- Faculty of Veterinary Medicine, University of Calgary, Calgary, AB, Canada
Digital Dermatitis (DD) is a polymicrobial disease characterized by ulcerative lesions on the heel bulb of cattle and for which, despite being reported almost 50 years ago, information on the causative agent is still lacking. Tissue biopsies are regularly collected to identify bacterial presence-absence and their relative abundance in the microbiome, with sufficient evidence for the high abundance of species of Treponema spp. and other anaerobes in lesions. However, it is unclear what the potential of less-invasive sampling methods is for bacterial detection and quantification. This study aimed to test whether less-invasive sampling techniques, such as swabs and fine-needle aspiration (FNA), can be a convenient alternative to tissue biopsies in detecting and quantifying seven DD-associated bacteria in active, ulcerative DD lesions by qPCR. Twenty-two M2 DD lesions were collected using corresponding swabs, aspirates, and biopsies from dairy cows. Presence/absence and quantities of Treponema phagedenis, Treponema medium, Treponema pedis, Porphryromonas levii, Bacteroides pyogenes, Fusobacterium necrophorum, and Fusobacterium mortiferum were correlated, and Bland-Altman plot, McNemar's test, and Cohen's kappa coefficient were used to calculate the agreement among the methods. The quantities of all species were larger in swabs and smaller in aspirates compared to biopsies; however, the differences in bacterial enumeration observed between biopsies and swabs were smaller than in biopsies and aspirates. A strong correlation was observed between the quantity of T. pedis, T. medium, P. levii, and F. mortiferum in biopsies, swabs, and FNA. Yet, T. phagedenis presented the smallest difference between biopsies and swabs, followed by T. pedis and T. medium. In conclusion, swabs, aspirates, and biopsies were equal in their capacity to detect Treponema species based on the good agreement for bacteria presence/absence, with a more limited agreement for the other anaerobes, which were more often present in M2 lesions swabs by qPCR. Bacterial numbers were higher in swabs and lower in aspirates compared to biopsies, with the amounts of treponemes in swabs being closer to biopsies than in aspirates to biopsies. Therefore, aspirates were less suitable for bacterial quantification in DD lesions compared to the other methods.
Introduction
Digital Dermatitis (DD) is a polymicrobial infectious disease that causes skin erosion on the heel bulbs of cattle, but its precise etiology is still unclear. It is one of the main causes of lameness in dairy cattle, compromising animal welfare and leading to direct and indirect economic losses (1–3). DD can be presented either as an acute and ulcerative lesion or as chronic and proliferative. Due to the fastidious nature of the organisms involved in DD, culture-independent molecular approaches have been used to identify certain DD-associated bacteria as important constituents of the DD microbiome that are not as abundant in healthy skin. Treponema spp. are suggested to play a crucial role in this disease because of their dominance in the bacterial population in DD lesions, with the most common species being T. phagedenis, T. medium, and T. pedis (4). Other anaerobes are frequently identified in lesions alongside Treponema spp., such as Porphyromonas, Mycoplasma, Fusobacterium, and Bacteroides spp. (5–7). However, the exact contribution of those bacterial species to lesion development and progression is not fully understood.
DD microbiome studies mostly report bacterial relative abundance and community structure and changes; however, few investigations have used quantitative methods and applied this knowledge to assess the role of different microorganisms in disease initiation and progression at the species level. Recently, real-time polymerase chain reactions (qPCR) assays have been developed to detect and quantify DD-associated bacteria (4, 5). Therefore, the density of microorganisms, patterns of prevalence, and variation can be determined and related to lesion initiation, development, progression, and chronicity, targeting specific species. For this, tissue biopsies have been commonly used. As an invasive sampling method, biopsies require local anesthesia and can lead to bleeding, scarring, and secondary infections (8), making it difficult to sample multiple sites on the same animal or the same site in long-term repeated-sampling studies. Although some Treponema phylotypes were detected more deeply in DD lesions based on fluorescent in situ hybridization (FISH) (9) and Steiner silver staining (10), they were also detected in stratum spinosum by Steiner silver stain (11), on hoof trimmers gloves for up to 3 days in the air by PCR and culture (12), and in superficial biopsies (top 2 mm) by 16S rRNA gene sequencing (13), demonstrating also more superficial presence. Thus, less-invasive sampling techniques, such as swabs and fine-needle aspiration (FNA), might be an alternative to sampling DD lesions helping to answer remaining questions about DD microbiota.
There is sufficient evidence for the presence and high abundance of species of Treponema spp. and other anaerobes in DD lesions; however, hitherto, is unclear what is the potential of less-invasive sampling methods for bacterial detection and quantification. Therefore, this study aimed to test whether swabs and FNA can be a convenient and reliable alternative to tissue biopsies in detecting and quantifying seven DD-associated bacteria at the species level by qPCR.
Materials and methods
Sampling collection and processing
Holstein dairy cattle from three commercial dairy farms in central and southern Alberta, Canada, with active ulcerative DD lesions that were more than 2 cm in diameter (described as stage M2) (11) were sampled. Milking cows were screened for M2 lesions in the milking parlor using a small mirror glued to a plastic kitchen spatula for foot inspection (14, 15) and their lesions were confirmed in the trimming chute. Corresponding biopsies, aspirates, and swabs from a total of 22 M2 lesions were collected. All animal use was approved by the University of Calgary Veterinary Services Animal Care Committee (VSACC) under animal care protocol #AC21-0146. Written informed consent was obtained from the owners for the participation of their animals in this study.
A swab followed by a FNA and a biopsy were collected from the same M2 lesion at the same time point. Once the cow was restrained in the trimming chute, the lesion was cleaned with tap water and dried off with paper towels to confirm the M2 stage based on lesion appearance and size. Local anesthesia was performed before sampling with 3 mL of lidocaine subcutaneously (Lidocaine HCl 2%, Zoetis Canada Inc., Kirkland, QC). The lesion was checked to be dried before sampling to avoid water absorption during the procedure, ensuring that the absorbed material was from the lesion itself. First, the tip of a dry sterile cotton swab was positioned in the center of the lesion at 90 degrees from the lesion surface and rotated for 10 sec with sufficient pressure to absorb fluid from the lesion and immediately placed into an anaerobic transport medium (ATM, Starswab Anaerobic Transport System, Etobicoke, ON). Then, FNA was performed by carefully introducing a 22G needle attached to an empty 10 mL syringe into the lesion to a depth of around 5 mm. Once the needle was inserted, 2 to 5 mL of negative pressure was applied by briskly withdrawing the plunger multiple times, until fluid appeared at the needle hub. The aspirate was immediately injected into ATM. Finally, one biopsy was obtained using a disposable 4 mm biopsy punch (Integra LifeSciences, Princeton, NJ) from the outer margin of the lesion, as regularly performed in previous studies (4, 6). Extra biopsies (n = 11) were collected from the center of the lesions, in addition to the edge, from the same lesion, to test which would be more coherent with swabs. All samples were transported in ATM, which supports bacteria viability, preventing replication during transportation at ambient temperature.
In the lab, specimens were removed from the anaerobic media tubes and were processed under anaerobic conditions (5% CO2, 5% H2, 90% N2) in an anaerobic chamber (Bactron3000, Sheldon Manufacturing, Inc.; Cornelius, OR). Tissue biopsies were sectioned longitudinally, without removing the outer layer of the epidermal skin, and up to 25 mg were processed. Bacterial DNA was extracted using the Qiagen DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) with minor modifications. Up to 25 mg of biopsy, up to 25 mg of aspirates, recovered from ATM using a sterile disposable inoculating loop, and swab cotton heads were placed in a 1.5 mL microcentrifuge tube with 40 μL of proteinase K and 180 μL of tissue lysis (ATL) buffer and incubated at 56°C overnight. Then, the manufacturer's recommendation was followed, with the swabs being processed in the same manner as the biopsies and aspirates except for their removal prior to the ethanol addition step, with the DNA being eluted in 100 μL of DNase/RNase-free water. Extraction controls were performed by following every step of the Qiagen kit without samples. DNA was stored at−20°C until their use in qPCR.
Quantitative polymerase chain reaction (qPCR)
Quantitative real-time PCR (CFX96 real-time system; Rio-Rad Laboratories Inc., Hercules, CA) was carried out for absolute quantification of species-specific gene copy numbers targeting seven clinically relevant DD-associated bacteria. Three different qPCR assays were conducted in this study: a multiplex qPCR developed by Beninger, et al. (4) targeting T. phagedenis, T. medium, and T. pedis, a multiplex qPCR developed by Caddey, et al. (5) for non-Treponema species targeting P. levii, B. pyogenes, and Fusobacterium sp. (97% identity with Fusobacterium mortiferum) (c), and a singleplex qPCR developed by Witcomb, et al. (16) targeting F. necrophorum. Those bacterial species were chosen because their identification as commonly detected organisms in bovine DD lesions in Alberta (4, 5). Primers and fluorescent probes for each reaction are shown in Table 1. For DNA quantification, sample threshold cycle (Ct) values were compared to a standard curve produced from 10-fold serial dilutions of known concentrations of template DNA measured with the Qubit dsDNA HS kit (Life Technologies, Carlsbad, CA, United States) before each reaction. For T. phagedenis, T. pedis, and T. medium, plasmids containing the species-specific regions as described by Beninger, et al. (4) were used as standards, whereas genomic DNAs were used for the non-Treponema qPCR assays as described by Caddey, et al. (5). After obtaining the copy numbers per μL of sample in the reaction, the value was multiplied by the elution volume to generate the copy numbers per sampling method. Negative controls were included in each reaction (extraction controls and non-template controls).
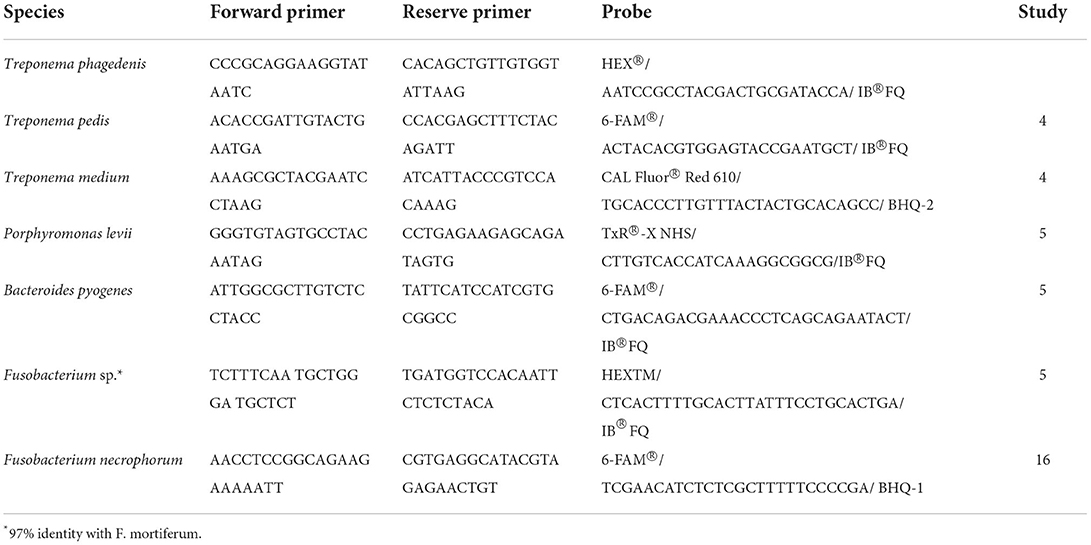
Table 1. Primer and probe sequences for Treponema and non-Treponema species-specific multiplex and singleplex qPCRs.
Statistical analysis
Copy numbers of bacterial DNA in M2 DD lesions collected by biopsy, FNA, and swab were log10-transformed and compared for each species using the Kruskal-Wallis test, followed by Dunn's post–hoc test to evaluate which method is significantly different from each other. To evaluate whether the comparison between samples collected from the center of the lesion with swabs was coherent with the biopsy collection on the edge of the lesion, log copy numbers per mg of tissue in each paired biopsy (edge vs. center) were compared using the Wilcoxon rank test. To investigate the relationship between methods, Spearman's correlation coefficient was used. To assess the agreement between the sampling methods using quantitative data, Lin's Concordance Correlation (CCC) coefficient was calculated, and Bland-Altman plots were produced on continuous data (log copy numbers of bacterial DNA). Lin's CCC measures agreement and ranges from−1 to 1, with 1 being a perfect agreement and−1 a perfect disagreement, and 0 no agreement between the predicted and observed values (17). Bland-Altman plots depict the difference found between the methods plotted against the mean, and a good agreement means that 95% of the data points are within ±2 standard deviations (SD) of the mean difference, representing the upper and lower limits of agreement (LoA) (18). For categorical data (bacterial presence or absence), contingency tables were created to test reliability among the sampling methods agreement using Cohen's Kappa coefficient, which ranges from 0 to 1. The following interpretation was used: κ = 0 indicated no agreement, κ = 0.01–0.2 as slight, κ = 0.21–0.40 as fair, κ = 0.41–0.60 as moderate, κ = 0.61–0.80 as substantial, and κ = 0.81–1.00 as almost perfect agreement (19). All the analyses were performed using R v2022.02.3 (RStudio, Boston, MA, United States) and were considered statistically significant when p < 0.05.
Results
Bacterial quantification was compared between two paired biopsies (edge vs. center, n = 11), and no significant difference in log copy numbers was observed for all species. The average bacterial DNA log copies and standard deviations recovered from edge biopsies was 8.1±0.7, 6.6 ±2.4, 5.3 ±2.9, 2.5 ±1.7, 2.3 ±1.8, 0.5 ±0.8, and 1.1 ±1.6, T. phagedenis, T. medium, T. pedis, P. levii, B. pyogenes, F. necrophorum, and F. mortiferum, respectively, whereas from center biopsies was 8.3 ±0.7, 6.9 ±1.8, 5.6 ±3.0, 2.6 ±1.7, 2.2 ±1.9, 0.7±1.1, and 0.4 ±0.9 for T. phagedenis, T. medium, T. pedis, P. levii, B. pyogenes, F. necrophorum, and F. mortiferum, respectively. The percentage of samples positive for the different target bacteria in DD M2 lesions collected using a punch biopsy, FNA, or swabs based on qPCR is shown in Figure 1. Swabs were more often positive than biopsies and FNA for all target species except for T. phagedenis and T. medium, in which the prevalence was 100% (22/22) in all sampling methods. Conversely, aspirates were more often negative for all target species. The frequency of detection of T. pedis, P. levii, B. pyogenes, F. necrophorum, and F. mortiferum was lower in biopsies than swabs, decreasing from 68% (15/22) to 64% (14/22), 95% (21/22) to 91% (20/22), 100% (22/22) to 64% (14/22), 64% (14/22) to 18% (4/22), and 68% (15/22) to 27% (6/22) in swabs and biopsies, respectively. Comparing FNA to biopsies, T. pedis was detected in the same proportion, whereas P. levii detection in biopsies increased from 50% (11/22) to 91% (20/22), B. pyogenes from 27% (6/22) to 64% (14/22), F. necrophorum from 9% (2/22) to 18% (4/22), and F. mortiferum from 5% (1/22) to 27% (6/22). Swabs of M2 lesions yielded more bacterial cells for all target species than biopsies, while fewer bacterial cells were recovered from aspirates (Figure 2). The average log copy numbers and standard deviations recovered from biopsies was 8.3 ±0.7, 6.5 ±1.5, 4.0 ±3.3, 2.4 ±1.5, 2.1 ±1.7, 0.6 ±1.3, and 0.8 ±1.3 for T. phagedenis, T. medium, T. pedis, P. levii, B. pyogenes, F. necrophorum, and F. mortiferum, respectively, whereas from swabs was 8.9 ±0.6, 7.6 ±1.1, 4.6 ±3.4, 4.2 ±1.5, 5.1 ±0.9, 2.2 ±1.8, and 2.7 ±2.1 and from FNA was 6.4 ±0.7, 4.9 ±0.9, 2.9 ±2.4, 1.0 ±1.2, 1.0 ±1.6, 0.3 ±1.0, and 0.1 ±0.5 for T. phagedenis, T. medium, T. pedis, P. levii, B. pyogenes, F. necrophorum, and F. mortiferum, respectively. The number of T. phagedenis, T. medium, P. levii, B. pyogenes, and F. mortiferum cells detected by qPCR was significantly different among all sampling methods; however, no difference was observed between the number of T. pedis recovered from lesion biopsies and swabs. When biopsies were compared to FNA, no difference was observed for T. pedis and F. necrophorum quantities. T. phagedenis was the most abundant species in all sampling techniques, whereas F. mortiferum was the least abundant in either biopsy or FNA, and F. necrophorum in swabs.
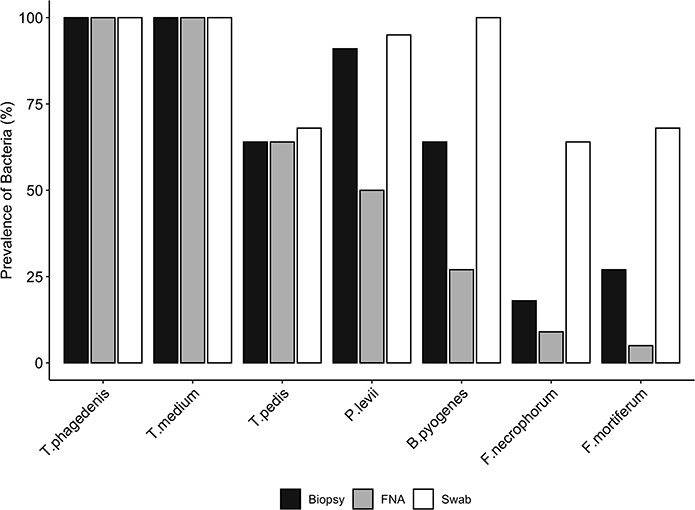
Figure 1. Prevalence (%) of each bacterial species presence in biopsies, swabs, and fine-needle aspiration (FNA) from M2 DD lesions detected by qPCR. A McNemar's test indicated significant differences in the proportions of the presence of Bacteroides pyogenes, Fusobacterium mortiferum, and Fusobacterium necrophorum between biopsies and swabs, and Porphyromonas levii, Bacteroides pyogenes, and Fusobacterium mortiferum between biopsies and aspirates (p < 0.01); however, no significant difference was observed for the other species.
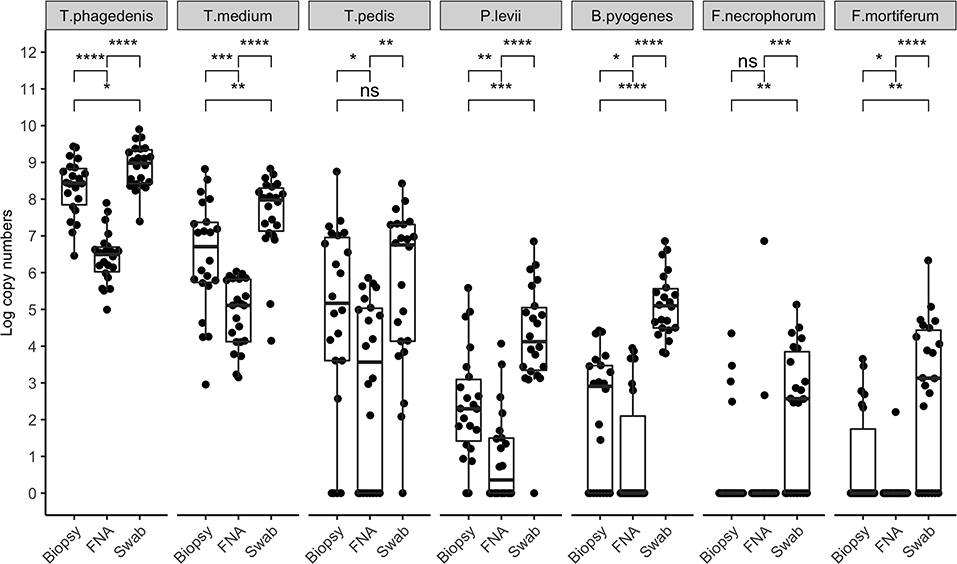
Figure 2. Bacterial quantification (log copy numbers) by species for each sampling method. Boxplot showing the distribution of data, with each dot representing one sample (n = 22). Related p-values for Dunn's post–hoc test among the sampling methods: biopsy, fine-needle aspiration (FNA), and swab for Treponema phagedenis, Treponema medium, Treponema pedis, Porphyromonas levii, Bacteroides pyogenes, Fusobacterium necrophorum, and F. mortiferum. No significant difference was observed for T. pedis between biopsy and swabs, and F. necrophorum in biopsy and aspirates only. Asterisks and horizontal bars indicate differences in bacterial quantification among the sampling methods: ns: p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Correlation coefficient and Lin's CCC among the sampling methods for each species are given in Table 2. Correlations between the number of the target bacterial species in swabs, aspirates, and biopsies were investigated by calculating Spearman's rho (ρ) correlation coefficient. Strong positive correlations were observed between the quantity of T. pedis and T. medium in biopsies and both less-invasive sampling methods. No significant correlations were observed for T. phagedenis when biopsies were compared to swabs and FNA (Figure 3). For the non-Treponema species, strong positive correlations between the number of P. levii and F. mortiferum in biopsies and both less-invasive sampling methods were observed and between the number of B. pyogenes in biopsies and swabs. No significant correlations were observed for F. necrophorum when biopsies were compared to swabs and FNA and for B. pyogenes in biopsies and FNA (Figure 4). Lin's CCC was also calculated to assess agreement between the log copy number of the target species in both sampling methods. A positive agreement was observed between bacterial quantification in biopsies and swabs, having T. pedis with the highest coefficient, followed by T. medium, P. levii, B. pyogenes, F. mortiferum, T. phagedenis, and F. necrophorum. When bacterial numbers were compared between biopsies and FNA, the highest coefficient was observed for T. pedis, followed by T. pedis, P. levii, B. pyogenes, T. medium, F. mortiferum, F. necrophorum, and T. phagedenis.
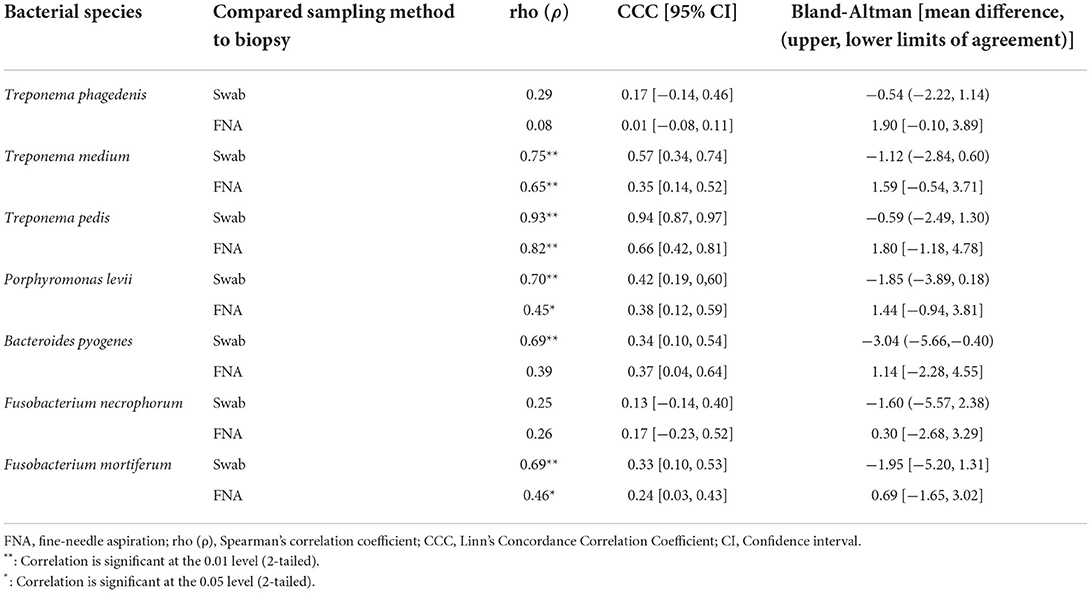
Table 2. Spearman's correlation coefficient, Lin's Concordance Correlation Coefficient, and Bland-Altman results of bacterial quantification of each species in DD M2 lesions (n = 22) among the sampling methods.
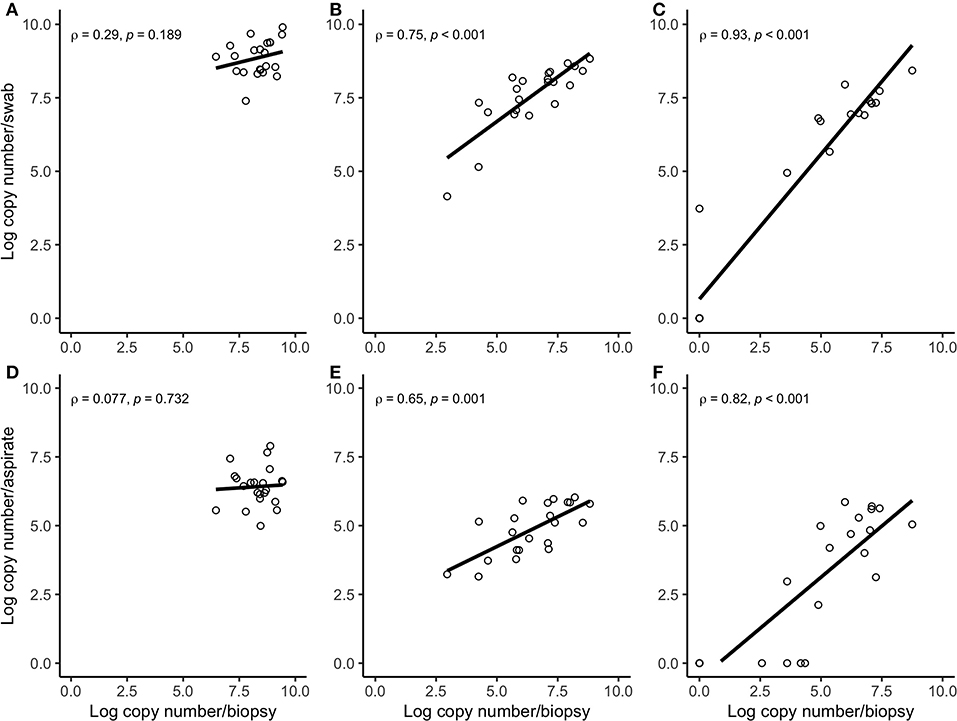
Figure 3. Spearman correlation plots between the amount (log-transformed copy numbers) of Treponema phagedenis (A), Treponema medium (B), Treponema pedis (C) in swabs and biopsies of bovine DD active M2 lesions, and T. phagedenis (D), T. medium (E), and T. pedis (F) in aspirates and biopsies of bovine DD active M2 lesions. There were significant correlations between the quantity of T. pedis and T. medium in biopsies and swabs and biopsies and aspirates. There were no significant correlations between the quantity of T. phagedenis, neither in biopsies and swabs nor in biopsies and aspirates.
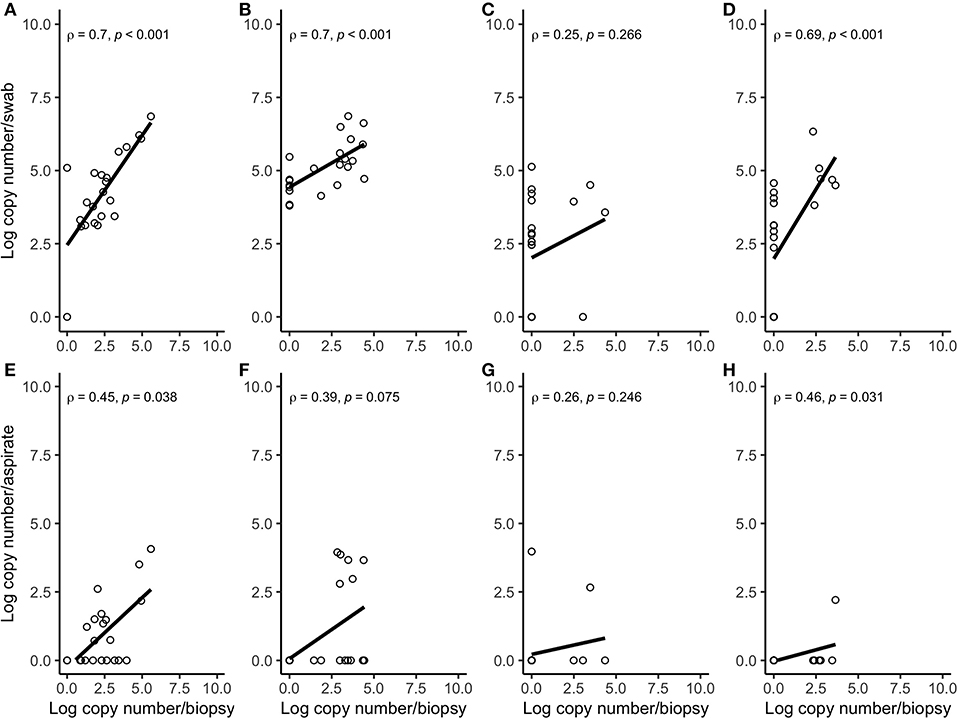
Figure 4. Spearman correlation plots between the amount (log-transformed copy numbers) of Porphyromonas levii (A), Bacteroides pyogenes (B), Fusobacterium necrophorum (C), and Fusobacterium mortiferum (D) in swabs and biopsies of bovine DD active M2 lesions, and P. levii (E), B. pyogenes (F), F. necrophorum (G), and F. mortiferum (H) in aspirates and biopsies of bovine DD active M2 lesions. There were significant correlations between the quantity of P. levii, B. pyogenes, and F. mortiferum in biopsies and swabs and P. levii, F. mortiferum in biopsies and aspirates. There were no significant correlations between the quantity of F. necrophorum, neither in biopsies and swabs nor in biopsies and aspirates and B. pyogenes in biopsies and aspirates.
The difference in bacterial enumeration depending on the sampling method was plotted against the mean using a Bland-Altman plot (Figures 5, 6). The difference in log copy numbers comparing biopsies with swabs for each target bacterial species was−0.5 for T. phagedenis,−0.6 for T. pedis,−1.1 for T. medium,−1.6 for F. necrophorum,−1.9 for P. levii,−2.0 for F. mortiferum, and−3.0 for B. pyogenes, whereas the comparison between the log copy numbers in biopsies and FNA resulted in the difference of 1.9 for T. phagedenis, 1.6 for T. medium, 1.8 for T. pedis, 1.4 for P. levii, 1.1 for B. pyogenes, 0.3 for F. necrophorum, and 0.7 for F. mortiferum (Supplementary material 1). B. pyogenes quantification in biopsies and swabs presented the greatest bias from zero, indicating the highest discrepancy between those methods. In contrast, T. phagedenis presented the smallest bias from zero. In the biopsies and FNA comparison, T. phagedenis presented the greatest bias from zero, and F. necrophorum the smallest. Furthermore, based on the LoA width, the widest LoA between biopsies and swabs quantification was observed for F. necrophorum followed by F. mortiferum, B. pyogenes, P. levii, T. pedis, T. medium, and T.phagedenis, in with decreasing order (Table 2). Regarding biopsies and FNA, the widest LoA was detected for B. pyogenes followed by F. necrophorum, T. pedis, P. levii, F. mortiferum, T. medium and T. phagedenis, in with decreasing order.
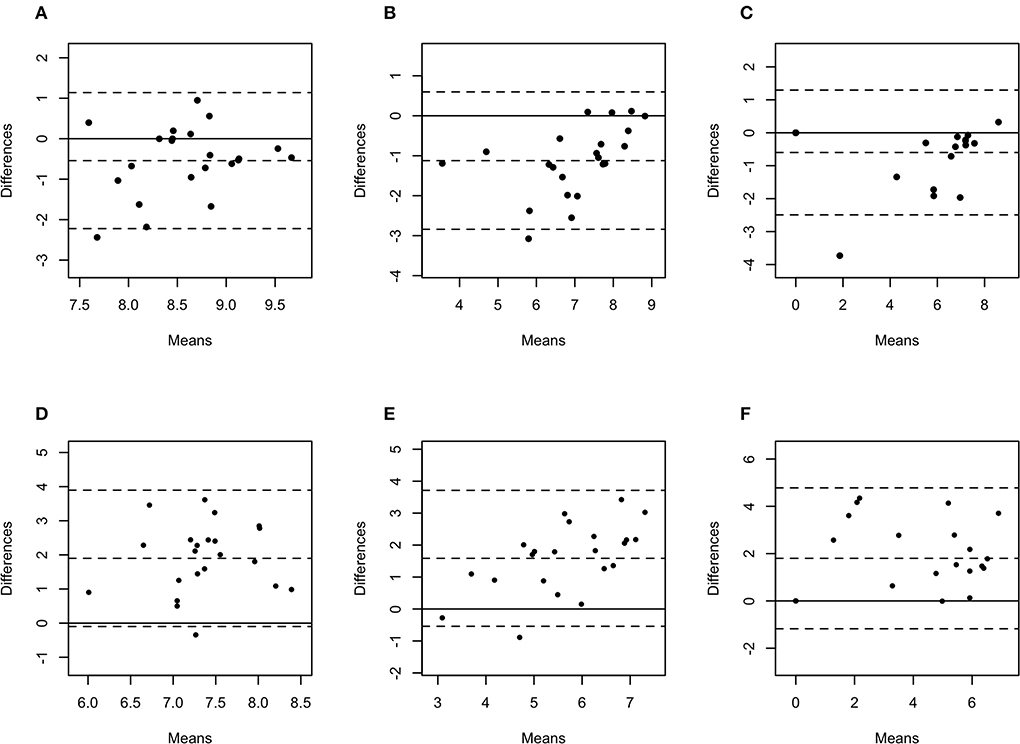
Figure 5. Bland-Altman plots representing the agreement between the quantity of Treponema phagedenis (A), Treponema medium (B), and Treponema pedis (C) in biopsies and swabs, and T. phagedenis (D), T. medium (E), and T. pedis (F) in biopsies and aspirates. The center dotted represents the mean of difference (bias), and the two contiguous dotted lines represent the upper and lower limit of 95% of agreement. The solid line crossing the axis at zero represents no difference between the sampling methods (perfect agreement; bias = 0).
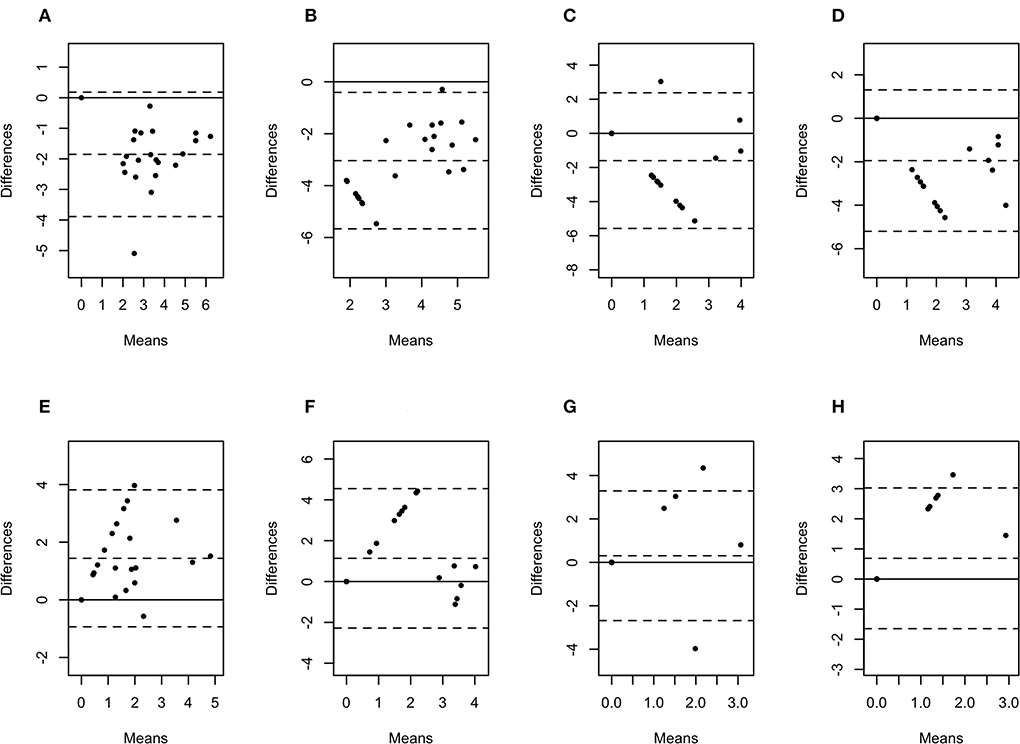
Figure 6. Bland-Altman plots representing the agreement between the quantity of Porphyromonas levii (A), Bacteroides pyogenes (B), Fusobacterium necrophorum (C), and Fusobacterium mortiferum (D) in biopsies and swabs, and P. levii (E), B. pyogenes (F), F. necrophorum (G), and F. mortiferum (H) in biopsies and aspirates. The center dotted represents the mean of difference (bias), and the two contiguous dotted lines represent the upper and lower limit of 95% of agreement. The solid line crossing the axis at zero represents no difference between the sampling methods (perfect agreement; bias = 0).
The qualitative outcomes for the presence/absence of target bacterial species were calculated using Cohen's kappa coefficient of agreement and reliability between biopsies and both less-invasive sampling methods (Table 3). The comparison between biopsies and both less-invasive methods resulted in a perfect agreement for T. phagedenis and T. medium. For T. pedis, the agreement was almost perfect between biopsies and swabs, and substantial between biopsies and FNA. The agreement of the presence in biopsies and swabs was substantial for P. levii, fair for F. mortiferum, slight for F. necrophorum, and null for B. pyogenes. In biopsies and FNA, the agreement was fair for B. pyogenes, F. mortiferum, and F. necrophorum, and slight for P. levii. Based on McNemar's test to assess whether the proportions of the presence of the target bacteria in the samples differed between biopsies and the less-invasive methods, a difference was observed for B. pyogenes, F. necrophorum, and F. mortiferum presence detection in comparison with swabs and observed for P. levii and B. pyogenes presence detection in comparison with FNA; however, no significant difference was observed for the other bacterial species among the sampling methods.

Table 3. Cohen's kappa coefficient and McNemar's p-value results of bacterial detection of the presence of each species in DD M2 lesions (n = 22) among the sampling methods.
Discussion
Using species-specific qPCR, swabs and aspirates of M2 DD lesions efficiently detect the presence of T. phagedenis, T. pedis, and T. medium at a similar frequency and in good agreement with lesion biopsies, but little agreement was observed for the detection of P. levii, B. pyogenes, F. necrophorum, F. mortiferum presence, which were more often present in swabs. Swabs collected higher bacterial loads than biopsies, with the smallest difference in quantity observed for the target Treponema species and the largest discrepancy for the non-Treponema anaerobes. The quantity of T. phagedenis, T. medium, and T. pedis was larger in swabs and smaller in aspirates when compared to biopsies; however, the differences in bacterial enumeration observed between biopsies and swabs are smaller than in biopsies and aspirates. Hence, the results from this study suggest that swabs provide a simpler and less-invasive technique for detecting the presence of treponemes in DD M2 lesions using qPCR. Because of the greater discrepancies in the number of bacteria recovered from biopsies and FNA, only swabs were effective in quantifying amounts of treponemes in DD lesions similar to biopsies, and aspirates were less suitable for bacterial quantification compared to other methods. However, taking into account that there is not enough evidence to attribute specific roles to each of the DD-associated bacteria as secondary or primary invaders, the greater disagreement among sampling methods might indicate different niches for Treponema and non-Treponema species in the lesion. T. phagedenis was the most prevalent species, matching the results observed in earlier studies in M2 lesions in dairy (4) and beef cattle (5). However, T. medium was more often detected than the previously reported quantities in lesions of the same stage (M2) in beef cattle, while T. pedis was detected in similar quantities, suggesting there may be differences in microbial community structure due to distinct housing and management practices between dairy and beef cattle operations.
Previous investigations of treponemes involved in DD using swabs detected them on hoof knifes (20, 21), DD lesions in captive European bison (22), and Contagious Ovine Digital Dermatitis (CODD) lesions experimentally induced in sheep (23); however, those results were based on relative abundance and disregarded total bacterial load, providing information about the presence/absence of bacterial species rather the amount of particular bacterial species. Real-time qPCR enables the amplification of species-specific genes, which are detected by specific fluorescent probes, and the DNA copies enumeration can be calculated using a standard curve produced with serial dilutions of known concentrations of template DNA, converting threshold cycles to the absolute quantity of target gene (24). With this molecular technique, we can increase the knowledge about bacterial quantifies and thereby further investigate their roles in the disease.
Biopsies sampled a more focused area at the lesion edge, capturing the three-dimensional structure of the skin, while swabs sampled only the surface area of lesions, but with a greater exudate absorption capacity. When the microbiome of CODD lesions in sheep was compared between swabs and biopsies, researchers observed a distinct bacterial diversity, with Spirochetaceae being more abundant in biopsies and Porphyromonadaceae being abundant in both methods (23). Similarly, Spirochetes were identified in DD deep biopsies, and Firmicutes in superficial strata using 16S rRNA gene sequencing, demonstrating the higher presence of opportunistic pathogens in a superficial skin layer (25). Additional studies investigating the microorganisms involved in udder cleft dermatitis in dairy cows failed to culture treponemes from swab (26), but successfully detected them by nested PCR (27). Since treponemes are difficult to cultivate, a cultured-based method may result in false negative outcomes. Furthermore, previous studies have demonstrated that Treponema species are present in different tissue layers in DD lesions based on fluorescent in situ hybridization (FISH) (9, 28) and Steiner silver staining (10). T. phagedenis has been detected in all skin layers, whereas T. medium was detected deeper. Our results have shown the presence of T. phagedenis and T. medium in 100% of samples using all sampling methods, and T. pedis in 68% of the swabs, 64% of aspirates, and 64% of biopsies demonstrating that, although those species are considered invasive, their DNA could be recovered by surface swabs. The M2 DD lesions assessed in this study are ulcerative, hemorrhagic, open wounds that lost completely the stratum corneum (11), which along with the application of sufficient pressure to obtain wound exudate using the swab allowed a greater detection of Treponema species DNA in this study. Although lidocaine has been suggested to have antimicrobial activities (29, 30), it can affect microbial activities but not bacterial DNA detection by qPCR (31). Therefore, the use of lidocaine in this study prior to sampling collection is unlikely to affect our results. In addition, active DD lesions can be painful, but not in all cases (32). As a consequence of local anesthesia typically not being required for swabbing DD lesions for research purposes, animals may experience some transient discomfort and pain during the swab sampling. However, the non-invasive nature of this method and the prevention of an open wound after the procedure in the heel bulb of cattle give preference to sampling by swabs rather than biopsies.
Despite the similar frequency of T. phagedenis, T. pedis, and T. medium in biopsies and FNA, and the usefulness of this technique to detect their presence in DD lesions, their numbers were significantly lower in aspirates. Fluids obtained from FNA are from deeper layers of tissue, being useful for microbiological studies. This method has been used to diagnose soft tissue infections (33–35) and the rate of bacterial DNA recovery differs among these studies depending on the disease characteristics. Because it penetrates deeper into the tissue compared to swab, FNA is considered equivalent to tissue biopsy without being more invasive and may recover more deeply invaded treponemes. However, our results demonstrate that the amount of bacterial DNA recovered from aspirates was significantly lower than that from biopsies and swabs, with a large difference in bacterial enumeration present in the lesion. This may be explained by the small fluid volume recovered from aspirates, and the challenge of recapturing the aspirate from the semi-solid anaerobic transport media prior to DNA extraction.
There was a positive correlation between the amount detected by FNA, swabs, and biopsies for all bacterial species, but this correlation was strong for T. pedis, T. medium, P. levii, F. mortiferum, and B. pyogenes, and weak for T. phagedenis and F. necrophorum in swabs and for T. phagedenis, F. necrophorum, and B. pyogenes in FNA. One important point is that correlation reveals the relationship between the outcomes but does not measure the agreement between sampling methods, meaning that as the log copy numbers of an organism increase in swabs or aspirates, there is an increase in log copy numbers of the same organism in biopsies. As shown in Figures 3, 4, T. phagedenis, F. mortiferum, and F. necrophorum presented a small variability in quantities, which results in a lower apparent correlation. However, T. phagedenis data is distinct from F. mortiferum and F. necrophorum. Whereas T. phagedenis in each sample (100%, 22/22) presented a small range of high log copy numbers in both methods, F. necrophorum and F. mortiferum presented an equal variability (homoscedasticity) since their log copy numbers in 81% (18/22) and 73% (16/22) of the biopsies, 36% (8/22) and 32% (7/22) of the swabs, and 91% (20/22) and 95% (21/22) of the aspirates was zero. For Lin's CCC, which measures how far the best-fit line deviates from the 45°line (perfect agreement), a small CCC between biopsies and swabs was observed for T. phagedenis, F. mortiferum, and F. necrophorum and between biopsies and FNA for those three species, suggesting a poorer agreement. Given that CCC also depends on the range of data, these correlation coefficients also need to be interpreted with caution.
Bland-Altman plot analysis was used to measure agreement between biopsies, swabs, and FNA bacterial quantification. Although the correlation observed for T. phagedenis was weak, this organism presented the lowest bias between biopsies and swabs through the Bland-Altman plot analysis, suggesting that on average, swabs measured amounts of T. phagedenis close to those by biopsies. Conversely, the lowest bias between biopsies and FNA was observed for F. necrophorum. A bias equal to zero means that both methods obtained the same bacterial quantity. The observed bias for each target species suggested that on average swabs measured 0.5, 0.6, 1.1, 1.6, 1.9, 2.0, and 3.0 log copies more than biopsies for T. phagedenis, T. pedis, T. medium, F. necrophorum, P. levii, F. mortiferum, and B. pyogenes, respectively. In contrast, FNA measured on average 1.9, 1.6, 1.8, 1.4, 1.1, 0.3, and 0.7 log copies less than biopsies for T. phagedenis, T. pedis, T. medium, F. necrophorum, P. levii, F. mortiferum, and B. pyogenes, respectively. Those results emphasize the larger amounts of bacterial DNA recovered from swabs and the smaller numbers recovered from FNA. The LoA representing 95% of the data that were within ±2 SD of the mean difference between biopsies and swabs and biopsies and FNA were wider for non-Treponema species than for treponemes, which had the narrowest LoA. Therefore, a better equivalence and agreement can be assumed for treponemes between biopsies and swabs, given their lower bias and LoA. Regarding the comparison between biopsies and FNA, treponemes presented the highest bias but the lowest LoA, emphasizing again the bigger difference in the number of bacterial DNA, but a better agreement than non-Treponema species in biopsies and FNA. The main reason for this finding is the high percentage of samples with negative detection results for the non-Treponema target species, given by bacterial counts of zero. As a consequence, the difference is zero in the Bland-Altman plot. However, the larger range of 95% of data represents the low agreement between the methods. The question remains if one or two log copy numbers can be considered clinically relevant because the minimum infective dose for DD-associated species and the importance of bacterial doses are unknown. Further research should be undertaken to investigate DD transmission and infective dose to help the interpretation of differences and similarities in sample type agreement.
Bacterial presence or absence analyses between the sampling methods detected agreement and reliability of results for each organism. There was a full agreement for the presence of T. phagedenis and T. medium in swabs, aspirates, and biopsies since all samples were positive. Although T. pedis and P. levii had the same proportion of agreement between biopsies and swabs (95.5%), the reliability of the T. pedis results was 90%, whereas only 64% for P. levii. Therefore, the agreement observed for P. levii detection was deemed less than for T. pedis. F. mortiferum, F. necrophorum, and B. pyogenes presented fair (30%), slight (7%), or null (0%) reliability results, respectively, despite some degree of agreement. A worse agreement was detected between biopsies and FNA, with reliability being substantial for T. pedis (56%), fair for B. pyogenes (35%), F. necrophorum (24%), F. mortiferum (23%), and slight for P. levii (18%). B. pyogenes, F. necrophorum, and F. mortiferum demonstrated the greatest discrepancies between the sampling methods, being more frequently detected in swabs than biopsies. Higher levels of non-Treponema organisms in swabs than biopsies and FNA might reflect the high density of those organisms on the lesion surface and their limited involvement in DD as secondary invaders. This argument has been supported by other researchers assessing the spatial distribution in lesions from FISH (7, 36) and the microbiome of DD lesions (6). P. levii, F. necrophorum and B. pyogenes are considered opportunistic pathogens and normal inhabitants of the rumen, and they have been previously associated with footrot, hepatic abscesses, and uterine infections in cattle (37, 38). However, a study assessing bacterial gene expression in DD lesions reinforces the involvement of Porphyromonas and Bacteroides in DD initiation, but not Fusobacterium (39). Indeed, the lowest detection frequencies and agreements in this study were related to F. necrophorum and F. mortiferum. Although it is possible that these non-Treponema anaerobes were contaminants and found in association with the lesions because of the lack of antiseptic cleaning before sampling, they have been frequently found in DD lesions (5, 6, 28), and they might have a distinct spatial distribution within the lesion, with their niche being more superficial than treponemes. Given our results, the high abundance of non-Treponema anaerobes detected from swabs compared to biopsies needs to be taken into consideration. Longitudinal studies into different sampling methods might reveal more differences in the quantification and detection of DD-associated bacterial species.
Some limitations regarding this study should be mentioned. First, we collected swab samples from the center of the lesion without covering a precise surface area; therefore, the comparison was made based on the total DNA yield from the swab cotton head, and the results could not be normalized and expressed in log copy numbers per cm2. Further investigation might be required to standardize the sampling collection in a known surface area. Second, the DNA detected by qPCR was not differentiated between viable and dead cells. Next studies might test the use of propidium monoazide (PMA) to treat the samples prior to DNA extraction for live/dead distinction by qPCR. Finally, no healthy tissue samples were obtained in the present study; we targeted only ulcerative, active lesions for comparison. This approach could potentially reveal bias in the results since bacteria from healthy skin or environment could also be detected by qPCR, especially by using surface swabs. Future surveys of healthy skin (stage M0) and other DD lesion stages, such as chronic (stage M4) and early stage (M1), will be necessary to expand the swab/FNA/biopsy comparison and agreement among different stages of the disease. Especially the collection of M1 lesions could be targeted to have a greater range of T. phagedenis, which had a lower apparent correlation due to limited variation between the samples.
In conclusion, it seems reasonable to employ swabs and FNA and real-time qPCR to screen for the presence of treponemes in M2 DD lesions. T. phagedenis, T. medium, and T. pedis were detected similarly in swabs, aspirates, and biopsies, and P. levii, F. mortiferum, F. necrophorum, and B. pyogenes were detected more frequently in swabs compared to aspirates and biopsies. More bacterial DNA was recovered from swabs, followed by biopsies and aspirates. A good and reliable agreement for bacterial quantification among the methods was observed only when T. phagedenis, T. medium, and T. pedis were targeted. Their quantities were larger in swabs and smaller in aspirates in comparison to biopsies; however, the differences in bacterial enumeration observed between biopsies and swabs were smaller than between biopsies and aspirates. The less-invasive sampling methods offer the opportunity to sample multiple places in the same animal or monitor DD initiation, treatment effectiveness, and progression in a long-term repeat-sampling study targeting treponemes involved in this disease.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by the University of Calgary Veterinary Services Animal Care Committee (VSACC) under animal care protocol #AC21-0146. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
APD and JDB contributed to the conception and design of the study. APD collected and processed the dairy cattle lesion samples, performed all statistical analyses, and wrote the manuscript. JDB edited and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Collaborative Research and Development grants (CRD) to JDB (CRDPJ/536202-2018).
Acknowledgments
We thank the Alberta dairy farmers who were involved in the study as well as the hooftrimmers Elbert Koster from No Tilt Hoof Trimming and Mark and Henri van Verk from Chinook Hoof Care for their assistance in identifying potential study participants. We thank Dr. Karin Orsel, Colton Scott, Susan Pyakurel, and Derrick Zhang for their assistance in the sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1040988/full#supplementary-material
References
1. Solano L, Barkema HW, Pajor EA, Mason S, LeBlanc SJ, Zaffino Heyerhoff JC, et al. Prevalence of lameness and associated risk factors in Canadian Holstein-Friesian cows housed in freestall barns. J Dairy Sci. (2015) 98:6978–91. doi: 10.3168/jds.2015-9652
2. Solano L, Barkema HW, Mason S, Pajor EA, LeBlanc SJ, Orsel K. Prevalence and distribution of foot lesions in dairy cattle in Alberta, Canada. J Dairy Sci. (2016) 99:6828–41. doi: 10.3168/jds.2016-10941
3. Bruijnis MRN, Hogeveen H, Stassen EN. Assessing economic consequences of foot disorders in dairy cattle using a dynamic stochastic simulation model. J Dairy Sci. (2010) 93:2419–32. doi: 10.3168/jds.2009-2721
4. Beninger C, Naqvi SA, Naushad S, Orsel K, Luby C, Derakhshani H. Associations between digital dermatitis lesion grades in dairy cattle and the quantities of four Treponema species. Vet Res. (2018) 49:111. doi: 10.1186/s13567-018-0605-z
5. Caddey B, Orsel K, Naushad S, Derakhshani H, De Buck J. Identification and quantification of bovine digital dermatitis-associated microbiota across lesion stages in feedlot beef cattle. mSystems. (2021) 6:e0070821. doi: 10.1128/mSystems.00708-21
6. Krull AC, Shearer JK, Gorden PJ, Cooper VL, Phillips GJ, Plummer PJ. Deep sequencing analysis reveals temporal microbiota changes associated with development of bovine digital dermatitis. Infect Immun. (2014) 82:3359–73. doi: 10.1128/IAI.02077-14
7. Moreira TF, Facury Filho EJ, Carvalho AU, Strube ML, Nielsen MW, Klitgaard K, et al. Pathology and bacteria related to digital dermatitis in dairy cattle in all year round grazing system in Brazil. PLoS ONE. (2018) 13:e0193870. doi: 10.1371/journal.pone.0193870
8. Nischal U, Nischal K, Khopkar U. Techniques of skin biopsy and practical considerations. J Cutan Aesthet Surg. (2008) 1:107. doi: 10.4103/0974-2077.44174
9. Moter A, Leist G, Rudolph R, Shrank K. Fluorescence in situ hybridization shows spatial distribution of as yet uncultured treponemes in biopsies from digital dermatitis lesions. Microbiology ((Reading). (1998) 144:2459–67. doi: 10.1099/00221287-144-9-2459
10. Berry SL, Read DH, Famula TR, Mongini A, Döpfer D. Long-term observations on the dynamics of bovine digital dermatitis lesions on a California dairy after topical treatment with lincomycin HCl. Vet J. (2012) 193:654–8. doi: 10.1016/j.tvjl.2012.06.048
11. Döpfer D, ter Huurne AAHM, Cornelisse JL, van Asten AJAM, Koopmans A, Meijer FA, et al. Histological and bacteriological evaluation of digital dermatitis in cattle, with special reference to spirochaetes and Campylobacter faecalis. Vet Rec. (1997) 140:620–3. doi: 10.1136/vr.140.24.620
12. Angell JW, Clegg SR, Grove-White DH, Blowey RW, Carter SD, Duncan JS, et al. Survival of contagious ovine digital dermatitis (CODD)-associated treponemes on disposable gloves after handling CODD-affected feet. Vet Rec. (2017) 181:89. doi: 10.1136/vr.104228
13. Zinicola M, Lima F, Lima S, Machado V, Gomez M, Döpfer D, et al. Altered microbiomes in bovine digital dermatitis lesions, and the gut as a pathogen reservoir. PLoS ONE. (2015) 10:e0120504. doi: 10.1371/journal.pone.0120504
14. Relun A, Guatteo R, Roussel P, Bareille N, A. simple method to score digital dermatitis in dairy cows in the milking parlor. J Dairy Sci. (2011) 94:5424–34. doi: 10.3168/jds.2010-4054
15. Solano L, Barkema HW, Jacobs C, Orsel K. Validation of the M-stage scoring system for digital dermatitis on dairy cows in the milking parlor. J Dairy Sci. (2017) 100:1592–603. doi: 10.3168/jds.2016-11365
16. Witcomb LA, Green LE, Kaler J, Ul-Hassan A, Calvo-Bado LA, Medley GF, et al. A longitudinal study of the role of Dichelobacter nodosus and Fusobacterium necrophorum load in initiation and severity of footrot in sheep. Prevent Vet Med. (2014) 115:48–55. doi: 10.1016/j.prevetmed.2014.03.004
17. Lin LI-K, A. concordance correlation coefficient to evaluate reproducibility. Biometrics. (1989) 45:255. doi: 10.2307/2532051
18. Bland, Altman. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. (1986) 327:307–10. doi: 10.1016/S0140-6736(86)90837-8
19. Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. (1977) 33:159. doi: 10.2307/2529310
20. Gillespie A, Carter SD, Blowey RW, Evans N. Survival of bovine digital dermatitis treponemes on hoof knife blades and the effects of various disinfectants. Vet Rec. (2020) 186:67. doi: 10.1136/vr.105406
21. Sullivan LE, Blowey RW, Carter SD, Duncan JS, Grove-White DH, Page P, et al. Presence of digital dermatitis treponemes on cattle and sheep hoof trimming equipment. Vet Rec. (2014) 175:201. doi: 10.1136/vr.102269
22. Hoby S, Jensen TK, Brodard I, Gurtner C, Eicher R, Steiner A, et al. Detection of treponemes in digital dermatitis lesions of captive European bison (Bison bonasus). PLoS ONE. (2021) 16:e0255921. doi: 10.1371/journal.pone.0255921
23. Duncan JS, Angell JW, Richards P, Lenzi L, Staton GJ, Grove-White D, et al. The dysbiosis of ovine foot microbiome during the development and treatment of contagious ovine digital dermatitis. Anim Microbiome. (2021) 3:19. doi: 10.1186/s42523-021-00078-4
24. Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, et al. The real-time polymerase chain reaction. Mol Aspects Med. (2006) 27:95–125. doi: 10.1016/j.mam.2005.12.007
25. Santos TMA, Pereira RV, Caixeta LS, Guard CL, Bicalho RC. Microbial diversity in bovine papillomatous digital dermatitis in Holstein dairy cows from upstate New York. FEMS Microbiol Ecol. (2012) 79:518–29. doi: 10.1111/j.1574-6941.2011.01234.x
26. van Engelen E, Dijkstra T, Meertens NM, van Werven T. Bacterial flora associated with udder cleft dermatitis in Dutch dairy cows. J Dairy Sci. (2021) 104:728–35. doi: 10.3168/jds.2020-18414
27. Sobhy NM, Mahmmod YS, Refaai W, Awad A. Molecular detection of Treponema species organisms in foremilk and udder cleft skin of dairy cows with digital dermatitis. Trop Anim Health Prod. (2020) 52:815–21. doi: 10.1007/s11250-019-02072-0
28. Klitgaard K, Foix Bretó A, Boye M, Jensen TK. Targeting the treponemal microbiome of digital dermatitis infections by high-resolution phylogenetic analyses and comparison with fluorescent in situ hybridization. J Clin Microbiol. (2013) 51:2212–9. doi: 10.1128/JCM.00320-13
29. Johnson SM, Saint John BE, Dine AP. Local anesthetics as antimicrobial agents: a review. Surg Infect. (2008) 9:205–13. doi: 10.1089/sur.2007.036
30. Razavi BM, Fazly Bazzaz BS. A review and new insights to antimicrobial action of local anesthetics. Eur J Clin Microbiol Infect Dis. (2019) 38:991–1002. doi: 10.1007/s10096-018-03460-4
31. Lefeuvre B, Cantero P, Ehret-Sabatier L, Lenormand C, Barthel C, Po C, et al. Effects of topical corticosteroids and lidocaine on Borrelia burgdorferi sensu lato in mouse skin: potential impact to human clinical trials. Sci Rep. (2020) 10:10552. doi: 10.1038/s41598-020-67440-5
32. Cutler JHH, Cramer G, Walter JJ, Millman ST, Kelton DF. Randomized clinical trial of tetracycline hydrochloride bandage and paste treatments for resolution of lesions and pain associated with digital dermatitis in dairy cattle. J Dairy Sci. (2013) 96:7550–7. doi: 10.3168/jds.2012-6384
33. Abdulbasith K, Bhaskar M, Munisamy M, Nagarajan R. Study of fine-needle aspiration microbiology versus wound swab for bacterial isolation in diabetic foot infections. Indian J Med Res. (2020) 152:312. doi: 10.4103/ijmr.IJMR_1151_18
34. Lee PC, Turnidge J, McDonald PJ. Fine-needle aspiration biopsy in diagnosis of soft tissue infections. J Clin Microbiol. (1985) 22:80–3. doi: 10.1128/jcm.22.1.80-83.1985
35. Parikh A, Hamilton S, Sivarajan V, Withey S, Butler P. Diagnostic fine-needle aspiration in postoperative wound infections is more accurate at predicting causative organisms than wound swabs. Annals. (2007) 89:166–7. doi: 10.1308/003588407X155761
36. Nielsen MW, Strube ML, Isbrand A, Al-Medrasi WDHM, Boye M, Jensen TK, et al. Potential bacterial core species associated with digital dermatitis in cattle herds identified by molecular profiling of interdigital skin samples. Vet Microbiol. (2016) 186:139–49. doi: 10.1016/j.vetmic.2016.03.003
37. Jeon SJ, Vieira-Neto A, Gobikrushanth M, Daetz R, Mingoti RD, Parize ACB, et al. Uterine microbiota progression from calving until establishment of metritis in dairy cows. Appl Environ Microbiol. (2015) 81:6324–32. doi: 10.1128/AEM.01753-15
38. Nagaraja TG, Narayanan SK, Stewart GC, Chengappa MM. Fusobacterium necrophorum infections in animals: pathogenesis and pathogenic mechanisms. Anaerobe. (2005) 11:239–46. doi: 10.1016/j.anaerobe.2005.01.007
Keywords: hairy heel warts, cattle, qPCR (quantitative PCR), Treponema, test agreement, sampling techniques, dairy cows
Citation: Dias AP and De Buck J (2022) Detection and quantification of bacterial species DNA in bovine digital dermatitis lesions in swabs and fine-needle aspiration versus biopsies. Front. Vet. Sci. 9:1040988. doi: 10.3389/fvets.2022.1040988
Received: 09 September 2022; Accepted: 07 November 2022;
Published: 18 November 2022.
Edited by:
Ameer Megahed, University of Illinois at Urbana–Champaign, United StatesReviewed by:
Paul Plummer, Iowa State University, United StatesNicholas James Evans, University of Liverpool, United Kingdom
Copyright © 2022 Dias and De Buck. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeroen De Buck, amRlYnVja0B1Y2FsZ2FyeS5jYQ==
 Angelica Petersen Dias
Angelica Petersen Dias Jeroen De Buck
Jeroen De Buck