- MOE Joint International Research Laboratory of Animal Health and Food Safety, College of Veterinary Medicine, Nanjing Agricultural University, Nanjing, China
Bovine coronavirus (BCoV) is a major cause of infectious disease in cattle, causing huge economic losses to the beef and dairy industries worldwide. BCoV can infect humans and multiple other species of animals. A rapid, reliable, and simple test is needed to detect BCoV infection in suspected farms. In this study, we developed a novel multienzyme isothermal rapid amplification (MIRA) and lateral flow dipstick (LFD) combination assay, targeting a highly conserved region of the viral nucleocapsid (N) gene for BCoV detection. The MIRA-LFD assay was highly specific and sensitive, comparable to a published reverse transcription quantitative PCR (RT-qPCR) assay for BCoV detection. Compared with the published RT-qPCR assay, the κ value of the MIRA-LFD assay in the detection of 192 cattle clinical samples was 0.982. The MIRA-LFD assay did not require sophisticated instruments and the results could be observed with eyes. Our results showed that the MIRA-LFD assay was a useful diagnostic tool for rapid on-site detection of BCoV.
Introduction
Coronaviruses are lipid-enveloped and include severe acute respiratory syndrome-associated coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and Middle East respiratory syndrome-associated coronavirus. They exhibit variable tissue tropism and the ability to infect various species of hosts, as exemplified by SARS-CoV-2 (1). Bovine coronavirus (BCoV) belongs to the genus Betacoronavirus of the family Coronaviridae, with a single-stranded and positive-sense RNA genome (2). BCoV has been linked with several clinical syndromes, including neonatal calf diarrhea (3, 4), winter dysentery (5), bovine respiratory disease, and shipping fever in adult cattle (6–8). The morbidity of BCoV ranged from 2 to 84%, and the mortality varied among cattle of different ages, and was approximately 8% in cattle with 5–90 days old (9, 10). BCoV is prevalent worldwide and spreads via the fecal-oral and respiratory routes (11, 12). Infections of BCoV and BCoV-like viruses have been found in goats, dromedary camels, deers, dogs, and humans (13–16).
Several diagnostic methods have been developed for detecting and identifying BCoV infections, including direct electron microscopy, protein A-gold immunoelectron microscopy (17), hybridization with cDNA probes (18), direct immunofluorescence (19), immunofluorescent staining, immune electron microscopy (20), immunohistochemistry (21), indirect ELISA (22), DNA microarray (23), rapid immunochromatographic assay (24), reverse transcription-polymerase chain reaction (RT-PCR) (25), semi-nested PCR assay (26), multiplex RT-PCR (27), and quantitative RT-PCR (RT-qPCR) (28, 29). These assays play a vital role in the detection of BCoV infections, but their applications are restricted because they require trained technicians, complex and expensive instruments, and/or long turn-over time. Therefore, these methods are unsuitable to be used at farms. In this study, we aimed to develop a rapid, reliable, simple, and isothermal amplification-based assay to fill the technological gap in BCoV detection at farms.
To date, several isothermal amplification-based techniques, such as the loop-mediated isothermal amplification assay (LAMP), recombinase polymerase amplification (RPA), and multienzyme isothermal rapid amplification (MIRA) (30, 31) have been developed for detecting pathogens. MIRA, whose principle is similar to that of RPA, is a new rapid isothermal amplification technique for the detection of nucleic acids. As shown in Figure 1A, at the first step of MIRA reverse transcriptase (moloney murine leukemia virus reverse transcriptase, M-MLV RT) reversely transcribes RNA into dsDNA. Then the complex of recombinase (the product of the recA locus in E. coli) and the primer binds to the dsDNA, which has been opened by single-stranded DNA-binding (SSB) protein (i.e., T4 phage gene 32 protein, gp32). After the binding, the recombinase is removed and the DNA polymerase (E. coli DNA polymerase I) binds to the 3' end of the primer and catalyzes the chain extension, which doubles the dsDNA. The synthesized dsDNA shall be opened by SSB and bound by the complex of recombinase and the primer, which begets a new round of dsDNA synthesis. The MIRA reaction can be completed within 20 min at 35–42°C (30, 31). It is fast and sensitive, and can be combined with a lateral flow dipstick (LFD) to visually evaluate the results (Figure 1B). The results of MIRA can be read out with the LFD step, which, like MIRA, does not require expensive instruments or trained technicians. The target RNA is amplified using biotin-labeled primers through MIRA, and the specific amplicons are bound by specific FAM-labeled probes, so the specific amplicons carry both FAM and biotin haptens. When the specific amplicons move from one terminal to another terminal of the test strip, the specific amplicons bind through their FAM haptens to FAM-antibodies conjugated with golden nanoparticles, and then the specific amplicons bind through their biotin haptens to the T-line where the biotin-antibodies have been immobilized, so the golden nanoparticles show red color there. The remaining FAM-antibodies conjugated with golden nanoparticles bind to the control (C)-line where the secondary antibodies against the FAM-antibodies have been immobilized, and so the golden nanoparticles show red color there. Therefore, red T-lines suggest positive MIRA results, and red C-lines suggest the FAM-antibodies conjugated with golden nanoparticles work well. Therefore, the MIRA-LFD combination assay has obvious advantages for virus detection, particularly at farms with limited resources. In this study, we developed a MIRA-LFD combination assay for BCoV detection and evaluated its sensitivity and specificity.
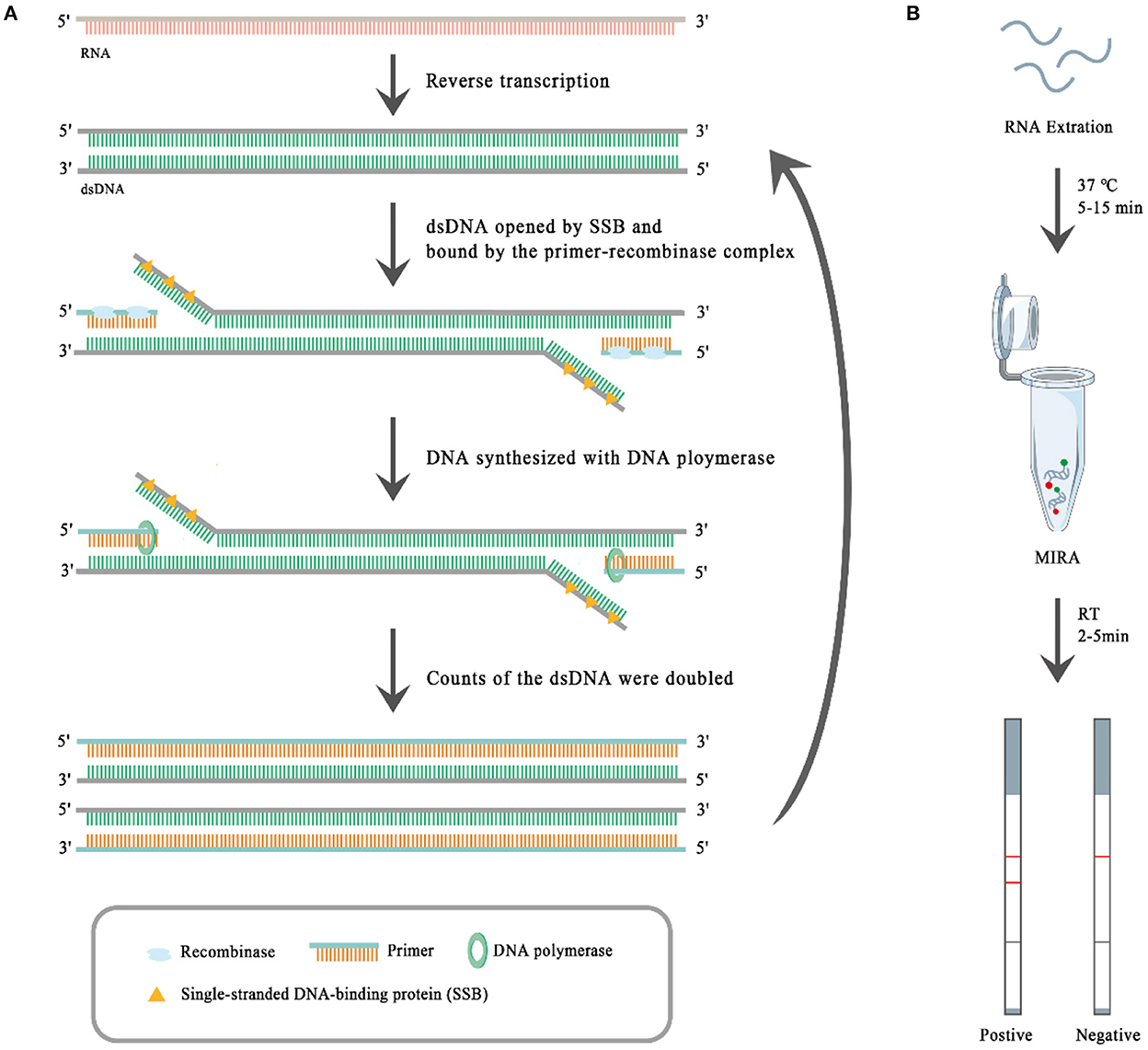
Figure 1. The principle of multienzyme isothermal rapid amplification (MIRA) (A) and the workflow of the MIRA-lateral flow dipstick (MIRA-LFD) assay (B). The presence of two red lines indicates a positive result and one red line a negative result.
2. Materials and methods
2.1. Viruses and plasmids
Field strains of bovine enterovirus (BEV), bovine herpesvirus-1 (BHV-1), bovine viral diarrhea virus 1 (BVDV-1) were cultured using MDBK (Madin-Darby bovine kidney) cells. Bovine clinical samples previously detected by RT-PCR and sequencing as positive in the detection of BCoV, bovine astrovirus (BAstV), or bovine rotavirus (BRV) were stored in our laboratory. The BCoV N gene was cloned into the pUC57 plasmid by Tsingke Biotechnology Company (Beijing, China).
2.2. Design and synthesis of primers and probes
To obtain the best diagnostic target for BCoV, we compared the full-length N gene sequences of BCoVs available in the GenBank. The N-terminal region of the gene is highly conserved and was hence selected as the target for the MIRA-LFD combination assay. The MIRA primers targeting the BCoV N gene were designed following the guidelines of AMP-Future Biotech company (Weifang, China). The sequences of the MIRA primers and the primers and probe of a RT-qPCR assay for BCoV detection, which was reported previously (28), were listed in Table 1. They were synthesized by Sangon Biotech Company (Shanghai, China) and purified with high-performance liquid chromatography (HPLC).
2.3. Extraction of RNA and DNA
Each feces or swab sample was homogenized in fivefold volume of phosphate-buffered saline (PBS) (pH 7.2). Viral DNA/RNA was extracted using the Vazyme FastPure Viral DNA/RNA Mini Kit from Vazyme Company (Nanjing, China) from 200 μL of the virus-infected cell culture supernatant or the homogenized clinical samples. The extracted DNA/RNA was dissolved in 50 μL nuclease-free water and stored at −80°C until the relevant assays were performed.
2.4. The MIRA reactions
The MIRA reactions were performed using the AMP-Future Biotech company kit (#WLRB8204KIT), according to the manufacturer's instructions. The reaction mixture contained 10 μL A buffer, 4 μL ddH2O, 1 μL of each primer (10 μM) 2 μL B buffer, and 2 μL RNA template. The mixture was incubated in a 42°C water bath for 30 min, after which, each mixture (5 μL) was subjected to electrophoresis with a 2% (g/g) agarose gel for 10 min at 160 V. Finally, the samples were visualized using an automated digital gel image analysis system (Bio-Rad).
2.5. The MIRA-LFD assay
MIRA-LFD reactions were performed using a kit (# WLRN8206KIT) from AMP-Future Biotech Company according to the manufacturer's instructions. The reaction mixture contained 29.4 μL A buffer, 11.5 μL ddH2O, 2 μL of each primer (10 μM), 2.5 μL B buffer, 0.6 μL probe, and 2 μL RNA template. After the 10 min reaction in a water bath at 37–39°C, the product was 1:10,000 diluted with H2O. The diluted product (100 μL) was added to the LFD strip (Gu'An Beiji Biotech Company, Hebei, China), and the color was observed 5 min later.
2.6. The specificity evaluation
The RNA of Nucleic acid templates prepared from other bovine pathogens, including BVDV-1, BHV-1, BRV, BEV, and BAstV, were detected using the MIRA-LFD assay to evaluate the specificity of this assay.
2.7. The sensitivity evaluation
To determine the analytical sensitivity of the MIRA-LFD assay, the BCoV-N-pUC57 plasmid was diluted to 101-108 copies per reaction and detected with the MIRA-LFD assay with three replicates for each dilution, using ribozyme-free water as the negative control. The extracted RNA of three BCoV positive samples was diluted serially from 1:5, and the diluted RNA was detected with the MIRA-LFD assay and the RT-qPCR assay reported previously (28).
2.8. Evaluation with clinical samples
A total of 132 feces samples and 60 nasal swab samples of bovine clinical samples were collected from cattle in Hebei, Inner Mongolia, Heilongjiang, and Ningxia provinces or autonomous regions of China between 2021 and 2022. The samples were collected and submitted to our laboratory by the relevant animal owners. These samples were detected in triplicate with the MIRA-LFD assay and the RT-qPCR assay reported previously (28).
2.9. Statistical analyses
The Kappa (κ) and possibility (p) values were used to compare the detection results of the MIRA-LFD assay and the RT-qPCR assay. The confidence level for statistical analyses was set as 95% (p < 0.05). Statistical analysis was performed using SPSS (version 21.0).
3. Results
3.1. Selection of the primer pair and the reaction temperature
Alignment of some representative BCoV genome sequences suggested that the viral N gene sequences were relatively conserved, and hence the gene was selected as the target gene for the MIRA-LFD assay. Then, a suitable fluorescent N-Probe was designed for the assay, and two forward primers (N-1F, N-3F) and three reverse primers (N-1R, N-2R, and N-3R) were designed to match N-Probe. The MIRA reactions were performed to compare six combinations of the upstream and downstream primers using 104 copies of the BCoV-N-pUC57 plasmid. As shown in Figure 2A, the combination of N-1F and N-2R was rated as one of the best-performing primer pairs and hence employed for the MIRA-LFD assay.
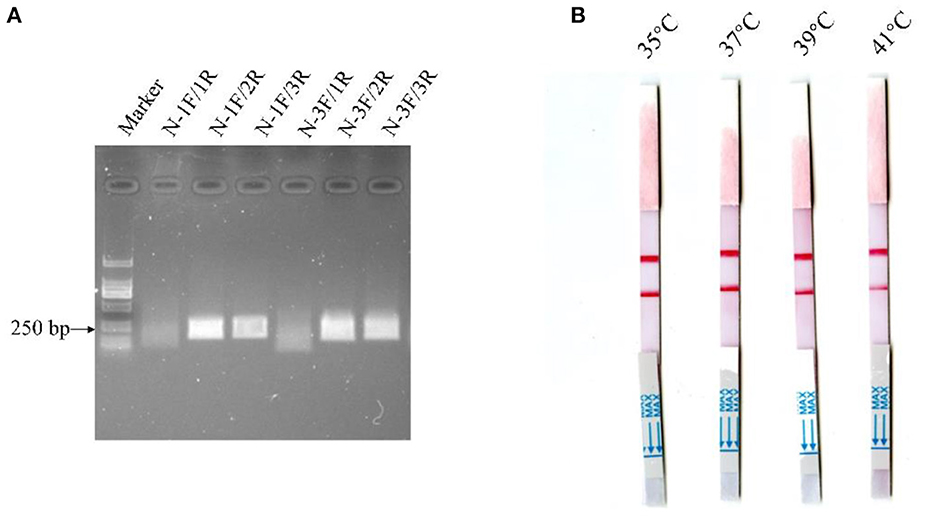
Figure 2. Detection of the diluted BCoV-N-pUC57 plasmid with the MIRA reactions using six pairs of primers (A) and comparison of the reaction temperatures for the MIRA-LFD assay (B).
The MIRA-LFD assay was performed at different temperatures (35, 37, 39, and 41°C) using the manufacturer's instructions. As shown in Figure 2B, the red line was the strongest and was produced in the shortest time at 37°C, which was therefore selected as the reaction temperature for the MIRA-LFD assay.
3.2. Evaluation with known viruses or controls
The viral RNA molecules of BCoV, BVDV-1, BHV-1, BRV, BEV, and BAstV were tested using the MIRA-LFD assay, and none of them were positive except the viral RNA molecules of BCoV (Figure 3A). The BCoV-N-pUC57 plasmid was tested using the MIRA-LFD assay after serial dilution, and Figure 3B showed that the assay could detect 100 copies of the plasmid. The RT-qPCR could detect 100 copies of the plasmid as well. The extracted RNA of three BCoV positive samples (samples 21, 46, and 78) was diluted serially from 1:5, and both the MIRA-LFD and the RT-qPCR assay could detect the RNA after 1:5 dilution (samples 21 and 78) or 1:25 dilution (sample 46).
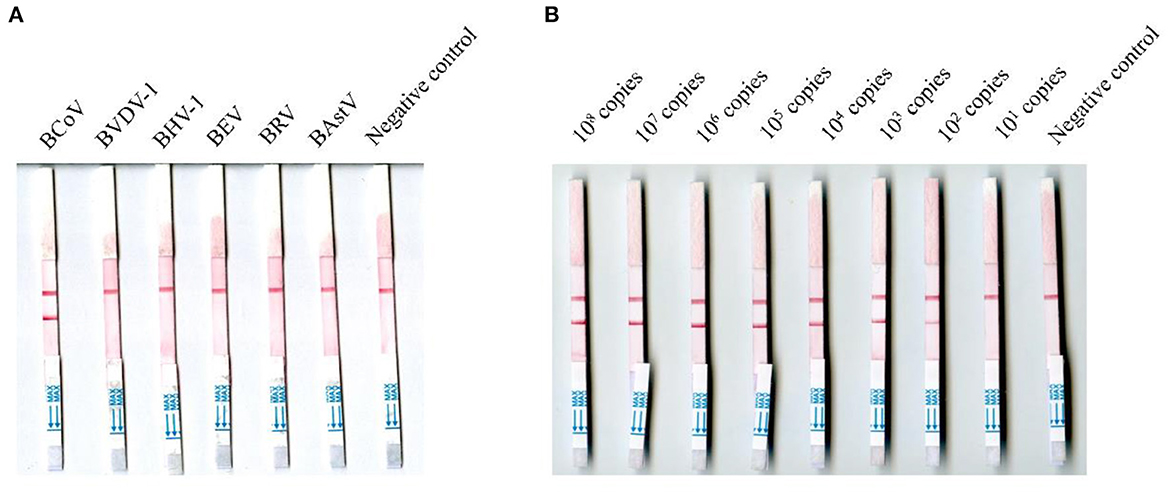
Figure 3. Detection of the RNA of six bovine viruses and the negative control (A) and different copies of the BCoV-N-pUC57 plasmid using the MIRA-LFD assay (B).
3.3. Evaluation with clinical samples
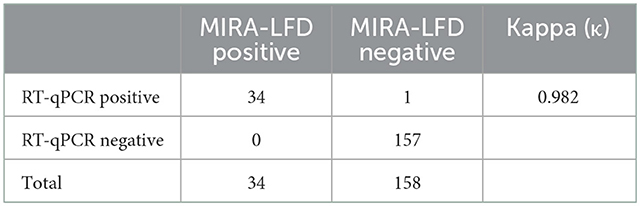
We detected 192 clinical samples submitted from several provinces or autonomous regions in China using the MIRA-LFD assay and the RT-qPCR assay reported previously. The RT-qPCR assay identified 35 (18.23%) BCoV positive samples, of which 34 were detected positive by the MIRA-LFD assay. The RT-qPCR assay identified 157 (81.77%) BCoV negative samples, which were all detected negative by the MIRA-LFD assay. The results of these two assays were highly consistent with each other (κ = 0.982) (Table 2).
4. Discussion
BcoV causes significant economic losses in the beef and dairy industries worldwide and can infect humans and multiple other species of animals (32). Rapid and accurate detection of BCoV is vital for the control of the virus infection. In this study, we established a new rapid assay for the BCoV detection through the combination of the new techniques of MIRA and LFD. The detection of known viruses suggests that this assay is highly specific, and the detection of the positive control of the plasmid and the serially diluted RNA of BCoV-positive samples suggests that this assay is almost as sensitive and specific as the RT-qPCR reported previously.
Compared with RT-qPCR assays, MIRA-LFD assays do not require expensive instruments. Compared with other isothermal amplification-based assays, such as LAMP (33, 34), MIRA-LFD assays are more easy in design. Moreover, MIRA-LFD assays require less time than RT-qPCR and LAMP assays. Notably, the collected clinical samples included both respiratory and feces samples, indicating that our MIRA-LFD assay applies to both types of samples. Therefore, MIRA-LFD assays are more suitable for on-site detection, which has advantages in taking prompt control measure or specific treatment and preventing the spread of infectious diseases.
MIRA-LFD assays have been employed in the detection of some human pathogens, such as SARS-CoV-2 and hepatitis C virus (35–37), but they have been rarely employed in the veterinary field. To our knowledge, this is the first study to report the detection of BCoV using this method, and this study shall inspire the development of MIRA-LFD assays for the detection of other human and animal viruses.
In conclusion, we developed a novel, specific, and sensitive MIRA-LFD assay targeting the viral N gene for the rapid and on-site detection of BCoVs, which can be applied for the diagnosis, surveillance, and control of BCoV infections in cattle and other animals as well as in humans.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
HY and ZP contributed to the design of the experimental work and revised the manuscript. CJ contributed to the experimental work and drafted the manuscript. YF, QG, and YZ performed the statistical analysis. JM and RS revised the manuscript. All authors reviewed the manuscript.
Funding
This study was supported by a grant from the Qingdao Lijian Diagnostic Technology Development Center (HMSY21001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Vlasova AN, Saif LJ. Bovine coronavirus and the associated diseases. Front Vet Sci. (2021) 8:643220. doi: 10.3389/fvets.2021.643220
2. Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB. Virus taxonomy: the database of the international committee on taxonomy of viruses (ICTV). Nucl Acids Res. (2018) 46:D708–17. doi: 10.1093/nar/gkx932
3. Tsunemitsu H, Saif LJ. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch Virol. (1995) 140:1303–11. doi: 10.1007/BF01322757
5. David D, Storm N, Ilan W, Sol A. Characterization of winter dysentery bovine coronavirus isolated from cattle in Israel. Viruses. (2021) 13:70. doi: 10.3390/v13061070
6. Lathrop SL, Wittum TE, Brock KV, Loerch SC, Perino LJ, Bingham HR, et al. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am J Vet Res. (2000) 61:1062–6. doi: 10.2460/ajvr.2000.61.1062
7. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract. (2010) 26:349–64. doi: 10.1016/j.cvfa.2010.04.005
8. Ellis J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can Vet J. (2019) 60:147–52.
9. Zhu Q, Li B, Sun D. Advances in bovine coronavirus epidemiology. Viruses. (2022) 14:109. doi: 10.3390/v14051109
10. Alfieri AA, Ribeiro J, de Carvalho BL, Lorenzetti E, Alfieri AF. Dairy calf rearing unit and infectious diseases: diarrhea outbreak by bovine coronavirus as a model for the dispersion of pathogenic microorganisms. Trop Anim Health Prod. (2018) 50:1937–40. doi: 10.1007/s11250-018-1592-9
11. Lotfollahzadeh S, Madadgar O, Reza MM, Reza MDM, George WD. Bovine coronavirus in neonatal calf diarrhoea in Iran. Vet Med Sci. (2020) 6:686–94. doi: 10.1002/vms3.277
12. Shin J, Tark D, Le VP, Choe S, Cha RM, Park GN, et al. Genetic characterization of bovine coronavirus in Vietnam. Virus Genes. (2019) 55:415–20. doi: 10.1007/s11262-019-01647-1
13. Munoz M, Alvarez M, Lanza I, Carmenes P. Role of enteric pathogens in the aetiology of neonatal diarrhoea in lambs and goat kids in Spain. Epidemiol Infect. (1996) 117:203–11. doi: 10.1017/S0950268800001321
14. Woo PC, Lau SK, Wernery U, Wong EY, Tsang AK, Johnson B, et al. Novel betacoronavirus in dromedaries of the Middle East, 2013. Emerg Infect Dis. (2014) 20:560–72. doi: 10.3201/eid2004.131769
15. Alekseev KP, Vlasova AN, Jung K, Hasoksuz M, Zhang X, Halpin R, et al. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol. (2008) 82:12422–31. doi: 10.1128/JVI.01586-08
16. Zhang XM, Herbst W, Kousoulas KG, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. (1994) 44:152–61. doi: 10.1002/jmv.1890440207
17. Athanassious R, Marsolais G, Assaf R, Dea S, Descoteaux JP, Dulude S, et al. Detection of bovine coronavirus and type A rotavirus in neonatal calf diarrhea and winter dysentery of cattle in Quebec: evaluation of three diagnostic methods. Can Vet J. (1994) 35:163–9.
18. Verbeek A, Dea S, Tijssen P. Detection of bovine enteric coronavirus in clinical specimens by hybridization with cDNA probes. Mol Cell Probes. (1990) 4:107–20. doi: 10.1016/0890-8508(90)90012-O
19. Heckert RA, Saif LJ, Myers GW. Development of protein A-gold immunoelectron microscopy for detection of bovine coronavirus in calves: comparison with ELISA and direct immunofluorescence of nasal epithelial cells. Vet Microbiol. (1989) 19:217–31. doi: 10.1016/0378-1135(89)90068-0
20. Sato K, Inaba Y, Tokuhisa S, Miura Y, Kaneko N, Asagi M, et al. Detection of bovine coronavirus in feces by reversed passive hemagglutination. Arch Virol. (1984) 80:23–31. doi: 10.1007/BF01315291
21. Singh S, Singh R, Singh KP, Singh V, Malik Y, Kamdi B, et al. Immunohistochemical and molecular detection of natural cases of bovine rotavirus and coronavirus infection causing enteritis in dairy calves. Microb Pathog. (2020) 138:103814. doi: 10.1016/j.micpath.2019.103814
22. Ven S, Arunvipas P, Lertwatcharasarakul P, Ratanapob N. Seroprevalence of bovine coronavirus and factors associated with the serological status in dairy cattle in the western region of Thailand. Vet World. (2021) 14:2041–7. doi: 10.14202/vetworld.2021.2041-2047
23. Thanthrige-Don N, Lung O, Furukawa-Stoffer T, Buchanan C, Joseph T, Godson DL, et al. A novel multiplex PCR-electronic microarray assay for rapid and simultaneous detection of bovine respiratory and enteric pathogens. J Virol Methods. (2018) 261:51–62. doi: 10.1016/j.jviromet.2018.08.010
24. Klein D, Kern A, Lapan G, Benetka V, Mostl K, Hassl A, et al. Evaluation of rapid assays for the detection of bovine coronavirus, rotavirus A and Cryptosporidium parvum in faecal samples of calves. Vet J. (2009) 182:484–6. doi: 10.1016/j.tvjl.2008.07.016
25. Symes SJ, Allen JL, Mansell PD, Woodward KL, Bailey KE, Gilkerson JR, et al. First detection of bovine noroviruses and detection of bovine coronavirus in Australian dairy cattle. Aust Vet J. (2018) 96:203–8. doi: 10.1111/avj.12695
26. Takiuchi E, Stipp DT, Alfieri AF, Alfieri AA. Improved detection of bovine coronavirus N gene in faeces of calves infected naturally by a semi-nested PCR assay and an internal control. J Virol Methods. (2006) 131:148–54. doi: 10.1016/j.jviromet.2005.08.005
27. Loa CC, Lin TL, Wu CC, Bryan TA, Hooper TA, Schrader DL. Differential detection of turkey coronavirus, infectious bronchitis virus, and bovine coronavirus by a multiplex polymerase chain reaction. J Virol Methods. (2006) 131:86–91. doi: 10.1016/j.jviromet.2005.07.007
28. Decaro N, Elia G, Campolo M, Desario C, Mari V, Radogna A, et al. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J Virol Methods. (2008) 151:167–71. doi: 10.1016/j.jviromet.2008.05.016
29. Tsuchiaka S, Masuda T, Sugimura S, Kobayashi S, Komatsu N, Nagai M, et al. Development of a novel detection system for microbes from bovine diarrhea by real-time PCR. J Vet Med Sci. (2016) 78:383–9. doi: 10.1292/jvms.15-0552
30. Li Q, Zhou QF, Xue CY, Ma JY, Zhu DZ, Cao YC. Rapid detection of porcine reproductive and respiratory syndrome virus by reverse transcription loop-mediated isothermal amplification assay. J Virol Methods. (2009) 155:55–60. doi: 10.1016/j.jviromet.2008.09.012
31. Xue G, Li S, Zhang W, Du B, Cui J, Yan C, et al. Reverse-transcription recombinase-aided amplification assay for rapid detection of the 2019 novel coronavirus (SARS-CoV-2). Anal Chem. (2020) 92:9699–705. doi: 10.1021/acs.analchem.0c01032
32. Oma VS, Klem T, Traven M, Alenius S, Gjerset B, Myrmel M, et al. Temporary carriage of bovine coronavirus and bovine respiratory syncytial virus by fomites and human nasal mucosa after exposure to infected calves. BMC Vet Res. (2018) 14:22. doi: 10.1186/s12917-018-1335-1
33. Qiao J, Meng Q, Cai X, Chen C, Zhang Z, Tian Z. Rapid detection of Betacoronavirus 1 from clinical fecal specimens by a novel reverse transcription loop-mediated isothermal amplification assay. J Vet Diagn Invest. (2012) 24:174–7. doi: 10.1177/1040638711425937
34. Amer HM, Almajhdi FM. Development of a SYBR green I based real-time RT-PCR assay for detection and quantification of bovine coronavirus. Mol Cell Probes. (2011) 25:101–7. doi: 10.1016/j.mcp.2011.03.001
35. Wang H, Zhang Y, Zhou J, Li M, Chen Y, Liu Y, et al. Rapid visual detection of hepatitis C virus using reverse transcription recombinase-aided amplification-lateral flow dipstick. Front Cell Infect Microbiol. (2022) 12:816238. doi: 10.3389/fcimb.2022.816238
36. Ghosh P, Chowdhury R, Hossain ME, Hossain F, Miah M, Rashid MU, et al. Evaluation of recombinase-based isothermal amplification assays for point-of-need detection of SARS-CoV-2 in resource-limited settings. Int J Infect Dis. (2022) 114:105–11. doi: 10.1016/j.ijid.2021.11.007
Keywords: bovine coronavirus, detection, multienzyme isothermal rapid amplification, lateral flow dipstick, diagnostic
Citation: Ji C, Feng Y, Sun R, Gu Q, Zhang Y, Ma J, Pan Z and Yao H (2023) Development of a multienzyme isothermal rapid amplification and lateral flow dipstick combination assay for bovine coronavirus detection. Front. Vet. Sci. 9:1059934. doi: 10.3389/fvets.2022.1059934
Received: 02 October 2022; Accepted: 08 December 2022;
Published: 04 January 2023.
Edited by:
Ji-Ming Chen, Foshan University, ChinaReviewed by:
Wenliang Li, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaKaicheng Wang, China Animal Health and Epidemiology Center, China
Copyright © 2023 Ji, Feng, Sun, Gu, Zhang, Ma, Pan and Yao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zihao Pan,  cGFuemloYW9AbmphdS5lZHUuY24=
cGFuemloYW9AbmphdS5lZHUuY24=
 Chengyuan Ji
Chengyuan Ji Yiqiu Feng
Yiqiu Feng Ruini Sun
Ruini Sun Qibing Gu
Qibing Gu Jiale Ma
Jiale Ma Zihao Pan
Zihao Pan Huochun Yao
Huochun Yao