
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 13 December 2022
Sec. Comparative and Clinical Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1036757
This article is part of the Research TopicRising Stars in Comparative and Clinical Medicine: 2022View all 29 articles
 Rokshana Parvin1*
Rokshana Parvin1* Congriev Kumar Kabiraj1
Congriev Kumar Kabiraj1 Ismail Hossain1
Ismail Hossain1 Alamgir Hassan1
Alamgir Hassan1 Jahan Ara Begum1
Jahan Ara Begum1 Mohammed Nooruzzaman1
Mohammed Nooruzzaman1 Md. Taohidul Islam2
Md. Taohidul Islam2 Emdadul Haque Chowdhury1
Emdadul Haque Chowdhury1For rapid and sensitive pathogen screening from field outbreaks, molecular techniques such as qPCR-based simultaneous detections are efficient. Respiratory diseases are the most detrimental diseases to the poultry industry and need to be addressed because of their major economic losses. In the current study, we have applied two different detection assays: one for simultaneous detection of avian influenza virus (AIV; M gene) and subtyping (H5, N1, H9, N2) using TaqMan probe chemistry (TaqMan multitarget) and another for simultaneous detection of Newcastle disease virus (NDV), infectious bronchitis virus (IBV), and infectious laryngotracheitis virus (ILTV) using SYBR Green chemistry (SYBR Green multitarget). Two individual qPCRs were conducted for the detection of four pathogens. Surveillance of tissue (n = 158) and oropharyngeal swab (206) samples from multiple poultry flocks during the years April 2020–July 2022 applying the TaqMan and SYBR Green multitarget qPCRs revealed that 48.9% of samples were positive for respiratory infections, of which 17.2% were positive for NDV, 25.5% were positive for AIV, 9.9% were positive for IBV, and only a single positive (0.3%) for ILTV. Among the AIV, 35% were highly pathogenic subtype H5N1 and 65% were low pathogenic subtype H9N2. Co-infections of 2–3 respiratory viruses were also accurately detected. Respiratory viral pathogens are quite common in Bangladeshi poultry and can be successfully detected using multitarget simultaneous real-time quantitative polymerase chain reaction (RT-qPCR) assays like those adopted in the current study. Increased mass surveillance, along with the molecular characterization of the circulating respiratory viruses, is crucial to control the epidemic and subsequently save the Bangladeshi poultry industry.
The worldwide poultry industry is expanding rapidly in response to the rising demand for animal-derived protein. In Bangladesh, poultry is reared both in the backyard and commercial settings where more than 50% of the households keep chickens and almost 30% of the households rear ducks (http://www.bbs.gov.bd/). The Bangladeshi commercial poultry industry comprises broilers, layers, and slow-growing colored meat-type chickens (Sonali) that produce ~0.2 million metric tons of poultry meat and 5,210 million table eggs per year (1). However, 60–70% of commercial hens are kept in small-scale facilities with limited biosecurity and management methods, which enhances disease susceptibility (2).
Intensification of commercial farming leads to the accumulation of multiple pathogens at a single site, in particular, a wide range of respiratory viral pathogens cause huge economic losses (3). The presence of more than one pathogen in hosts at the same time causes the complexity of diseases (3–8). Globally, poultry production encounters more viral infections compared to infections with other pathogens (9). Among different viral pathogens of poultry, avian influenza virus (AIV), Newcastle disease virus (NDV), infectious bronchitis virus (IBV), and infectious laryngotracheitis virus (ILTV) are the most prevalent and can produce disease independently, concurrently, or in association with bacterial agents and mycoplasmas (8, 10–13). Furthermore, the biosecurity of poultry farms in Bangladesh is quite unsatisfactory, particularly in small- to medium-scale poultry farms (14, 15), allowing pathogens to enter the farms. Early diagnosis of respiratory pathogens can minimize mortality in the infected flocks as well as could suggest effective supportive treatment options. The success of vaccination depends on the genetic background of the circulating strains factor and the selection of broadly protective vaccines against avian viral diseases (16, 17).
Respiratory infections are very common in commercial poultry and duck populations of Bangladesh (18–21). Therefore, rapid detection is required, followed by prevention and control. The current study emphasizes the prevalence of respiratory viruses in poultry and their early, accurate detection. Several laboratory methods such as virus isolation in embryonated eggs and organ cultures and serological tests are being used for decades for detecting and differentiating avian respiratory viral infections (3, 22–24). These methods are time-consuming and laborious, particularly when used separately for a single pathogen (25–27). Nowadays, rapid diagnostic tests have been developed for the detection of either viral nucleic acids or viral antigens, including rapid chromatographic and, molecular techniques (28–33). However, those techniques detect only one specific pathogen at a time. The duplex or multiplex PCR has the ability to amplify and differentiate multiple specific nucleic acids. Many countries have implemented respiratory disease surveillance, and the use of multiplex real-time PCR assays has aided in detecting circulating viruses and outbreaks, as well as estimating disease burden (29–32). Recently, a PCR-based panel assay for the detection of multiple poultry respiratory pathogens has been established using a real-time PCR system in a single reaction with SYBR Green reagent (3). A mini version of the Riems Influenza a Typing Array (RITA) assay (34, 35) has also been used in the subtype detection of AIVs and identification of other avian viral pathogens in Bangladesh (20, 36). The RITA assay is very useful for detecting the AIV subtype in a single run, but it is not cost-effective for a country with limited resources. As a result, selecting particular primers and probes for a specific target run may reduce PCR costs while simultaneously enabling rapid diagnosis. The aim of the study was to detect respiratory viral pathogens (AIV, NDV, IBV, and ILT) and selective subtyping of AIVs using two multitarget real-time quantitative polymerase chain reaction (RT-qPCR) simultaneous detection assays from outbreak samples. We adopt previously established assays (34, 35) in a format of multitarget simultaneous runs for the detection of respiratory viral pathogens that are common and endemic in Bangladesh.
Ten (10) of each positive AIV– subtype H5N1 (n = 5) and H9N2 (n = 5) along with NDV, IBV, five (5) ILTV, and 10 mycoplasma-positive (referred to as negative here) samples were collected from virus repositories at the Department of Pathology and of which three vaccine strains were purchased from commercial sources (Nobilis ND LaSota, NOBILIS® MA5 + CLONE 30, and Nobilis® ILT, MSD animal health) to adopt and validate the simultaneous RT-qPCR assays.
A total of 364 poultry flocks, including layer (n = 97), broiler and broiler breeder (n = 115), Sonali (n = 63), duck (n = 79), pigeon (n = 8), and turkey (n = 2) with a history of respiratory signs from different commercial poultry flocks in the Mymensingh division of Bangladesh were investigated from April 2020 to June 2022. Dead birds from those natural outbreaks were submitted to the Department of Pathology, necropsy was performed, and gross pathological changes were recorded. A total of 158 tissue samples (lung and trachea from each individual animal were pooled) were collected in sterile tubes. Tissues were homogenized to prepare a 20% w/v suspension in minimum essential medium (MEM, Thermo Fisher Scientific, USA) containing a mixture of streptomycin and penicillin (50 μg/ml), and the supernatant was collected following centrifugation at 3,000 rpm for 10 min. In addition, 206 oropharyngeal swabs were collected from different poultry flocks with a similar history. Five individual birds were randomly chosen, and swabs were pooled in the tube containing MEM and antibiotics and considered single-swab samples. Samples were kept at −70°C until further analysis.
Viral DNA or RNA was extracted using a Monarch nucleic acid purification kit (New England Biolabs Inc., USA). One-step RT-qPCR was performed using TaqMan probe-based and SYBR Green chemistry. The current study used target primers developed for specific diseases in previous investigations (28, 35, 37–39), which are listed in Supplementary Table 1.
The TaqMan probe-based single-step RT-qPCR assay (referred to as “TaqMan multitarget” here) was optimized using a Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs Inc., USA) and AIV-specific oligonucleotides as used previously (Supplementary Table 1). The final reaction volume was 12.5 μl, containing 2.5 μl of DNA or RNA template, 6 μl of 2× RT-PCR reaction mix, 1 μl of RT- PCR Enzyme Mix, 1 μl of nuclease-free water, and 2 μl of primer probe mix (10 pmol each). The RT-qPCR thermocycling conditions were 55°C for 10 min (reverse transcription) and 95°C for 1 min (initial denaturation), followed by 45 cycles at 95°C for 10 s (denaturation) and 60°C for 30 s (annealing and elongation) with the reading of fluorescence in this step. The Optimized TaqMan Multiplex is used for the simultaneous detection of generic AIV (M gene) and subtype of AIV (H5, H9, N1, and N2 genes).
The detection of NDV, IBV, and infectious laryngotracheitis (ILT) using single-step RT-qPCR SYBR Green assay (referred to as “SYBR Green multitarget” here) was carried out by specific target primers against avian paramyxovirus 1 NP gene, IBV UTR region, and ILT glycoprotein gene-specific oligonucleotides, respectively (Supplementary Table 1). The assay was carried out using a Luna Universal One-Step RT-qPCR Kit (New England Biolabs Inc., USA) containing SYBR Green reagents following a previously established protocol (23). The final reaction volume was 12.5 μl, including 2.5 μl of the RNA template, 5 μl of 2× RT-PCR SYBR reaction mix, 0.5 μl of Luna WarmStart RT Enzyme Mix, 2.5 μl of nuclease-free water, and 2 μl of primer mix (10 pmol each). The RT-qPCR thermocycling conditions were 55°C for 10 min (reverse transcription) and 95°C for 2 min (initial denaturation), followed by 45 cycles at 95°C for 10 s (denaturation) and 60°C for 30 s (annealing and elongation) with a reading of fluorescence in this step. Immediately after PCR, a melting curve analysis was performed with a continuous temperature increment of 0.5°C/s between 65 and 95°C. Samples yielding cycle threshold (Ct) values ≤ 32 with an appropriate melting temperature (Tm) were considered positive.
In total, 45 reference samples, AIV (n = 10), NDV (n = 10), IBV (n = 10), ILTV (n = 5)-positive samples (virus from the repository and vaccine strains), and avian mycoplasma-positive here considered as negative sample (n = 10), were tested using single-target single-run and multitarget (TaqMan multitarget and SYBR Green multitarget) RT- qPCR in parallel. The threshold cycle (Ct) values of each individual run were compared with the Ct obtained at multitarget runs. The reproducibility of the multitarget RT-qPCRs was recorded by three independent runs. The sensitivity and specificity of the applied multitarget RT-qPCRs tested in the reference samples were verified, and true-positive and negative results were calculated using the web-based tool (https://www.medcalc.org/calc/diagnostic_test.php). The false-negative and false-positive results were verified using the positive and negative control within each sample panel.
A total of 364 poultry flocks, including layer, broiler and broiler breeder, Sonali, duck, pigeon, and turkey flocks, were investigated. Each type of bird has varied flock sizes (Table 1), and the vaccination schedule for respiratory pathogens was also varied, however, in most cases vaccination was not done. In general, the affected flocks reported clinical signs of off-feed, gasping, sneezing, coughing, drowsiness, decreasing output, and varying degree of mortality. Table 1 displays the outbreak overview for various bird species. The outbreak mostly occurred from pre- to post-winter seasons (October–May) in Bangladesh when the average temperature ranges from 15 to 30°C. At necropsy, the dead or affected birds showed congested and hemorrhagic trachea, cloudy air sac, hemorrhages in the intestine, brain, blackish, fibrinous, edematous, hemorrhagic, and congested and consolidated lung as shown in Supplementary Figure 1. Following the acquisition of respiratory clinical characteristics, samples from several commercial poultry flocks were examined using simultaneous RT-qPCRs.
Two multitarget parallel RT-qPCR assays (TaqMan multitarget and SYBR Green multitarget) were applied, which were originally based on the “Riems Influenza a Typing Array” (34, 35). Each multitarget parallel assay's PCR plate or strip (depending on the number of samples to be analyzed in each RT-qPCR) was set so that each column can include six unknown samples, one positive and one negative control (Supplementary Table 2).
As all target viruses have RNA genome except ILTV (DNA genome), we applied RT-qPCR methods for SYBR Green multitarget parallel assay to facilitate simultaneous run. TaqMan multitarget RT-qPCR exclusively amplified the M, H5, H9, N1, and N2 genes of AIV in a single run (Figure 1A). Other respiratory viruses, such as NDV, IBV, and ILTV, were detected as well using SYBR Green multitarget RT-qPCR but in a separate run (Figure 1B). The construction of a dissociation melt curve (Tm) in SYBR Green multitarget RT-qPCR at temperatures between 83 and 84°C further aided the accurate identification of specific target viruses as the negative control produced an unspecific Tm at 75°C (Figure 1C). The final interpretation for multitarget RT-qPCRs was based on the Ct (cycle threshold) values on the sigmoid curve and the Tm values on the melt curve (SYBR Green multitarget only), and it was fairly good for detecting pathogen target genes (Figure 1). Ct values above 35 were considered for negative interpretation of the target pathogens.
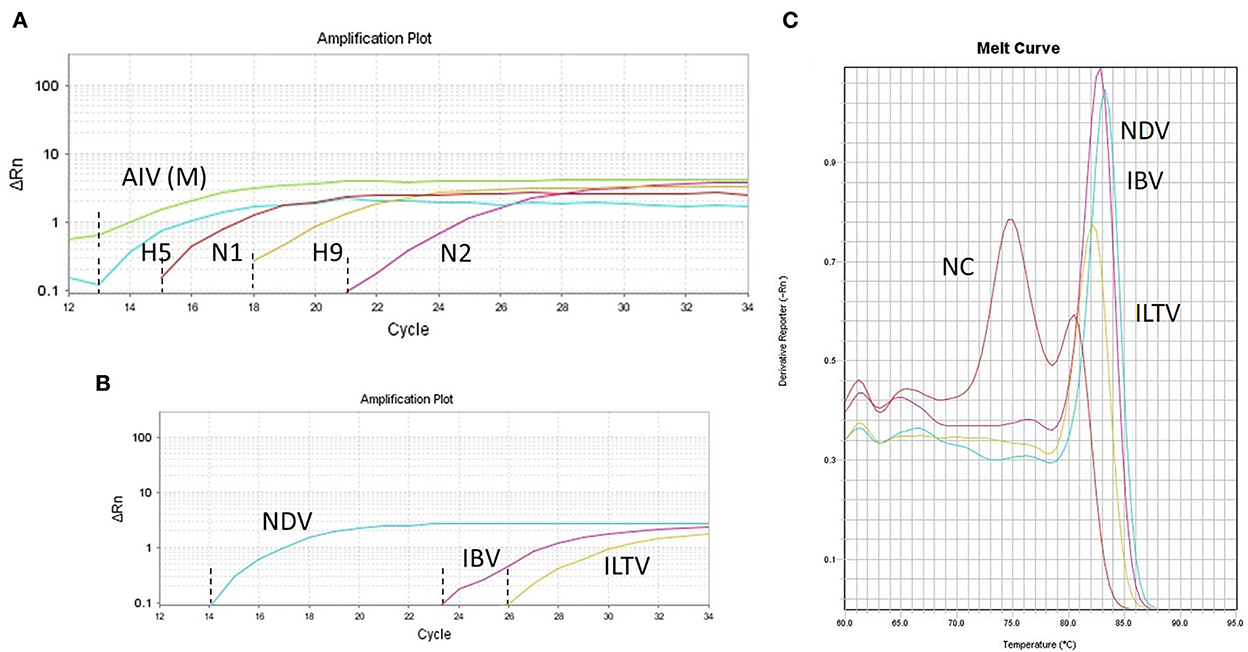
Figure 1. RT-qPCRs amplification plots for specific target viral pathogens. (A) Simultaneous detection of generic avian influenza virus (AIV-M) and subtyping of AIV (H5, H9, N1, and N2) in a single run. (B) Simultaneous detection of Newcastle disease virus (ND), infectious bronchitis virus (IBV), and infectious laryngotracheitis virus (ILT) along with the melt curve (C) in a single run. NC denotes a negative control that shows a melt curve at a different temperature (~75°C). The Ct values of individual gene is indicated by dash lines (A,B) generated from the number of cycles.
All reference (positive and negative) viruses (n = 45) were tested blindly against all target genes using multitarget parallel RT-qPCR assays and compared with single-target RT-qPCRs. The resulting Ct and Tm values in specific targets at positive controls were then used as a baseline for the evaluation of other samples. Triplicate runs of the selected reference samples were employed in both single-target and multitarget parallel RT-qPCR assays. Each test performed using single- or multiple-target assays in two different thermal cyclers on two different days yielded almost similar Ct values (Figure 2). The mean Ct value for each target in two independent assays did not differ considerably, indicating that the applied simultaneous TaqMan and SYBR Green multitarget detection approaches are as specific as single-target RT-qPCR runs.
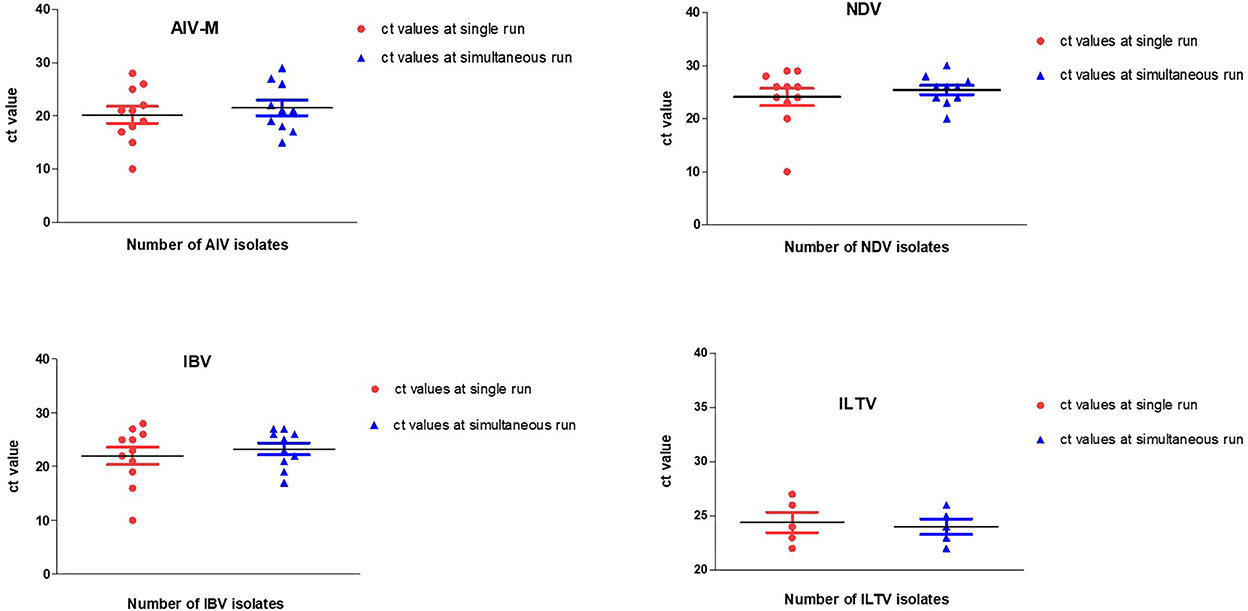
Figure 2. Scatter plots representing Ct values at two independent RT-qPCR runs. There were no significant variations in Ct values generated using single-target single run and multitarget parallel RT-qPCR simultaneous run.
The semi-quantitation and detection limits for the multitarget simultaneous assays are given in Table 2. The applied TaqMan multitarget and SYBR Green multitarget simultaneous RT-qPCR assays maintained linearity of magnitude and, using the slope from the linear equation, the overall efficiency was estimated to be 99% as of single-target single RT-qPCR run. The three replicates also do not differ with the generation of Ct values and thereafter have shown the less standard error of the average Ct values (Table 2).
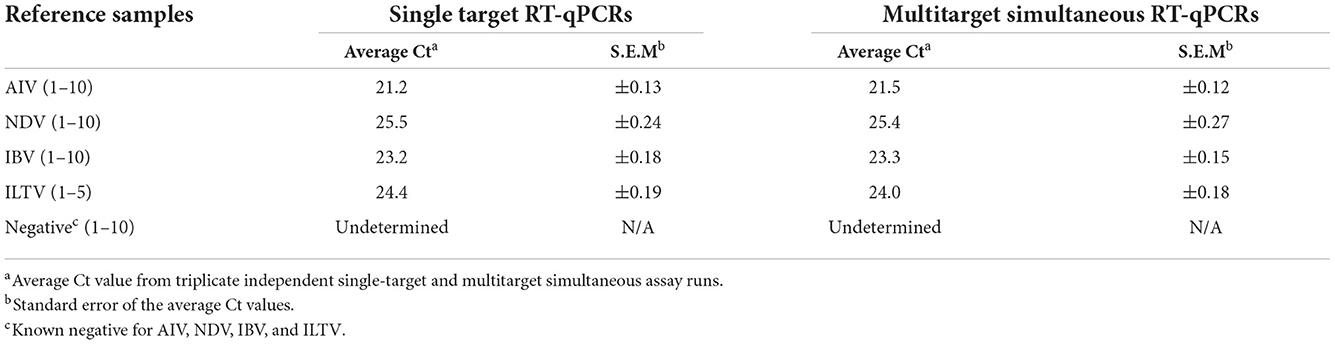
Table 2. Detection limit (Ct) and reproducibility of reference samples in applied multitarget simultaneous RT-qPCR assays.
Among the 158 tissue samples, there were 26 positives (16.5%) for NDV, 54 positives (34.2%) for AIV, 19 positives (12%) for IBV, and only a single positive (0.8%) for ILTV. On the other hand, 206 oropharyngeal swabs were detected 18 NDV (8.7%), 39 AIV (18.9%), and 17 IBV (8.3%) with no ILTV detection. Figure 3 shows the percentage of flocks afflicted by viral infections by bird type from tissue (Figure 3A) and swab (Figure 3B) samples.
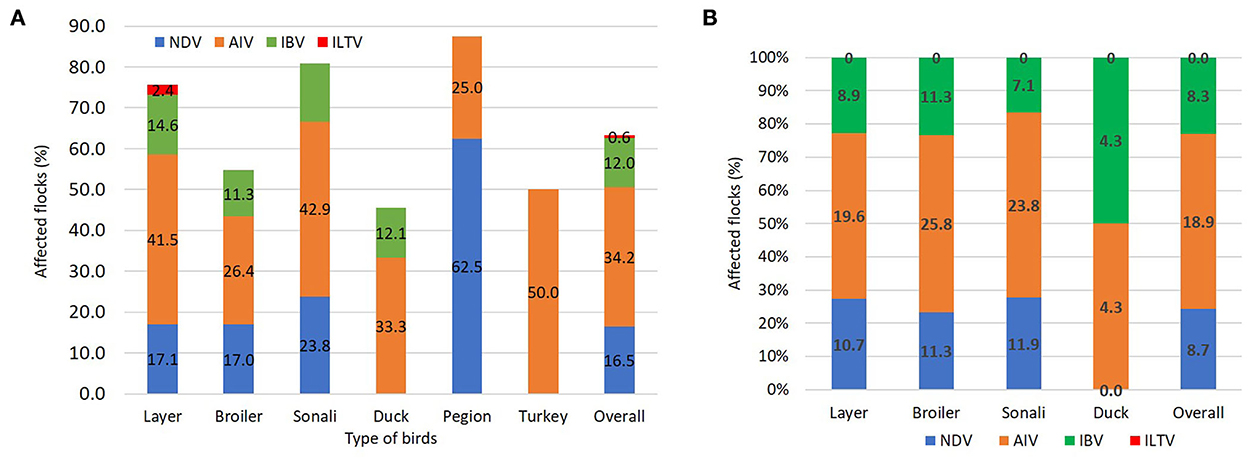
Figure 3. Detection of respiratory pathogens by applying the multitarget simultaneous RT-qPCRs. (A) The stacked column showing the percentage of respiratory pathogens in tissue samples among different bird types. (B) The stacked column showing the percentage of respiratory pathogens in oropharyngeal swab samples among different bird types.
Altogether, 48.9% of samples carried respiratory infections of which the highest detected (16.5%) pathogen was low pathogenic avian influenza (LPAI) subtype H9N2, followed by NDV (13.2%), IBV (9.9%), highly pathogenic avian influenza (HPAI) subtype H5N1 (9.1%) with only 0.3% ILTV (Figure 4A). Thus, within the AIV, 35% belonged to the HPAI subtype H5N1 and 65% were LPAI subtype H9N2. In addition, mixed viral infection was detected using the multiplex simultaneous RT-qPCRs, and the positive virus co-infection was found within the AIV, NDV, and IBV as depicted in Figure 4B. Briefly, 13 samples carried co-infections of which 7 were positives for both AIV and NDV, 2 positives for NDV and IBV, 3 positives for AIV and IBV while a single flock was positive for triple infection with AIV H9N2, NDV, and IBV. The 13 positive co-infected samples were further reconfirmed using single RT-qPCRs for each target independently, yielding almost identical amplification results.
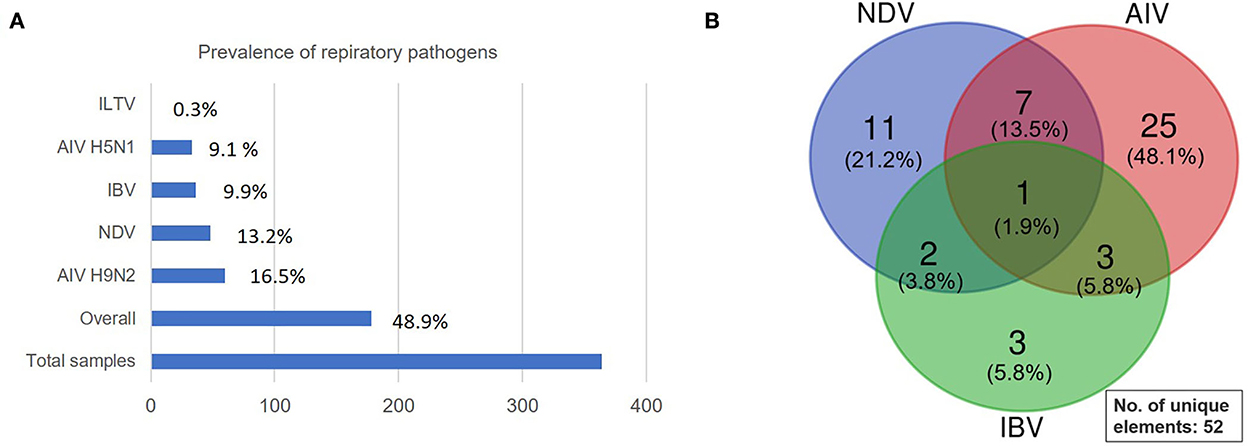
Figure 4. Overall prevalence of respiratory pathogens. (A) A clustered bar showing the overall prevalence of respiratory pathogens identified in the current study. (B) Venn diagram showing the presence of co-infections of 2–3 different respiratory viruses detected by multitarget simultaneous RT-qPCR assays.
The ability to detect and differentiate causative viruses from surveillance or outbreak samples quickly helps in controlling the spread of viruses. The current study focuses on the detection of respiratory viral infections in poultry and the circulation of either high or low pathogenic subtypes of AIV in Bangladesh. Another important aim is the application of rapid and sensitive real-time PCR-based multitarget simultaneous assays (TaqMan multitarget and SYBR Green multitarget) for the successful detection of respiratory viral pathogens. In this study, we used tissue (pooled lung and trachea) and oropharyngeal swab samples from dead or sick animals when flock morbidity and mortality were noticed. The detection and identification of avian respiratory pathogens, particularly AIV, IBV, and NDV, have been the subject of numerous research studies due to the significance of these viruses in commercial poultry.
Riems influenza a typing array (RITA) is an RT-qPCR-based low-density array diagnostic approach for the detection of 14 HA and 9 NA subtypes of AIVs (34), and an improved version of this method has been developed recently (35). The assay uses a hydrolysis probe technique (TaqMan), one of the most powerful and widespread methodologies in diagnostic microbiology (40). The RITA represents a competitive, fast, and sensitive subtyping tool that requires neither new machinery nor additional training of staff in a lab where RT-qPCR is already established (34). Researchers have used PCR extensively as an indispensable diagnostic technology to detect viruses since its introduction because of its high sensitivity and specificity. Based on this assay, we applied two multitarget simultaneous assays, one is TaqMan probe-based RT-qPCR for AIV detection and subtyping in a single run. The sample gives positive results against the target generic gene of AIV (M) and simultaneously detects the AIV subtypes (H5, H9, N1, and N2) within a short period of time. Multiplex PCR is the simultaneous detection of more than one virus in a single-tube reaction, whereas our approach is the uniplex PCR in multiwell layout. Due to the endemic nature of H5N1 and H9N2 in Bangladesh (19), the current study concentrated on subtyping these viruses. Recently circulated global havoc of the HPAI H5N8 subtype is not yet detected in Bangladesh; therefore, it is opted out of current multitarget assays. However, it is quite possible to incorporate N8 detection into the same TaqMan multitarget assays to detect the N8 subtype simultaneously in a future study. Another assay is SYBR Green-based RT-qPCR that can detect target genes against three other respiratory pathogens (NDV, IBV, and ILT) simultaneously in a single run. Such molecular approaches have been employed in the past with various target pathogen combinations and have proven to be highly useful (34, 39, 41).
Before applying the assays in field samples, we evaluated the assays with reference (n = 45) samples from the laboratory repositories. As those target primers have already been established and sensitivity and specificity were evaluated, sensitized (34, 37–39), and are using for many years, we further combined those targets as per the requirement of the current study. Further specificity of the multitarget assays was ensured when negative commercial flocks were turned out negative by applying the simultaneous RT-qPCR assays. The most commonly determined respiratory diseases in different poultry farms during the study period were avian influenza (AI), with a prevalence of 25.5% (HPAI H5N1 and LPAI H9N2), followed by Newcastle disease (ND) and infectious bronchitis (IB), each with a prevalence of 13.2 and 9.9%, respectively. ILT, on the other hand, was identified only in a layer flock, indicating an occasional infection. In the current study, AI and ND offered considerable challenges in the layer, broiler, Sonali, and pigeon flocks, whereas AI and IB had a significant impact on duck flocks. The death of the two turkeys examined here may not have been caused by any of the viral diseases tested here; however, one turkey was found to be positive for the LPAI subtype H9N2. These similar fundamental respiratory viral diseases have been identified previously in many other countries in Asia and Africa, including Bangladesh (4, 24, 31–33, 42, 43). For many years, Bangladesh has reported ~30–50% prevalence of respiratory infections in commercial poultry (22, 23, 42, 43), also in line with our observation, which has led to slow growth and productivity, generating substantial economic losses in the poultry industry. The overall mortality shown in the investigated flocks, however, may not solely be the result of the following four viral infections. Other co-infections and poor biosecurity might have accelerated the infectivity and death rate (41, 44, 45). In the current investigation, using the multitarget simultaneous RT-qPCR assays a total of 13 farms successfully detected co-infections of either AIV and NDV, IBV and NDV, or AIV and IBV, as well as a triple infection with AIV H9N2, NDV, and IBV.
Despite the fact that all the flocks evaluated in this study were suffering from respiratory diseases, none of them tested for Mycoplasma gallisepticum (MG) infections. The gross pathological observation, on the other hand, suggested that there was a good chance of obtaining MG-positive pathogens alone or in combination with other detected viral pathogens. As a result, a new investigation is needed to detect mycoplasma and other bacterial infections as a single or mixed pathogen. According to our current investigation, respiratory disease outbreak is one of the most challenging problems in the poultry sector of Bangladesh, which could be due to minimum biosecurity practices in the farms and inadequate vaccination programs. For the reduction of respiratory diseases in poultry, rapid and sensitive detection is required. Continuous monitoring of circulating viruses and reviewing farm vaccination programs using vaccines similar to the circulating strains in addition to effective vaccination are the strategies that need to be implemented.
Respiratory pathogens are a major threat to the poultry industry in Bangladesh. AIV (HPAI H5N1 and LPAI H9N2), NDV, and IBV are the common respiratory pathogens circulating among poultry flocks. The TaqMan and SYBR Green multitarget simultaneous RT-qPCRs assays were successfully employed for screening the outbreak samples (tissue and swabs), and the interpretation was accurate, rapid, and reproducible. The specific laboratories having RT-PCR facilities could easily adopt the assays for simultaneous detection of respiratory viral pathogens within a shorter period of time as well as be able to do selective subtyping for AIVs that are endemic in the respective countries.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
The current study has been approved by the Ethical Committee of Bangladesh Agricultural University Research System under the approval number BAURES/ESRC /ET_28122. Written informed consent was obtained from the owners for the participation of their animals in this study.
RP and CK wrote the first draft of the manuscript. RP, CK, IH, and AH organized samples and data collection. RP, CK, IH, AH, JB, MN, and MI analyzed formally and interpreted the results. RP, CK, IH, AH, JB, MN, MI, and EC edited and critically reviewed the manuscript. RP and EC designed the study and supervised the study. All authors have read and agreed to the published version of the manuscript.
This research was funded by the Bangladesh Agricultural University Research System (BAURES), Mymensingh, Grant Number (2021/3/Emergency-Fund).
The authors thank the farm owners for bringing their animals for necropsy and technician Md. Shafiqul Islam for his excellent support at the laboratory.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1036757/full#supplementary-material
1. Ali MZ, Rahman MM, Sultana S. Seroprevalence of Mycoplasma gallisepticum antibody by elisa and serum plate agglutination test of laying chicken. Vet World. (2015) 8:9. doi: 10.14202/vetworld.2015.9-14
2. Poultry Sector Study Bangladesh; the Hague the Netharlands. (2020). Available online at: https://Www.Rvo.Nl/Sites/Default/Files/2020/12/Poultry%20sector%20study%20bangladesh.Pdf (accessed March 3, 2022).
3. Rahman MM, Nooruzzaman M, Kabiraj CK, Mumu TT, Das PM, Chowdhury EH, et al. Surveillance on respiratory diseases reveals enzootic circulation of both H5 and H9 avian influenza viruses in small-scale commercial layer farms of Bangladesh. Zoonoses Public Health. (2021) 68:896–907. doi: 10.1111/zph.12879
4. Roussan D, Haddad R, Khawaldeh G. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult Sci. (2008) 87:444–8. doi: 10.3382/ps.2007-00415
5. Sid H, Benachour K, Rautenschlein S. Co-infection with multiple respiratory pathogens contributes to increased mortality rates in algerian poultry flocks. Avian Dis. (2015) 59:440–6. doi: 10.1637/11063-031615-Case.1
6. Umar S, Guerin JL, Ducatez MF. Low pathogenic avian influenza and coinfecting pathogens: a review of experimental infections in avian models. Avian Dis. (2017) 61:3–15. doi: 10.1637/11514-101316-Review
7. Ball C, Forrester A, Ganapathy K. Co-circulation of genetically diverse population of vaccine related and unrelated respiratory mycoplasmas and viruses in UK poultry flocks with health or production problems. Veterinary Microbiol. (2018) 225:132–8. doi: 10.1016/j.vetmic.2018.09.009
8. Broomand Z, Jafari R, Mayahi M. Detection of newcastle disease, H9n2 avian influenza, and infectious bronchitis viruses in respiratory diseases in backyard chickens in Ahvaz, Iran, in 2014-2015. Arch Razi Inst. (2018) 73:19–25. doi: 10.22092/ARI.2018.114056
9. Bertran K, Cortey M, Díaz I. The use of H-index to assess research priorities in poultry diseases. Poultry Sci. (2020) 99:6503–12. doi: 10.1016/j.psj.2020.09.017
10. Croville G, Foret C, Heuillard P, Senet A, Delpont M, Mouahid M, et al. Disclosing respiratory co-infections: a broad-range panel assay for avian respiratory pathogens on a nanofluidic PCR platform. Avian Pathol. (2018) 47:253–60. doi: 10.1080/03079457.2018.1430891
11. Clothier KA, Torain A, Reinl S. Surveillance for avibacterium paragallinarum in autopsy cases of birds from small chicken flocks using a real-time PCR assay. J Vet Diagn Invest. (2019) 31:364–7. doi: 10.1177/1040638719844297
12. Hasni MS, Chaudhry M, Mushtaq MH, Durrani AZ, Bin Rashid H, Gill SS, et al. Prevalence and associated risk factors of avian influenza h9 in backyard poultry populations of two agroecological zones of Pakistan. Pak Vet J. (2021) 41:132–6. doi: 10.29261/pakvetj/2020.085
13. Ul-Rahman A, Shabbir MAB, Mehmood A, Shabbir MZ. Genotypic and sub-genotypic diversity of avian paramyxoviruses 2, 4 and 6. Pak Vet J. (2021) 41:156–9. doi: 10.29261/pakvetj/2020.088
14. Rimi N, Sultana R, Muhsina M, Uddin B, Haider N, Nahar N, et al. Biosecurity conditions in small commercial chicken farms, Bangladesh 2011–2012. Ecohealth. (2017) 14:244–58. doi: 10.1007/s10393-017-1224-2
15. Rahman M, Chowdhury E, Parvin R. Small-scale poultry production in Bangladesh: challenges and impact of COVID-19 on sustainability. Ger J Vet Res. (2021) 1:19–27. doi: 10.51585/gjvr.2021.0004
16. Javadov E, Vikhreva I, Sukhanova V, Kozyrenko O, Kisil A. Efficacy of live vaccines and specific prevention in chicken infectious bronchitis. Int Int J Vet Sci. (2020) 9:470–2. doi: 10.37422/IJVS/042
17. Ravikumar R, Chan J, Prabakaran M. Vaccines against major poultry viral diseases: strategies to improve the breadth and protective efficacy. Viruses. (2022) 14:1195. doi: 10.3390/v14061195
18. Islam M, Khan M, Islam M, Hassan J. Isolation and characterization of infectious laryngotracheitis virus in layer chickens. Bangladesh J Vet Med. (2010) 8:123–30. doi: 10.3329/bjvm.v8i2.11194
19. Parvin R, Begum JA, Nooruzzaman M, Chowdhury EH, Islam MR, Vahlenkamp TW. Review analysis and impact of co-circulating H5N1 and H9N2 avian influenza viruses in Bangladesh. Epidemiol Infect. (2018) 146:1259–66. doi: 10.1017/S0950268818001292
20. Parvin R, Kabiraj CK, Mumu TT, Chowdhury EH, Islam MR, Beer M, et al. Active virological surveillance in backyard ducks in Bangladesh: detection of avian influenza and gammacoronaviruses. Avian Pathol. (2020) 49:361–8. doi: 10.1080/03079457.2020.1753654
21. Bhuiyan ZA, Ali MZ, Moula MM, Bary MA, Arefin N, Giasuddin M, et al. Seroprevalence of major avian respiratory diseases in broiler and sonali chicken in selected areas of Bangladesh. J Adv Vet Anim Res. (2019) 6:561. doi: 10.5455/javar.2019.f383
22. Ganapathy K, Ball C, Kabiraj CK, Nooruzzaman M, Chowdhury EH, Islam MR. Mycoplasma gallisepticum detection in bangladesh table egg laying chicken flocks. Pak Vet J. (2021) 41:306–8. doi: 10.29261/pakvetj/2021.024
23. Nooruzzaman M, Mumu TT, Kabiraj CK, Hasnat A, Rahman MM, Chowdhury EH, et al. Genetic and biological characterization of newcastle disease viruses circulating in Bangladesh during 2010–2017: further genetic diversification of class II genotype XIII in Southcentral Asia. J Gen Virol. (2021) 102:001554. doi: 10.1099/jgv.0.001554
24. Parvin R, Begum JA, Nooruzzaman M, Kabiraj CK, Chowdhury EH. Circulation of three genotypes and identification of unique mutations in neutralizing epitopes of infectious bronchitis virus in chickens in Bangladesh. Arch Virol. (2021) 166:3093–103. doi: 10.1007/s00705-021-05227-3
25. Adi A, Astawa NM, Putra KSA, Hayashi Y, Matsumoto Y. Isolation and characterization of a pathogenic newcastle disease virus from a natural case in Indonesia. J Vet Med Sci. (2010) 72:313. doi: 10.1292/jvms.09-0303
26. Sato T, Sugimori T, Ishii S. Infectious bronchitis of chickens in Japan. I epidemiological, clinical, and pathological studies and isolation of the causative virus in embryonated eggs. Jap J Exp Med. (1955) 25:115–31.
27. Suarez DL, Das A, Ellis E. Review of rapid molecular diagnostic tools for avian influenza virus. Avian Dis. (2007) 51:201–8. doi: 10.1637/7732-101006-REGR.1
28. Spackman E, Senne DA, Myers T, Bulaga LL, Garber LP, Perdue ML, et al. Development of a real-time reverse transcriptase pcr assay for type a influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol. (2002) 40:3256–60. doi: 10.1128/JCM.40.9.3256-3260.2002
29. Meir R, Maharat O, Farnushi Y, Simanov L. Development of a real-time TaqMan RT-PCR assay for the detection of infectious bronchitis virus in chickens, and comparison of RT-PCR and virus isolation. J Virol Methods. (2010) 163:190–4. doi: 10.1016/j.jviromet.2009.09.014
30. Shirvan AS, Mardani K. Molecular detection of infectious bronchitis and newcastle disease viruses in broiler chickens with respiratory signs using duplex Rt-Pcr. Vet Res Forum. (2014) 5:319–23.
31. Haji-Abdolvahab H, Ghalyanchilangeroudi A, Bahonar A, Ghafouri SA, Vasfi Marandi M, Mehrabadi MHF, et al. Prevalence of avian influenza, newcastle disease, and infectious bronchitis viruses in broiler flocks infected with multifactorial respiratory diseases in Iran, 2015–2016. Trop Anim Health Prod. (2019) 51:689–95. doi: 10.1007/s11250-018-1743-z
32. Hassan KE, Shany SA, Ali A, Dahshan A-HM, Azza A, El-Kady MF. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult Sci. (2016) 95:1271–80. doi: 10.3382/ps/pew068
33. Umar S, Teillaud A, Aslam HB, Guerin J-L, Ducatez MF. Molecular epidemiology of respiratory viruses in commercial chicken flocks in Pakistan from 2014 through to 2016. BMC Vet Res. (2019) 15:1–12. doi: 10.1186/s12917-019-2103-6
34. Hoffmann B, Hoffmann D, Henritzi D, Beer M, Harder TC. Riems influenza a typing array (rita): an Rt-QPCR-based low density array for subtyping avian and mammalian influenza a viruses. Sci Rep. (2016) 6:1–10. doi: 10.1038/srep27211
35. Hassan KE, Ahrens AK, Ali A, El-Kady MF, Hafez HM, Mettenleiter TC, et al. Improved subtyping of avian influenza viruses using an Rt-Qpcr-based low density array: “riems influenza a typing array”, version 2 (Rita-2). Viruses. (2022) 14:415. doi: 10.3390/v14020415
36. Parvin R, Begum JA, Chowdhury EH, Islam MR, Beer M, Harder T. Co-Subsistence of avian influenza virus subtypes of low and high pathogenicity in bangladesh: challenges for diagnosis, risk assessment and control. Sci Rep. (2019) 9:1–10. doi: 10.1038/s41598-019-44220-4
37. Wäckerlin R. Untersuchungen Zu Infektionsverlauf Und Ausbildung Einer Protektiven Immunität Im Haushuhn (Gallus Gallus) Nach in Ovo Infektion Mit Einem Apathogenen Aviären Paramyxovirusisolat Vom Serotyp 1: lmu (Ph.D. thesis). (2009). Available online at: https://edoc.ub.uni-muenchen.de/10339/1/Waeckerlin_Regula.pdf
38. Chamings A, Nelson TM, Vibin J, Wille M, Klaassen M, Alexandersen S. Detection and characterisation of coronaviruses in migratory and non-migratory Australian wild birds. Sci Rep. (2018) 8:1–10. doi: 10.1038/s41598-018-24407-x
39. Callison S, Riblet S, Oldoni I, Sun S, Zavala G, Williams S, et al. Development and validation of a real-time taqman® PCR assay for the detection and quantitation of infectious laryngotracheitis virus in poultry. J Virol Methods. (2007) 139:31–8. doi: 10.1016/j.jviromet.2006.09.001
40. Nagy A, Cerníková L, Vitásková E, Krivda V, Dán Á, Dirbáková Z, et al. MeltMan: optimization, evaluation, and universal application of a qPCR system integrating the TaqMan qPCR and melting analysis into a single assay. PLoS ONE. (2016) 11:e0151204. doi: 10.1371/journal.pone.0151204
41. Yadav JP, Singh Y, Batra K, Khurana SK, Mahajan NK, Jindal N. Molecular detection of respiratory avian mycoplasmosis associated bacterial and viral concurrent infections in the poultry flocks. Anim Biotechnol. (2022) 33:1–9. doi: 10.1080/10495398.2022.2032725
42. Parvin R, Heenemann K, Halami MY, Chowdhury EH, Islam MR, Vahlenkamp TW. Full-genome analysis of avian influenza virus H9N2 from Bangladesh reveals internal gene reassortments with two distinct highly pathogenic avian influenza viruses. Arch Virol. (2014) 159:1651–61. doi: 10.1007/s00705-014-1976-8
43. Islam MR, Haque ME, Giasuddin M, Chowdhury EH, Samad MA, Parvin R, et al. New introduction of clade 2321 avian influenza virus (H5n1) into Bangladesh. Transbound Emerg Dis. (2012) 59:460–3. doi: 10.1111/j.1865-1682.2011.01297.x
44. Parvin R, Nooruzzaman M, Kabiraj CK, Begum JA, Chowdhury EH, Islam MR, et al. Controlling avian influenza virus in bangladesh: challenges and recommendations. Viruses. (2020) 12:751. doi: 10.3390/v12070751
45. Hassan KE, El-Kady MF, El-Sawah AAA, Luttermann C, Parvin R, Shany S, et al. Respiratory disease due to mixed viral infections in poultry flocks in Egypt between 2017 and 2018: upsurge of highly pathogenic avian influenza virus subtype H5n8 since 2018. Transbound Emerg Dis. (2021) 68:21–36. doi: 10.1111/tbed.13281
Keywords: outbreak investigation, RT-qPCR assays, simultaneous detection, respiratory viral pathogens, mixed infection
Citation: Parvin R, Kabiraj CK, Hossain I, Hassan A, Begum JA, Nooruzzaman M, Islam MT and Chowdhury EH (2022) Investigation of respiratory disease outbreaks of poultry in Bangladesh using two real-time PCR-based simultaneous detection assays. Front. Vet. Sci. 9:1036757. doi: 10.3389/fvets.2022.1036757
Received: 05 September 2022; Accepted: 16 November 2022;
Published: 13 December 2022.
Edited by:
Khalid Mehmood, Islamia University of Bahawalpur, PakistanReviewed by:
Ali Raza Jahejo, Shanxi Agricultural University, ChinaCopyright © 2022 Parvin, Kabiraj, Hossain, Hassan, Begum, Nooruzzaman, Islam and Chowdhury. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rokshana Parvin, cm9rc2hhbmEucGFydmluQGJhdS5lZHUuYmQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.