- 1School of Science, RMIT University, Bundoora, VIC, Australia
- 2Scolexia Pty Ltd., Moonee Ponds, VIC, Australia
- 3Institute for Future Farming Systems, Central Queensland University, Rockhampton, QLD, Australia
Spotty Liver Disease (SLD) is a serious infectious disease which occurs mainly in laying chickens in free range production systems. SLD outbreaks can increase mortality and decrease egg production of chickens, adversely impact welfare and cause economic hardship for poultry producers. The bacterium Campylobacter hepaticus is the primary cause of the disease. This study aimed to identify the effects of C. hepaticus on chicken gut microbiota and gut structure. Three C. hepaticus strains (HV10T, NSW44L and QLD19L), isolated from different states of Australia, were used in the study. Chickens at 26-weeks post-hatch were orally dosed with one of the C. hepaticus strains (challenged groups) or Brucella broth (unchallenged or control group). Six days after the challenge, birds were necropsied to assess liver damage, and caecal content and tissue samples were collected for histology, microbiology, and 16S rRNA gene amplicon sequencing to characterize the composition of the bacterial microbiota. Strain C. hepaticus NSW44L produced significantly more disease compared to the other C. hepaticus strains and this coincided with more adverse changes observed in the caecal microbiota of the birds challenged with this strain compared to the control group. Microbial diversity determined by Shannon and Simpson alpha diversity indices was lower in the NSW44L challenged groups compared to the control group (p = 0.009 and 0.0233 respectively, at genus level). Short-chain fatty acids (SCFAs) producing bacteria Faecalibacterium, Bifidobacterium and Megamonas were significantly reduced in the challenged groups compared to the unchallenged control group. Although SLD-induction affected the gut microbiota of chickens, their small intestine morphology was not noticeably affected as there were no significant differences in the villus height or ratio of villus height and crypt depth. As gut health plays a pivotal role in the overall health and productivity of chickens, approaches to improve the gut health of the birds during SLD outbreaks such as through diet and keeping the causes of stress to a minimum, may represent significant ways to alleviate the impact of SLD.
Introduction
Spotty Liver Disease (SLD), causes small whitish-gray miliary spots on the surface of the liver, and is a significant disease in poultry. It causes significant loss to the poultry industry as it can cause up to 10% flock mortalities and 25% reduction in egg production in flocks experiencing a disease outbreak (1, 2). Even though SLD was noted decades earlier, its specific etiology was recognized only in 2015, when a novel Campylobacter was isolated from SLD-affected chickens (3). Following this discovery, Van et al. (4) isolated, characterized, and formally named a new bacterium isolated from SLD affected birds in Australia (4). This specific bacterium was named Campylobacter hepaticus. C. hepaticus was confirmed as the cause of SLD after experiments showing that the Koch's Postulates were fulfilled (5). Unlike C. jejuni and C. coli, which are mostly commensal in chickens but pathogenic in human, C. hepaticus causes spotty liver disease in chickens but pathogenicity to humans has not been reported.
C. hepaticus is a Gram-negative, S-shaped bacterium which grows under microaerobic conditions. It has singular bipolar flagella and ranges in length between 1 and 1.2 um and width of 0.3–0.4 um (4). Compared to other Campylobacter species, C. hepaticus has a small genome size of ~1.5 Mb and a lower G+C content, between 27.9 and 28.5%, depending on the strain (6). C. hepaticus colonizes the small intestine of infected chickens, increasing in population from duodenum to ileum, and is at the highest levels within the caeca. They were also readily detected in cloacal swabs, indicating it is likely transmitted via a fecal-oral infection (7). The bacterium can be cultured from the liver and bile of infected birds. Isolation of C. hepaticus from complex primary microbial sources has proven to be difficult due to the absence of a fully selective media and growth of faster growing contaminating microorganisms (4, 8). Chickens entering the peak lay period, between 22 and 27 weeks, were found to be highly susceptible to SLD outbreaks. They can be infected with the responsible bacterium as young as 12 weeks old and up to 8 weeks before clinical SLD is manifested (8). C. hepaticus has shown the potential to survive for long periods in the farm environment by entering viable but non-culturable (VBNC) states (9).
The gastrointestinal tract (GI) of chickens contains diverse and complex microbiota which contribute to digestion and absorption of nutrients, immune system development, and pathogen exclusion (10). The functionality and the health of chicken gut is dependent on the surrounding environment, feed, and the GI microbiota. Chicken GI microbiota plays a pivotal role in the maintenance of intestinal health and can form a protective barrier by attaching to the GI tissues to reduce the opportunity for colonization of pathogenic bacteria (11). A healthy structure and function of chicken intestinal microbiota is crucial for the positive production performance of poultry (12, 13).
This study aimed to investigate the gut health of chickens by determining whether the SLD pathogenesis resulted in changes in the caecal microbiota by comparing the microbiota of an unchallenged group with C. hepaticus infected groups which were challenged with three different C. hepaticus strains. The effect of SLD on gut morpho-histology was also studied. The understanding of changes in gut microbiota and structure during SLD will help to direct the development of intervention strategies, such as feed modifications or probiotic application, to improve the gut health of the chickens.
Materials and methods
Bacterial strains and growth conditions
C. hepaticus HV10T, C. hepaticus NSW44L and C. hepaticus QLD19L were grown on Brucella agar (Becton Dickinson) with 5% horse blood (HBA) and incubated at 37 °C in microaerobic conditions using CampyGen gas packs (Oxoid) for 3 days (6). To prepare the inoculum for the chicken challenge, the culture was grown in Brucella broth supplemented with l-cysteine (0.4 mM), and l-glutamine (4 mM) and sodium pyruvate (10 mM) (modified Brucella broth) in 75 cm2 tissue culture flasks at 37°C for 48 h in microaerophilic conditions and used directly for the challenge, as previously described (14).
Chicken challenge experiments
A C. hepaticus challenge trial of 26-week old Hy-Line layer hens was carried out to investigate the changes in gut microbiota and gut structure of the C. hepaticus unchallenged group (control group) compared to challenged groups, where birds were challenged with HV10T, NSW44L or QLD19L strains. Birds from each group were housed in 4 cages, 3 birds per cage. Birds were sourced from a farm that had not observed any SLD in their flocks for several years and birds were confirmed as C. hepaticus-negative by specific PCR of fecal material collected by cloacal swabbing, using the previously published protocol (5). The animal experiment was approved by the Wildlife and Small Institutions Animal Ethics Committee of the Victorian Department of Economic Development, Jobs, Transport and Resources (approval number 14.16). The challenges were carried out as described in (14). In brief, the control chickens were given 1 mL of modified Brucella broth, the challenged groups were dosed with 1 x 109 CFU of the relevant C. hepaticus strain in 1 mL of modified Brucella broth. The birds were fed ad libitum with a standard, antibiotic-free layer diet. There were 12 birds each in the negative control and C. hepaticus HV10T groups, and 8 birds for the C. hepaticus NSW44L and C. hepaticus QLD19L groups.
At 6 days post infection, the birds were euthanized by cervical dislocation and the livers were examined for lesions. Liver lesion score was measured by the number of spots on a liver: a score of 0 indicated no spots; 1 indicated 1–5 spots; 2 indicated 6–20 spots; 3 indicated 21–100 spots; 4 indicated 101–1,000 spots; and 5 indicated more than 1,000 spots. Caecal content samples were collected and stored at −20°C until further analysis. Bile samples were taken aseptically from the gall bladder, kept on ice and processed within 3 h. Isolation from bile samples was performed as previously described (5), briefly, 20 μL of bile was spread directly onto horse blood agar plates and incubated at 37°C in microaerobic conditions for 3 days and confirmed by MALDI-TOF. Tissue samples were taken from the distal end of the jejunum of each bird and preserved in 10% neutral phosphate-buffered formalin for histology.
Histology
The tissue samples were embedded, sectioned, and hematoxylin and eosin stained by Gribbles Pathology (Australia). The control group, HV10T, and NSW44L groups were investigated. The mounted tissue sections were examined and scored for villus height and crypt depth. Ten villus/crypt measurements were made for each bird. The histology data were analyzed using one-way ANOVA in Minitab 21. The Dunnett test was used to compare every mean to a control mean.
16S rRNA gene amplicon sequencing and data analysis
DNA from the caecal samples was extracted using a Bioline Isolate Fecal DNA kit (Meridian, cat.no#BIO-52082). Primers were selected to amplify the V3–V4 region of 16S rRNA genes: forward ACTCCTACGGGAGGCAGCAG and reverse GGACTACHVGGGTWTCTAAT, and also contained barcodes, spacer and Illumina sequencing linker sequences as detailed previously (15). PCR was carried out in a final volume of 20 μL using Q5 High-Fidelity 2× Master Mix (New England Biolabs), primers at a final concentration of 250 nM each and 1 μL of template DNA. An Eppendorf Mastercycler Pro PCR instrument was used for amplification with cycling conditions of; 98°C for 1 min, 35 cycles of 98°C for 10 s; 49°C for 30 s and 72 °C for 30 s, and final extension at 72 °C for 10 min. Negative controls (water) were added in the sample preparation process and no amplicons were produced from the negative controls. Sequencing was performed on an Illumina MiSeq platform using 2 x 300 bp paired-end sequencing.
Sequence data were trimmed with Trimmomatic (v 0.39) and then fastq files were analyzed using DADA2 in QIIME2 v2020.6 (16) to denoise and produce Amplicon Sequence Variants (ASVs). Quality filtering was applied with the default option. ASVs were clustered at 99% identity using the VSEARCH plugin (17). Taxonomy was assigned using the SILVA database (v138). A total of 940,924 high-quality paired-end reads were obtained, with an average of 23,523 reads per sample, with a minimum of 10,023 and a maximum of 36,955. Obtaining feature table was further filtered (features that were present in only a single sample and samples with a total frequency <1,000). A total of 9,624 OTUs were found. The downstream statistical microbial data analyses and visualizations were done using MicrobiomeAnalyst (18), where features with mean values of <4 were filtered. The community profiling was based on the R Phyloseq and Vegan packages. Principal coordinates analysis (PCoA) (Jaccard Index and PERMANOVA) was used to visualize the clustering of samples based on their genus-level compositional profiles. Associations of specific bacteria within each group were identified using the linear discriminant analysis effect size and the multivariate analysis implemented in the MicrobiomeAnalyst software (18). The sequence data used for analysis is available in NCBI under BioProject accession number PRJNA877767.
Results
C. hepaticus NSW44L produced significant more SLD clinical disease compared to other C. hepaticus strains
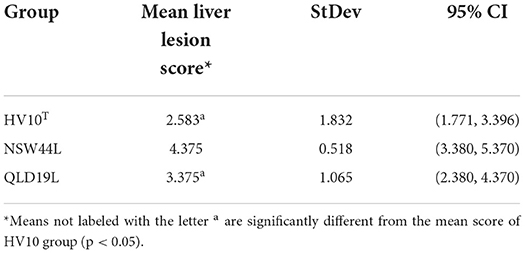
At 6-days post challenge, the birds were euthanized and the livers were accessed for typical SLD lesion. As expected, all unchallenged birds had normal healthy livers with no SLD lesions. All birds from the NSW44L challenged group had typical SLD liver lesions, with liver scores of 4 and 5 for all birds (mean liver score of 4.375), whereas 2 birds in the HV10T groups didn't have lesions on the livers and the average liver score for the group was 2.57, all birds from QLD19L had SLD liver lesions, but both HV10T and QLD19L challenged groups had significantly lower liver score than the NSW44L group (p < 0.05, Table 1). C. hepaticus was isolated from all challenged birds, except one from the QLD19L challenged group.
SLD had negative effects on the gut microbiota of the C. hepaticus NSW44L challenged group
The C. hepaticus NSW44L challenged group had significantly lower Shannon and Simpson alpha diversity values (p = 0.009 and 0.0233, respectively, at genus level) compared to the unchallenged control group. No significant difference was found in the Chao1 richness index between the control and NSW44 challenged groups (Figure 1).
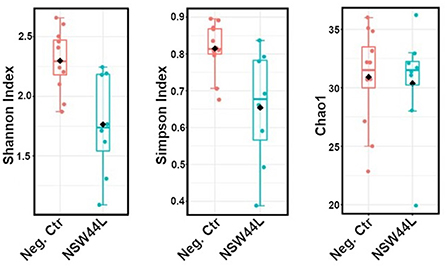
Figure 1. Alpha-diversity plots showed the C. hepaticus NSW44L challenged group has significantly lower Shannon and Simpson alpha diversity [p = 0.009 and 0.0233, respectively, (left and middle panels)] compared to the control group. No significant difference was found in the Chao1 richness index (right panel).
Principal Coordinate Analysis (PCoA) was used to explore and visualize similarities/dissimilarities in the overall microbiota compositions of the C. hepaticus NSW44L challenged group and the unchallenged control group. Gut microbiota compositions of the challenged birds were distinct from the non-challenged group (p < 0.001) (Figure 2).
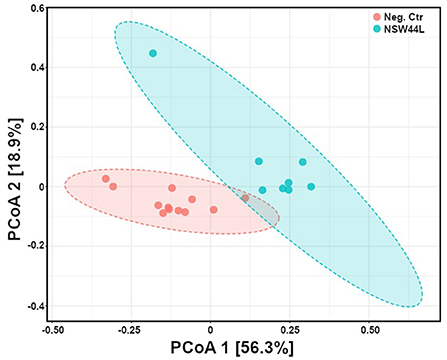
Figure 2. Principal coordinates analysis (PCoA) of gut microbiota composition of C. hepaticus challenged groups compared with non-challenged birds (at phylum level). The group challenged with the NSW44L strain was clustered away from the control group.
The relative abundance of the top 10 genera is visualized in a stacked bar chart and it can be seen that members of the Faecalibacterium, Bifidobacterium and Megamonas genera have reduced relative abundance in the C. hepaticus NSW44L challenged group compared to the unchallenged control group (presented in red, orange and purple respectively in Figure 3).
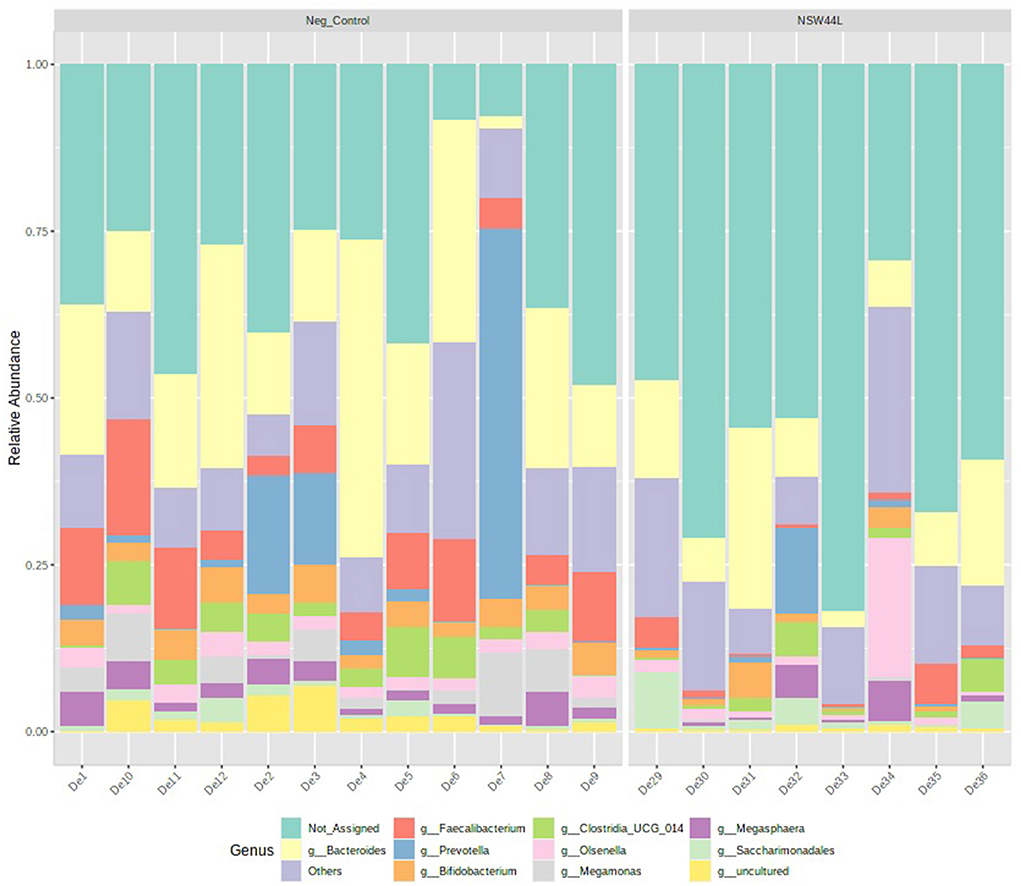
Figure 3. Relative abundance of top 10 genera detected in caecal content of unchallenged and C. hepaticus NSW44L challenged birds.
Univariate analysis showed significant reductions in the relative abundances of Faecalibacterium, Bifidobacterium, Ruminococcus torques, Megamonas and Enorma genera in the challenged group compared to the unchallenged group (p < 0.05) (Figure 4).
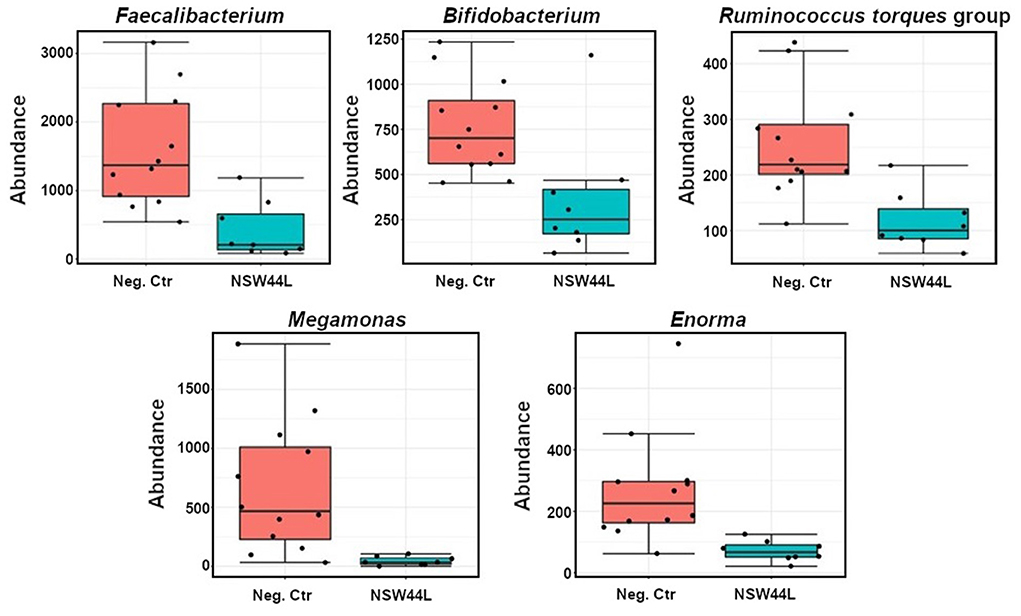
Figure 4. Classical univariate statistical comparisons showed significant reductions of Faecalibacterium, Bifidobacterium, Ruminococcus torques group, Megamonas and Enorma genera in the NSW44L challenged group.
SLD also adversely affected the microbiota of birds challenged with different strains of C. hepaticus
To determine whether the changes in microbiota composition seen following infection with C. hepaticus NSW44L were unique to that particular treatment group or indicated a more general response to the challenge, two other pathogenic strains of C. hepaticus, QLD19L and the type strain HV10T, were used to infect layer birds. There was no significant difference in the bacteria diversity (Shannon and Simpson indexes) and Chao1 richness index among the control, HV10T and QLD19L groups (data not shown).
Principal Coordinates Analysis (PCoA) using a Jaccard distance matrix index showed gut microbiota composition of challenged NSW44 was distinct from the non-challenged group, but the separation was not apparent for the other two challenged groups. This result is consistent with disease severity as this group had significantly more liver lesions than the other groups (Figure 5).
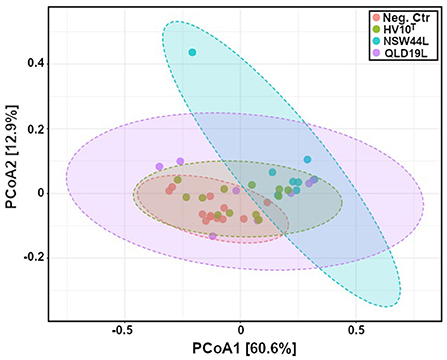
Figure 5. Principal coordinates analysis (PCoA) of gut microbiota composition of C. hepaticus challenged groups compared with non-challenged birds (p < 0.001). The group challenged with the NSW44L strain was clustered away from the control group, but the separation was not obvious for the other two challenged groups.
The relative abundance of the top 10 genera presented in Figure 6 showed that members of the Faecalibacterium and Megamonas genera had reduced relative abundance in the C. hepaticus HV10T and QLD19L challenged groups compared to the unchallenged control group (presented in blue and purple respectively in Figure 6).
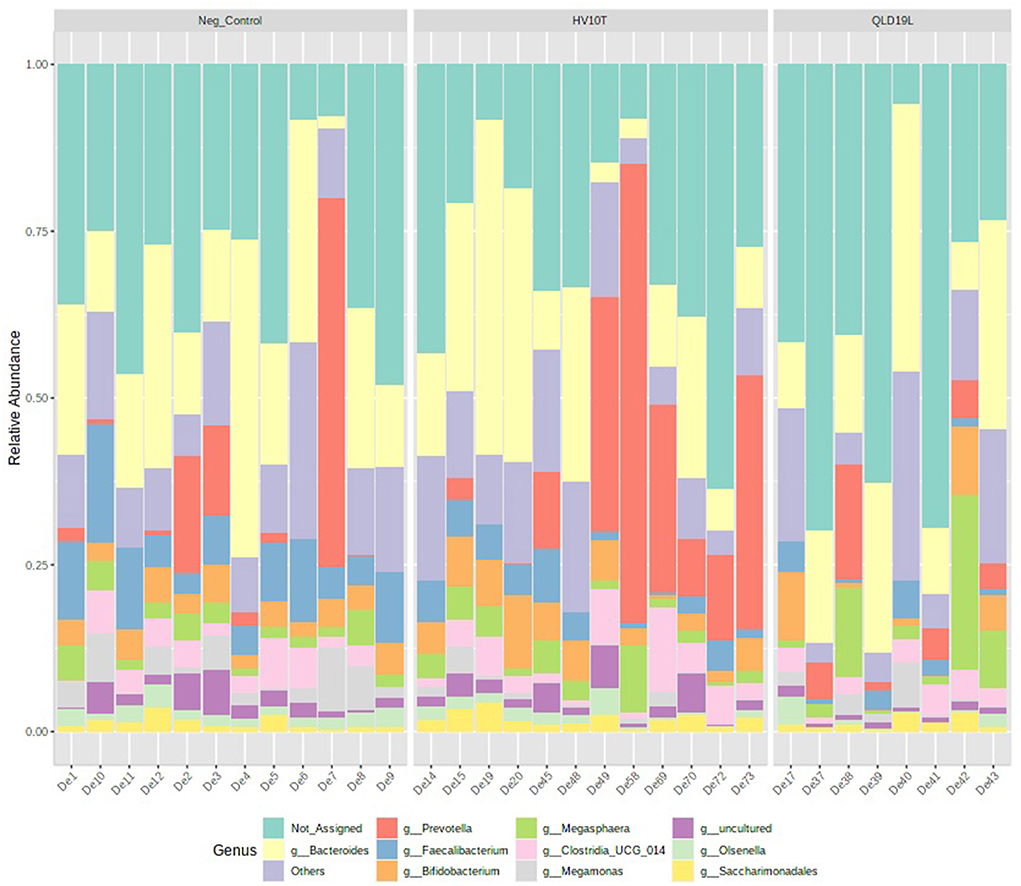
Figure 6. Relative abundance of top 10 genera detected in caecal content of unchallenged and C. hepaticus HV10T and QLD19L challenged birds.
Furthermore, the univariate statistical comparisons showed significantly reduced relative abundances of Faecalibacterium, and Megamonas in all challenged groups compared to the unchallenged control group (p < 0.05) (Figure 7).
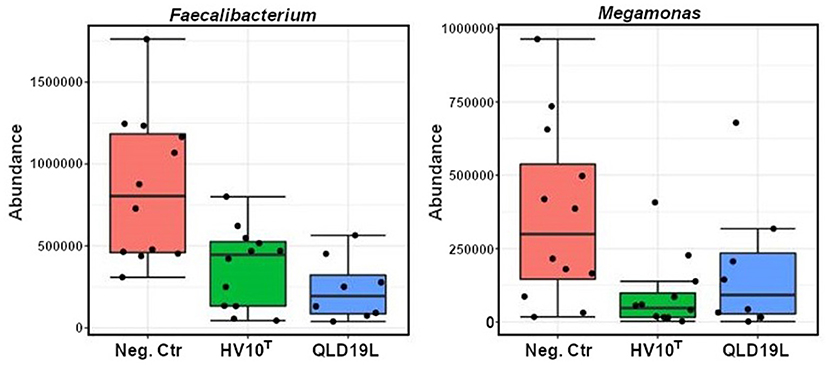
Figure 7. Classical Univariate Statistical Comparisons showed significant reductions of Faecalibacterium and Megamonas in the HV10T and QLD19L groups (p < 0.05).
Gut histology
A typical gut histological section used to measure the villus height and crypt depth is shown in Figure 8.
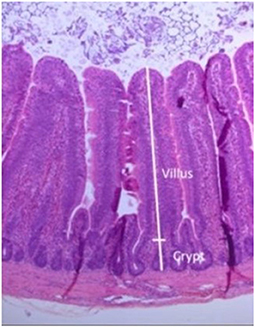
Figure 8. Gut section stained with hematoxylin and eosin, showing measurements taken of villus and crypt.
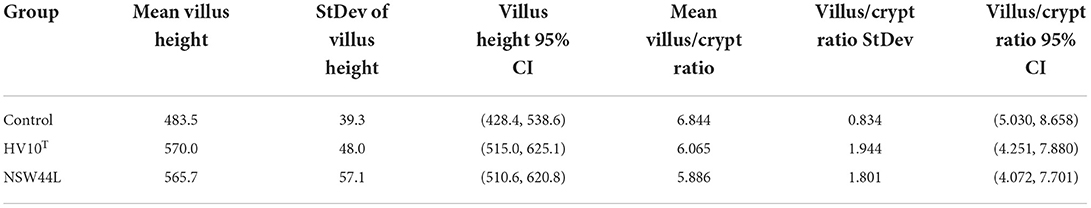
The group mean measurements of villus heights and villus/crypt height ratios are shown in Table 2. No statistically significant differences between groups were observed for either villus height or villus/crypt ratios.
Discussions
C. hepaticus strains isolated from different states in Australia, including HV10T (Victoria), NSW44L (New South Wales) and QLD19L (Queensland) were used to challenge birds at peak lay to examine the effect of SLD on gut health. The NSW44L strain produced significantly more liver lesions than the other two strains (p < 0.05). The genetic differences between different strains may play a role in disease pathogenesis, although genome analysis showed that the HV10T and NSW44L strains belong to the same lineage, whereas QLD19L was in another lineage (6).
As gut microbiota has important functions of food digestion and production of essential nutrients, regulation of immune, metabolic and nervous systems, diverse gut bacteria are necessary to support these functions (11). The adverse effects of C. hepaticus on alpha diversity were observed in C. hepaticus NSW44L challenged group, as the Shannon and Simpson alpha diversities were significantly reduced in the challenged groups compared to the control group (p < 0.05). Beta diversity analysis indicated a significant difference (p < 0.001) in community structure between the control and NSW44_ challenged groups with clear separation of the clustered samples of each group. There was considerable overlap in the coordination of the samples in beta diversity between the unchallenged, HV10T and QLD19L groups, indicating little systematic difference in the overall compositions of these caecal microbiotas. This apparent difference in the overall structure of the microbiota of the NSW44L infected group compared to the other two infection groups is consistent with the degree of pathogenicity of the C. hepaticus strains, as NSW44L strains produced significantly more disease than the other two strains.
When the microbiota communities of the challenged and unchallenged birds were investigated in more detail, it was observed that Faecalibacterium and Megamonas genera were reduced in the challenged groups compared to the non-challenged group. Faecalibacterium prausnitzii is the most well-characterized species of the Faecalibacterium genus, and other species have recently been identified and reported, including F. moorei (19), F. butyricigenerans, F. hominis, F. longum (20), F. duncaniae, F. gallinarum, and F. hattorii (21). Most Faecalibacterium bacteria are butyric acid producers, therefore they can exhibit anti-inflammatory effects and are potential probiotics for the treatment of gut inflammation (20). Comparing to the non-challenged group, the Megamonas genus was significantly reduced in all challenged groups. Megamonas is a major propionate producer via methylmalonyl-CoA mutase, epimerase, and decarboxylase (22). Propionate is one of three (acetate, propionate, and butyrate) important short-chain fatty acids. SCFAs are essential for host metabolism, induce the differentiation of regulatory T cells to enhance the host health, and serve as energy and carbon sources for poultry (23–25).
Bifidobacterium genus was another beneficial taxa reduced in abundance in the C. hepaticus NSW44L challenged group. Bifidobacterium play a role in gut health promotion by increasing intestinal immunostimulation and producing host beneficial volatile fatty acids (26, 27). Faecalibacterium and Bifidobacterium are both anaerobic bacteria and have shown potential use as probiotics to improve human and animal health. Administration of strains of B. breve and B. infantis species showed body weight gain and prevented the deleterious effects and mortality due to Salmonella infection in chickens through competitive exclusion and the release of cytokines (28). Therefore, the reduction of SCFAs producer Faecalibacterium, Bifidobacterium and Megamonas in the SLD-challenged groups further demonstrates the negative effect of C. hepaticus on chickens. It will be interesting in future work to assess the SCFA levels in SLD affected birds.
Ruminococcus torques group and Enorma genera were also reduced in the NSW44L challenged group. A high abundance of these bacteria has previously been associated with better performance in poultry (29, 30). The role of Enorma in human and animal health has not been studied widely, however, Khan and Chousalkar have shown that the Enorma genus was also reduced from Salmonella Typhimurium-challenged group (31).
The small intestine is important for the digestion and absorption of nutrients, and histological alterations have been found to be related to changes in intestinal function (32). Longer intestinal villi increase the absorptive surface, resulting in increased absorption capacity of the intestine in chickens. Similarly, a higher villus height and crypt depth ratio are associated with a greater capacity for nutrient digestibility and absorption (33, 34). Although SLD-induction affected the gut microbiota of chickens, their small intestine structure was not significantly different in the villus height and ratio of villus height and crypt depth. This is in contrast to the effects induced by some pathogens that cause specific damage to the small intestine as part of the pathogenic processes that they initiate, for example, by Clostridium perfringens infections, where birds inoculated with 1 mL of 2.0 × 108 cfu/mL showed decreased intestinal villus height and a lower ratio of villus height to crypt depth ratio (35).
In conclusion, C. hepaticus infection impacted the microbiota of chickens. They were most affected in the C. hepaticus NSW44L challenged group, which also had the highest lesion score amongst the challenged groups. The Shannon and Simpson diversity indexes were decreased in NSW44L challenged group, indicating lower bacterial diversity in infected birds. Some SCFAs producing bacteria were significantly reduced in all challenged groups. C. hepaticus might activate the host's innate immune responses, such as antimicrobial peptides, that could lead to the changes in the composition of the gut microbiota of the host. The change in caecal microbiota in an unhealthy direction may be one of the reasons for production loss during SLD outbreaks. Approaches to improve the birds' gut health during SLD outbreaks, such as through diet, probiotic supplementation, SCFA addition, and keeping the causes of stress to a minimum, may assist in the management of SLD.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA877767.
Ethics statement
The animal study was reviewed and approved by the Wildlife and Small Institutions Animal Ethics Committee of the Victorian Department of Economic Development, Jobs, Transport and Resources (Approval number 14.16).
Author contributions
TV and RM conceived the study. TV and LL performed the experiments. TW, AA, PS, TV, and RM carried out the chicken trial. TV and DS performed bioinformatics analysis of the sequencing data. TV and RP drafted the manuscript. RM and DS edited the manuscript. All authors have edited and approved the final manuscript.
Conflict of interest
Authors AA, TW, and PS were employed by Scolexia Pty Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Crawshaw T, Irvine R. Spotty liver syndrome in poultry in Great Britain. Vet Rec. (2012) 170:317–8. doi: 10.1136/vr.e2201
2. Grimes T, Reece R. (2011). Spotty liver disease – an emerging disease in free-range egg layers in Australia. In: 60th Western Poultry Disease Conference. Sacremento, CA, 53–56.
3. Crawshaw TR, Chanter JI, Young SC, Cawthraw S, Whatmore AM, Koylass MS, et al. Isolation of a novel thermophilic Campylobacter from cases of spotty liver disease in laying hens and experimental reproduction of infection and microscopic pathology. Vet Microbiol. (2015) 179:315–21. doi: 10.1016/j.vetmic.2015.06.008
4. Van TTH, Elshagmani E, Gor MC, Scott PC, Moore RJ. Campylobacter hepaticus sp. nov, isolated from chickens with spotty liver disease. Int J Syst Evol Microbiol. (2016) 66:4518–24. doi: 10.1099/ijsem.0.001383
5. Van TT, Elshagmani E, Gor MC, Anwar A, Scott PC, Moore RJ. Induction of spotty liver disease in layer hens by infection with Campylobacter hepaticus. Vet Microbiol. (2017) 199:85–90. doi: 10.1016/j.vetmic.2016.12.033
6. Van TTH, Lacey JA, Vezina B, Phung C, Anwar A, Scott PC, et al. Survival mechanisms of Campylobacter hepaticus identified by genomic analysis and comparative transcriptomic analysis of in vivo and in vitro derived bacteria. Front Microbiol. (2019) 10:107. doi: 10.3389/fmicb.2019.00107
7. Van TTH, Gor MC, Anwar A, Scott PC, Moore RJ. Campylobacter hepaticus, the cause of spotty liver disease in chickens, is present throughout the small intestine and caeca of infected birds. Vet Microbiol. (2017) 207:226–30. doi: 10.1016/j.vetmic.2017.06.022
8. Phung C, Vezina B, Anwar A, Wilson T, Scott PC, Moore RJ, et al. Campylobacter hepaticus, the cause of spotty liver disease in chickens: transmission and routes of infection. Front Vet Sci. (2019) 6:505. doi: 10.3389/fvets.2019.00505
9. Phung C, Moore RJ, Van TTH. Campylobacter hepaticus, the cause of spotty liver disease in chickens, can enter a viable but nonculturable state. Vet Microbiol. (2022) 266:109341. doi: 10.1016/j.vetmic.2022.109341
10. Shang Y, Kumar S, Oakley B, Kim WK. Chicken gut microbiota: importance and detection technology. Front Vet Sci. (2018) 5:254. doi: 10.3389/fvets.2018.00254
11. Diaz Carrasco JM, Casanova NA, Fernández Miyakawa ME. Microbiota, gut health and chicken productivity: what is the connection? Microorganisms. (2019) 7:374. doi: 10.3390/microorganisms7100374
12. Rubio LA, Peinado MJ, Ruiz R, Suárez-Pereira E, Ortiz Mellet C, García Fernández JM. Correlations between changes in intestinal microbiota composition and performance parameters in broiler chickens. J Anim Physiol Anim Nutr (Berl). (2015) 99:418–23. doi: 10.1111/jpn.12256
13. Stanley D, Hughes RJ, Geier MS, Moore RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: Challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. (2016) 7:187. doi: 10.3389/fmicb.2016.00187
14. Phung C, Wilson TB, Quinteros JA, Scott PC, Moore RJ, Van TTH. Enhancement of Campylobacter hepaticus culturing to facilitate downstream applications. Sci Rep. (2021) 11:20802. doi: 10.1038/s41598-021-00277-8
15. Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. (2014) 2:1–7. doi: 10.1186/2049-2618-2-6
16. Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. (2019) 37:852–7. doi: 10.1038/s41587-019-0209-9
17. Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. (2016) 4:e2584. doi: 10.7717/peerj.2584
18. Dhariwal A, Chong J, Habib S, King IL, Agellon LB, Xia J. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. (2017) 45:W180–8. doi: 10.1093/nar/gkx295
19. Fitzgerald CB, Shkoporov AN, Sutton TDS, Chaplin AV, Velayudhan V, Ross RP, et al. Comparative analysis of Faecalibacterium prausnitzii genomes shows a high level of genome plasticity and warrants separation into new species-level taxa. BMC Genomics. (2018) 19:931. doi: 10.1186/s12864-018-5313-6
20. Zou Y, Lin X, Xue W, Tuo L, Chen M-S, Chen X-H, et al. Characterization and description of Faecalibacterium butyricigenerans sp. nov and F longum sp nov, isolated from human faeces. Sci Rep. (2021) 11:11340. doi: 10.1038/s41598-021-90786-3
21. Sakamoto M, Sakurai N, Tanno H, Iino T, Ohkuma M, Endo A. Genome-based, phenotypic and chemotaxonomic classification of Faecalibacterium strains: proposal of three novel species Faecalibacterium duncaniae sp. nov., Faecalibacterium hattorii sp. nov. and Faecalibacterium gallinarum sp. nov'. Int J Syst Evol Microbiol. (2022) 72. doi: 10.1099/ijsem.0.005379
22. Polansky O, Sekelova Z, Faldynova M, Sebkova A, Sisak F, Rychlik I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. (2015) 82:1569–76. doi: 10.1128/AEM.03473-15
23. Deleu S, Machiels K, Raes J, Verbeke K, Vermeire S. Short chain fatty acids and its producing organisms: an overlooked therapy for IBD? EBioMedicine. (2021) 66:103293. doi: 10.1016/j.ebiom.2021.103293
24. Józefiak D, Rutkowski A, Martin SA. Carbohydrate fermentation in the avian ceca: a review. Anim Feed Sci Technol. (2004) 113:1–15. doi: 10.1016/j.anifeedsci.2003.09.007
25. Sun B, Hou L, Yang Y. The development of the gut microbiota and short-chain fatty acids of layer chickens in different growth periods. Front Vet Sci. (2021) 8:666535. doi: 10.3389/fvets.2021.666535
26. Grant A, Gay CG, Lillehoj HS. Bacillus s:as direct-fed microbial antibiotic alternatives to enhance growth, immunity, and gut health in poultry. Avian Pathol. (2018) 47:339–51. doi: 10.1080/03079457.2018.1464117
27. Haghighi HR, Gong J, Gyles CL, Hayes MA, Sanei B, Parvizi P, et al. Modulation of antibody-mediated immune response by probiotics in chickens. Clin Diagn Lab Immunol. (2005) 12:1387–92. doi: 10.1128/CDLI.12.12.1387-1392.2005
28. El-Sharkawy H, Tahoun A, Rizk AM, Suzuki T, Elmonir W, Nassef E, et al. Evaluation of bifidobacteria and lactobacillus probiotics as alternative therapy for salmonella typhimurium infection in broiler chickens. Animals (Basel). (2020) 10. doi: 10.3390/ani10061023
29. Crisol-Martínez E, Stanley D, Geier MS, Hughes RJ, Moore RJ. Understanding the mechanisms of zinc bacitracin and avilamycin on animal production: linking gut microbiota and growth performance in chickens. Appl Microbiol Biotechnol. (2017) 101:4547–59. doi: 10.1007/s00253-017-8193-9
30. Torok VA, Hughes RJ, Mikkelsen LL, Perez-Maldonado R, Balding K, MacAlpine R, et al. Identification and characterization of potential performance-related gut microbiotas in broiler chickens across various feeding trials. Appl Environ Microbiol. (2011) 77:5868–78. doi: 10.1128/AEM.00165-11
31. Khan S, Chousalkar KK. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J Anim Sci Biotechnol. (2020) 11:29. doi: 10.1186/s40104-020-0433-7
32. Yamauchi K. Review on chicken intestinal villus histological alterations related with intestinal function. J Poult Sci. (2002) 39:229–42. doi: 10.2141/jpsa.39.229
33. Prakatur, I., Miskulin, M., Pavic, M., Marjanovic, K., Blazicevic, V., Miskulin, I., et al. Intestinal morphology in broiler chickens supplemented with propolis and bee pollen. Animals. (2019) 9:301doi: 10.3390/ani9060301
34. Silva MA, Pessotti BMS, Zanini SF, Colnago GL, Rodrigues MRA, Nunes LC, et al. Intestinal mucosa structure of broiler chickens infected experimentally with Eimeria tenella and treated with essential oil of oregano. Cienc Rural. (2009) 39:1471–7. doi: 10.1590/S0103-84782009005000135
Keywords: spotty liver disease, microbiota, layers, chickens, Campylobacter, gut health
Citation: Van TTH, Lee Nen That LFM, Perera R, Anwar A, Wilson TB, Scott PC, Stanley D and Moore RJ (2022) Spotty liver disease adversely affect the gut microbiota of layers hen. Front. Vet. Sci. 9:1039774. doi: 10.3389/fvets.2022.1039774
Received: 08 September 2022; Accepted: 10 October 2022;
Published: 28 October 2022.
Edited by:
Dirk Werling, Royal Veterinary College (RVC), United KingdomReviewed by:
Ashenafi Feyisa Beyi, Iowa State University, United StatesAlois Cizek, University of Veterinary and Pharmaceutical Sciences Brno, Czechia
Laura Glendinning, University of Edinburgh, United Kingdom
Copyright © 2022 Van, Lee Nen That, Perera, Anwar, Wilson, Scott, Stanley and Moore. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thi Thu Hao Van, dGhpdGh1aGFvLnZhbkBybWl0LmVkdS5hdQ==
 Thi Thu Hao Van
Thi Thu Hao Van Lisa F. M. Lee Nen That
Lisa F. M. Lee Nen That Rachelle Perera
Rachelle Perera Arif Anwar2
Arif Anwar2 Timothy B. Wilson
Timothy B. Wilson Peter C. Scott
Peter C. Scott Dragana Stanley
Dragana Stanley Robert J. Moore
Robert J. Moore