
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Vet. Sci. , 13 December 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1059054
 Remous Ocloo1*
Remous Ocloo1* Justin Nyasinga2,3,4
Justin Nyasinga2,3,4 Zubair Munshi2
Zubair Munshi2 Aisha Hamdy5
Aisha Hamdy5 Tessa Marciniak6
Tessa Marciniak6 Manonmani Soundararajan6
Manonmani Soundararajan6 Mae Newton-Foot1,7
Mae Newton-Foot1,7 Wilma Ziebuhr6
Wilma Ziebuhr6 Adebayo Shittu8
Adebayo Shittu8 Gunturu Revathi2
Gunturu Revathi2 Alaa Abouelfetouh5,9
Alaa Abouelfetouh5,9 Andrew Whitelaw1,7
Andrew Whitelaw1,7Introduction: Staphylococci other than Staphylococcus aureus (SOSA) in animals are becoming more pathogenic and antibiotic resistant and can potentially disseminate to humans. However, there is little synthesized information regarding SOSA from animals in Africa. This systematic review provides a comprehensive overview of the epidemiology and antimicrobial resistance of SOSA in companion animals (pets) and livestock in Africa.
Method: This systematic review (PROSPERO-CRD42021252303) was conducted according to the PRISMA guidelines, and 75 eligible studies from 13 countries were identified until August 2022. Three electronic databases (Pubmed, Scopus and Web of Science) were employed.
Results: The frequently isolated SOSA were S. epidermidis, S. intermedius, S. pseudintermedius, S. xylosus, S. chromogenes, S. hyicus, M. sciuri, S. hominis, and S. haemolyticus. Thirty (40%) studies performed antibiotic susceptibility testing (AST). Penicillin (58%) and tetracycline (28%) resistance were most common across all SOSA with high rates of resistance to aminoglycosides, fluoroquinolones, and macrolides in some species. Resistance to last-resort antibiotics such as linezolid and fusidic acid were also reported. Limited data on strain typing and molecular resistance mechanisms precluded analysis of the clonal diversity of SOSA on the continent.
Conclusion: The findings of this review indicate that research on livestock-associated SOSA in Africa is lacking in some regions such as Central and Western Africa, furthermore, research on companion animals and more advanced methods for identification and strain typing of SOSA need to be encouraged.
Systematic review registration: https://www.crd.york.ac.uk/prospero/, identifier: CRD42021252303.
Staphylococci colonize the skin and mucous membranes of a wide range of vertebrate hosts and account for several human and animal diseases (1–4). Staphylococci other than Staphylococcus aureus (SOSA) is a relatively new term, encompassing largely coagulase-negative staphylococci, including two coagulase-positive staphylococcal species (S. pseudintermedius and S. schleiferi) (2). There are increasing reports of SOSA infections associated with implantable foreign bodies and sepsis, particularly among newborns and preterm neonates (5). Moreover, some SOSA (e.g., S. pseudintermedius, S. schleiferi, and S. felis) are important pathogens in veterinary medicine (6). SOSA infections are a global economic problem in the animal production sector (7–11). SOSA are largely responsible for mastitis in milk-producing animals, leading to reduced milk production (12).
Antimicrobials are administered to animals to enhance growth and prevent infection. However, the doses of antimicrobials dispensed by practitioners in animal husbandry are mostly sub-therapeutic and contribute to the emergence of antimicrobial resistance (AMR) (13–15). The spread of AMR genes may be exacerbated by trading with animals between farms and other animal husbandry practices (16, 17).
High resistance rates have been described in SOSA from animals. Tetracycline resistance has been described in SOSA isolates from normal and subclinical mastitic buffalo milk in Egypt: 100% in S. lugdunensis and S. hominis, and 66.6% in S. epidermidis (18). SOSA isolates recovered from turkey farms in Egypt were also resistant to tetracycline (100%), oxacillin (92.3%) and daptomycin (89.7%) (19). In Turkey, 75% of SOSA recovered from raw milk were resistant to erythromycin (20) and in South Africa, 51% of SOSA isolates from subclinical mastitis cow milk samples were multidrug resistant (MDR) (21). Antimicrobial-resistant SOSA (AMRSOSA) in livestock and companion animals pose an economic threat to the livestock industry and a risk for spill-over into the human environment, threatening public health (22–24).
SOSA are considered less pathogenic than S. aureus and this perception has led to limited knowledge on the virulence, prevalence, and dissemination of SOSA, particularly in animals (25, 26). However, in recent years, resistance to last-resort antibiotics has been described in SOSA (19, 27, 28). There is also evidence that SOSA and S. aureus exchange virulence and resistance genes continuously in the natural environment, which is a potential risk to the empiric treatment of S. aureus infections (29–33). For example, Mammaliicoccus sciuri, formerly known as Staphylococcus sciuri, is postulated to be the origin of mecA and the precursor to the evolution of the staphylococcal cassette chromosome mec (SCCmec) element (34, 35). The methicillin resistance gene (mecA) encodes an alternative penicillin-binding protein (PBP2A) (36, 37), which confers resistance to the β-lactam class of antibiotics such as penicillins and cephalosporins.
Systematic reviews on S. aureus in animals have observed that both animal clonal complex (CC) 398, CC130, CC133 and human-associated lineages (CC1, CC15, CC72, CC80, CC10, and CC152) are common in Africa. Methicillin resistant S. aureus (MRSA) prevalence ranges from 0 to 3 % (38, 39). However, the epidemiology of SOSA in companion animals and livestock in Africa is not well-described, despite the potential for transfer of staphylococci and their resistance mechanisms from these animals to humans. This study aims to provide a comprehensive overview of the published data describing the epidemiology and antimicrobial resistance of SOSA in companion animals and livestock in Africa.
This systematic review was registered on https://www.crd.york.ac.uk/prospero/ (PROSPERO-CRD42021252303) and conducted following the preferred reporting items for systematic reviews and meta-analyses (PRISMA) guidelines (40).
A literature search of relevant articles in PubMed, Scopus, and Web of Science, published from 1991 to 2022, was conducted and retrieved until August 2022 using the search terms described in Supplementary material 1. The results were transferred onto Microsoft Excel, and duplicates were removed. Titles and abstracts were screened, and full-text articles were assessed for eligibility. All searches and screenings were done by two independent reviewers each, and discrepancies were resolved by consensus.
Eligible studies included reports on SOSA isolated from companion animals and livestock in Africa and published in peer-reviewed English journals. Other criteria for inclusion were descriptions of population size and laboratory methods. Mammaliicoccus sciuri was included in this study as it was previously regarded as S. sciuri.
Studies not published in English journals, published in predatory journals (according to Beall's list, 2021), which did not report on primary data, and those which reported on S. aureus only or did not perform speciation of SOSA were excluded. Review articles, notes, e-mails, editorials, articles without original data and studies that only described animal products and wild animals were also excluded.
The following data were extracted onto Microsoft Excel: first author, year of publication, study design, country/region, type of animals, number of animals, type of samples, number of samples, and year of sample collection. Other information included species isolated, number of isolates, susceptibility data for various classes of antibiotics (fluoroquinolones, cephalosporins, penicillins, aminoglycosides, macrolides, and polymyxins), resistance genes, laboratory methods, and strain types. Data extraction was performed by two independent reviewers and discrepancies were resolved by consensus. The data were stratified using the United Nations geoscheme subregion classification (https://unstats.un.org/unsd/methodology/m49/).
Antibiotic resistance rates were only reported for a species when a minimum of 30 isolates were tested (41). Antibiotic resistance rates for each species were calculated using the number of resistant isolates (NR) and total number of isolates tested (NT).
Differences between antibiotic rates in carriage and pathogenic SOSA were calculated using Fisher exact and Chi squared tests where appropriate.
The systematic search of the three databases yielded 9,160 articles. After de-duplication and exclusion by title and abstract screening, 395 full-text articles were screened, of which 75 were considered eligible based on our inclusion criteria (Figure 1). The majority of the studies (97%; n = 73), were published between 2000 and 2022, with 69% of the studies (n = 52) published between 2012 and 2022. According to regions, Northern Africa (n = 28) had the highest number of eligible studies, followed by Eastern Africa (n = 26), Western Africa (n = 11), and Southern Africa (n = 10) (Figure 2). No articles were available from Central Africa.
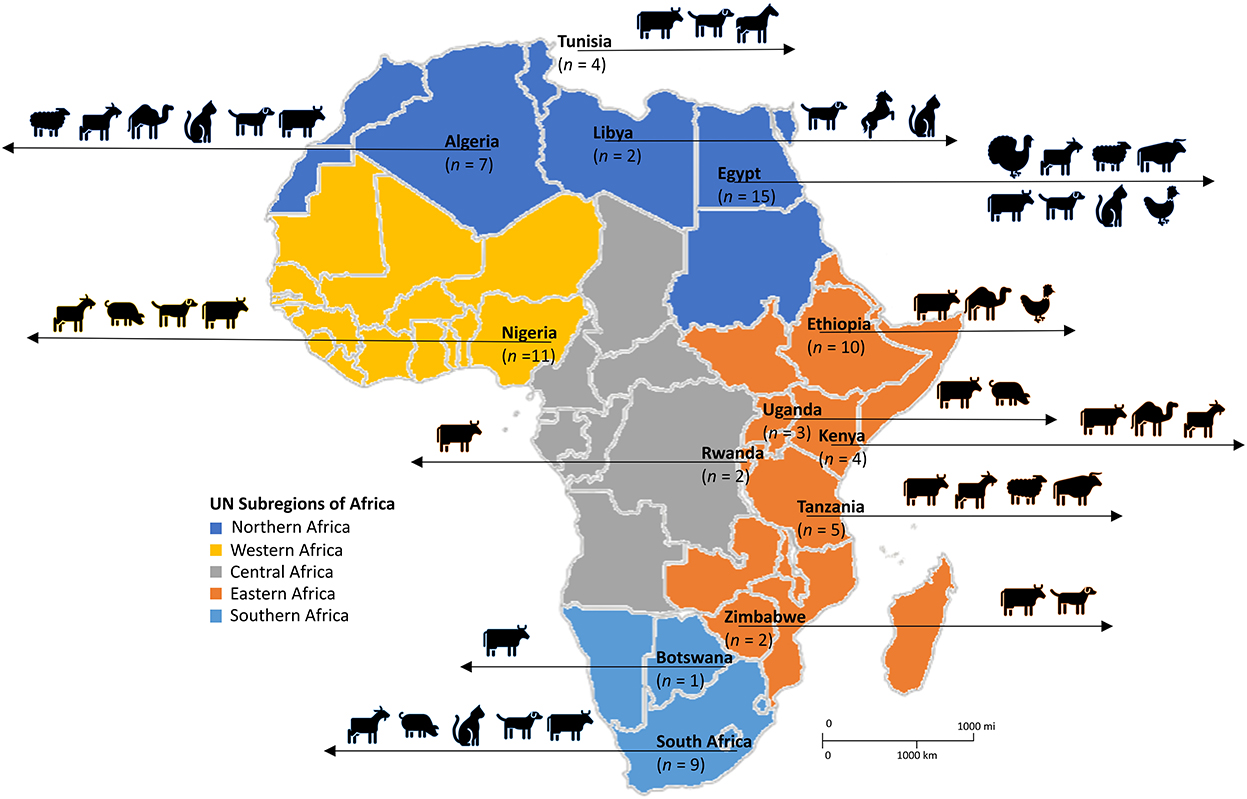
Figure 2. Diversity of livestock and domestic animals in which SOSA was described in Africa. Adapted from the United Nations geoscheme subregions African Map. n, number of articles.
Of the 75 eligible studies, 62 described SOSA in livestock, 12 in companion animals, and one included both livestock and companion animal. Of the reports from livestock, 17 were performed on subsistence farms, nine on commercial farms, and the rest (n = 49) did not provide information on farm type. Overall, 46 reports were from diseased animals, 17 from healthy animals, and ten from diseased and healthy animals; while 2 did not provide information. Dogs were the most common companion animal investigated, in 12 studies, while cows and goats were the main livestock investigated, in 32 and 13 studies, respectively. Figure 2 represents the diversity of animals in which SOSA were described in Africa. Milk was the leading sample type screened, representing 65% (n = 49) of the eligible studies. Most SOSA were isolated from cows with mastitis (Table 1).
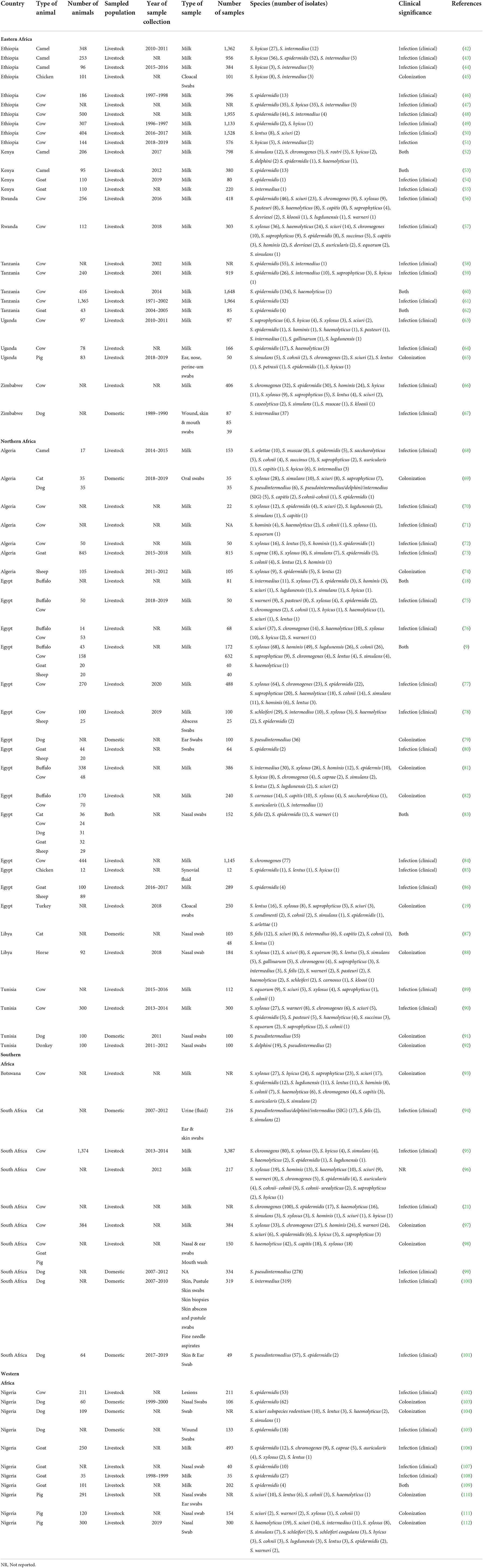
Table 1. Sources, clinical significance & geographical distribution of Staphylococci other than S. aureus (SOSA) in Africa.
The frequently isolated SOSA in Africa were S. epidermidis (23%; n = 784), S. intermedius (14%; n = 446), S. pseudintermedius (11%; n = 371), S. xylosus (10%; n = 346), S. chromogenes (9.0%; n = 303), S. hyicus (4.7%; n = 157), M. sciuri (4.5%; n = 151), S. hominis (3.9%; n = 130), and S. haemolyticus (3.7%; n = 123) (Figure 3). S. xylosus was most frequently reported in Northern Africa, S. intermedius/S. pseudintermedius in Southern Africa, while S. epidermidis was predominant in Eastern and Western Africa (Table 1). API-Staph and BD Phoenix were the common biochemical tests employed to identify SOSA, in 64% (n = 48) studies (Supplementary material 2).
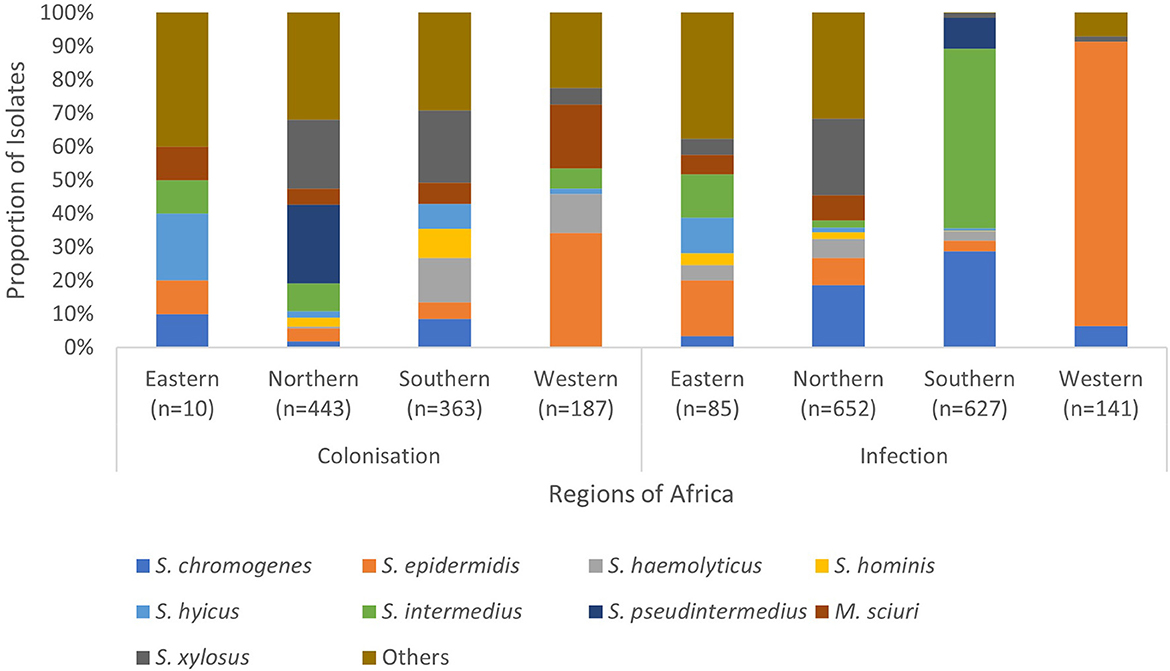
Figure 3. Regional distribution of SOSA in Africa. SOSA isolates were classified as “Others” when the total number included in the review was <100. Isolates (n = 177) in which colonization and infection were not distinguished were excluded. Eastern Africa (Ethiopia, Kenya, Rwanda, Tanzania, Uganda & Zimbabwe), Northern Africa (Algeria, Egypt, Libya & Tunisia), Southern Africa (Botswana & South Africa), Western Africa (Nigeria).
Thirty (40%) studies identified SOSA to the species level and performed antibiotic susceptibility testing (AST), of these only five (13%) were published prior to 2012. The Kirby Bauer disc diffusion method was widely utilized for AST (87%; n = 26) and the zone of inhibition was mostly interpreted using Clinical and Laboratory Standards Institute (CLSI) guidelines (81%; n = 21) (Supplementary material 2a).
Generally, data on methicillin resistance in SOSA was lacking. However, methicillin resistance rates were high among S. pseudintermedius (89%; n = 83) and M. sciuri (54%; n = 37), S. xylosus (45%; n = 37), and S. epidermidis (44%; n = 14) isolates. The typical human-associated SOSA showed high rates of penicillin resistance, S. epidermidis (64%), S. haemolyticus (63%), and S. hominis (96%), however, there was less resistance to glycopeptides and rifampicin. High rates of fluoroquinolone resistance were also observed in S. epidermidis (69%), S. hyicus (56%), S. xylosus (42%), and M. sciuri (36%). Aminoglycoside resistance was high in S. xylosus (31%), M. sciuri (36%), and S. hominis (33%) compared to the other SOSA. Lincosamide resistance was also high in M. sciuri (54%) and S. xylosus (31%) and M. sciuri demonstrated high rates of macrolide resistance (56%; n = 32). Overall, in SOSA, resistance to penicillin (58%) and tetracycline (28%) were most common (Supplementary material 2b). A few studies (n = 5) reported on susceptibility to last-resort antibiotics. Linezolid resistance was described in S. intermedius (n = 5), S. xylosus (n = 3), S. equorum (n = 1) and S. epidermidis (n =1) and resistance to fusidic acid was observed in S. pseudintermedius (n = 4), M. sciuri (n = 5), S. xylosus (n = 3), and S. hominis (n = 1) (Supplementary material 3). Figure 4 shows the rate of antibiotic resistance in SOSA species commonly encountered in the clinical setting. However, we were unable to stratify data geographically due to insufficient data from other regions.
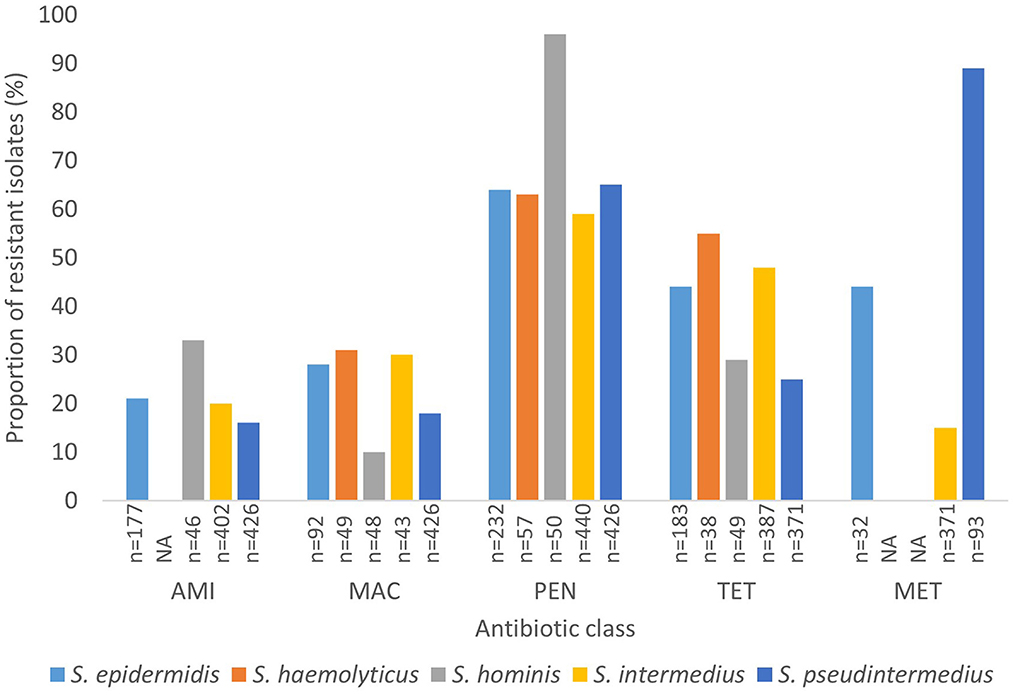
Figure 4. Rate of antibiotic resistance of clinically significant SOSA. AMI, Aminoglycoside; MAC, Macrolide; PEN, Penicillin; TET, Tetracycline; MET, Methicillin. “n” Denotes the number of isolates tested against a particular antibiotic. “NA” denotes not applicable.
Higher rates of aminoglycoside, fluoroquinolone, lincosamide, macrolide, rifampicin and methicillin resistance have been seen in carriage SOSA compared to pathogenic SOSA (Supplementary material 4).
Fifteen (20%) studies performed molecular screening for AMR genes and reported the presence of the mecA gene in at least one member of the SOSA. The mecA was detected in S. pseudintermedius (n = 49), S. sciuri (n = 31), S. intermedius (n = 27), S. xylosus (n = 23), S. epidermidis (n = 12), S. schleiferi (n = 10), S. haemolyticus (n = 8), S. hominis (n = 3), S. hyicus (n = 2), and S. chromogenes (n = 1) (Supplementary material 5). Only three studies reported on SCCmec typing: SCCmec type V was noted in S. warneri, and SCCmec types I and IVa were detected in S. epidermidis. Only one study reported on sequence types (STs), diverse STs were described in S. delphini and S. pseudintermedius (Supplementary material 6).
Diverse SOSA were recovered from clinical and non-clinical samples from livestock and companion animals in Africa. The most common species include S. epidermidis, S. pseudintermedius, S. xylosus, and S. chromogenes. S. pseudintermedius was most commonly isolated in pets and S. epidermidis in livestock. However, there is lack of data on certain species in some countries such Botswana, Nigeria and Libya. We recommend that studies focus more on the following SOSA organisms which can also help understand the burden of SOSA infections in animals in Africa: S. intermedius, S. xylosus and S. chromogenes. S. pseudintermedius is considered as typical zoonotic commensal organism in the ears and skin of dogs and birds (79, 91). It is responsible for skin and soft tissue infections in dogs (67, 100), however in humans it is predominantly associated with infected dog bites and bacteraemia (113, 114). Surprisingly, none of the dogs from Western Africa (Nigeria) harbored S. pseudintermedius (104, 105, 108). S. xylosus is ubiquitous bacterium which is naturally occurring in the soil, food and on surfaces (115–117). Some strains are associated with opportunistic infections in humans (118, 119). There were no livestock associated infections/colonisations due to S. xylosus reported in Botswana, Ethiopia, Libya or Tanzania (42, 46, 58–62, 87, 93). S. chromogenes is a major cause of mastitis and subclinical mastitis in animals as seen in this review (21, 56, 57, 66, 75, 76, 84, 90, 106). However, no S. chromogenes infections were recorded in Botswana, Algeria, Ethiopia, Libya and Tanzania (42, 46, 58–62, 64, 70, 73, 87, 93, 120). The strain distribution could not be described due to the insufficient published data, but we were able to describe epidemiology of SOSA in terms of species distribution.
We observed that about 50% of the eligible studies utilized semi-automated methods (API-Staph and BD Phoenix) based on biochemical reactions. However, speciation of SOSA through these methods is unreliable as various species exhibit similar characteristics, hence molecular and spectrometric methods such as polymerase chain reaction (PCR) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) have been advocated (121, 122). Investigators pay little attention to SOSA as they are often considered contaminants, leading to the lack of speciation and strain typing. Therefore, there is sparse genotyping data on members of the SOSA, and it is difficult to understand the clonal diversity of SOSA on the continent and we could not describe clonal relationships with human-associated isolates.
The use of antibiotics in animal production and its consequent impact on AMR is a major challenge globally, particularly in Africa (15, 123, 124). We observed high rates of penicillin (58%) and tetracycline (28%) resistance in SOSA generally. The high rates (≥50%) of lincosamide, macrolide, and methicillin resistance observed in M. sciuri are note-worthy. Genes mediating resistance to these antibiotics could be transferred via mobile genetic elements to pathogenic bacteria such as S. aureus, as has previously been described for the SCCmec element (125, 126). High resistance to aminoglycosides, macrolides, tetracyclines, and methicillin was also observed in SOSA species (15–89%) which are commonly encountered in human medicine, such as S. epidermidis, S. haemolyticus, S. hominis, S. intermedius, and S. pseudintermedius, which might be due to the overuse of clinical antibiotics, particularly penicillin and tetracycline, in veterinary medicine (15). This could result in empiric treatment failures in clinical settings since these antibiotics are commonly used in human medicine. This again highlights the need for policymakers to enforce regulations on the use of antibiotics in animal husbandry in Africa.
Resistance to last resort antibiotics such as linezolid was also reported. This raises the risk of antibiotic resistance transfer to commensal S. aureus in animals, in which low rates of resistance to methicillin and other antibiotics have generally been observed in Africa (0–3%) (39). Interactions at the human-animal interface also raise the risk of antibiotic resistance transfer to humans. Studies have demonstrated that SOSA are becoming more resistant than S. aureus in humans (127, 128), although rates of methicillin resistance across Africa vary widely, from 12 to over 80% (129). The lack of methicillin resistance detection seen in this review might be due to the challenges associated with using cefoxitin and oxacillin for methicillin resistance screening in SOSA as discussed by Yang et al. (130) and Humphries et al. (2). Although, mecA or PBP2a PCR detection is the gold standard for assessment of methicillin resistance in staphylococci, PBP2a phenotypic testing may be an efficient, labor- and cost-saving approach (130, 131). The uneven distribution of studies has made it difficult to compare antibiotic resistant rates across the different regions. Furthermore, the high rates of antibiotic resistance seen in carriage SOSA can be attributed to the small sample size and the differences in species distribution but needs to be investigated further.
The findings from this review revealed that the epidemiology of SOSA is described mainly in cows, with insufficient data on companion animals such as dogs and cats in Africa. The very heterogeneous nature of the livestock and companion animals sampled in different regions does make it difficult to draw firm conclusions about the geographic distribution of different species and AMR rates among SOSA. We suggest that other livestock and companion animals should be investigated in future studies to better understand the problem of AMR in animals on the continent. Studies in Eastern Africa are more focused on infected livestock. Ethiopia particularly is home to Africa's largest livestock population and human interaction with healthy livestock is more frequent (132). We recommend that research in this region focuses on healthy animals to help address the problem of zoonotic transmissions. There is also a lack of data particularly in Central and Western Africa. It is important that research in these regions of Africa is encouraged to help understand the burden of AMR in animals in Africa. Few studies performed strain typing and molecular screening of antibiotic resistance genes. With the decrease in the cost of next-generation sequencing, researchers in Africa should consider employing whole genome sequencing (WGS), which can also provide additional data such as resistance, virulence, pathogenicity, and genetic composition of the organism (133).
Many studies did not indicate the sample collection period, and there was inadequate longitudinal data, describing SOSA or AMR in specific countries or regions; for example, from West Africa only Nigeria reported on SOSA in animals. Therefore, it was not possible to stratify data which is a limitation of the review. However, findings from this review shows that SOSA species are becoming more resistant to antibiotics, and this requires immediate attention.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author/s.
RO contributed to the conceptualization and design of the systematic review, performed literature searches, data extraction and synthesis, data interpretation, and wrote the manuscript. JN, ZM, AH, TM, and MS performed literature searches, data extraction and synthesis, and provided critical feedback on the manuscript. MN-F, AW, WZ, GR, AA, and AS contributed to the conceptualization and design of the systematic review and provided critical feedback on the manuscript. All authors contributed to the article and approved the submitted version.
This study was supported by the German Research Council (DFG) through grant ZI665/3-1 and the German Federal Ministry of Education and Research (BMBF) through grant number 01KI1727E.
Thanks to Dinah Seligsohn, National Veterinary Institute, Sweden for providing us additional data on speciation of SOSA from her publication (52).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1059054/full#supplementary-material
1. Shoen HRC, Rose SJ, Ramsey SA, de Morais H, Bermudez LE. Analysis of Staphylococcus infections in a veterinary teaching hospital from 2012 to 2015. Comp Immunol Microbiol Infect Dis. (2019) 66:101332. doi: 10.1016/j.cimid.2019.101332
2. Humphries RM, Magnano P, Burnham CAD, Bard JD, Dingle TC, Callan K, et al. Evaluation of surrogate tests for the presence of mecA- mediated methicillin resistance in Staphylococcus capitis, Staphylococcus haemolyticus, Staphylococcus hominis, and Staphylococcus warneri. J Clin Microbiol. (2021) 59:1–11. doi: 10.1128/JCM.02290-20
3. Marincola G, Liong O, Schoen C, Abouelfetouh A, Hamdy A, Wencker FDR, et al. Antimicrobial resistance profiles of coagulase-negative Staphylococci in community-based healthy individuals in Germany. Front public Heal. (2021) 9:684456. doi: 10.3389/fpubh.2021.684456
4. Ma GC, Worthing KA, Ward MP, Norris JM. Commensal Staphylococci including methicillin-resistant Staphylococcus aureus from dogs and cats in remote New South Wales, Australia. Microb Ecol. (2020) 79:164–74. doi: 10.1007/s00248-019-01382-y
5. Michels R, Last K, Becker SL, Papan C. Update on coagulase-negative staphylococci—what the clinician should know. Microorganisms. (2021) 9:830. doi: 10.3390/microorganisms9040830
6. Thomson P, García P, Miles J, Isla D, Yáñez C, Santibáñez R, et al. Isolation and identification of Staphylococcus species obtained from healthy companion animals and humans. Vet Sci. (2022) 9:79. doi: 10.3390/vetsci9020079
7. Huijps K, Lam TJGM, Hogeveen H. Costs of mastitis: facts and perception. J Dairy Res. (2008) 75:113–20. doi: 10.1017/S0022029907002932
8. Stepień-Pyśniak D, Wilczyński J, Marek A, Smiech A, Kosikowska U, Hauschild T. Staphylococcus simulans associated with endocarditis in broiler chickens. Avian Pathol. (2017) 46:44–51. doi: 10.1080/03079457.2016.1203392
9. El-Jakee JK, Aref NE, Gomaa A, El-Hariri MD, Galal HM, Omar SA, et al. Emerging of coagulase negative staphylococci as a cause of mastitis in dairy animals: an environmental hazard. Int J Vet Sci Med. (2013) 1:74–8. doi: 10.1016/j.ijvsm.2013.05.006
10. Bennett RM, Christiansen K, Clifton-Hadley RS. Estimating the costs associated with endemic diseases of dairy cattle. J Dairy Res. (1999) 66:455–9. doi: 10.1017/S0022029999003684
11. Romero J, Benavides E, Meza C. Assessing financial impacts of subclinical mastitis on Colombian Dairy Farms. Front Vet Sci. (2018) 5:273. doi: 10.3389/fvets.2018.00273
12. Schepers JA, Dijkhuizen AA. The economics of mastitis and mastitis control in dairy cattle: a critical analysis of estimates published since 1970. Prev Vet Med. (1991) 10:213–24. doi: 10.1016/0167-5877(91)90005-M
13. Manyi-Loh C, Mamphweli S, Meyer E, Okoh A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules. (2018) 23:795. doi: 10.3390/molecules23040795
14. Lekagul A, Tangcharoensathien V, Liverani M, Mills A, Rushton J, Yeung S. Understanding antibiotic use for pig farming in Thailand: a qualitative study. Antimicrob Resist Infect Control. (2021) 10:1–11. doi: 10.1186/s13756-020-00865-9
15. Van TTH, Yidana Z, Smooker PM, Coloe PJ. Antibiotic use in food animals worldwide, with a focus on Africa: pluses and minuses. J Glob Antimicrob Resist. (2020) 20:170–7. doi: 10.1016/j.jgar.2019.07.031
16. Founou LL, Founou RC, Allam M, Ismail A, Finyom Djoko C, Essack SY. Genome analysis of methicillin-resistant Staphylococcus aureus isolated from pigs: detection of the clonal lineage ST398 in Cameroon and South Africa. Zoonoses Public Health. (2019) 66:512–25. doi: 10.1111/zph.12586
17. Park S, Ronholm J. Staphylococcus aureus in agriculture: lessons in evolution from a multispecies pathogen. Clin Microbiol Rev. (2021) 34:1–27. doi: 10.1128/CMR.00182-20
18. El-Razik KAA, Arafa AA, Hedia RH, Ibrahim ES. Tetracycline resistance phenotypes and genotypes of coagulase-negative staphylococcal isolates from bubaline mastitis in Egypt. Vet World. (2017) 10:702–10. doi: 10.14202/vetworld.2017.702-710
19. Moawad AA, Hotzel H, Awad O, Roesler U, Hafez HM, Tomaso H, et al. Evolution of antibiotic resistance of coagulase-negative staphylococci isolated from healthy Turkeys in Egypt: first report of linezolid resistance. Microorganisms. (2019) 7:476. doi: 10.3390/microorganisms7100476
20. Eker E, Yildirim Y. Antibiotic resistance of coagulase negative Staphylococci isolated from buffalo milk and some milk products. Kocatepe Vet J. (2010) 3:7–12. Available online at: https://dergipark.org.tr/en/download/article-file/108810 (accessed August 22, 2021).
21. Phophi L, Petzer IM, Qekwana DN. Antimicrobial resistance patterns and biofilm formation of coagulase-negative Staphylococcus species isolated from subclinical mastitis cow milk samples submitted to the Onderstepoort Milk Laboratory. BMC Vet Res. (2019) 15:420. doi: 10.1186/s12917-019-2175-3
22. Woolhouse M, Ward M, Van Bunnik B, Farrar J. Antimicrobial resistance in humans, livestock and the wider environment. Philos Trans R Soc B Biol Sci. (2015) 370:20140083. doi: 10.1098/rstb.2014.0083
23. Hosain MZ, Lutful Kabir SM, Kamal MM. Antimicrobial uses for livestock production in developing countries. Vet World. (2021) 14:210–21. doi: 10.14202/vetworld.2021.210-221
24. Ikhimiukor OO, Odih EE, Donado-Godoy P, Okeke IN. A bottom-up view of antimicrobial resistance transmission in developing countries. Nat Microbiol. (2022) 7:757–65. doi: 10.1038/s41564-022-01124-w
25. Hischebeth GT, Randau TM, Ploeger MM, Friedrich MJ, Kaup E, Jacobs C, et al. Staphylococcus aureus versus Staphylococcus epidermidis in periprosthetic joint infection—outcome analysis of methicillin-resistant versus methicillin-susceptible strains. Diagn Microbiol Infect Dis. (2019) 93:125–30. doi: 10.1016/j.diagmicrobio.2018.08.012
26. Bonvegna M, Grego E, Sona B, Stella MC, Nebbia P, Mannelli A, et al. Occurrence of Methicillin-Resistant Coagulase-Negative Staphylococci (MRCoNS) and Methicillin-Resistant Staphylococcus aureus (MRSA) from pigs and farm environment in Northwestern Italy. Antibiotics. (2021) 10:676. doi: 10.3390/antibiotics10060676
27. Kosecka-Strojek M, Sadowy E, Gawryszewska I, Klepacka J, Tomasik T, Michalik M, et al. Emergence of linezolid-resistant Staphylococcus epidermidis in the tertiary children's hospital in Cracow, Poland. Eur J Clin Microbiol Infect Dis. (2020) 39:1717–25. doi: 10.1007/s10096-020-03893-w
28. Gupta V, Garg S, Jain R, Garg S, Chander J. Linezolid resistant Staphylococcus haemolyticus: first case report from India. Asian Pac J Trop Med. (2012) 5:837–8. doi: 10.1016/S1995-7645(12)60155-2
29. França A, Gaio V, Lopes N, Melo LDR. Virulence factors in coagulase-negative staphylococci. Pathogens. (2021) 10:1–46. doi: 10.3390/pathogens10020170
30. Haaber J, Penadés JR, Ingmer H. Transfer of antibiotic resistance in Staphylococcus aureus. Trends Microbiol. (2017) 25:893–905. doi: 10.1016/j.tim.2017.05.011
31. Othman AA, Hiblu MA, Abbassi MS, Abouzeed YM, Ahmed MO. Nasal colonization and antibiotic resistance patterns of staphylococcus species isolated from healthy horses in tripoli, libya. J Equine Sci. (2021) 32:61–5. doi: 10.1294/jes.32.61
32. Phumthanakorn N, Prapasarakul N, Yindee J, Gronsang D. Frequency, distribution, and antimicrobial resistance of coagulase-negative staphylococci isolated from clinical samples in dogs and cats. Microb Drug Resist. (2022) 28:236–43. doi: 10.1089/mdr.2020.0586
33. Suwito W, Nugroho WS, Wahyuni AETH, Sumiarto B. Antimicrobial resistance in coagulase-negative staphylococci isolated from subclinical mastitis in Ettawa crossbred goat (Pe) in Yogyakarta, Indonesia. Biodiversitas. (2021) 22:3418–22. doi: 10.13057/biodiv/d220650
34. Miragaia M. Factors contributing to the evolution of Meca-mediated β-lactam resistance in staphylococci: update and new insights from whole genome sequencing (WGS). Front Microbiol. (2018) 9:2723. doi: 10.3389/fmicb.2018.02723
35. Madhaiyan M, Wirth JS, Saravanan VS. Phylogenomic analyses of the staphylococcaceae family suggest the reclassification of five species within the genus staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five staphylococcu. Int J Syst Evol Microbiol. (2020) 70:5926–36. doi: 10.1099/ijsem.0.004498
36. Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother. (2010) 54:4352–9. doi: 10.1128/AAC.00356-10
37. Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. (2001) 9:486–93. doi: 10.1016/S0966-842X(01)02175-8
38. Samutela MT, Kwenda G, Simulundu E, Nkhoma P, Higashi H, Frey A, et al. Pigs as a potential source of emerging livestock-associated Staphylococcus aureus in Africa: a systematic review. Int J Infect Dis. (2021) 109:38–49. doi: 10.1016/j.ijid.2021.06.023
39. Lozano C, Gharsa H, Ben Slama K, Zarazaga M, Torres C. Staphylococcus aureus in animals and food: methicillin resistance, prevalence and population structure. A review in the African continent. Microorganisms. (2016) 4:12. doi: 10.3390/microorganisms4010012
40. Tawfik GM, Dila KAS, Mohamed MYF, Tam DNH, Kien ND, Ahmed AM, et al. A step by step guide for conducting a systematic review and meta-analysis with simulation data. Trop Med Health. (2019) 47:1–9. doi: 10.1186/s41182-019-0165-6
41. CLSI. CLSI M100-ED29: 2021 Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. Pittsburgh (2020). p. 50–1.
42. Regassa A, Golicha G, Tesfaye D, Abunna F, Megersa B. Prevalence, risk factors, and major bacterial causes of camel mastitis in Borana Zone, Oromia Regional State, Ethiopia. Trop Anim Health Prod. (2013) 45:1589–95. doi: 10.1007/s11250-013-0403-6
43. Abel Gadir AE, Hildebrandt G, Kleer J, Molla, Kyule MB. Prevalence and risk factors of camel (Camelus dromedarius) mastitis based on bacteriological examinations in selected regions of Ethiopia. (2005) 12:33–6. Available online at: https://www.researchgate.net/publication/283052883_Prevalence_and_risk_factors_of_camel (accessed August 22, 2021).
44. Alebie A, Molla A, Adugna W, Tesfaye A, Ejo M. Prevalence, isolation, identification, and risk factors of major bacterial cause of camel subclinical mastitis. Biomed Res Int. (2021) 2021:5522331. doi: 10.1155/2021/5522331
45. Abunna F, Adugna B, Tufa TB, Ayana D, Gutema FD, Waktole H, et al. Detection and antimicrobial resistance of Staphylococcus species from chicken, chicken litter, and humans in Addis Ababa, Ethiopia. Vet Med Int. (2022) 2022:1–8. doi: 10.1155/2022/9084334
46. Workineh S, Bayleyegn M, Mekonnen H, Potgieter LND. Prevalence and aetiology of Mastitis in cows from two major Ethiopian dairies. Trop Anim Health Prod. (2002) 34:19–25. doi: 10.1023/A:1013729626377
47. Abdel Gadir AE, Hildebrandt G, Kleer JBBM. Susceptibility of bacteria isolated from camel (Camelus dromedarius) mastitis to commonly used antimicrobials. (2005) 12:33–6. Available online at: https://www.researchgate.net/publication/283052883_Prevalence_and_risk_factors_of_camel (accessed August 22, 2021).
48. Getahun K, Kelay B, Bekana M, Lobago F. Bovine mastitis and antibiotic resistance patterns in Selalle smallholder dairy farms, central Ethiopia. Trop Anim Health Prod. (2008) 40:261–8. doi: 10.1007/s11250-007-9090-5
49. Kerro Dego O, Tareke F. Bovine mastitis in selected areas of southern Ethiopia. Trop Anim Health Prod. (2003) 35:197–205. doi: 10.1023/a:1023352811751
50. Dabele DT, Borena BM, Admasu P, Gebremedhin EZ, Marami LM. Prevalence and risk factors of mastitis and isolation, identification and antibiogram of staphylococcus species from mastitis positive zebu cows in toke kutaye, cheliya, and dendi districts, west shewa zone, oromia, ethiopia. Infect Drug Resist. (2021) 14:987–98. doi: 10.2147/IDR.S295257
51. Fesseha H, Mathewos M, Aliye S, Wolde A. Study on prevalence of bovine mastitis and associated risk factors in dairy farms of Modjo Town and Suburbs, Central Oromia, Ethiopia. Vet Med Res Rep. (2021) 12:271–83. doi: 10.2147/VMRR.S323460
52. Seligsohn D, Nyman AK, Younan M, Sake W, Persson Y, Bornstein S, et al. Subclinical mastitis in pastoralist dairy camel herds in Isiolo, Kenya: prevalence, risk factors, and antimicrobial susceptibility. J Dairy Sci. (2020) 103:4717–31. doi: 10.3168/jds.2019-17701
53. Toroitich KC, Gitau GK, Kitala PM, Gitao GC. The Prevalence and Causes of Mastitis in Lactating Traditionally Managed One-Humped Camels (Camelus dromedarius) in West Pokot County, Kenya. Livestock Research for Rural Development (2015) 1–13. Available online at: http://www.lrrd.org/lrrd29/4/gita29062.html (accessed August 22, 2021).
54. Okoko IM, Maina N, Kiboi D, Kagira J. β-lactam resistance in bacteria associated with subclinical mastitis in goats in Thika Subcounty, Kenya. Vet World. (2020) 13:1448–56. doi: 10.14202/vetworld.2020.1448-1456
55. Mahlangu P, Maina N, Kagira J. Prevalence, risk factors, and antibiogram of bacteria isolated from milk of goats with subclinical mastitis in Thika East Subcounty, Kenya. J Vet Med. (2018) 2018:1–8. doi: 10.1155/2018/3801479
56. Ndahetuye JB, Persson Y, Nyman AK, Tukei M, Ongol MP, Båge R. Aetiology and prevalence of subclinical mastitis in dairy herds in peri-urban areas of Kigali in Rwanda. Trop Anim Health Prod. (2019) 51:2037–44. doi: 10.1007/s11250-019-01905-2
57. Antók FI, Mayrhofer R, Marbach H, Masengesho JC, Keinprecht H, Nyirimbuga V, et al. Characterization of antibiotic and biocide resistance genes and virulence factors of staphylococcus species associated with bovine mastitis in Rwanda. Antibiotics. (2020) 9:1. doi: 10.3390/antibiotics9010001
58. Mdegela RH, Karimuribo E, Kusiluka LJM, Kabula B, Manjurano A, Kapaga AM, et al. Mastitis in smallholder dairy and pastoral cattle herds in the urban and peri-urban areas of the Dodoma municipality in Central Tanzania. Livest Res Rural Dev. (2005) 17:1–9. Available online at: http://www.lrrd.org/lrrd17/11/mdeg17123.htm (accessed August 22, 2021).
59. Mdegela RH, Kusiluka LJM, Kapaga AM, Karimuribo ED, Turuka FM, Bundala A, et al. Prevalence and determinants of mastitis and milk-borne zoonoses in smallholder dairy farming sector in Kibaha and Morogoro districts in eastern Tanzania. J Vet Med Ser B Infect Dis Vet Public Heal. (2004) 51:123–8. doi: 10.1111/j.1439-0450.2004.00735.x
60. Suleiman TS, Karimuribo ED, Mdegela RH. Prevalence of bovine subclinical mastitis and antibiotic susceptibility patterns of major mastitis pathogens isolated in Unguja island of Zanzibar, Tanzania. Trop Anim Health Prod. (2018) 50:259–66. doi: 10.1007/s11250-017-1424-3
61. Kivaria FM, Noordhuizen JPTM. A retrospective study of the aetiology and temporal distribution of bovine clinical mastitis in smallholder dairy herds in the Dar es Salaam region of Tanzania. Vet J. (2007) 173:617–22. doi: 10.1016/j.tvjl.2006.01.008
62. Mbilu TJNK. Status of mastitis in lactating goats at Sokoine University of agriculture and neighbouring smallholder farms in Morogoro Municipality, Tanzania. Livest Res Rural Dev. (2007) 19:1–8. Available online at: http://www.lrrd.org/lrrd19/3/mbil19040.htm
63. Kateete DP, Kabugo U, Baluku H, Nyakarahuka L, Kyobe S, Okee M, et al. Prevalence and antimicrobial susceptibility patterns of bacteria from milkmen and cows with clinical mastitis in and around Kampala, Uganda. PLoS ONE. (2013) 8:e63413. doi: 10.1371/journal.pone.0063413
64. Björk S, Båge R, Kanyima BM, André S, Nassuna-Musoke MG, Owiny DO, et al. Characterization of coagulase negative staphylococci from cases of subclinical mastitis in dairy cattle in Kampala, Uganda. Ir Vet J. (2014) 67:1–3. doi: 10.1186/2046-0481-67-12
65. Ikwap K, Gertzell E, Hansson I, Dahlin L, Selling K, Magnusson U, et al. The presence of antibiotic-resistant Staphylococcus spp. and Escherichia coli in smallholder pig farms in Uganda. BMC Vet Res. (2021) 17:31. doi: 10.1186/s12917-020-02727-3
66. Kudinha T, Simango C. Prevalence of coagulase-negative staphylococci in bovine mastitis in Zimbabwe. J S Afr Vet Assoc. (2002) 73:62–5. doi: 10.4102/jsava.v73i2.557
67. Kelly PJ, Mason PR, Els J, Matthewman LA. Pathogens in dog bite wounds in dogs in Harare, Zimbabwe. Vet Rec. (1992) 131:464–6. doi: 10.1136/vr.131.20.464
68. Hadef L, Aggad H, Hamad B. Bacterial causative agents associated with subclinical mastitic in dromedary she-camels in southeastern Algeria. Jordan J Biol Sci. (2018) 11:209–14.
69. Razali K, Kaidi R, Giarratana F, Ait-Oudhia KH. Staphylococci and zoonotic potential: oral carriage and antibiotic susceptibility in healthy dogs and cats in Algeria. J Hell Vet Med Soc. (2022) 73:3621–8. doi: 10.12681/jhvms.25239
70. Zaatout N, Ayachi A, Kecha M. Interaction of primary mammary bovine epithelial cells with biofilm-forming staphylococci associated with subclinical bovine mastitis. Iran J Vet Res. (2019) 20:27–32.
71. Guendouze A, Djeghader A, Oucheriah Y, Hamidechi MA. Identification of bacteria causing subclinical cow mastitis: comparative 16S DNA sequence analysis and biotyping using MALDI-TOF MS. Res J Pharm Biol Chem Sci. (2017) 8:1249–54. Available online at: https://www.rjpbcs.com/pdf/2017_8(1)/%5b158%5d.pdf (accessed August 22, 2021).
72. Bakir M, Sabrina R, Toufik M. Antibacterial susceptibility profiles of sub-clinical mastitis pathogens isolated from cows in batna and setif governorates (east of algeria). Vet World. (2011) 4:537–41. doi: 10.5455/vetworld.2011.537-541
73. Gabli Z, Djerrou Z, Gabli AE, Bensalem M. Prevalence of mastitis in dairy goat farms in Eastern Algeria. Vet World. (2019) 12:1563–72. doi: 10.14202/vetworld.2019.1563-1572
74. Smaali S, Boukazoula F, Saadi L, Brahem I. Prevalence of methicillin-resistant staphylococcal mastitis in ruminants. Adv Anim Vet Sci. (2020) 8:428–32. doi: 10.17582/journal.aavs/2020/8.4.428.432
75. Ahmed W, Neubauer H, Tomaso H, El Hofy FI, Monecke S, Abdeltawab AA, et al. Characterization of staphylococci and streptococci isolated from milk of bovides with mastitis in Egypt. Pathogens. (2020) 9:381. doi: 10.3390/pathogens9050381
76. El-Ashker M, Gwida M, Monecke S, Ehricht R, Elsayed M, El-Gohary F, et al. Microarray-based detection of resistance genes in coagulase-negative staphylococci isolated from cattle and buffalo with mastitis in Egypt. Trop Anim Health Prod. (2020) 52:3855–62. doi: 10.1007/s11250-020-02424-1
77. Abed AH, Menshawy AMS, Zeinhom MMA, Hossain D, Khalifa E, Wareth G, et al. Subclinical mastitis in selected bovine dairy herds in north upper egypt: assessment of prevalence, causative bacterial pathogens, antimicrobial resistance and virulence-associated genes. Microorganisms. (2021) 9:1175. doi: 10.3390/microorganisms9061175
78. Abed AH, Hegazy EF, Omar SA, Abd El-Baky RM, El-Beih AA, et al. Carvacrol essential oil: a natural antibiotic against zoonotic multidrug-resistant staphylococcus species isolated from diseased livestock and humans. Antibiotics. (2021) 10:1328. doi: 10.3390/antibiotics10111328
79. Fahmy HA, Mahrous E. Detection of multidrug resistant strains in some pathogenic bacteria and fungi caused otitis in pet animals. Int J Vet Sci. (2020) 9:453–7. doi: 10.37422/IJVS/20.059
80. Hatem ME, Arab RH, Nagwa AS, Abd El-Moez SI, Khairy EA, Fouad EA. Bacterial abscessationin sheep and goat in Giza governorate with full antibiogram screening. Glob Vet. (2013) 10:372–81. doi: 10.5829/idosi.gv.2013.10.4.72112
81. Osman KM, Abd El-Razik KA, Marie HSH, Arafa A. Coagulase-negative staphylococci collected from bovine milk: species and antimicrobial gene diversity. J Food Saf. (2016) 36:89–99. doi: 10.1111/jfs.12216
82. Osman KM, Pires ÁDS, Franco OL, Orabi A, Hanafy MH, Marzouk E, et al. enterotoxigenicity and antibiotic resistance of coagulase-negative staphylococci isolated from raw buffalo and cow milk. Microb Drug Resist. (2020) 26:520–30. doi: 10.1089/mdr.2019.0114
83. Abdel-Moein KA, Zaher HM. The nasal carriage of coagulase-negative staphylococci among animals and its public health implication. Vector-Borne Zoonotic Dis. (2020) 20:897–902. doi: 10.1089/vbz.2020.2656
84. Youssif Ismail Badr NH, Hafiz NM, Halawa MA, Aziz HM. Genes conferring antimicrobial resistance in cattle with subclinical mastitis. Bulg J Vet Med. (2021) 24:67–85. doi: 10.15547/bjvm.2019-0028
85. Amer MM, Mekky HM, Fedawy HS. Molecular identification of Mycoplasma synoviae from breeder chicken flock showing arthritis in Egypt. Vet World. (2019) 12:535–41. doi: 10.14202/vetworld.2019.535-541
86. Abdalhamed AM, Zeedan GSG, Zeina HAAA. Isolation and identification of bacteria causing mastitis in small ruminants and their susceptibility to antibiotics, honey, essential oils, and plant extracts. Vet World. (2018) 11:355–62. doi: 10.14202/vetworld.2018.355-362
87. Elnageh HR, Hiblu MA, Abbassi MS, Abouzeed YM, Ahmed MO. Prevalence and antimicrobial resistance of staphylococcus species isolated from cats and dogs. Open Vet J. (2020) 10:452–6. doi: 10.4314/ovj.v10i4.13
88. Othman AM, Al-Huraibi BS, Assayaghi RM, Al-Shami HZ. Nasal carriage and methicillin resistance of Staphylococcus aureus among Schoolchildren in Sana'a City, Yemen. Int J Microbiol. (2021) 2021:5518317. doi: 10.1155/2021/5518317
89. Dhaouadi S, Soufi L, Campanile F, Dhaouadi F, Sociale M, Lazzaro L, et al. Prevalence of meticillin-resistant and -susceptible coagulase-negative staphylococci with the first detection of the mecC gene among cows, humans and manure in Tunisia. Int J Antimicrob Agents. (2020) 55:105826. doi: 10.1016/j.ijantimicag.2019.10.007
90. Klibi A, Maaroufi A, Torres C, Jouini A. Detection and characterization of methicillin-resistant and susceptible coagulase-negative staphylococci in milk from cows with clinical mastitis in Tunisia. Int J Antimicrob Agents. (2018) 52:930–5. doi: 10.1016/j.ijantimicag.2018.07.026
91. Gharsa H, Ben Slama K, Gómez-Sanz E, Lozano C, Klibi N, Jouini A, et al. Antimicrobial resistance, virulence genes, and genetic lineages of Staphylococcus pseudintermedius in healthy dogs in Tunisia. Microb Ecol. (2013) 66:363–8. doi: 10.1007/s00248-013-0243-y
92. Gharsa H, Ben SK, Gómez-Sanz E, Gómez P, Klibi N, Zarazaga M, et al. Characterisation of nasal Staphylococcus delphini and Staphylococcus pseudintermedius isolates from healthy donkeys in Tunisia. Equine Vet J. (2015) 47:463–6. doi: 10.1111/evj.12305
93. Guta C, Sebunya TK, Gashe BA. Antimicrobial susceptibility of staphylococci species from cow foremilk originating from dairy farms around Gaborone, Botswana. East Afr Med J. (2002) 79:45–8. doi: 10.4314/eamj.v79i1.8925
94. Qekwana DN, Sebola D, Oguttu JW, Odoi A. Antimicrobial resistance patterns of Staphylococcus species isolated from cats presented at a veterinary academic hospital in South Africa. BMC Vet Res. (2017) 13:286. doi: 10.1186/s12917-017-1204-3
95. Schmidt T, Kock MM, Ehlers MM. Diversity and antimicrobial susceptibility profiling of staphylococci isolated from bovine mastitis cases and close human contacts. J Dairy Sci. (2015) 98:6256–69. doi: 10.3168/jds.2015-9715
96. Monday IE, Hosu YS. Seasonal distribution, antimicrobial activity, and risk factors associated with the incidence of staphylococcus species in bovine milk from a dairy farm in Eastern Cape, South Africa. Asian J Agric Rural Dev. (2022) 12:10–9. doi: 10.18488/5005.v12i1.4412
97. Pekana A, Nwodo UU, Okoh AI, Green E. Distribution and antibiotic susceptibility profiles of Staphylococcus spp isolated from unpasteurized cow milk locally consumed in Nkonkobe local municipality, South Africa. Int J Appl Res Vet Med. (2017) 15:75–84. doi: 10.3390/ijerph15102223
98. Adegoke AA, Okoh AI. Species diversity and antibiotic resistance properties of Staphylococcus of farm animal origin in Nkonkobe Municipality, South Africa. Folia Microbiol. (2014) 59:133–40. doi: 10.1007/s12223-013-0275-1
99. Qekwana DN, Oguttu JW, Sithole F, Odoi A. Patterns and predictors of antimicrobial resistance among Staphylococcus spp. from canine clinical cases presented at a veterinary academic hospital in South Africa. BMC Vet Res. (2017) 13:116. doi: 10.1186/s12917-017-1034-3
100. Blunt CA, van Vuuren M, Picard J. Antimicrobial susceptibility profiles of staphylococcus intermedius isolates from clinical cases of canine pyoderma in South Africa. J S Afr Vet Assoc. (2013) 84:E1–6. doi: 10.4102/jsava.v84i1.276
101. Prior CD, Moodley A, Karama M, Malahlela MN, Leisewitz A. Prevalence of methicillin resistance in Staphylococcus pseudintermedius isolates from dogs with skin and ear infections in South Africa. J S Afr Vet Assoc. (2022) 93:10–7. doi: 10.36303/JSAVA.2022.93.1.505
102. Dalis JS, Kazeem HM, Makinde AA, Fatihu MY. Bacteria associated with bovine dermatophilosis in Zaria, Nigeria. African J Microbiol Res. (2010) 4:1475–9. Available online at: https://academicjournals.org/journal/AJMR/article-full-text-pdf/751548C13843
103. Ajuwape ATP, Roberts AA, Solarin OO, Adetosoye AI. Bacteriological and haematological studies of clinical mastitis in goats in Ibadan, OYO State, Nigeria. Small Rumin Res. (2005) 60:307–10. doi: 10.1016/j.smallrumres.2004.10.009
104. Chah KF, Gómez-Sanz E, Nwanta JA, Asadu B, Agbo IC, Lozano C, et al. Methicillin-resistant coagulase-negative staphylococci from healthy dogs in Nsukka, Nigeria. Brazilian J Microbiol. (2014) 45:215–20. doi: 10.1590/S1517-83822014005000034
105. Chukwu CO, Ogo NI, Jimoh A, Chukwu DI. Pathogenic bacteria associated with cutaneous canine myiasis due to Cordylobia anthropophaga. Vet World. (2012) 5:617–20. doi: 10.5455/vetworld.2012.617-620
106. Danmallam FA, Pimenov NV. Study on prevalence, clinical presentation, and associated bacterial pathogens of goat mastitis in bauchi, plateau, and edo states, nigeria. Vet World. (2019) 12:638–45. doi: 10.14202/vetworld.2019.638-645
107. Ugochukwu EI, Agwu CO. Aerobic bacteria from nasal discharge of goats suffering from clinical PPR: isolation and identification. Microbios. (1991) 65:81–5.
108. Ajuwape ATP, Oyebanji MO, Adetosoye AI. Bacteriological examination of normal upper respiratory tract of puppies with particular reference to staphylococci. Vet Arh. (2006) 76:179–84.
109. Alawa JP, Ngele MB, Ogwu D. Chronic caprine mastitis in Nigerian goat breeds: microbiological flora and histopathological findings. Small Rumin Res. (2000) 35:203–7. doi: 10.1016/S0921-4488(99)00099-1
110. Ugwu CC, Gomez-Sanz E, Agbo IC, Torres C, Chah KF. Characterization of mannitol-fermenting methicillin-resistant staphylococci isolated from pigs in Nigeria. Brazilian J Microbiol. (2015) 46:885–92. doi: 10.1590/S1517-838246320140644
111. Lawal OU, Adekanmbi AO, Adelowo OO. Occurrence of methicillin-resistant staphylococci in the pig-production chain in Ibadan, Nigeria. Onderstepoort J Vet Res. (2021) 88:1–4. doi: 10.4102/ojvr.v88i1.1959
112. Mamfe LM, Akwuobu CA, Ngbede EO. Phenotypic detection, antimicrobial susceptibility and virulence profile of staphylococci in the pig production setting, Makurdi, Nigeria. Access Microbiol. (2021) 3:000293. doi: 10.1099/acmi.0.000293
113. Talan DA, Goldstein E, Staatz D, Overturf GD. Staphylococcus intermedius: clinical presentation of a new human dog bite pathogen. Ann Emerg Med. (1989) 18:410–3. doi: 10.1016/S0196-0644(89)80582-7
114. Viau R, Hujer AM, Hujer KM, Bonomo RA, Jump RLP. Are Staphylococcus intermedius infections in humans cases of mistaken identity? A case series and literature review. Open Forum Infect Dis. (2015) 2:ofv110. doi: 10.1093/ofid/ofv110
115. Kessie G, Ettayebi M, Haddad AM, Shibl AM, Al-Shammary FJ, Tawfik AF, et al. Plasmid profile and antibiotic resistance in coagulase-negative staphylococci isolated from polluted water. J Appl Microbiol. (1998) 84:417–22. doi: 10.1046/j.1365-2672.1998.00358.x
116. Nimrat S, Siriboonlamom S, Zhang S, Xu Y, Vuthiphandchai V. Chilled storage of white shrimp (Litopenaeus vannamei) spermatophores. Aquaculture. (2006) 261:944–51. doi: 10.1016/j.aquaculture.2006.08.018
117. Shale K, Lues JFR, Venter P, Buys EM. The distribution of Staphylococcus sp. on bovine meat from abattoir deboning rooms. Food Microbiol. (2005) 22:433–8. doi: 10.1016/j.fm.2004.09.007
118. Siqueira JE, Lima KC. Staphylococcus epidermidis and Staphylococcus xylosus. in a secondary root canal infection with persistent symptoms: a case report. Austral Endodontic J. (2002) 28:61–3. doi: 10.1111/j.1747-4477.2002.tb00382.x
119. Pinna A, Zanetti S, Sotgiu M, Sechi LA, Fadda G, Carta F. Identification and antibiotic susceptibility of coagulase negative staphylococci isolated in corneal/external infections. Br J Ophthalmol. (1999) 83:771–3. doi: 10.1136/bjo.83.7.771
120. Kateete DP, Asiimwe BB, Mayanja R, Najjuka CF, Rutebemberwa E. Species and drug susceptibility profiles of staphylococci isolated from healthy children in Eastern Uganda. PLoS ONE. (2020) 15:e0229026. doi: 10.1371/journal.pone.0229026
121. Rudkjøbing VB, Thomsen TR, Xu Y, Melton-Kreft R, Ahmed A, Eickhardt S, et al. Comparing culture and molecular methods for the identification of microorganisms involved in necrotizing soft tissue infections. BMC Infect Dis. (2016) 16:652. doi: 10.1186/s12879-016-1976-2
122. Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. (2015) 6:791. doi: 10.3389/fmicb.2015.00791
123. Kimera ZI, Mshana SE, Rweyemamu MM, Mboera LEG, Matee MIN. Antimicrobial use and resistance in food-producing animals and the environment: an African perspective. Antimicrob Resist Infect Control. (2020) 9:1–12. doi: 10.1186/s13756-020-0697-x
124. Landers TF, Cohen B, Wittum TE, Larson EL. A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. (2012) 127:4–22. doi: 10.1177/003335491212700103
125. Barbier F, Ruppé E, Hernandez D, Lebeaux D, Francois P, Felix B, et al. Methicillin-resistant coagulase-negative staphylococci in the community: high homology of SCCmec IVa between Staphylococcus epidermidis and major clones of methicillin-resistant Staphylococcus aureus. J Infect Dis. (2010) 202:270–81. doi: 10.1086/653483
126. Bloemendaal ALA, Brouwer EC, Fluit AC. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS ONE. (2010) 5:e11841. doi: 10.1371/journal.pone.0011841
127. Cui J, Liang Z, Mo Z, Zhang J. The species distribution, antimicrobial resistance and risk factors for poor outcome of coagulase-negative staphylococci bacteraemia in China. Antimicrob Resist Infect Control. (2019) 8:1–10. doi: 10.1186/s13756-019-0523-5
128. Ehlers MM, Strasheim W, Lowe M, Ueckermann V, Kock MM. Molecular epidemiology of staphylococcus epidermidis implicated in catheter-related bloodstream infections at an academic hospital in Pretoria, South Africa. Front Microbiol. (2018) 9:417. doi: 10.3389/fmicb.2018.00417
129. Falagas ME, Karageorgopoulos DE, Leptidis J, Korbila IP. MRSA in Africa: filling the Global Map of Antimicrobial Resistance. PLoS ONE. (2013) 8:68024. doi: 10.1371/journal.pone.0068024
130. Yang C, Anahtar MN, Pierce VM. It's not you, it's SOSA: a case study on breaking up with an FDA-cleared susceptibility testing system's oxacillin results for Staphylococcus spp. other than S. aureus and S. lugdunensis. Open Forum Infect Dis. (2022) 9:ofac421. doi: 10.1093/ofid/ofac421
131. Canver MC, Gonzalez MD, Ford BA, Arnold AR, Lawhon SD, Burnham CAD, et al. Improved performance of a rapid immunochromatographic assay for detection of PBP2a in non-staphylococcus aureus staphylococcal species. J Clin Microbiol. (2018) 57:4–8. doi: 10.1128/JCM.01417-18
132. Food and Agriculture Organization of the United Nations. Livestock Production Systems spotlight Ethiopia. Rome, Italy: FAO (2018). Available online at: http://www.fao.org/3/CA2149EN/ca2149en.pdf (accessed November 16, 2022).
Keywords: Africa, animals, antibiotic resistance, coagulase-negative staphylococci, non-aureus staphylococci, Staphylococci other than S. aureus
Citation: Ocloo R, Nyasinga J, Munshi Z, Hamdy A, Marciniak T, Soundararajan M, Newton-Foot M, Ziebuhr W, Shittu A, Revathi G, Abouelfetouh A and Whitelaw A (2022) Epidemiology and antimicrobial resistance of staphylococci other than Staphylococcus aureus from domestic animals and livestock in Africa: a systematic review. Front. Vet. Sci. 9:1059054. doi: 10.3389/fvets.2022.1059054
Received: 30 September 2022; Accepted: 16 November 2022;
Published: 13 December 2022.
Edited by:
Faham Khamesipour, Shahid Beheshti University of Medical Sciences, IranReviewed by:
Newton Valerio Verbisck, Brazilian Agricultural Research Corporation (EMBRAPA), BrazilCopyright © 2022 Ocloo, Nyasinga, Munshi, Hamdy, Marciniak, Soundararajan, Newton-Foot, Ziebuhr, Shittu, Revathi, Abouelfetouh and Whitelaw. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Remous Ocloo, cmVtb3VzQHN1bi5hYy56YQ==; cmVtb3Vzb0BnbWFpbC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.