
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 22 December 2022
Sec. Veterinary Infectious Diseases
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1058115
This article is part of the Research Topic Bacterial Diseases In Poultry: Biology, Virulence And Prevention In The Age Of Reduced Antibiotic Use View all 9 articles
 Chake Keerqin1
Chake Keerqin1 Katherine McGlashan2
Katherine McGlashan2 Thi Thu Hao Van3
Thi Thu Hao Van3 Helene N. Chinivasagam2
Helene N. Chinivasagam2 Robert J. Moore3
Robert J. Moore3 Mingan Choct1
Mingan Choct1 Shu-Biao Wu1*
Shu-Biao Wu1*Background: Bacteriophages are viral predators of bacteria and are common in nature. Their host-specific infections against specific bacteria make them an attractive natural agent to control bacterial pathogens. Interest in the potential of bacteriophages as antibacterial agents in the production animal industries has increased.
Methods: A total of 18 bacteriophages were isolated from Australian commercial poultry environments, from which three highly active phages were chosen for enrichment. Sequencing libraries were prepared using a Nextera XT kit (Illumina) and sequenced on an Illumina MiSeq instrument using 2 × 300 bp paired-end chemistry. The sequence data were then assembled and aligned with a2 bacteriophage as the reference. An animal trial was performed by oral gavaging Clostridium perfringens netB containing strain EHE-NE18 to the Ross 308 broiler chickens prior inoculation with Eimeria species. The chickens were raised following the management guide for Ross 308 from d 0 to d 21 and fed with starter and grower diets met the specific breed nutrient requirements. Body weight gain and feed intake were measured on d 9 and d 21 and FCR adjusted with mortality was calculated.
Results: The isolated bacteriophages only had only 96.7% similarity to the most closely related, previously characterized, Clostridium bacteriophage indicated that they might represent a novel strain of bacteriophage. A “cocktail” containing the three bacteriophages was capable of lysing four known disease-inducing C. perfringens strains in vitro. Oral administration of the bacteriophages cocktail to broilers challenged with necrotic enteritis markedly alleviated intestinal necrotic lesions in the duodenum and jejunum on day 16 post-hatch. The phage treatment significantly reduced the lesion scores of birds challenged with NE (P < 0.01), and the lesion scores between birds treated with the bacteriophages and the unchallenged birds were not statistically different (P > 0.05). However, no effect on the growth performance was observed during the recorded period of days 9-21.
Conclusion: These findings suggest that bacteriophage treatment is a promising approach to protect intestinal health from C. perfringens induced necrotic enteritis. Further research will be required on the dosing, route of administration, and large scale validation studies to further advance this approach to pathogen control.
Necrotic enteritis (NE) is a bacterial enteric disease in poultry accountable for over US$6 billion profit loss annually to the industry worldwide in the treatments and containment measures (1). The etiology of NE has been well characterized and it is mainly due to the rapid proliferation of C. perfringens Type G (2) strains that express the NetB toxin (3), albeit NetB negative strains have also been reported to cause NE (4). Outbreaks of NE also depend on a number of predisposing factors that result in degradation of intestinal health and compromise of the host immune status (3, 5, 6). It is reported that change of feed, use of different cereal bases in diet formulation, i.e., non-viscous grains to viscous grains, the inclusion of low digestibility protein sources, Eimeria infection, and compromise of to the host immune status, all exacerbate the chances of NE outbreaks (7). Flocks affected by acute NE exhibit clinical symptoms coupled with a rapid increase in flock mortality and distress in the affected bird populations, but subclinical (mild) disease are more frequent with little mortality rate if any, and suboptimal growth performance. The subclinical form of necrotic enteritis leads to more significant economic consequences with prolonged deterioration of growth performance resulting from loss of feed efficiency over time (8).
For decades, poultry and other animal production systems across the globe have depended on in-feed antibiotics to sustain growth performance and control enteric pathogens as they can effectively maintain intestinal microbiota uniformity and suppress bacterial infection (9). However, the emergence of antibiotic-resistant bacteria and the possibility of horizontal transfer of antibiotic resistance genes to human pathogens resulted in the ban of in-feed antibiotics and has ushered in antibiotic-free practices that require alternatives to conventional in-feed antibiotics to maintain flock performance and health. The suppression of specific intestinal bacterial populations is essential to manage opportunistic pathogens and stop them from reaching a disease-causing threshold (10). Following the 2006 EU's legislative ban on antibiotic use in animal feed, numerous countries across the globe have followed suit and are dealing with the challenges of running antibiotic-free production systems. Early efforts to overcome these challenges focused on the adoption of antibiotic-free measures that provided some protection against pathogens (11), but the end results have been inconsistent.
The lack of consistency in the efficacy of the current alternatives for maintaining performance means that novel solutions continue to be explored. The interest in bacteriophages stems from the fact that they naturally target specific bacteria and have narrow host ranges. On exposure to specific bacteriophages, a host bacterium undergoes lysis resulting in the death of the bacterium and the release of bacteriophages that can continue to infect other host bacteria; this self-replicating process is a very different process compared to other alternatives to antibiotics. Hence, bacteriophages are an attractive and feasible option for pathogen control. The use of phage therapy is not new. In fact, the clinical application of bacteriophages was first implemented in the early 1920s, with extensive therapeutic application in human medicine across the eastern European countries, and it has continued to the present time (12). Many known attributes of bacteriophages fit the criteria for an effective antibacterial alternative, including automatic dosing capability through lytic propagation of bacteriophages progeny, minimal inherent toxicity, low disruption to commensal microbiota, biofilm clearing, and variable routes of administration (13). Bacteriophage absorption on the surface of susceptible bacteria is mediated by specific receptors on the bacteria cell wall (14). Different types of bacteriophages that infect any one species of bacteria may recognize different receptors. Hence, the combination of several bacteriophages in the form of a cocktail may improve their efficacy against a target bacteria. Indeed, the use of multivalent bacteriophage cocktails was reported to be successful in reducing mortality and improving performance in chickens infected with C. perfringens resulting in clinical NE (15).
However, only limited research has been reported on phage prophylactic treatment against C. perfringens induced subclinical necrotic enteritis in chickens. This study was conducted to isolate and purify bacteriophages from poultry production environments that could infect and lyse C. perfringens, determine their lytic profile against a range of known virulent C. perfringens strains capable of inducing NE, and then the isolated bacteriophage was evaluated for prophylactic properties in vivo using a necrotic enteritis challenge model.
Samples from various sources within poultry environments were aseptically collected for bacteriophage isolation, which included soil samples from backyard chicken sheds and yards located in Armidale and Tamworth, NSW, Australia, sewage and offal wash from seven individual chicken farms, and intestinal content of chickens from experiments conducted at UNE (Armidale, New South Wales, Australia).
All collected samples were kept on ice until processed. C. perfringens were isolated by plating serial dilutions in 0.1% peptone onto Perfringens Agar Base (Oxoid, Hampshire, UK) supplemented with Tryptose Sulphite Cycloserine (TSC) (Oxoid, Hampshire, UK), followed by incubation under anaerobic conditions using an anaerobic sachet (Oxoid Australia) at 39oC for 48 h. Putative C. perfringens colonies were re-streaked twice (to ensure purity) on fresh Horse Blood Agar (HBA) (Oxoid, Hampshire, UK) and incubated at 39oC for 18 h. Bacteria isolates were picked and suspended in the Brain Heart Infusion (BHI, Oxoid, Hampshire, UK) broth with sterile glycerol addition to 40% (v/v) and stored at −20oC until used.
Samples for bacteriophage isolation were prepared by gently shaking 1 g of solid samples (i.e., soil, gut content or litter) or 1 ml liquid sample (effluent) in 9 ml of SM buffer (0.1 M NaCl, 1 mM MgSO4, 0.2 M Tris-HCl, pH 7.5) for 12 h. The supernatant was then centrifuged at 11,000 × g at 4oC for 5 mins. The supernatant was then filtered through 0.22-μm-pore-size Millipore filters (MF-Millipore™, MERCK, Australia). Bacteriophages were isolated as per the methods of Smith (16) using the isolated C. perfringens host strains. In brief, overnight growth of C. perfringens from HBA agar was initially mixed in 3 ml brain-heart infusion (BHI) broth, and then further BHI was added to adjust the turbidity to 220 ± 15 Nephelometric Turbidity Unit (NTU). Serial dilutions of bacteriophage filtrates were prepared in SM buffer and 100 μl of each bacteriophage dilution was mixed with 200 μl of C. perfringens and incubated at 37oC for 30 min. Each set of the bacteriophages-host mixture was mixed into 7 ml of 0.5% BHI soft agar (supplemented with 1 mM MgCl2 and 1 mM Ca Cl2) and plated as an overlay on 1% BHI base agar. Plates were incubated anaerobically at 39oC for 48 h. Well separated plaques were picked and resuspended in 1 ml of SM buffer, then stored at 4°C overnight. Serial dilutions of the homogenized plaques were made and cultured with the susceptible host in three cycles of plaque purification (16, 17).
Phage stocks were prepared from three to five plates with confluent plaques from the third round of phage purification. SM buffer (5 ml) was added to confluent plates and shaken slowly at 10 rpm for 12 h to resuspend the bacteriophage. The supernatants were then filtrated through 0.22-μm-pore-size Millipore filters (MF-Millipore™, MERCK, Australia). The resulting filtrates were centrifuged at 40,000 × g at 4°C for 2h. The resulting bacteriophage pellets were resuspended in SM buffer to form 1 × 109 pfu/ml bacteriophage stock and stored in the dark at 4°C.
The bacteriophages were tested for lytic ability against four pathogenic C. perfringens strains (NE14, NE18, NE21, and NE36) to investigate the bacteriophage's virulence against target hosts in vitro. The characteristics of the tested C. perfringens strains are detailed in Table 1. Bacterial lawns were prepared using turbidity-adjusted C. perfringens strains (220 ± 15 NTU) mixed with 7 ml of 0.5% BHI soft agar (supplemented with 5mM MgSO4 and 10mM CaCl2) and set on the prepared 1% BHI base agar. The bacteriophage lysates were titered, and all were within the range of 1–7 × 109pfu/ml. A volume of 20 μl of each bacteriophage preparation was aseptically placed in duplicate on marked positions of the bacterial lawns and set to dry with minimal disturbance to retain the bacteriophage droplets at the designated positions. Plates were incubated anaerobically at 39°C up to 48 h and inspected for plaque formation. All positive tests were replicated three times with the relevant C. perfringens host.
The bacteriophage stocks were concentrated by precipitation with polyethylene glycol, and DNA was isolated using a phenol/chloroform extraction method. Sequencing libraries of 5 bacteriophages from different sources (A2, C2, D1, E1, and H1) were prepared using a Nextera XT kit (Illumina). The libraries were sequenced on an Illumina MiSeq instrument using 2 × 300 bp paired-end chemistry. The sequence data generated from the MiSeq were assembled using the A5-miseq assembly pipeline (21). The bacteriophage DNA sequences were aligned using Clone Manager Suite V8 (Scientific and Educational Software, NC) with a2 bacteriophage as the reference. Homology analysis of the bacteriophage a2 sequence was performed against the NCBI database using the BLASTn program. Gene prediction was performed by submitting the DNA sequence of bacteriophage a2 to the RAST server (https://rast.nmpdr.org/) for RAST annotations. The whole genome sequence data has been deposited in the NCBI database with the accesion numbers OP753449–OP753453.
Eighteen bacteriophage isolates that had lytic activity against the C. perfringens EHE-NE18 strain were enriched for the preparation of a bacteriophage inoculate. Three selected bacteriophages were prepared in larger volumes using the procedure adopted for purification as described above. Pelleted bacteriophages were resuspended in SM buffer to a final concentration of 1 × 106 pfu/ml for the in vivo experiment.
Day-old birds (Ross 308 broilers) were obtained from a commercial hatchery (Baiada Hatchery, Tamworth, NSW, Australia). Birds were housed in 18 pens, each containing 11 birds. The pens (0.9 × 0.95 m) were equipped with feeders and drinkers and hardwood shavings were used as bedding materials. The temperature and lighting were controlled according to the Ross 308 broiler guideline. A standard wheat-soybean starter diet formulated to meet the 2014 Ross 308 nutrient specifications was fed to all birds from day 0 to 10 (Table 2). A grower diet consisting of the wheat-based formulation was fed from day 10 to the end of the trial on day 21 (Table 2). Feed and water were available ad libitum. Pen weight and feed intake were measured on days 9 and 21, and the feed conversion ratio (FCR) was calculated by pen feed intake divided by pen bird weight gain and adjusted with mortality during the rearing period of d 9 to d 21.
The experiment consisted of a control untreated and unchallenged group, and necrotic enteritis challenge groups with or without bacteriophage treatment. On day 9, the NE challenge groups were given 1 ml of a mixture of field strains of Eimeria species containing 5,000 oocysts each of E. acervulina and E. maxima, and 2,500 oocysts of E. brunetti (Eimeria Pty Ltd, Werribee, VIC, Australia) by oral gavage, and unchallenged control birds were orally gavaged with 1 ml phosphate buffered saline as a sham treatment. On days 14-15, birds in the challenged groups were inoculated by oral gavage with 1 × 108 CFU of C. perfringens EHE-NE18 strain as described by Wu et al. (22). The bacteriophage treatment birds were orally gavaged with 1 × 106 units of the C. perfringens specific bacteriophage inoculate approximately 1 h following C. perfringens gavage. On day 16, two birds per pen from all the groups were randomly selected, weighed, and euthanized by cervical dislocation to perform post-mortem analysis and intestinal lesion scoring at a 0–6 scale according to Keyburn et al. (19). On day 21, the experiment was completed, and birds were euthanized and disposed of according to the approved ethics protocol.
Data were analyzed by one-way ANOVA with the pen as the experimental unit. Paired comparisons of means were performed using Tukey's test when differences among treatments were detected by ANOVA. Significance at P < 0.05 was declared for paired comparison.
The current study isolated and purified eighteen C. perfringens bacteriophages isolates. Each bacteriophage isolate was capable of producing plaques (Figure 1A) on each of the C. perfringens hosts obtained from the same poultry environments (soil, sewage, offal wash or intestinal content) and corresponding geographic locations. Among 46 field C. perfringens isolates from the environment, only three C. perfringens isolates were susceptible to all the bacteriophages isolates and thus used as the hosts for the bacteriophages. Therefore, all 18 bacteriophages were subsequently enriched with these three common hosts for bacteriophage purification, and the resulting bacteriophage preparations were used to perform lytic tests individually against pathogenic C. perfringens strains derived from chickens with necrotic enteritis. Positive lytic activities were observed for all these bacteriophage strains against four previously described virulent C. perfringens strains (NE14, NE18, NE21, and NE36) in our spot-testing assay. However, other strains of C. perfringens (NE1, NE4, NE7, NE16, NE31, and NE38) were not susceptible to these bacteriophages (Table 1). As the bacteriophage preparation was lytic against NE18 strain, which has been reported to produce NetB toxin (3) and has been used in NE experimental disease induction models (23), this preparation was used as an inoculate in the subsequent in vivo experiment with EHE-NE18 as the challenge strain.
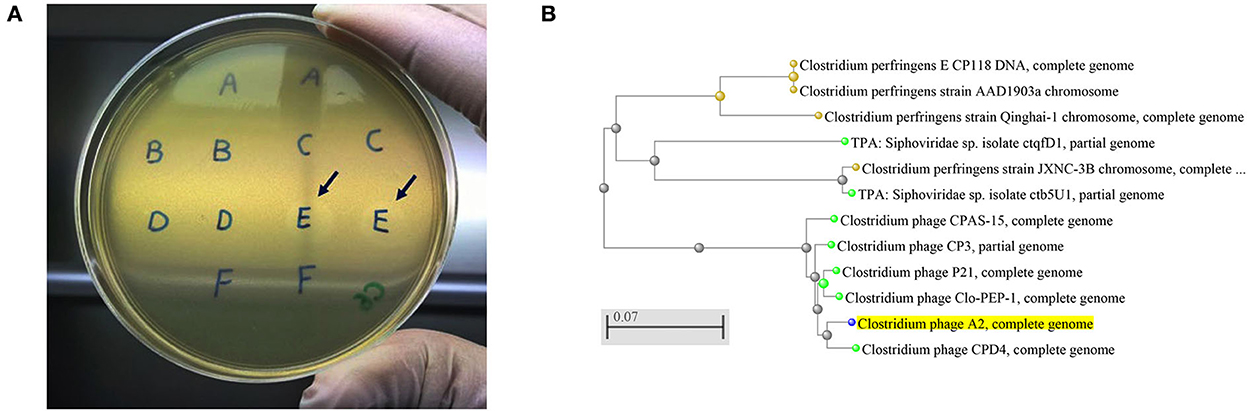
Figure 1. (A) Clostridium perfringens bacteriophage plaques. Arrows showing the plaques produced by the strains from sample E that was collected from chicken caecal content, (B) An evolutionary tree of Clostridium bacteriophage strains produced by NCBI online blast tools with the Fast Minimum Evolution tree method. The Clostridium phage A2 isolated in the current study is highlighted in yellow showing its close relation with Clostridium phage CPD4. The scale bar corresponds to 0.07 substitutions per nucleotide position.
A narrow spectrum of infectiousness against C. perfringens strains also presents the possibility of a controlled elimination of pathogenic C. perfringens strains but minimal harm to commensal C. perfringens. It has been demonstrated that C. perfringens exists as normal gut commensals with low abundance in the intestinal tract of healthy chickens (6).
Five bacteriophages genomes were randomly chosen for sequencing, and the BLAST analysis against the NCBI database indicated the bacteriophages isolated aligned with the Clostridium bacteriophages CPD4 with a homology of 97.7% which clustered closely as shown in Figure 1B and 77 genes were predicted, of which 19 had specific bacteriophage functions. The degree of homology to previously characterized bacteriophages indicates that the bacteriophages isolated in this study might represent a novel bacteriophage strain. C. perfringens specific bacteriophage isolates have been reported to belong to the family Siphoviridae or Podviridare, and bacteriophages endolysin has been shown to be responsible for the lytic activity (17, 24). The sequences of the bacteriophage isolates produced in the current study generated a single contig with the same GC content (34%) and showed a similar genome size with 99.98–100.00% identities (Table 3). The genomes of several C. perfringens specific bacteriophages have been characterized as reported previously, providing essential information for understanding the virulence of phages to their hosts and for downstream applications (17, 25).
No mortality of birds due to NE and low lesion scores (0.8 out of a maximum of 6) were observed in the in vivo challenge experiment, indicating that subclinical NE had been induced. The lesion score results are shown in Table 4. Higher lesion scores were recorded in both the duodenum (P < 0.01) and jejunum (P < 0.01) of the challenged birds than those unchallenged. The phage treatment significantly reduced the lesion scores of birds challenged with NE (P < 0.01), such that the lesion scores between birds treated with the bacteriophages and the control unchallenged birds were not statistically different (P > 0.05). However, this significant effect of the bacteriophages on lesion scores did not fully manifest in the measures of bird performance. In the current study, the birds under NE challenge were significantly affected, showing reduced BWG, FI, and increased FCR in the challenged treatments compared to the control (Table 5). NE challenged birds with or without bacteriophage treatment had reduced BWG and FI and increased FCR during days 9 to 21 or 0 to 21 (P < 0.001 for all except FCR during d 0–21 being P = 0.002). The challenge model used in the current study experimentally reproduced the conditions of possible NE outbreaks in the field, of which coccidiosis serves as a predisposing factor to damage intestinal epithelium before C. perfringens infection (7, 22). The current study was not set out to apportion the impact of the Eimeria per se, and therefore, it is difficult to determine with any certainty to what extent the lack of performance improvement by the bacteriophage therapy could be due to its inability to counter any damage that resulted from coccidiosis. Eimeria negatively affects nutrient utilization by interfering with the integrity of epithelium cells, leading to compromised growth of the animals (6, 26).
Although bacteriophage treatment did not translate into improved bird performance, less severity of intestinal lesions in the treated birds suggests that bacteriophages appropriately selected against the NetB producing strain of C. perfringens may be worthy of further exploration.
The current study focused on the performance of the birds during days 9–21, when the NE challenge was applied, and the NE effect was expected. However, a prolonged rearing period might be useful to observe the bacteriophage treatment effect on later stage performance through improved intestinal health. In addition, the dosage and the route of bacteriophage administration may also be important factors for the effectiveness of the bacteriophage treatment to protect birds from NE infection. These will need to be addressed in future studies to maximize the protective effect of bacteriophages, such as water and feed delivery and higher doses. Further, more bacteriophage strains may be needed to control C. perfringens strains in chickens through the administration of a wider spectrum cocktail.
In conclusion, 18 isolated C. perfringens specific bacteriophage strains were isolated. The finding that they had 96.7% similarity to the most closely related, previously characterized, Clostridium bacteriophage indicated that they represent a similar cluster of Clostridium bacteriophage. The result showed an encouraging protective effect of the bacteriophage application with alleviated lesions in the gut of NE-challenged birds despite the fact that no performance improvement was seen. Whether bacteriophage can be used in the poultry production to combat NE is yet to be assured. Further investigations are warranted to optimize the dosage and administration protocol and to isolate more NetB positive and even negative bacteriophage strains for a wider spectrum control of different C. perfringens pathogens.
The data presented in the study are deposited in the NCBI GenBank repository, accession numbers OP753449 – OP753453.
The animal study was reviewed and approved by the Animal Ethics Committee of the University of New England (UNE), Armidale, Australia.
CK performed data collection and analysis, interpreted results, and wrote the manuscript. KM performed isolation of the bacteriophages and analysis. TV sequenced the bacteriophage genome, analyzed data, and read the manuscript critically. HC participated the design of the project, isolated bacteriophages, and reviewed the manuscript critically. RM participated the design of the project, data analysis, and reviewed the manuscript critically. MC participated the conceptualization of the project and reviewed the manuscript critically. S-BW conceptualized, designed the project, performed the animal experiment, analyzed data, and reviewed the manuscript critically. All authors contributed to the article and approved the submitted version.
This study was supported by the Poultry CRC (#1-3-4).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
2. Rood, JI, Adams V, Lacey J, Lyras D, McClane BA, et al. Expansion of the Clostridium perfringens toxin-based typing scheme. Anaerobe. (2018) 53:5–10. doi: 10.1016/j.anaerobe.2018.04.011
3. Keyburn AL, Boyce JD, Vaz P, Bannam TL, Ford ME, Parker D, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS Pathogens. (2008) 4:e26. doi: 10.1371/journal.ppat.0040026
4. Cooper KK, Songer JG. Virulence of Clostridium perfringens in an experimental model of poultry necrotic enteritis. Vet Microbiol. (2010) 142:323–8. doi: 10.1016/j.vetmic.2009.09.065
5. Keyburn AL, Bannam TL, Moore RJ, Rood JI. NetB, a pore-forming toxin from necrotic enteritis strains of Clostridium perfringens. Toxins. (2010) 2:1913–27. doi: 10.3390/toxins2071913
6. Van Immerseel F, Rood JI, Moore RJ, Titball RW. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends Microbiol. (2009) 17:32–6. doi: 10.1016/j.tim.2008.09.005
7. Moore, RJ. Necrotic enteritis predisposing factors in broiler chickens. Avian Pathol. (2016) 45:1–22. doi: 10.1080/03079457.2016.1150587
8. Skinner JT, Bauer S, Young V, Pauling G, Wilson J. An economic analysis of the impact of subclinical (mild) necrotic enteritis in broiler chickens. Avian Dis. (2010) 54:1237–40. doi: 10.1637/9399-052110-Reg.1
9. Lanckriet A, Timbermont L, De Gussem M, Marien M, Vancraeynest D, Haesebrouck F, et al. The effect of commonly used anticoccidials and antibiotics in a subclinical necrotic enteritis model. Avian Pathol. (2010) 39:63–8. doi: 10.1080/03079450903505771
10. Eeckhaut V, Wang J, Van Parys A, Haesebrouck F, Joossens M, Falony G, et al. The probiotic Butyricicoccus pullicaecorum reduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front Microbiol. (2016) 7:1416. doi: 10.3389/fmicb.2016.01416
11. Cogliani C, Goossens H, Greko C. Restricting antimicrobial use in food animals: lessons from Europe. Microbe. (2011) 6:274.
12. Sulakvelidze A, Alavidze Z, Morris JG. Bacteriophage therapy. Antimicrob Agents Chemother. (2001) 45:649–59. doi: 10.1128/AAC.45.3.649-659.2001
13. Karthik K, Muneeswaran NS, Manjunathachar HV, Gopi M, Elamurugan A, Kalaiyarasu S. Bacteriophages: effective alternative to antibiotics. Adv Anim Vet Sci. (2014) 2:1–7. doi: 10.14737/journal.aavs/2014/2.3s.1.7
14. Pleteneva EA, Shaburova OV, Sykilinda NN, Miroshnikov KA, Krylov SV, Mesianzhinov VV, Krylov, VN. Study of the diversity in a group of phages of Pseudomonas aeruginosa species PB1 (Myoviridae) and their behavior in adsorbtion-resistant bacterial mutants. Genetika. (2008) 44:185–94. doi: 10.1134/S1022795408020051
15. Miller RW, Skinner J, Sulakvelidze A, Mathis GF, Hofacre, CL. Bacteriophage therapy for control of necrotic enteritis of broiler chickens experimentally infected with Clostridium perfringens. Avian Dis. (2010) 54:33–40. doi: 10.1637/8953-060509-Reg.1
16. Smith HW. The bacteriophages of Clostridium perfringens. J Gen Microbiol. (1959) 21:622–30. doi: 10.1099/00221287-21-3-622
17. Seal BS, Fouts DE, Simmons M, Garrish JK, Kuntz RL, Woolsey R, et al. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: genomic organization and proteomic analysis of the virions. Arch Virol. (2011) 156:25–35. doi: 10.1007/s00705-010-0812-z
18. Sheedy SA, Ingham AB, Rood JI, Moore RJ. Highly conserved alpha-toxin sequences of avian isolates of Clostridium perfringens. J Clin Microbiol. (2004) 42:1345–7. doi: 10.1128/JCM.42.3.1345-1347.2003
19. Keyburn AL, Sheedy SA, Ford ME, Williamson MM, Awad MM, Rood JI. Alpha-toxin of Clostridium perfringens is not an essential virulence factor in necrotic enteritis in chickens. Inf Immun. (2006) 74:6496–500. doi: 10.1128/IAI.00806-06
20. Keyburn AL, Portela RW, Sproat K, Ford ME, Bannam TL, Yan X, et al. Vaccination with recombinant NetB toxin partially protects broiler chickens from necrotic enteritis. Vet Res. (2013) 44:1. doi: 10.1186/1297-9716-44-54
21. Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. (2015) 31:587–9. doi: 10.1093/bioinformatics/btu661
22. Wu S-B, Rodgers N, Choct M. Optimized necrotic enteritis model producing clinical and subclinical infection of Clostridium perfringens in broiler chickens. Avian Dis. (2010) 54:1058–65. doi: 10.1637/9338-032910-Reg.1
23. Gharib-Naseri, K, Kheravii, S, Keerqin, C, Swick RA, Choct, M, Wu, S-B. Differential expression of intestinal genes in necrotic enteritis challenged broiler chickens with two different Clostridium perfringens strains. Poultry Sci. (2020) 100:100886. doi: 10.1016/j.psj.2020.11.063
24. Ha E, Son, B, Ryu, S. Clostridium perfringens virulent bacteriophage CPS2 and its thermostable endolysin LysCPS2. Viruses. (2018) 10:251. doi: 10.3390/v10050251
25. Morales CA, Oakley BB, Garrish JK, Siragusa GR, Ard MB, Seal BS. Complete genome sequence of the podoviral bacteriophage ΦCP24R, which is virulent for Clostridium perfringens. Arch Virol. (2012) 157:769–72. doi: 10.1007/s00705-011-1218-2
Keywords: bacteriophage, Clostridium perfringens, necrotic enteritis, lession, broiler
Citation: Keerqin C, McGlashan K, Van TTH, Chinivasagam HN, Moore RJ, Choct M and Wu S-B (2022) A lytic bacteriophage isolate reduced Clostridium perfringens induced lesions in necrotic enteritis challenged broilers. Front. Vet. Sci. 9:1058115. doi: 10.3389/fvets.2022.1058115
Received: 30 September 2022; Accepted: 06 December 2022;
Published: 22 December 2022.
Edited by:
Juan D. Latorre, University of Arkansas, United StatesReviewed by:
Thainá Landim de Barros, The Ohio State University, United StatesCopyright © 2022 Keerqin, McGlashan, Van, Chinivasagam, Moore, Choct and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shu-Biao Wu,  c2h1Ymlhby53dUB1bmUuZWR1LmF1
c2h1Ymlhby53dUB1bmUuZWR1LmF1
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.