-lactamase enzymes was observed in 141 (33.38%) isolates. The isolation rate of β-lactamase producing E. coli, Salmonella enterica, and Klebsiella pneumoniae was significantly higher (p < 0.05) in the birds reared in the South Andaman district (25.6, 17.5, and 18.7%, respectively) than in Nicobar (11.5, 7.6, 7.1%, respectively). Genotyping of the β-lactamase-producing isolates revealed the maximum possession of blaTEM, followed by blaSHV and blaCTX−M. The nucleotide sequences were found to be similar with blaCTX−M−15, blaSHV−11, blaSHV−27, blaSHV−228, blaTEM−1, and blaAmpC in BLAST search. Distribution of studied biofilm-associated genes in Enterobacteriaceae strains from different varieties of the birds revealed that the layer birds had the maximum possession, followed by Vanraja, Desi, broilers, and Nicobari fowls. The phylogenetic analysis of selected sequences revealed a partial clonal relationship with human clinical strains of the Indian subcontinent. Molecular docking depicted the Gibbs free energy release for 10 different macromolecules (proteins) and ligand (antibiotic) complexes, ranging from −8.1 (SHV-27 + cefotaxime) to −7 (TEM-1 + cefotaxime) kcal/mol.
Conclusion and relevance: The study revealed β-lactamase variants circulating in the fowl population of the Andaman and Nicobar Islands (India), even in remote places with low anthropogenic activity. Most of the strains possessed blaTEM−1, followed by blaCTX−M−15. Possession of blaSHV−11, blaSHV−27, and blaSHV−228 in poultry Enterobacteriaceae strains was not reported earlier from any part of the world. The phylogenetic analysis revealed a partial clonal relationship of β-lactamase sequences with the human clinical strains isolated from the Indian subcontinent.
Introduction
Antimicrobial resistance in livestock is a global challenge as the bacteria possessing the resistance genes can be disseminated into the human food chain through cross-contamination by means of occupational exposure, contaminated environment, and consumption of animal-origin foods. Extended spectrum-β-lactamase (ESBL) and AmpC-β-lactamase (ACBL) producing Enterobacteriaceae are the most reported antimicrobial-resistant bacteria in humans and livestock in the last two decades (1). Poultry was identified as the major reservoir of ESBL-producing Enterobacteriaceae in comparison to pigs, cattle, and other members of the livestock family (2). The poultry as a reservoir of ESBL-producing bacteria acts as a challenge for the farmers and slaughterhouse workers or meat vendors, as increased gut colonization of the resistant bacteria was detected in people who had more contact with the birds than the community (3). A recent whole-genome sequencing-based study also evidenced the transmission of ESBL-producing bacteria from poultry to the human population (4).
The ESBL enzyme generates resistance to β-lactam antibiotics, including higher-generation cephalosporins and monobactams. AmpC β-lactamase-producing bacteria (ACBL) can develop resistance against β-lactam antibiotics in addition to β-lactamase inhibitors like clavulanic acid. There are three classical ESBLs, i.e., TEM (except TEM-1, TEM-2, and TEM-13), SHV (except SHV-1, SHV-2, and SHV-11), and CTX-M (5). CTX-M-15 is the most common ESBL genotype prevalent currently among the human clinical isolates with a rising trend of CTX-M-1, frequently originating from livestock and poultry (6). Poultry acts as the major reservoir of CTX-M-1, SHV-12, TEM-52, and AmpC β-lactamases (7).
Anthropogenic activities were found to be associated with the development of an ESBL-“resistome” in the environment including aquatic settings either due to the dissemination of ESBL-determinants or the bacteria carrying the genes associated with direct human activities and/or the release of the antimicrobials in the sub-therapeutic level in the environment because of indirect human activities (8–10). Persistence of ESBL-producers on the abiotic or biotic surface, associated with the development of “resistome”, is dependent on the capacity to form biofilms, as they help in the survival of the bacterial colony against physical and chemical stresses, including disinfectants, host phagocytosis, and antibiotics (11). However, a recent study identified antimicrobial resistance genes in the commensals present in soil exposed to low anthropogenic activities (12).
Several studies found variants of ESBL in Enterobacteriaceae in healthy or diseased poultry birds (6, 13), but limited literature is available about the affinity of the β-lactamases for the precise class of cephalosporins. The present study was conducted to detect the presence of β-lactamase and biofilm-producing Escherichia coli, Salmonella, and Klebsiella in broilers and backyard or native fowl reared in the Andaman and Nicobar Islands (India), even in remote places with low anthropogenic activities. The study also included molecular docking experiments to confirm the nature of the catalytic domains in β-lactamase variants (blaCTX − M, blaSHV, and blaTEM) and phylogenetic analysis to reveal the clonal relationship of the poultry-origin Enterobacteriaceae isolates with human clinical strains from the GenBank database. Enterobacteriaceae was selected as the study bacteria as the family is included in the World Health Organization (WHO) global priority list under “critical” category as an indicator of antibiotic resistance.
Materials and methods
Sampling
During the period from November 2019 to January 2021, a total of 199 cloacal swabs (Table 1) were collected from five poultry breeds or varieties (broiler, Vanraja, Desi, Nicobari, and layer) irrespective of age and sex in three different districts of the Andaman and Nicobar Islands (India), i.e., South Andaman (S/A) (11.74°N/92.65°E), North and Middle Andaman (N & M/A) (12.65°N/92.89°E), and Nicobar (C/N) (7.12°N/93.78°E). The sample size varied between the districts depending on the accessibility and willingness of the farmers to join the study. The collected swabs taken from live birds were properly labeled and were aseptically transported, maintaining the cold chain, into the bacteriology laboratory of the Animal Science Division, ICAR-CIARI, Andaman and Nicobar Islands (India).
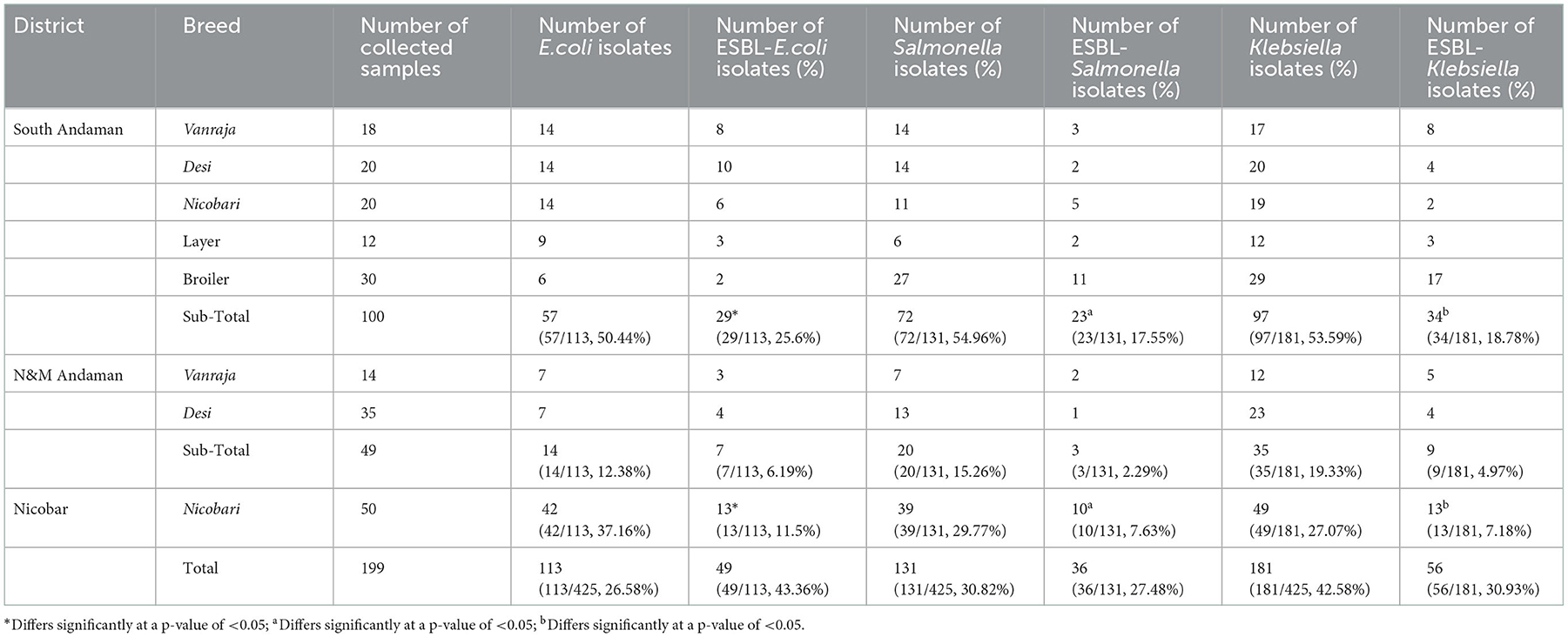
Table 1. Distribution of ESBL-producing E. coli, Salmonella, and Klebsiella in three districts of Andaman and Nicobar Islands (India).
No clinical symptoms were reported by the farmers during the collection of cloacal swabs from the birds. The contract farmers reared the broilers in medium-sized flocks (100–200 birds) with the guidelines, feed, vaccines, and medicines (including antibiotics like doxycycline, neomycin, and cephalexin) provided by the enterprise. The backyard farmers reared Vanraja, layer birds, and native fowls such as Desi and Nicobari in small flocks consisting of 15–20 birds per household with occasional exposure to tetracyclines, neomycin, and fluoroquinolones for therapy under the guidance of local veterinarians, para-veterinarians, and drug shop owners. The backyard farmers reared the birds under a semi-intensive system with daytime roaming around the houses and overnight shelter at the farmer's house. No commercial feed mixture was detected to have been used for feeding. The contract farmers prepared a separate bamboo or brick poultry shed and used feeders and waterers, with occasional cleaning and disinfection of the shed with formalin.
Isolation, identification, and PCR-based confirmation of Escherichia coli, Salmonella, and Klebsiella
The swab samples were transported in a sterile transport medium (transport liquid medium, HiMedia, India) and inoculated into the nutrient broth (HiMedia, India) and incubated at 37°C for 24 h. The loopful of overnight growth was streaked onto MacConkey agar (HiMedia, India) and incubated at 37°C for 24 h. The selected single pink colonies were transferred into eosin methylene blue (EMB) agar (HiMedia, India) and incubated at 37°C for 24 h. The single colonies with a greenish metallic sheen were selected for further morphological and biochemical identification (14). For the isolation of Salmonella, the swab samples collected were enriched with overnight growth in selenite broth (HiMedia, India) at 37°C. The loopful of growth was streaked onto brilliant green agar (BGA) (HiMedia, India) and incubated at 37°C. The single reddish colonies were selected for further morphological and biochemical identification (14). Similarly, for the isolation of Klebsiella, the growth in nutrient broth was streaked into Klebsiella selective agar (HiMedia, India) and incubated at 37°C. The single purple magenta colonies were considered for further morphological and biochemical identification (14). The tentatively identified E. coli, Salmonella, and Klebsiella isolates were confirmed by 16SrRNA gene-specific PCR (15, 16). Klebsiella pneumoniae was also identified by specific PCR with the Klebsiella species isolates (17). The PCR products were agarose gel electrophoresed containing ethidium bromide, and the gel was visualized and documented in a gel documentation system (Labmate Asia, India).
Antibiogram
All the E. coli, Salmonella enterica, and Klebsiella pneumoniae isolates were screened for antibiotic sensitivity with ceftazidime (CAZ) (30 μg), cefotaxime (CTX) (30 μg), ceftriaxone (CTR) (30 μg), cefpodoxime (CPD) (10 μg), cefoxitin (CX) (30 μg), aztreonam (AT) (30 μg), erythromycin (E) (15 μg), tetracycline (TE) (30 μg), chloramphenicol (C) (30 μg), amoxicillin/clavulanic acid (AMC) (20/10 μg), gentamicin (GEN) (10 μg), sulphafurazole (SF) (300 μg), ampicillin/cloxacillin (AX) (10 μg), ampicillin (AMP) (10 μg), co-trimoxazole (COT) (23.75/1.25 μg), amikacin (AK) (30 μg), ciprofloxacin (CIP) (5 μg), and oxytetracycline (O) (30 μg). The interpretation of the susceptibility or resistance was calculated as per the CLSI recommendation (18).
Double disc diffusion test
The bacterial isolates with a zone of inhibition (ZOI) diameter of ≤ 22 mm for ceftazidime, ≤ 27 mm for cefotaxime, ≤ 25 mm for ceftriaxone, ≤ 17 mm for cefpodoxime, and ≤ 27 mm for aztreonam were considered for disc diffusion testing to detect phenotypical ESBL or AmpC production. For confirmation of ESBL production, the isolates that showed an increase of ≥5 mm in ZOI diameter when tested with CTZ and CAZ alone and in combination with ceftazidime/clavulanic acid (CAC/CAZ) (30/10 μg) and cefotaxime/clavulanic acid (CEC/CTX) (30 /10 μg) (18).
Cefoxitin (CX) (30 μg) disc screening was used for the initial detection of AmpC producers, and the isolates with ZOI diameter ≥18 mm were considered for the cefoxitin-cloxacillin double disc test. For confirmation of AmpC production, the isolates showed an increase of ≥4 mm in ZOI diameter when tested with cefoxitin alone and in combination with cefoxitin/cloxacillin (19).
PCR-based detection of ESBL and chromosomal AmpC genes
All the isolates showing phenotypical β-lactamase production were screened for the presence of blaCTX − M, blaSHV, blaTEM, and blaAmpC genes by PCR (20, 21). The PCR products were electrophoresed with ethidium bromide (0.5 μg/ml) and the gel was visualized and documented in a gel documentation system (Labmate Asia, India). The commercial source (Xcelris Genomics, India) was used for the sequencing of selected PCR products as representative of each breed or variety of the birds or the districts studied. The sequence homology was detected by the standard nucleotide BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome). The sequences were submitted to the DNA Data Bank of Japan (DDBJ; www.ddbj.nig.ac.jp).
Detection of biofilm-associated genes
All the 425 isolates were subjected to PCR-based detection of biofilm-associated genes, namely, csgA, sdiA, and rpoS (22, 23). The commercial source (Xcelris Genomics, India) was used for the sequencing of selected PCR products. The sequence homology was detected by the standard nucleotide BLAST algorithm (https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome).
Homology modeling, model validation, and active site identification of different ESBL variants
Available PDB structures of CTX-M-15 (PDB id: 4HBU), SHV-11 (PDB id: 6NFD), and TEM-1 (PDB id: 1BTL) were pulled out from the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB-PDB) database (https://www.rcsb.org/). A position-specific iterated basic local alignment search tool (PSI-BLAST) was performed to find out suitable templates for SHV-28 (template PDB id: 3D4F) and SHV-228 (template PDB id: 3OPL) (https://www.ebi.ac.uk/Tools/sss/psiblast/). Protein homology modeling was performed by using the SWISS-MODEL server (https://swissmodel.expasy.org/). Other structural assessments and stereochemical qualities (Supplementary Figure 1) were verified by the PROCHECK server (https://www.ebi.ac.uk/thornton-srv/software/PROCHECK/). Catalytic active sites for the crystal structures and the modeled proteins were further deposited to DoGSiteScorer, a web server for automatic binding site detection, under the proteins plus (http://dogsite.zbh; uni-hamburg.de/) to get the potential pockets for molecular docking analyses (Supplementary Figure 2).
Docking of third-generation cephalosporins with the ESBL variants
Molecular docking was performed on the Autodock Vina Windows Desktop Suite (https://autodock.scripps.edu/download-autodock4/) as described earlier (24). Three-dimensional SDF file structures of cefotaxime (C16H17N5O7S2; PubChem id: 5742673) and cefpodoxime (C15H17N5O6S2; PubChem id: 6335986) were retrieved from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). Receptor energy minimization was done in Swiss-PdbViewer (https://spdbv.unil.ch/energy_tut.html), and the ligand structures were optimized by the Avogadro desktop suite (https://avogadro.cc/). Two-dimensional macromolecule + ligand complexes were visualized by LIGPLOT (https://www.ebi.ac.uk/thornton-srv/software/LigPlus/install.html) analysis, and 3D complexes were made in the PyMOL (https://www.schrodinger.com/products/pymol) desktop suite.
Partial clonal relationship of poultry origin β-lactamases producing Enterobacteriaceae strains with human clinical isolates
The selected β-lactamase sequences from the present study were compared with the ESBL sequences of clinical Enterobacteriaceae strains isolated from human patients in India and Indian subcontinents (Bangladesh, Myanmar, China, Thailand), available in the NCBI-Genbank database (National Centre for Biotechnology Information; https://www.ncbi.nlm.nih.gov/genbank/). The phylogenetic tree was constructed by the maximum likelihood (ML) method using molecular evolutionary genetics analysis (MEGA-X; https://www.megasoftware.net/) and analyzed in iTOL v6 (https://itol.embl.de/).
Statistical analysis
The chi-square test (SPSS Inc.) was applied to reveal the statistical differences in the occurrence of β-lactamase-producing E. coli, Salmonella, and Klebsiella strains among the studied fowl population reared in the South Andaman and Nicobar districts.
Results
In total, 425 Enterobacteriaceae strains were isolated from the collected samples (n = 199). K. pneumoniae (42.58%) was found to be the most prevalent, followed by Salmonella enterica (30.82%) and E. coli (26.58%) (Table 1). E. coli, Salmonella, and Klebsiella were tentatively identified by biochemical tests and confirmed with 16S-rRNA gene-specific PCR.
Phenotypical antibiotic resistance profiling of all 425 isolates showed the highest resistance against oxytetracycline (61–76%), amoxicillin/clavulanic acid (61–76%), and co-trimoxazole (60–72%), and the lowest resistance was observed against gentamicin (15–20%). E. coli (81.42%) and Salmonella (80.92%) showed the highest phenotypical resistance against oxytetracycline, whereas Klebsiella showed the highest resistance against ciprofloxacin (70.72%) (Table 2; Figure 1).
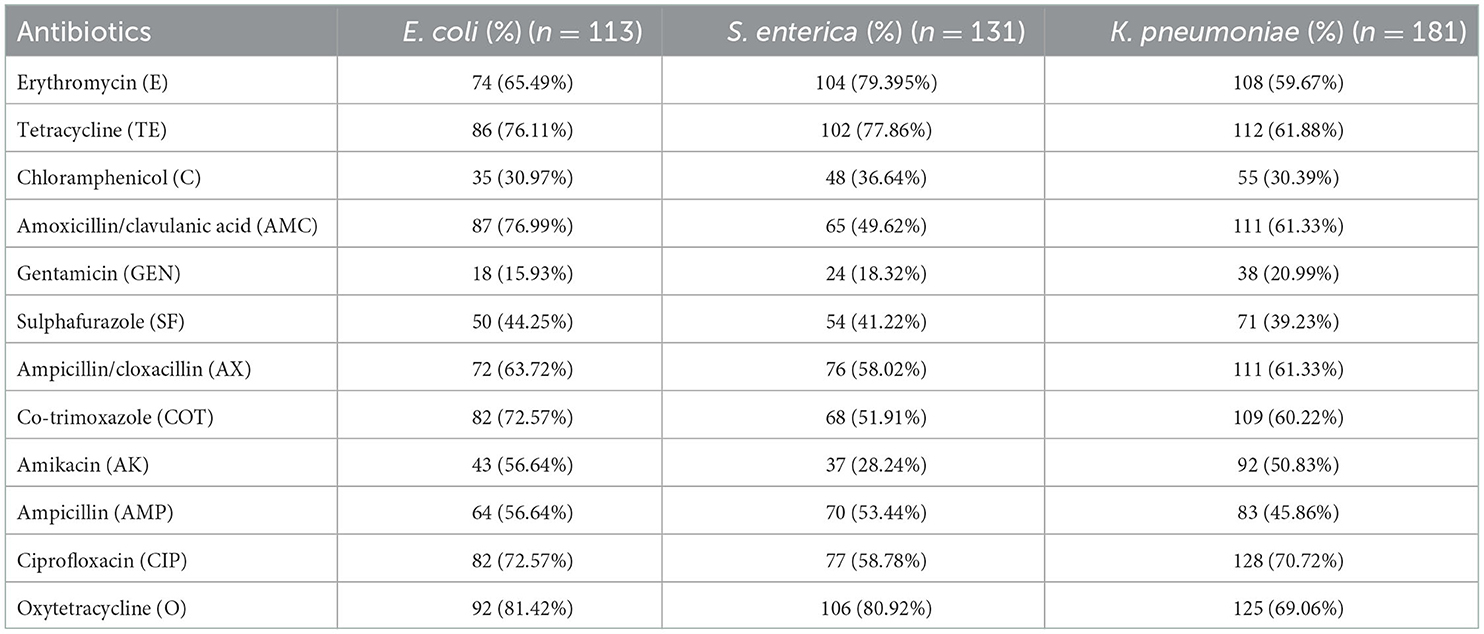
Table 2. Phenotypical antibiotic resistance in Enterobacteriaceae strains isolated from poultry in Andaman and Nicobar Islands (India).

Figure 1. Phenotypical antibiotic resistance profile of bacterial strains isolated from different birds reared in Andaman and Nicobar Islands (India).
Out of 425 isolates, phenotypical production of β-lactamase enzymes was observed by double disc test in 141 (33.38%) isolates. Production of β-lactamase enzymes was detected maximum in E. coli (43.36%) isolates, followed by the Salmonella (27.48%) and Klebsiella (30.93%) strains (Table 1). The isolation rate of β-lactamase-producing Enterobacteriaceae was significantly higher (p < 0.05) in the birds reared in the South Andaman district than in the Nicobar district (Table 1). Using the cefoxitin-cloxacillin double disc synergy (CC-DDS) test, phenotypical AmpC production was found in 28.24% (120/425) bacterial isolates. Klebsiella (51.33%) was the highest AmpC producer, followed by Salmonella (36.28%) and E. coli (18.58%).
Genotyping of the β-lactamase-producing isolates revealed maximum possession of blaTEM by E. coli (92.04%), Salmonella (78.63%), and Klebsiella (94.48%) isolates followed by blaSHV and blaCTX − M (Figure 2). None of the Salmonella isolates possessed blaCTX − M. Moreover, none of the E. coli, Salmonella, and Klebsiella isolates possessed all the studied ESBL genes (blaCTX − M, blaTEM, and blaSHV) together. Furthermore, blaTEM + blaSHV genotype was possessed by the maximum number of isolates, followed by the genotype blaTEM + blaCTX − M. All the phenotypical AmpC-producing isolates possessed blaAmpC in PCR. The nucleotide sequences of the PCR products were compared and found similar with blaCTX − M−15 (98.1% cognate), blaSHV − 11 (99.45% cognate), blaSHV − 27 (98.01–99.38% cognate), blaSHV − 228 (99.38% cognate), blaTEM − 1 (99.33% cognate), and blaAmpC (99.88% cognate) in the BLAST search. The sequences were published by DDBJ with accession numbers (https://getentry.ddbj.nig.ac.jp/) (Table 3).
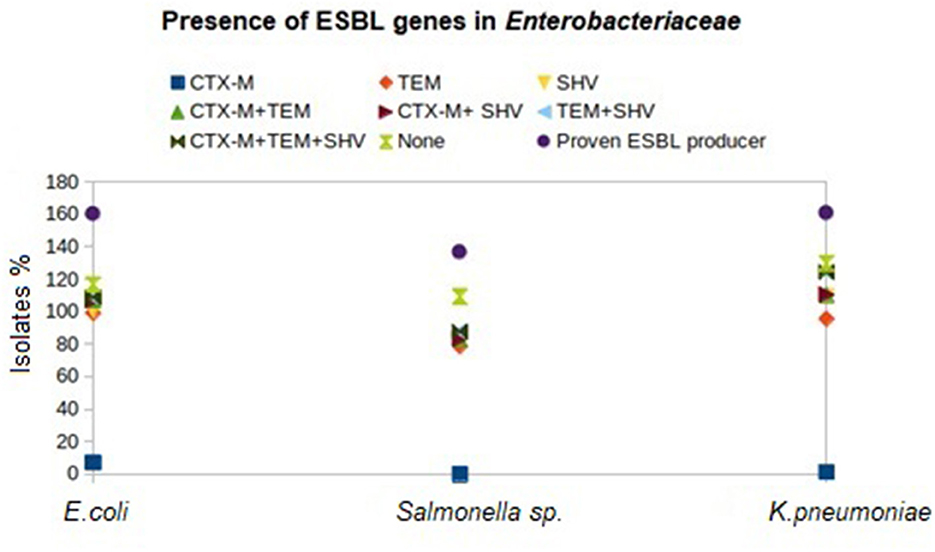
Figure 2. Distribution of β-lactamase genes in different birds reared in Andaman and Nicobar Islands (India).
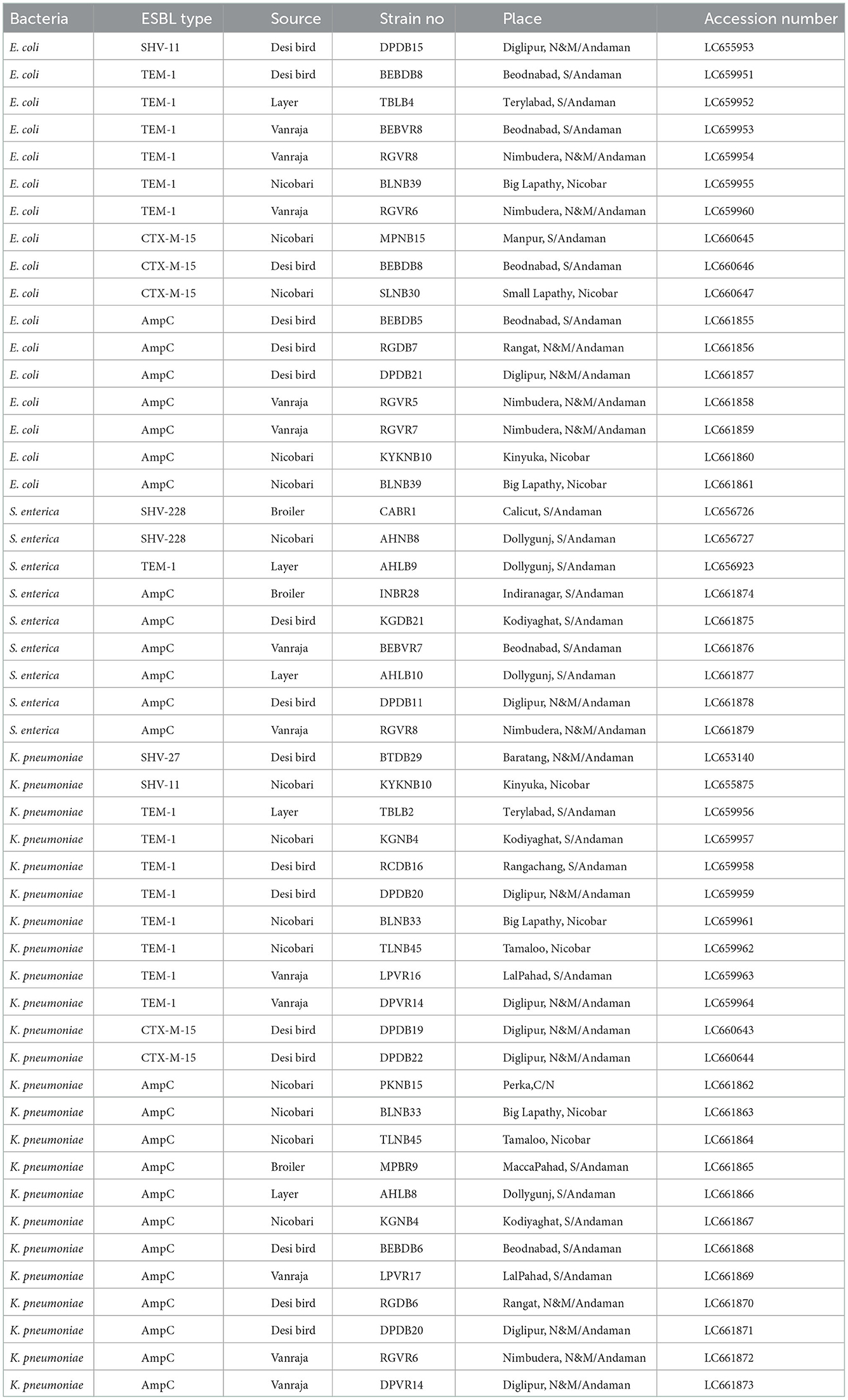
Table 3. Accession numbers of nucleotide sequences of ESBL/AmpC genes possessed by E. coli, Salmonella enterica, and Klebsiella pneumoniae strains isolated from different birds in Andaman and Nicobar Islands (India).
The distribution of biofilm-associated genes (csgA, rpoS, and sdiA) in the studied Enterobacteriaceae strains from different breeds or varieties of the birds revealed the maximum possession mostly by layer birds, followed by the other varieties of the studied birds (Figure 3). The csgA was detected with the highest frequency in the isolates from layer birds (92.5%), followed by Desi (84.6%), Vanraja (79%), Nicobari (66.6%), and broiler (53.2%). The sdiA was detected with the highest frequency in the isolates from layer birds (88.8%), followed by Desi (83.5%), Vanraja (79%), Nicobari (70.6%), and broiler (56.4%). The rpoS was detected with the highest frequency in the isolates from Vanraja (99%), followed by layer birds (96.3%), Desi (94.5%), Nicobari (78.7%), and broiler (77.4%).
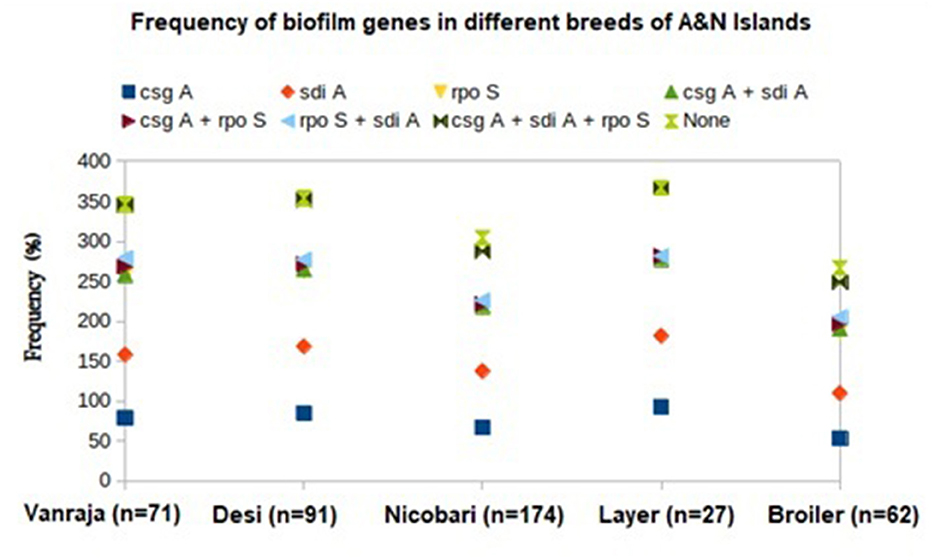
Figure 3. Distribution of biofilm-associated genes in different birds reared in Andaman and Nicobar Islands (India).
The phylogenetic analysis revealed a partial clonal relationship of β-lactamase sequences of the present study (Table 3), i.e., 15 blaTEM − 1 (LC659951-64 and LC656923), 2 blaSHV − 228 (LC656726-27), 5 blaCTX − M−15 (LC660643-47), 1 blaSHV − 27 (LC653140), and 2 blaSHV − 11 (LC655875 and LC655953) sequences with blaCTX − M−15, blaSHV − 11, and blaSHV − 27 and blaTEM − 1 sequences possessed by clinical strains isolated from human patients in India, Bangladesh, China, Myanmar, and Thailand (Figure 4).
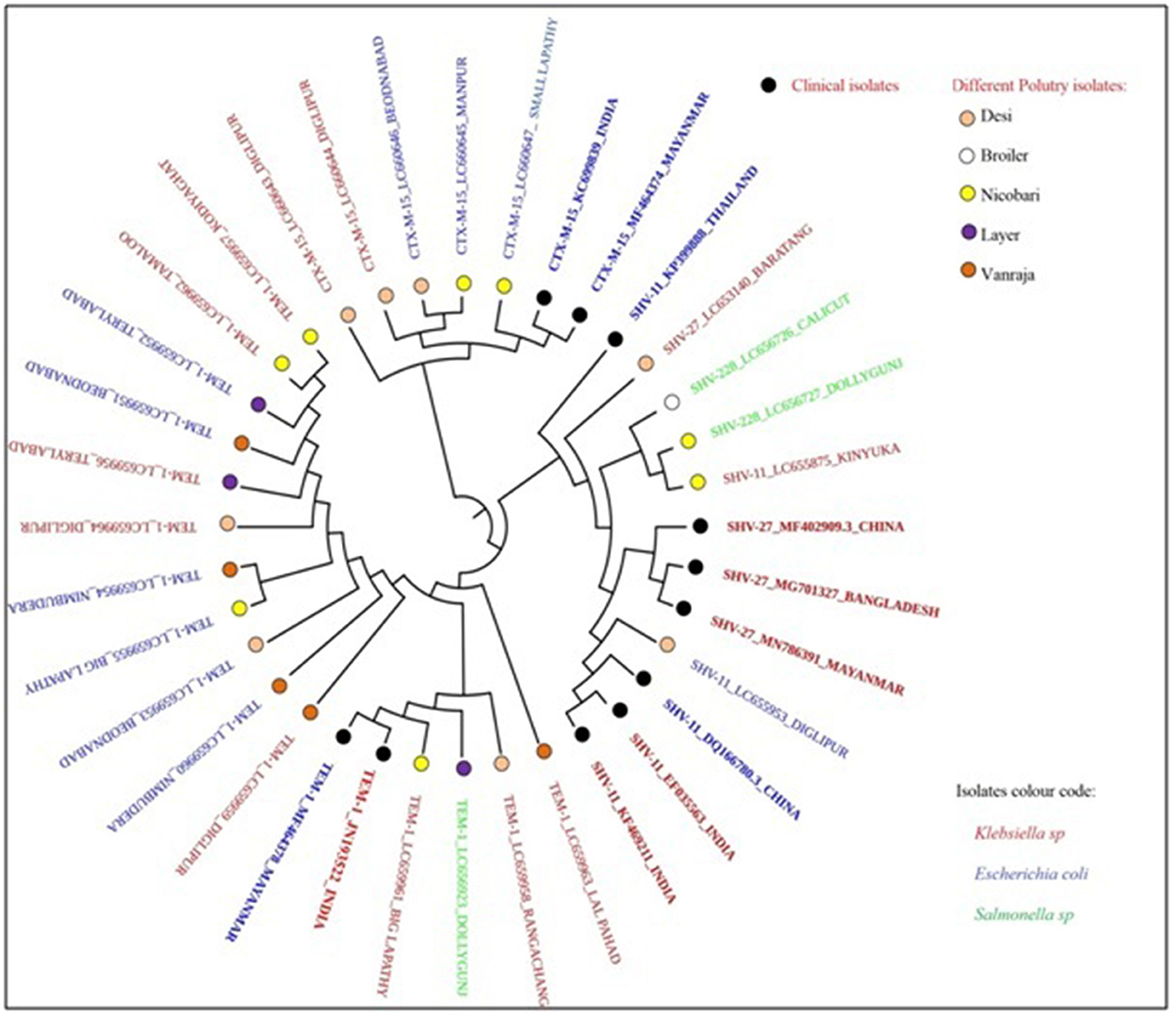
Figure 4. Clonal relationship of β-lactamase-producing Enterobacteriaceae isolated from locally reared fowls in Andaman and Nicobar Islands (India) with human clinical isolates.
Molecular docking depicted the Gibbs free energy release for 10 different macromolecules (proteins) and ligand (antibiotic) complexes, ranging from −8.1 (SHV-27+cefotaxime) to −7 (TEM-1+cefotaxime) kcal/mol. The color code of the drug and receptor molecules was maintained throughout the study (Figure 5). A summary of all the 10 complexes and participating amino acid residues in molecular interaction is described in Table 4. Different ligand + receptor complexes (2D Ligplot Plus and 3D PyMOL) are described in Figures 6–10.
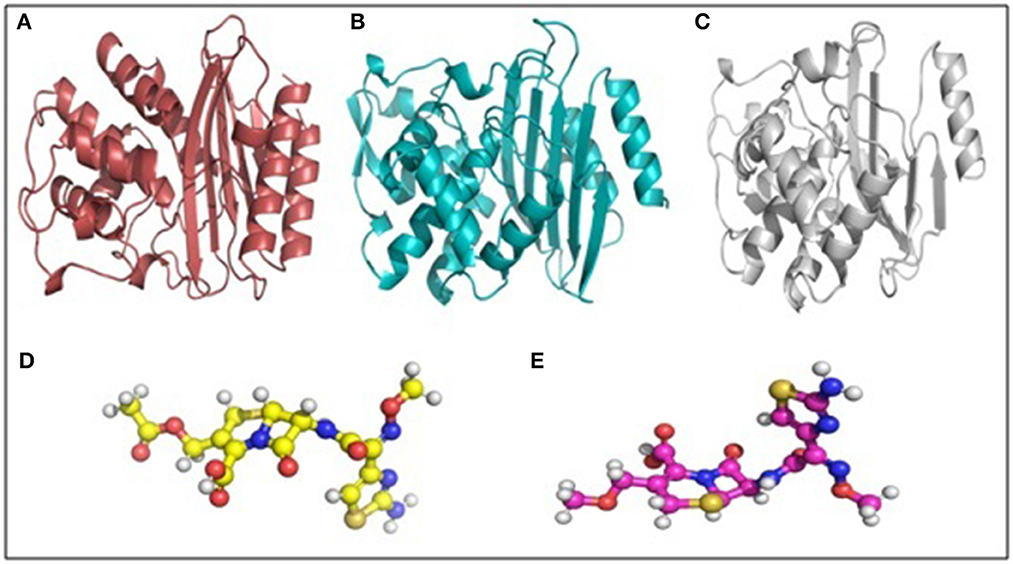
Figure 5. Macromolecule and ligand representation (3D) in PyMOL. These color codes have been mentioned throughout the study. (A) Cartoon representation of TEM-1 in ruby color. (B) Cartoon representation of CTX-M-15 in cyan color. (C) Cartoon representation of SHV in gray color. (D) Ball and stick representation of cefotaxime (ligand bonds are in golden color). (E) Ball and stick representation of cefpodoxime (ligand bonds are in lilac color).
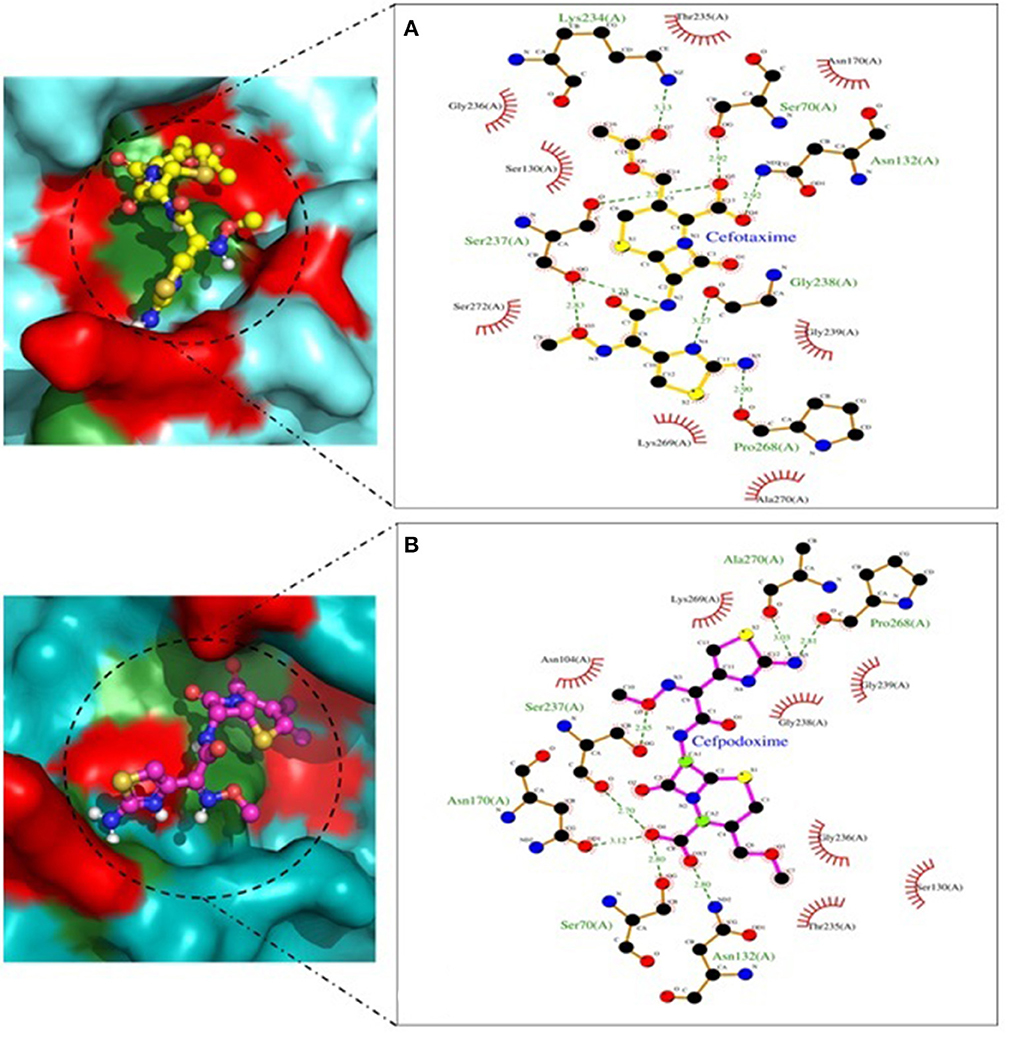
Figure 6. CTX-M-15 involved in interactions. (A) A binding map for cefotaxime+CTX-M-15. (B) A binding map for cefpodoxime+CTX-M-15.
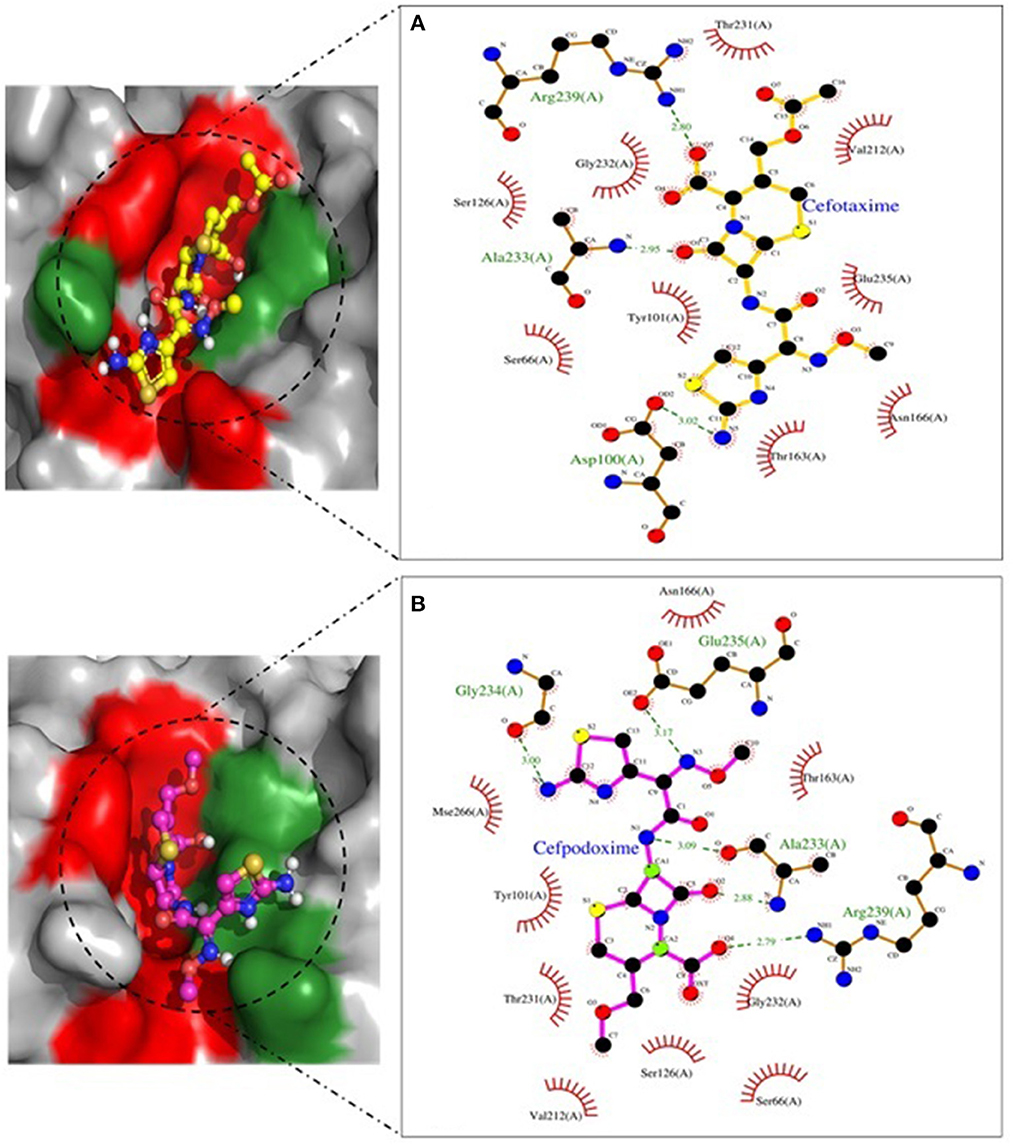
Figure 7. SHV-11 involved in interactions. (A) A binding map for cefotaxime+SHV-11. (B) A binding map for cefpodoxime+SHV-11.

Figure 8. SHV-27 involved in interactions. (A) A binding map for cefotaxime+SHV-27. (B) A binding map for cefpodoxime+SHV-27.
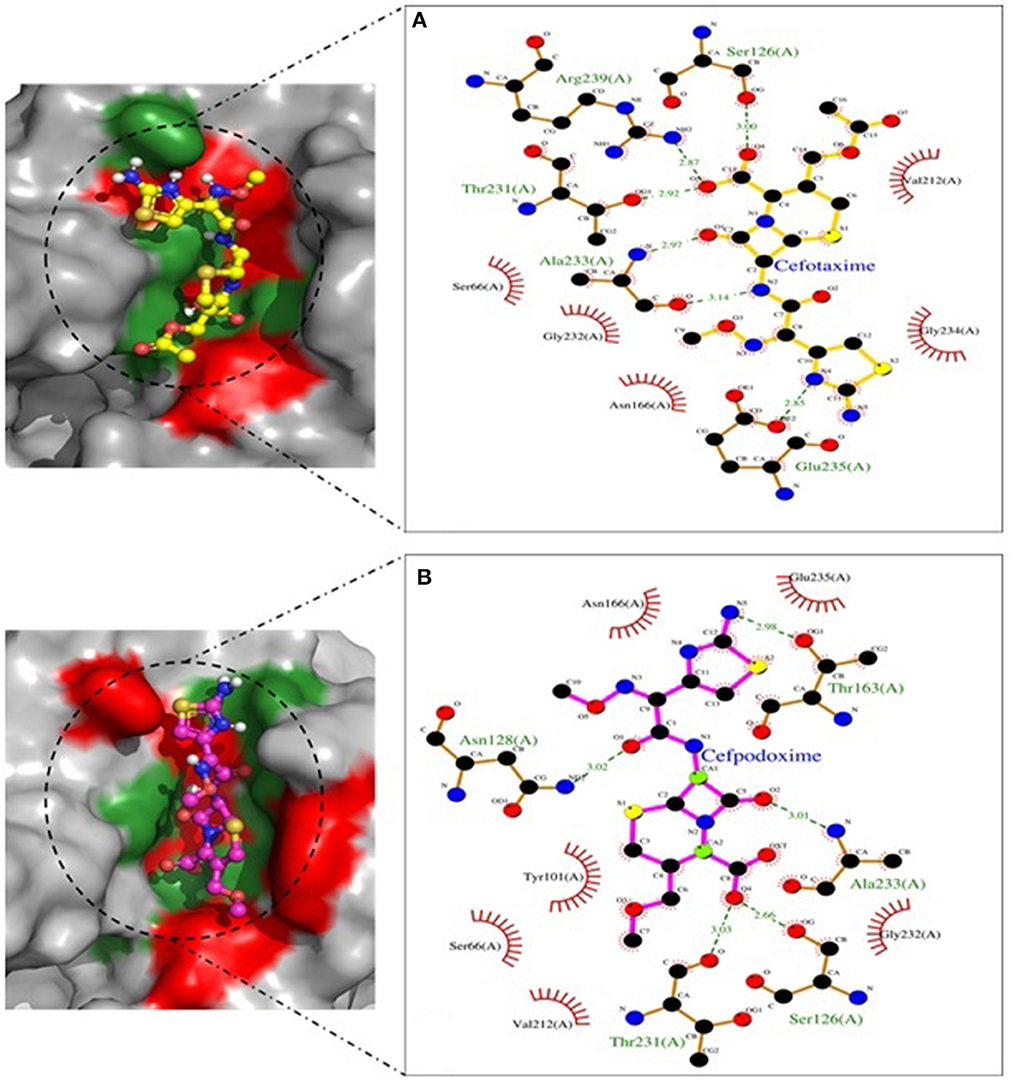
Figure 9. SHV-228 involved in interactions. (A) A binding map for cefotaxime+SHV-228. (B) A binding map for cefpodoxime+SHV-228.
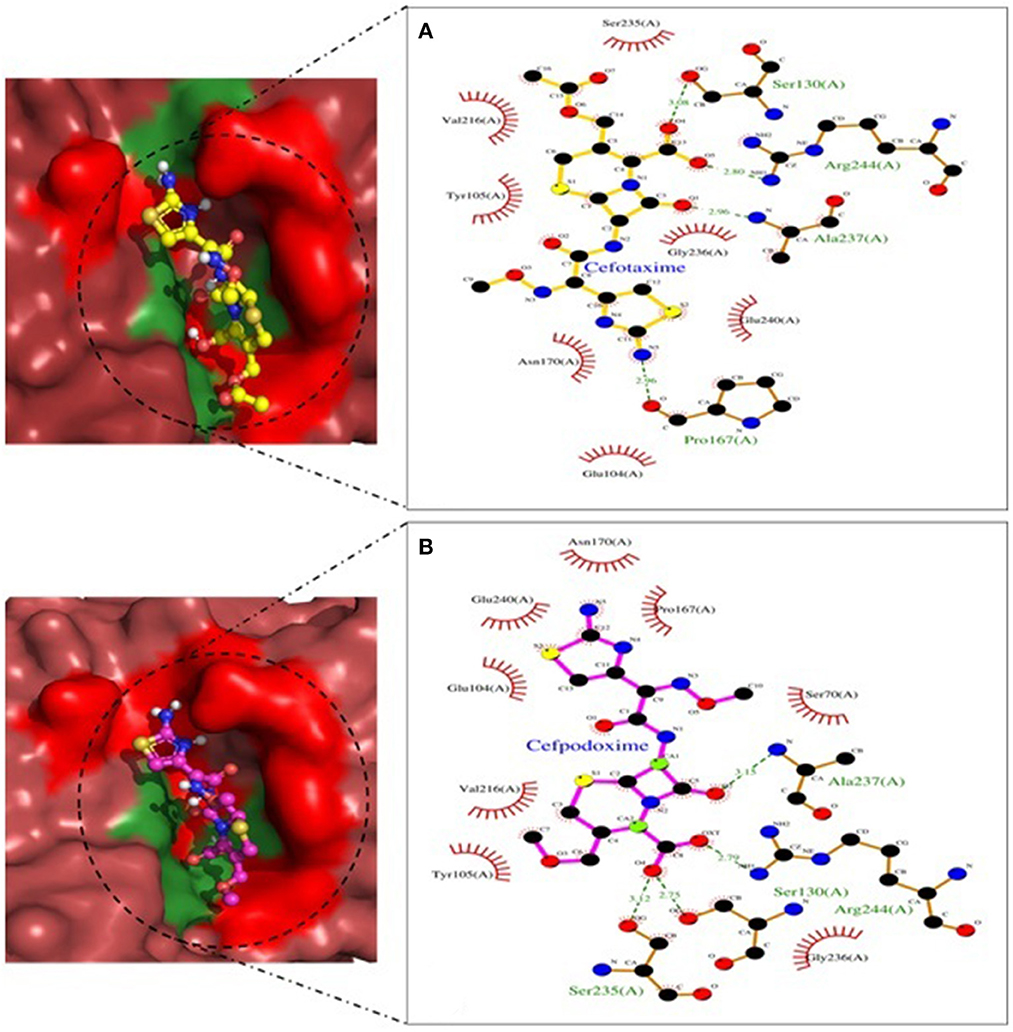
Figure 10. TEM-1 involved in interactions. (A) A binding map for cefotaxime+TEM-1. (B) A binding map for cefpodoxime+TEM-1.
Discussions
In the present study, K. pneumoniae was found to be the most prevalent in the cloacal swabs of the birds collected from different districts of A&N Islands (India), followed by Salmonella (30.82%) and E. coli (26.58%). Similar isolation rates of Klebsiella were reported earlier from poultry (43.8–72.3%) and bovine milk samples (45.2%) in other parts of India (25, 26). The isolation rate of Salmonella and E. coli in the present study was found to be corroborative with earlier reports (27, 28). However, the recovery of Salmonella, E. coli, and Klebsiella from poultry varied in different geographical regions depending on isolation protocol, sample size, and animal husbandry practices (29).
Antibiogram profiling of all 425 isolates showing maximum resistance against tetracycline is corroborative with the previous findings in Bangladesh (30), Iran (31), Malaysia (32), and Egypt (33). The resistance of poultry origin-Enterobacteriaceae to quinolone antibiotics (ciprofloxacin) from South China (34), Spain (35), and Egypt (36) was also reported, where co-resistance to ciprofloxacin and tigecycline was reported. Resistance to quinolones is often linked to tetracycline as the tetracycline molecule activates mutations in the mar operon, which results in more expression of the MarA protein increasing multidrug resistance (37). Most of the isolates in the present study showed resistance to three or more antibiotics and were considered multidrug resistant (38). The most common MDR pattern was found as E-TE-C-AMC-SF-COT-AMP-AX-CIP-O (9.7% in E. coli, 6.1% in Salmonella, and 5% in Klebsiella). All three studied bacterial strains (86.19%) were found susceptible to gentamicin, indicating the possible future usage of gentamicin for the treatment of bacterial infections in poultry in the A&N Islands.
Phenotypical β-lactamase production was detected maximum in E. coli (43.36%) isolates, followed by Salmonella (27.48%) and Klebsiella (30.93%). The majority of the ESBL producers in poultry belonged to the E. coli and Salmonella group of bacteria throughout the world (7). The occurrence of ESBL-producing Enterobacteriaceae was in accordance with those reported in Thailand (24.9%) (39), Lebanon (28%) (29), Ghana (29%) (4), and Denmark (27%) (40), lower than the prevalence rate reported in Germany (81–85%) (41) and Spain (79%) (42), and higher than Nicaragua (13%) (43) and Finland (14%) (44). The occurrence of ESBL-producing Enterobacteriaceae in poultry varies widely according to geographical location and antibiotic exposure, and the plasmids play a significant role in the clonal spread of ESBL genes in the poultry production system as the vertical route has less importance (44).
The isolation rate of β-lactamase producing Enterobacteriaceae was significantly higher (p < 0.05) in the birds reared in the South Andaman district than in Nicobar, which is correlated with more anthropogenic activities as the total human population and population density of South Andaman is significantly higher than the Nicobar (45). Anthropogenic activities were found to be directly correlated with the generation of ESBL-resistome in the environment either due to the dissemination of ESBL-producing bacteria or the release of the antimicrobials at the sub-therapeutic level in the environment (8–10). However, the occurrence of β-lactamase-producing Enterobacteriaceae in the birds reared in the Nicobar Islands with the minimum anthropogenic activities is an important finding as it may be correlated with increased soil salinity and high incidence of migratory birds in the islands after tsunami (46). Increased translation of multiple antibiotic resistance operons and transfer of ESBL gene containing plasmid was detected in soil bacteria to cope with the salinity stress as the stressors and the antimicrobials use the same bacterial cellular components or processes (47, 48). An increased presence of migratory birds after tsunami was associated with the generation of feeding habitats by the submergence of agricultural fields (49).
The nucleotide sequencing of the PCR products revealed that the variants of the β-lactamase circulating in the fowl population of A&N Islands were TEM-1 with the highest frequency, followed by CTX-M-15, SHV-11, SHV-27, and SHV-228. Similarly, TEM-1 was reported with a maximum frequency in E. coli strains isolated from diseased poultry in China (50) and in Salmonella strains isolated from poultry or poultry products in the Netherlands (51). Although TEM-1 is not considered as a classical ESBL, it is reported with high frequency in human clinical isolates throughout the world, and TEM-1-encoded enzyme was sometimes detected to demonstrate ESBL properties (52, 53). The high prevalence of TEM-1 in the fowl population of the present study also indicated the probable presence of subclinical bacterial infections, which was overlooked by the farmers who were not trained in poultry farming (13). The possession of blaCTX − M−15 is mostly associated with clinical Enterobacteriaceae isolates originated from both human and animal populations worldwide (54). CTX-M-15-producing Enterobacteriaceae were earlier reported in poultry from different parts of the globe (41, 42, 55). The SHV-27 was earlier reported in Klebsiella strains isolated from neonatal blood in Brazil, and the enzyme was found to show resistance against cefotaxime, ceftazidime and aztreonam (56). However, SHV-27, SHV-11, and SHV-228 were not reported from poultry in any part of the world.
Using the cefoxitin-cloxacillin double disc synergy (CC-DDS) test, phenotypical AmpC enzyme production was found to be 28.24% (120/425). In India, earlier studies revealed the occurrence of chromosomal AmpC (blaAmpC) in Enterobacteriaceae strains isolated from poultry, cattle with mastitis, pig and farm environments, and ducks (57). Other than therapeutic exposure to cefotaxime and ceftazidime, which was not detected in the present study, the occurrence of AmpC-producing bacteria might be associated with clonal transmission from the environment, as observed in a transmission dynamics study of ESBL-producing Enterobacteriaceae (39). The co-existence of ESBL and AmpC enzymes was detected in 10.82% (46/425) of the isolates, which is consistent with the earlier findings in the poultry production system (29, 58).
The generation of environmental resistomes is dependent on the persistence of ESBL/AmpC-producers on the abiotic or biotic surface with the capacity to form biofilms, as it helps in the survival of the bacterial colony against physical and chemical stresses (11). The present study detected a high prevalence (76%) of biofilm-associated genes in the Enterobacteriaceae strains isolated from the studied fowl population, indicating their possible environmental origin, although the soil microbial profile and the phenotypical biofilm-forming capacity of the strains were not validated.
The phylogenetic analysis revealed a partial clonal relationship between the fowl origin Enterobacteriaceae isolates and human clinical strains from the Indian subcontinent. Earlier studies revealed genetic relatedness of strains, similarity in types of β-lactamase genes, and/or associated plasmids in E. coli strains originating from animals and humans depicting the transmission probabilities (59).
Molecular docking interaction in the present study demonstrated the probable interactions among the different macromolecule-ligand complexes. The ligands with the minimum binding energy have the highest affinity of β-lactamases for cefotaxime and cefpodoxime. In our study, SHV-27 variants possessed the highest activity against cefotaxime. Improved docking scores were observed for the SHV variants because of the size and volume of its catalytic pocket and its druggability (Supplementary Figure 2). Antibiotic degradation by blaSHV in the present study has also revealed the participation of almost equivalent amino acids in terms of hydrophobic contacts (Ser 66, Tyr 101, Asn 166, and Val 212), which further emphasizes the structural homology of the other related variants. The study suffers from limitations related to sequencing, clonality analysis, and restricted numbers of isolates. The future characterization of this geographical location with the advent of next-generation sequencing can reveal the picture in detail.
The present study thus described the occurrence of β-lactamase/AmpC-producing Enterobacteriaceae in the local fowl population, even with the exposure of limited anthropogenic activities. Most of the strains possessed blaTEM − 1, followed by blaCTX − M−15. The possession of blaSHV − 11, blaSHV − 27, and blaSHV − 228 in poultry Enterobacteriaceae strains was not reported earlier. ESBL variants were modeled by the SWISS-MODEL and verified. Ligand with the minimum binding energy has the highest affinity of β-lactamases for cefotaxime and cefpodoxime. Phylogenetic analysis of the fowl origin ESBL-producing Enterobacteriaceae strains revealed a partial clonal relationship with the clinical isolates from human patients.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was reviewed and approved by Institutional Animal Ethics Committee, WBUAFS. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
SBh collected the samples and did all the laboratory works. SP conducted bioinformatics analysis. JS and IS supervised the study. TS, AD, SJ, KB, TD, SBa, and AT conceptualized the study. TM and AS helped in the analysis. IS, SBh, and AT wrote the primary and revised manuscripts. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors provide sincere gratitude to the Director, ICAR-Central Island Agricultural Research Institute, Port Blair-744105, Andaman and Nicobar Islands, India, for providing infrastructure and facilities. It was a collaborative work among WBUAFS (India), ICAR-CIARI (India), and the University of Helsinki (Finland).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1075133/full#supplementary-material
Supplementary Figure 1. RC plot analyses for the modeled SHV-27 protein structure.
Supplementary Figure 2. Summary of main chain parameters for the modeled SHV-27 protein structure.
Supplementary Figure 3. Summary of side chain parameters for the modeled SHV-27 protein structure.
Supplementary Figure 4. RMSD profile for the modeled SHV-27 protein structure.
Supplementary Figure 5. RC plot analyses for the modeled SHV-228 protein structure.
Supplementary Figure 6. Summary of main chain parameters for the modeled SHV-228 protein structure.
Supplementary Figure 7. Summary of side chain parameters for the modeled SHV-228 protein structure.
Supplementary Figure 8. RMSD profile for the modeled SHV-228 protein structure.
Supplementary Figure 9. Surface view of macromolecules. (A) TEM-1 (in ruby color) showing the opening of the catalytic pocket marked in yellow color. (B) CTX-M-15 (in cyan color) showing the opening of the catalytic pocket marked in yellow color. (C) SHV (in gray color) showing the opening of the catalytic pocket marked in yellow color.
References
1. Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. (2017) 72:2145–55. doi: 10.1093/jac/dkx146
2. Wadepohl K, Müller A, Seinige D, Rohn K, Blaha T, Meemken D, et al. Association of intestinal colonization of ESBL-producing Enterobacteriaceae in poultry slaughterhouse workers with occupational exposure-A German pilot study. PLoS ONE. (2020) 15:e0232326. doi: 10.1371/journal.pone.0232326
3. Price LB, Graham JP. Lackey LG, Roess A, Vailes R, Silbergeld E. Elevated risk of carrying gentamicin-resistant Escherichia coli among US poultry workers. Environ hlth Pers. (2007) 115:1738–42. doi: 10.1289/ehp.10191
4. Falgenhauer L, Imirzalioglu C, Oppong K, Akenten CW, Hogan B, Krumkamp R, et al. Detection and characterization of ESBL-producing Escherichia coli from humans and poultry in Ghana. Front Microbiol. (2019) 9:3358. doi: 10.3389/fmicb.2018.03358
5. EFSA Panel on Biological Hazards. Scientific opinion on the public health risks of bacterial strains producing extended-spectrum β-lactamases and/or AmpC β-lactamases in food and food-producing animals. EFSA J. (2011) 9:2322. doi: 10.2903/j.efsa.2011.2322
6. Valentin L, Sharp H, Hille K, Seibt U, Fischer J, Pfeifer Y, et al. Subgrouping of ESBL-producing Escherichia coli from animal and human sources: an approach to quantify the distribution of ESBL types between different reservoirs. Int J Med Microbiol. (2014) 304:805–16. doi: 10.1016/j.ijmm.2014.07.015
7. Saliu EM, Vahjen W, Zentek J. Types and prevalence of extended–spectrum beta–lactamase producing Enterobacteriaceae in poultry. Anim Hlth Res Rev. (2017) 18:46–57. doi: 10.1017/S1466252317000020
8. Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. (2011) 2:246. doi: 10.3389/fmicb.2011.00246
9. Huang X, Liu C, Li K, Liu F, Liao D, Liu L, et al. Occurrence and distribution of veterinary antibiotics and tetracycline resistance genes in farmland soils around swine feedlots in Fujian Province, China. Environ Sci Poltn Res. (2013) 20:9066–74. doi: 10.1007/s11356-013-1905-5
10. Al Salah DMM, Slaveykova VI. Otamonga JP, Poté J. Impact of anthropogenic activities on the occurrence and distribution of toxic metals, extending-spectra β-lactamases and carbapenem resistance in sub-Saharan African urban rivers. Sci Total Environ. (2020) 727:138129. doi: 10.1016/j.scitotenv.2020.138129
11. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. (2010) 8:623–33. doi: 10.1038/nrmicro2415
12. Tang X, Shen M, Zhang Y, Zhu D, Wang H, Zhao Y, et al. The changes in antibiotic resistance genes during 86 years of the soil ripening process without anthropogenic activities. Chemosphere. (2021) 266:128985. doi: 10.1016/j.chemosphere.2020.128985
13. Olsen RH, Bisgaard M, Löhren U, Robineau B, Christensen H. Extended-spectrum β-lactamase-producing Escherichia coli isolated from poultry: a review of current problems, illustrated with some laboratory findings. Avian Pathol. (2014) 43:199–208. doi: 10.1080/03079457.2014.907866
14. Quinn PJ, Carter ME, Markey B, Carter GR. Veterinary Clinical Microbiology. London, UK: Wolfe Publication (1994).
15. Wang RF, Cao WW, Cerniglia CE. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. (1996) 62:1242–7. doi: 10.1128/aem.62.4.1242-1247.1996
16. Brisse S, Verhoef J. Phylogenetic diversity of Klebsiella pneumoniae and Klebsiella oxytoca clinical isolates revealed by randomly amplified polymorphic DNA, gyrA and parC genes sequencing and automated ribotyping. Int J SystEvol Microbiol. (2001) 51:915–24. doi: 10.1099/00207713-51-3-915
17. Liu Y, Liu C, Zheng W, Zhang X, Yu J, Gao Q, et al. detection of Klebsiella pneumoniae in infant formula based on 16S−23S internal transcribed spacer. Int J Food Microbiol. (2008) 125:230–5. doi: 10.1016/j.ijfoodmicro.2008.03.005
18. Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement CLSI Document M100. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
19. Polsfuss S, Bloemberg GV, Giger J, Meyer V, Böttger EC, Hombach M. Practical approach for reliable detection of AmpC beta-lactamase-producing Enterobacteriaceae. J Clin Microbiol. (2011) 49:2798–803. doi: 10.1128/JCM.00404-11
20. Féria C, Ferreira E, Correia JD, Gonçalves J, Caniça M. Patterns and mechanisms of resistance to β-lactams and β-lactamase inhibitors in uropathogenic Escherichia coli isolated from dogs in Portugal. J Antimicrob Chemother. (2002) 49:77–85. doi: 10.1093/jac/49.1.77
21. Weill FX, Lailler R, Praud K, Kérouanton A, Fabre L, Brisabois A, et al. Emergence of extended-spectrum-β-lactamase (CTX-M-9)-producing multi resistant strains of Salmonella enterica serotype Virchow in poultry and humans in France. J Clin Microbiol. (2004) 42:5767–73. doi: 10.1128/JCM.42.12.5767-5773.2004
22. Silva VO, Espeschit IF, Moreira MA. Clonal relationship of Escherichia coli biofilm producer isolates obtained from mastitic milk. Canadian J Microbiol. (2013) 59:291–3. doi: 10.1139/cjm-2013-0053
23. Adamus-Białek W, Kubiak A, Czerwonka G. Analysis of uropathogenic Escherichia coli biofilm formation under different growth conditions. Acta Biochimica Polonica. (2015) 62. doi: 10.18388/abp.2015_1127
24. Banerjee A, Pal S, Goswami P, Batabyal K, Joardar SN, Dey S, et al. Docking analysis of circulating CTX-M variants in multi-drug resistant, beta-lactamase and biofilm-producing E. coli isolated from pet animals and backyard livestock. Microbial Patho. (2022) 170:105700. doi: 10.1016/j.micpath.2022.105700
25. Koovapra S, Bandyopadhyay S, Das G, Bhattacharyya D, Banerjee J, Mahanti A, et al. Molecular signature of extended spectrum β-lactamase producing Klebsiella pneumoniae isolated from bovine milk in eastern and north-eastern India. Infct Gen Evol. (2016) 44:395–402. doi: 10.1016/j.meegid.2016.07.032
26. Mahanti A, Ghosh P, Samanta I, Joardar SN, Bandyopadhyay S, Bhattacharyya D, et al. Prevalence of CTX-M-Producing Klebsiella spp. in broiler, kuroiler, and indigenous poultry in West Bengal State. India Microb Drug Resist. (2018) 24:299–306. doi: 10.1089/mdr.2016.0096
27. Karim MR, Giasuddin M, Samad MA, Mahmud MS, Islam MR, Rahman MH, et al. Prevalence of Salmonella spp. in poultry and poultry products in Dhaka, Bangladesh. Int J Anim Biol. (2017) 3:18–22.
28. Shecho M, Thomas N, Kemal J, Muktar Y. Cloacal Carriage and Multidrug Resistance Escherichia coli O157:H7 from Poultry Farms, Eastern Ethiopia. J Vet Med. (2017) 2017:8264583. doi: 10.1155/2017/8264583
29. Dandachi I, Sokhn ES, Dahdouh EA, Azar E, El-Bazzal B, Rolain JM, et al. Prevalence and characterization of multi-drug-resistant gram-negative bacilli isolated from Lebanese poultry: a nationwide study. Front Microbiol. (2018) 9:550. doi: 10.3389/fmicb.2018.00550
30. Das A, Dhar PK, Dutta A, Jalal MS, Ghosh P, Das T, et al. Circulation of oxytetracycline-and ciprofloxacin-resistant commensal Escherichia coli strains in broiler chickens and farm environments, Bangladesh. Vet World. (2020) 13:2395. doi: 10.14202/vetworld.2020.2395-2400
31. Jahantigh M, Samadi K, Dizaji RE, Salari S. Antimicrobial resistance and prevalence of tetracycline resistance genes in Escherichia coli isolated from lesions of colibacillosis in broiler chickens in Sistan, Iran. BMC Vet Res. (2020) 16:1–6. doi: 10.1186/s12917-020-02488-z
32. Ibrahim S, Wei Hoong L, Lai Siong Y, Mustapha Z, CW Zalati CS, Aklilu E, et al. Prevalence of antimicrobial resistance (AMR) Salmonella spp. and Escherichia coli isolated from broilers in the East Coast of Peninsular. Malaysia Antibiotics. (2021) 10:579. doi: 10.3390/antibiotics10050579
33. Elmonir W, Abd El-Aziz NK, Tartor YH, Moustafa SM, Abo Remela EM, Eissa R, et al. Emergence of colistin and carbapenem resistance in extended-spectrum β-lactamase producing Klebsiella pneumoniae isolated from chickens and humans in Egypt. Biol. (2021) 10:373. doi: 10.3390/biology10050373
34. Wang XM, Liao XP, Zhang WJ, Jiang HX, Sun J, Zhang MJ, et al. Prevalence of serogroups, virulence genotypes, antimicrobial resistance, and phylogenetic background of avian pathogenic Escherichia coli in south of China. Foodborne Patho Dis. (2010) 7:1099–106. doi: 10.1089/fpd.2010.0542
35. Sáenz Y, Zarazaga M, Briñas L, Lantero M, Ruiz-Larrea F, Torres C. Antibiotic resistance in Escherichia coli isolates obtained from animals, foods and humans in Spain. Int J Antimicrob Agents. (2001) 18:353–8. doi: 10.1016/S0924-8579(01)00422-8
36. El-Aziz A, Norhan K, Tartor YH, Gharieb R, Erfan AM, Khalifa E, et al. Extensive drug-resistant Salmonella enterica. isolated from poultry and humans: prevalence and molecular determinants behind the co-resistance to ciprofloxacin and tigecycline. Front Microbiol. (2021) 3 12:738784398. doi: 10.3389/fmicb.2021.738784
37. Ruiz C, Levy SB. Many chromosomal genes modulate MarA-mediated multidrug resistance in Escherichia coli. Antimicrob Agents Chemother. (2010) 54:2125–34. doi: 10.1128/AAC.01420-09
38. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pan drug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infct. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
39. Tansawai U, Walsh TR, Niumsup PR. Extended spectrum ß-lactamase-producing Escherichia coli among backyard poultry farms, farmers, and environments in Thailand. Poult Sci. (2019) 98:2622–31. doi: 10.3382/ps/pez009
40. Agersø Y, Jensen JD, Hasman H, Pedersen K. Spread of extended spectrum cephalosporinase-producing Escherichia coli clones and plasmids from parent animals to broilers and to broiler meat in a production without use of cephalosporins. Foodborne Patho Dis. (2014) 11:740–6. doi: 10.1089/fpd.2014.1742
41. Blaak H, van Hoek AH, Hamidjaja RA, van der Plaats RQ. Kerkhof-de Heer L, de RodaHusman AM, Schets FM. Distribution, numbers, and diversity of ESBL-producing E coli in the poultry farm environment. PloS ONE. (2015) 10:e0135402. doi: 10.1371/journal.pone.0135402
42. Blanc V, Mesa R, Saco M, Lavilla S, Prats G, Miró E, et al. ESBL-and plasmidic class C β-lactamase-producing E. coli strains isolated from poultry, pig and rabbit farms. Vet Microbiol. (2006) 118:299–304. doi: 10.1016/j.vetmic.2006.08.002
43. Hasan B, Laurell K, Rakib MM, Ahlstedt E, Hernandez J, Caceres M, et al. Fecal carriage of extended-spectrum β-lactamases in healthy humans, poultry, and wild birds in León, Nicaragua—a shared pool of bla CTX-M genes and possible interspecies clonal spread of extended-spectrum β-lactamases-producing Escherichia coli. Microbial Drug Resist. (2016) 22:682–7. doi: 10.1089/mdr.2015.0323
44. Oikarainen PE, Pohjola LK, Pietola ES, Heikinheimo A. Direct vertical transmission of ESBL/pAmpC-producing Escherichia coli limited in poultry production pyramid. Vet Microbiol. (2019) 231:100–6. doi: 10.1016/j.vetmic.2019.03.001
45. Census. Andaman and Nicobar Islands Population 2011–2022. (2011). Available online at: https://www.census2011.co.in/census/state/andaman+and+nicobar+islands.html (accessed on October 17, 2022).
46. Amaresan N, Kumar K, Madhuri K, Usharani GK. Isolation and characterization of salt tolerant plant growth promoting rhizobacteria from plants grown in tsunami affected regions of Andaman and Nicobar Islands. Geomicrobiol J. (2016) 33:942–7. doi: 10.1080/01490451.2015.1128994
47. McMahon MA, Xu J, Moore JE, Blair IS, McDowell DA. Environmental stress and antibiotic resistance in food-related pathogens. Appl Environ Microbiol. (2007) 73:211–7. doi: 10.1128/AEM.00578-06
48. Poole K. Stress responses as determinants of antimicrobial resistance in Gram-negative bacteria. Trends Microbiol. (2012) 20:227–34. doi: 10.1016/j.tim.2012.02.004
49. Purti N, Shankar VS, Narshimulu G, Halder S, Ramayya C, Singh RP. Study on the diversity of birds in the new abode of wetlands created by the 2004 tsunami in South Andaman. J Threatened Taxa. (2022) 14:20811–20. doi: 10.11609/jott.6804.14.4.20811-20820
50. Yuan L, Liu JH, Hu GZ, Pan YS, Liu ZM, Mo J, et al. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolates from chickens in Henan Province, China. J Med Microbiol. (2009) 58:1449–53. doi: 10.1099/jmm.0.012229-0
51. Hasman H, Mevius D, Veldman K, Olesen I, Aarestrup FM. β-Lactamases among extended-spectrum β-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J Antimicrob Chemother. (2005) 56:115–21. doi: 10.1093/jac/dki190
52. Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev. (2005) 18:657–86. doi: 10.1128/CMR.18.4.657-686.2005
53. Carattoli A, García-Fernández A, Varesi P, Fortini D, Gerardi S, Penni A, et al. Molecular epidemiology of Escherichia coli producing extended-spectrum β-lactamases isolated in Rome, Italy. J Clin Microbiol. (2008) 46:103–8. doi: 10.1128/JCM.01542-07
54. Haenni M, Ponsin C, Métayer V, Médaille C, Madec JY. Veterinary hospital-acquired infections in pets with a ciprofloxacin-resistant CTX-M-15-producing Klebsiella pneumoniae ST15 clone. J Antimicrob Chemother. (2012) 67:770–1. doi: 10.1093/jac/dkr527
55. Kawamura K, Goto K, Nakane K, Arakawa Y. Molecular epidemiology of extended-spectrum β-lactamases and Escherichia coli isolated from retail foods including chicken meat in Japan. Foodborne Patho Dis. (2014) 11:104–10. doi: 10.1089/fpd.2013.1608
56. Corkill JE, Cuevas LE, Gurgel RQ, Greensill J, Hart CA. SHV-27, a novel cefotaxime-hydrolysing β-lactamase, identified in Klebsiella pneumoniae isolates from a Brazilian hospital. J Antimicrob Chemother. (2001) 47:463–5. doi: 10.1093/jac/47.4.463
57. Banerjee A, Bardhan R, Chowdhury M, Joardar SN, Isore DP, Batabyal K, et al. Characterization of beta-lactamase and biofilm producing Enterobacteriaceae isolated from organized and backyard farm ducks. Lett Appl Microbiol. (2019) 69:110–5. doi: 10.1111/lam.13170
58. Castellanos LR, Donado-Godoy P, León M, Clavijo V, Arevalo A, Bernal JF, et al. High heterogeneity of Escherichia coli sequence types harbouring ESBL/AmpC genes on IncI1 plasmids in the Colombian poultry chain. PLoS ONE. (2017) 12:e0170777. doi: 10.1371/journal.pone.0170777
Keywords: Andaman and Nicobar, docking, clonal, ESBL, poultry
Citation: Bhowmick S, Pal S, Sunder J, Sujatha T, De AK, Mondal T, Singh AD, Joardar SN, Batabyal K, Dutta TK, Bandyopadhyay S, Tiwari A and Samanta I (2023) Exploring broilers and native fowls of Andaman and Nicobar Islands as a source of β-lactamase-producing Enterobacteriaceae even with limited anthropogenic activities and docking-based identification of catalytic domains in novel β-lactamase variants. Front. Vet. Sci. 9:1075133. doi: 10.3389/fvets.2022.1075133
Received: 20 October 2022; Accepted: 28 November 2022;
Published: 05 January 2023.
Edited by:
William Calero-Cáceres, Technical University of Ambato, EcuadorReviewed by:
Raoudha Dziri, Tunis El Manar University, TunisiaYasmine Hasanine Tartor, Zagazig University, Egypt
Copyright © 2023 Bhowmick, Pal, Sunder, Sujatha, De, Mondal, Singh, Joardar, Batabyal, Dutta, Bandyopadhyay, Tiwari and Samanta. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Indranil Samanta,  aXNhbWFudGE3NkBnbWFpbC5jb20=;
aXNhbWFudGE3NkBnbWFpbC5jb20=;  ZHJpc2FtYW50YUB3YnVhZnNjbC5hYy5pbg==; Ananda Tiwari,
ZHJpc2FtYW50YUB3YnVhZnNjbC5hYy5pbg==; Ananda Tiwari,  YW5hbmRhLnRpd2FyaUBoZWxzaW5raS5maQ==
YW5hbmRhLnRpd2FyaUBoZWxzaW5raS5maQ==
 Sneha Bhowmick1,2
Sneha Bhowmick1,2 Surajit Pal
Surajit Pal Arun Kumar De
Arun Kumar De Siddhartha Narayan Joardar
Siddhartha Narayan Joardar Kunal Batabyal
Kunal Batabyal Tapan Kumar Dutta
Tapan Kumar Dutta Samiran Bandyopadhyay
Samiran Bandyopadhyay Indranil Samanta
Indranil Samanta