- Key Lab of New Animal Drug Project of Gansu Province, Key Lab of Veterinary Pharmaceutical Development of Ministry of Agriculture and Rural Affairs, Lanzhou Institute of Husbandry and Pharmaceutical Sciences of Chinese Academy of Agricultural Sciences, Lanzhou, China
Clostridium difficile infection (CDI) in human and animals belonged usually to antibiotic-associated diarrhea, ranging in severity from mild to life-threatening intestinal tract illnesses. This study aimed to isolation and characterization, toxin genes test, molecular typing, and drug sensitivity of Clostridium difficile (C. difficile) which were isolated from clinical diseased dogs and cats. A total of 247 clinical samples were collected from five animal hospitals in Lanzhou City of Northwest China, of which dogs and cats accounted for 74.9% (185/247) and 25.1% (62/247), respectively. We successfully identified 24 C. difficile strains by 16S rRNA and Matrix-Assisted Laser Desorption/Ionization Time of Fight Mass Spectroscopy (MALDI-TOF-MS). 10.3% (19/185) of dogs and 8.1% (5/62) of cats were positive for C. difficile. Among them, 16 strains were toxic and 8 were non-toxic, with a toxic rate of 57.9% (11/19) in dogs and 100% (5/5) in cats. A total of 10 STs and 10 RTs were identified in this study. The percentages of ST42 (RT106) and ST2 (RT014/LW01) among 16 toxic strains were 41.7 and 12.5%, respectively. However, ST3 (RT001), ST1 (RT027), ST133 (LW04), and ST-UN (LW04) had only one strain. ST42 (RT106) was the most common genotype and RT027 strain was first isolated in China from pets. Antimicrobial susceptibility test showed that isolates were extremely sensitive to vancomycin and metronidazole but were resistant to erythromycin and ciprofloxacin. The drug resistant rates to clindamycin, levofloxacin, moxifloxacin and meropenem were 62.5, 20.8, 16.7, and 8.3%, respectively. In conclusion, C. difficile was quietly prevalent in dogs and cats in Lanzhou city with RT106 and RT014 as the main ribotypes. The CDI in pets should be paying more attention and further studies are needed.
Introduction
C. difficile is an obligate anaerobic spore-forming Gram-positive opportunistic pathogen isolated first from infants by Hall in 1935 (1). The toxin-producing bacteria mainly colonize in the intestinal tract of human and animals (2). However, C. difficile spores are widely distributed in the environment, including soil, water, plants and animals, particularly in hospital facilities (3).
C. difficile pathogenicity is primarily mediated by its toxin. Toxin-producing C. difficile proliferates in the intestine and releases toxins that cause C. difficile infection (CDI) in the intestinal mucosa, such as diarrhea, colitis, and even death in severe cases. C. difficile can produce a 304 kDa enterotoxin A (TcdA), a 270 kDa cytotoxin B (TcdB), and a C. difficile binary toxin (CDT) in the intestine (4–6). The genes encoding the toxins A and B are located in the same pathogenicity locus (PaLoc) of the chromosome (6, 7). This region also contains three additional accessory genes as the positively regulated gene tcdR, the negatively regulated gene tcdC, and the porin gene tcdE (5). Toxin A and toxin B have similar pathogenic mechanisms in that both of them can inactivate the Rho protein family in host cells via glycosylation modification (6). They cause intestinal cytoskeleton loss and structural disruption, resulting in strong inflammatory responses and multiple clinical symptoms, such as enteritis and diarrhea (7). In addition, some strains produce binary toxins encoded by cdtA and cdtB genes located outside the PaLoc. The binary toxins act as an ADP ribosyltransferase and break down action on the cell surface, causing intestinal epithelial cell variation and apoptosis (8). However, the pathogenic mechanism of CDT remains unknown (9).
The overall incidence and number of severe cases of CDI have increased recently due to the prevalence of highly virulent strains and the increasing rate of community-acquired infections (10). Based on molecular typing, CDI outbreaks in Europe and the United States have been linked to large-scale epidemics of highly virulent strains RT027 and RT078 (11). Nevertheless, CDI and CDI-related epidemics are not limited to these ribotypes (RTs). RTs 001, 002 and 014/020 are frequently associated with CDI clusters in the United States and Europe (12, 13). Recent study revealed that the isolation rate of C. difficile from hospitalized patients ranged from 9% to 14% in China (14, 15). The main epidemic strains were ST37(RT017), ST3(RT001), ST54(RT012) (15–18).
Community-acquired CDI is one of the main factors for the increased incidence of CDI. With the progress in research on community-acquired CDI, more cases of animals carrying C. difficile have been reported (19). The most common RTs isolated from dogs in Spain are RT106 and RT154 (20). In addition, virulent strains RT027 and RT078 are isolated in dogs in Canada and diarrheic calves in Germany (21, 22). Whole-genome sequencing revealed that C. difficile isolates causing CDI between animal and human were genetically related (23). Since companion animals such as dogs and cats live in close proximity with humans, susceptible people may become carriers of C. difficile when exposed to dogs and cats with CDI (24).
Antibiotics such as vancomycin, metronidazole, and fidaxomicin are the mainstays for CDI treatment with high cure rate (25). However, the high recurrence of CDI treated with these antibiotics cannot be neglected. The mortality and treatment costs of recurrent CDI was considerably high (26). On the other hand, C. difficile drug resistance is also increasing, leading to clinical CDI treatment failure and promoting CDI's occurrence and recurrence, thus bringing great difficulties to the treatment, prevention, and control of clinical CDI (27).
In recent years, with the rapid increase in pets and pet hospitals in China, the exposure of dogs and cats to individuals susceptible to CDI and environments contaminated with C. difficile and the use of antibiotics has led to increasingly severe risk factors for C. difficile colonization and transmission about CDI epidemiology on pets. However, there has been few studies in China on the clinical carriage of C. difficile in pets. Therefore, this study investigated the carriage of C. difficile in hospitalized dogs and cats using molecular epidemiology and resistance mechanisms to prevent large-scale transmission of C. difficile.
Materials and methods
Sample collection
Between March 2021 and December 2021, a total of 247 samples with anal swab were collected from dogs (n = 185) and cats (n = 62). There are five animal hospitals in Lanzhou City, Gansu Province: Qilihe District, Anning District, Xigu District, and Chengguan District. Five animal hospitals (denoted by “A,” “B,” “C,” “D,” “E”), “A” is a general practice hospital in Qilihe District, “B” and “C” are referred to the central referral hospital in Qilihe District and Anning District, and “D” and “E” are community hospitals in Chengguan District and Xigu District. “A” is the largest animal hospital with the most cases in Lanzhou City. “B” and “C,” as the referral center hospital, have a relatively small number of cases and treat severe diseases. “D” and “E”, as a community hospital, have a relatively small number of cases and treat mainly animals with vaccination and common diseases. All samples consisted of “A” (48.2%, 119), “B” (13.4%, 33), “C” (4.0%, 10), “D” (18.6%, 46), and “E” (15.8%, 39). In addition, all procedures were conducted following an approved protocol by the Institutional Animal Care and Use Committee at the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of the Chinese Academy of Agricultural Sciences.
Bacterial isolation and identification
Samples were collected with a disposable sampling anal swab from five animal hospitals in downtown Lanzhou city and inserted into Amies transport medium (Qingdao Haibo Biotechnology Co., Ltd, Qingdao, China). Samples were kept at 4°C before being sent to the laboratory within 12 h. Subsequently, samples were inoculated into a 5 mL cycloserine-cefoxitin fructose agar (CCFA) medium (an enriched, selective, and differential medium for the isolation of C. difficile) and placed in a Forma Anaerobic System (Thermo Scientific, USA) (28). The pretreated samples were incubated anaerobically (85% N2, 10% H2, and 5% CO2) at 37 °C for 7 d. Then 10 μL of the enriched culture was inoculated onto Chrome ID C. difficile agar medium (Merial, France) using the four-zone streak plate cultivation method and incubated anaerobically at 37°C for 48 h. Plates with growing colonies were placed under UV light at 365 nm to observe fluorescent spots. Single blue-fluorescent colony was inoculated onto an anaerobic agar medium (Beijing Land Bridge Technology Co., Ltd, China). Finally, the single colony morphology was observed and stained by Gram's method after 48 h incubation.
The isolates were identified using a matrix-assisted laser desorption–ionization time-of-flight mass spectrometry system (MALDI-TOF MS) (Bruker, Germany) (30). Briefly, colonies with blue fluorescence (a characteristic of C. difficile) and Gram-positive staining on the chromogenic medium were re-cultured for 48 h. A small number of fresh colonies were then evenly coated on the VITEK MS target plate. A 1 μL of α-Cyano-4-hydroxycinnamic acid solution was added dropwise to these colonies. The VITEK MS target plate was placed in MALDI–TOF-MS after the substrate had dried naturally. The mass spectral data were acquired between 2,000 and 20,000 Da. Finally, the strains were identified by comparing mass data to a database of standard C. difficile in data base through the Biotyper software.
Toxin gene test and TcdC gene sequencing
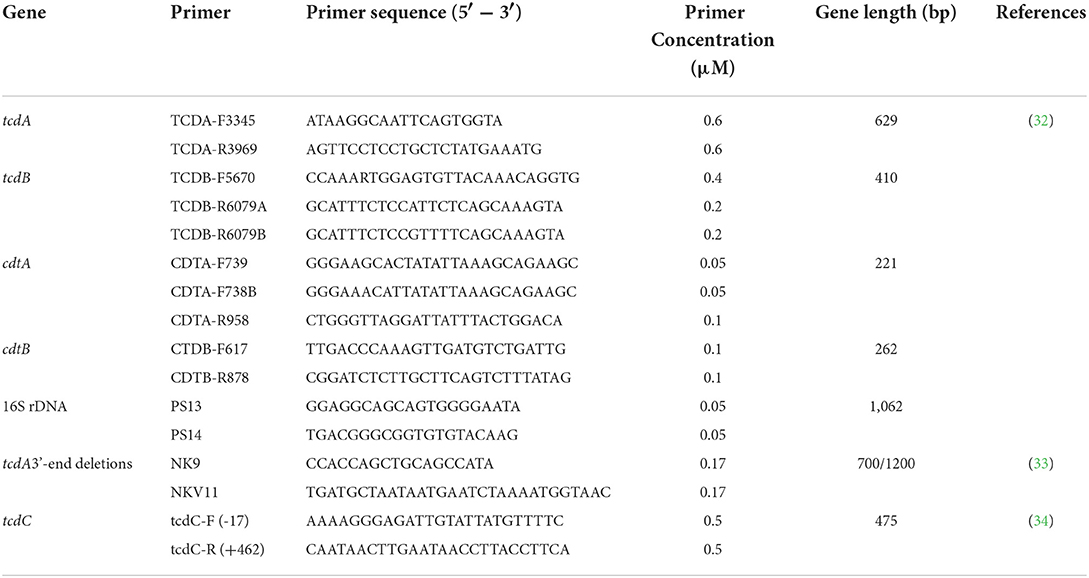
Single colonies were inoculated into an anaerobic liquid medium and incubated anaerobically overnight. Following the operation manual procedures, genomic DNA was extracted from the bacterial solution using the Bacterial DNA Extraction Kit (Solarbio Technology Co., Ltd., Beijing, China) and stored at−20 °C. The extracted DNA was used as a template for five-fold polymerase chain reaction (PCR) amplification of the toxin genes tcdA, tcdB, and CDT, and the 16S rRNA following the method recommended by Cheng et al. (31). PCR amplification primer sequences were seen in Table 1. Additionally, A 1.8 kb at the 3' terminal of the tcdA gene was examined for deletion. The tcdC gene was also amplified and detected using the method proposed by Curry et al. (34). The sequence results were compared to the tcdC gene standard sequence (NC_009089.1, a gene bank accession number for tcdC gene sequence of Clostridioides difficile 630) to identify any gene deletion.
Multilocus sequencing typing
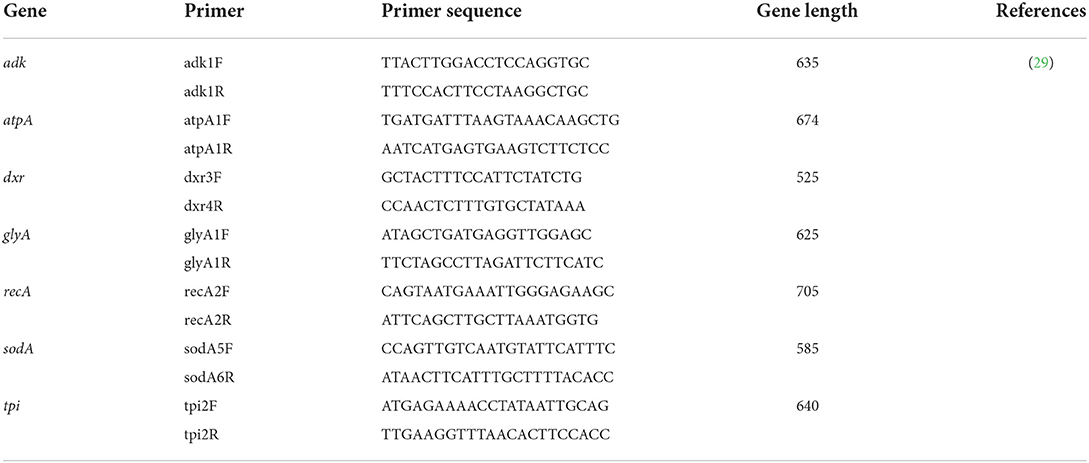
Seven C. difficile housekeeping genes were amplified according to the primers given on the official Multi-locus Sequencing Typing (MLST) website (http://pubMLST.org/cdifficile) using the method recommended by Griffiths et al. (29), followed by gene sequencing. The primers for MLST amplification and sequencing seen in Table 2. The sequencing results were submitted to the official website for comparison to obtain the allele type and the C. difficile sequence type (ST).
PCR-ribotyping
PCR-ribotyping was performed according to nucleotide sequence polymorphisms in the 16S-23S rRNA intergenic spacer region of C. difficile. The gene fragments in this region were amplified according to the method recommended by Indra et al. (35). The size and peak of the fragments read by Gene Marker were submitted to the WEBRIBO database (http://webribo.ages.at) to determine the RT of C. difficile. Novel RTs were named with “LW” plus two Arabic numbers, such as LW01.
Drug susceptibility test
The antimicrobial susceptibility test of the isolated C. difficile was performed using the agar dilution method recommended by the US Clinical and Laboratory Standards Institute (CLSI) M11-A8 (36). The following eight antibiotics were included in this study: vancomycin, clindamycin, erythromycin, levofloxacin, moxifloxacin, ciprofloxacin, meropenem, and metronidazole (Yuan ye Biotech., Shanghai, China). The results were interpreted with reference to the susceptibility breakpoints for anti-anaerobic drugs in CLSI 2017 version M100-S27 (37). Furthermore, for antibiotics with no susceptibility breakpoints in the CLSI document, the susceptibility breakpoints were proposed according to Huang's report (16).
Statistical analysis
Statistical significance was determined using the chi-square test and univariate logistic regression in SPSS Statistics (version 22; IBM Corporation). The level of significance was set at P < 0.05 and P < 0.01.
The genetic relationships of the isolates was determined by cluster analysis using the minimum-spanning tree available in the BioNumerics software V6.5 (Applied Maths).
Results
Prevalence of CDI in dogs and cats
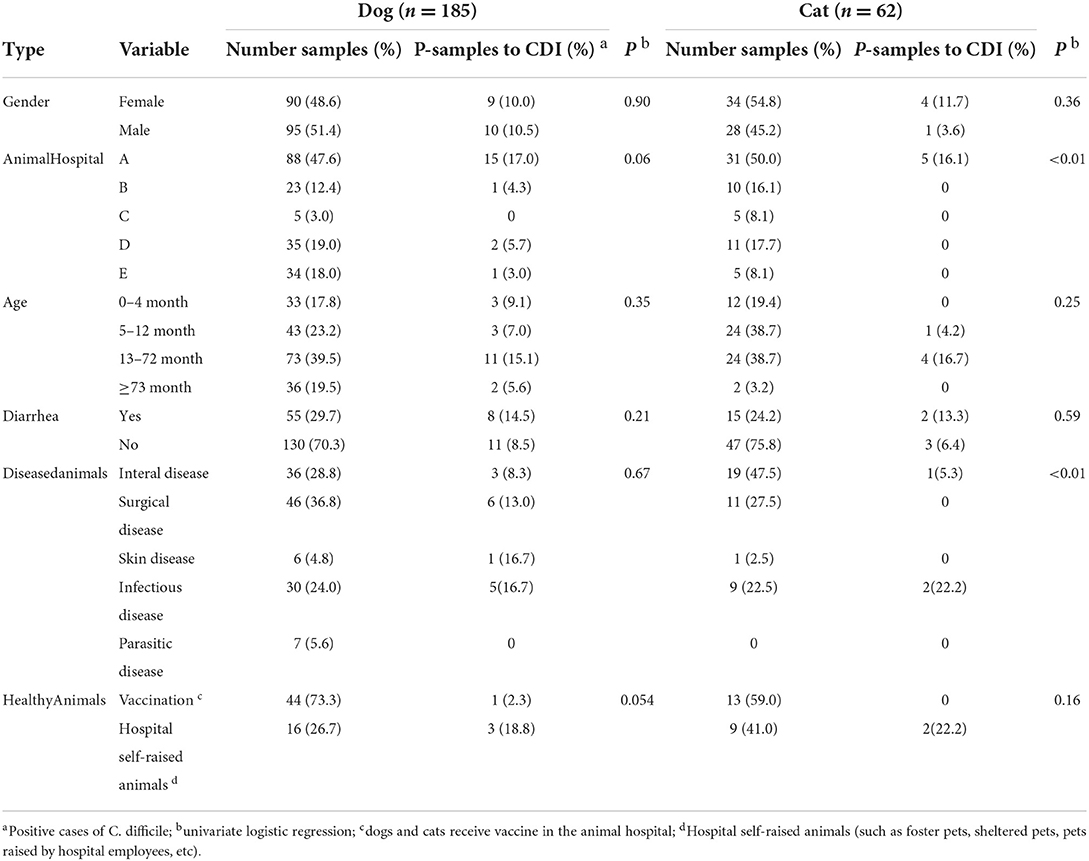
A total of 247 clinical samples were collected from five animal hospitals in Lanzhou City, of which dogs and cats accounted for 74.9% (185/247) and 25.1% (62/247), respectively. Among them, 10.3% (19/185) of dogs and 8.1% (5/62) of cats were positive for CDI. Five animal hospitals tested positive of CDI at rates of A (16.8%, 20/119), B (3.0%, 1/33), C (0%, 0/10), D (4.3%, 2/46) and E (2.6%, 1/39). Notably, in hospital A, 48.2% (119/247) of the samples were collected, but the positive samples account for 83.3% (20/24) of the total positive samples. The positive rate of hospital A was nearly 5 times higher than other hospitals. Treatment information and the health status of the dogs and cats were collected for statistical analysis (Table 3). Male and female dogs tested positive for CDI at rates of 10.5 and 10.0%, respectively. But male and female cats tested positive at rates of 3.6 and 11.7%, respectively. However, no significant difference in positive rates was observed in dogs and cats of different ages. Moreover, there was no significant correlation between diarrhea and isolated C. difficile. The positive rates for CDI in infectious, dermatological, and surgical diseases was diagnosed in 16.7, 16.7, and 13.0% of dogs, respectively, higher than its internal diseases, parasitic diseases, and immunizations. The rate of CDI positivity for infectious diseases in cats was 22.2%, higher than its other diseases' rates. The CDI positivity rate for vaccination in dogs and cats were 2.3 and 0.0%, which were extremely low. However, the positive rates for CDI in 25 hospital self-raised animals from five different animal hospitals were 18.8% (3/16) for dogs and 22.2% (2/9) for cats.
Toxin genotyping and TcdC gene test
We identified successfully 24 C. difficile strains by MALDI-TOF MS. There were 16 toxin-producing strains and 8 non-toxin-producing strains among them, with a toxic rate of 57.9% (11/19) in dogs and 100% (5/5) in cats. Five-fold PCR was used to amplify three toxin types: A+B+CDT+, A+B+CDT-, and A-B-CDT- (Figure 1). One of 16 toxic strains was A+B+CDT+ and other 15 were A+B+CDT-. Eight non-toxic strains all belonged to the A-B-CDT-. Next, the tcdA gene of 16 toxic strains was then amplified. Gel electrophoresis displayed typical tcdA bands at 1200 bp, meaning no gene deletion at 3' terminus of tcdA of all the 16 isolates (Figure 2). In other words, all the 16 isolates were normal toxic without toxic enhancement caused by gene deletion at 3' terminus of tcdA.
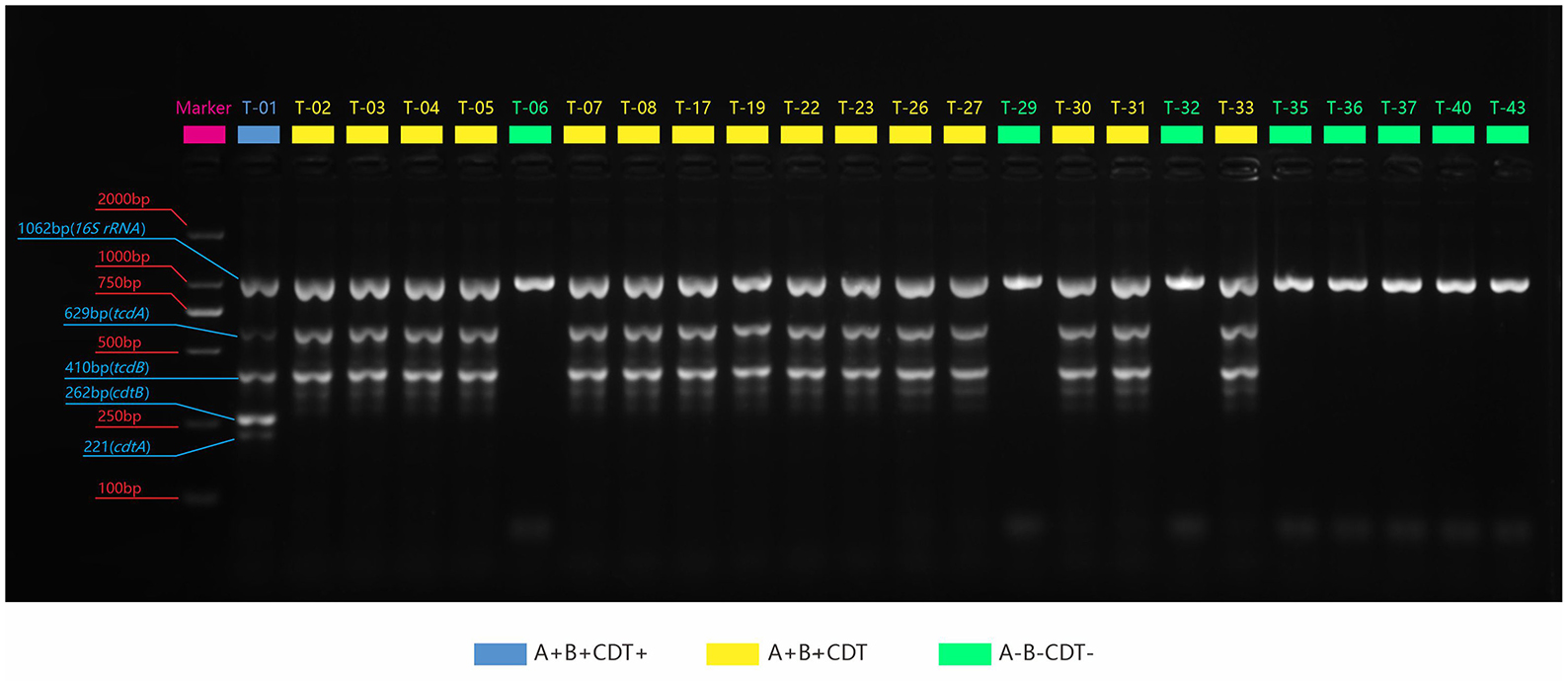
Figure 1. PCR amplification plots of the three toxin genes and the 16S rRNA genes. Blue: The strain contains four genes: tcdA, tcdB, cdtA, and cdtB; Yellow: It contains two genes: tcdA, tcdB; Green: It does not contain any toxin genes.
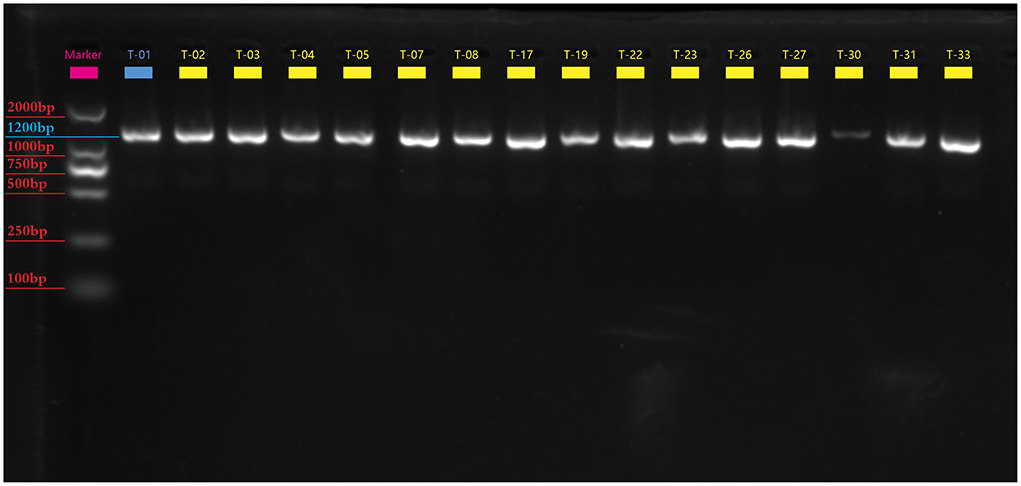
Figure 2. The electrophotogram of the tcdA gene PCR amplification. There are 16 toxin-producing Clostridium difficile tcdA gene bands at 1200bp.
The tcdC gene of 16 strains were amplified and sequenced. The obtained sequences were compared to the standard tcdC sequences (NC_009089.1) from NCBI. Only one strain (OP615994) had a single nucleotide deletion at position 117 and 18 nucleotide deletions at positions 330–348 in the tcdC gene (Figure 3).
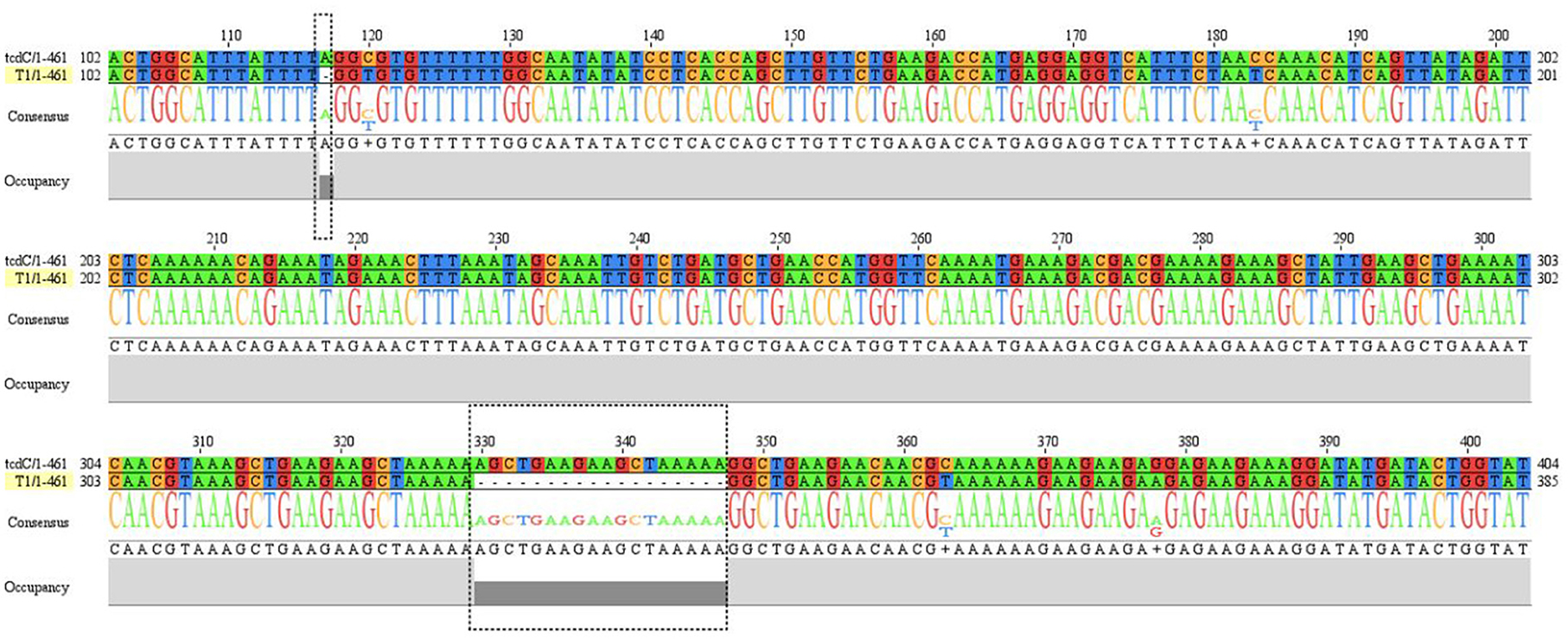
Figure 3. The sequence alignment between T1 strain (OP615994) and reference tcdC gene (NC_009089.1). tcdC gene of strain T1 has nucleotides deletions at position 117 and position 330-347 compared to reference sequence.
MLST
Ten ST types were identified from seven housekeeping genes of 24 C. difficile strains in 3 MLST clades by MLST database matching. Among them, ST2, ST3, ST42, ST15, ST26, ST76 and ST133 strains were located in MLST clade1. ST1 strain and ST39 strain were located in MLST clade2 and MLST clade 4, respectively. The most common strains in the isolated C. difficile were ST42 (41.7%, 10/24), ST39 (16.7%, 4/24), and ST2 (12.5%, 3/24). The remaining seven STs included ST1, ST3, ST15, ST26, ST76, ST133 and the newly identified strain ST-UN, all of which had only one strain with an isolation rate of <5% (Table 4).
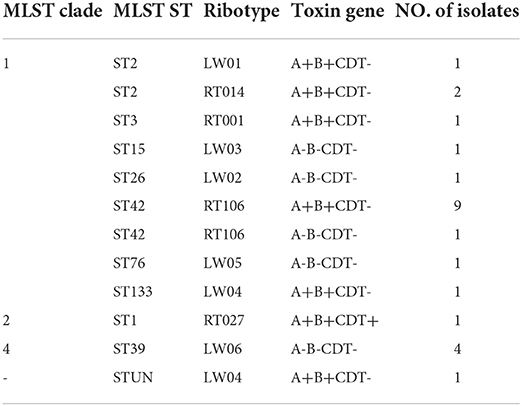
Table 4. Multilocus sequence type (MLST), ribotype, and toxin genotypes of 24 clostridium difficile clinical isolates.
RT
The data from 10 RTs were submitted to the WEBRIBO database for comparison. The RTs named RT027, RT014, RT106, and RT001 matched successfully with 14 strains. In addition, six new RTs were identified and named as LW “01–06”. Among these RTs, the most common type was RT106 (10/24), followed by LW06 (4/24) and RT014 (2/24) (Table 4).
The relationship between MLST, RT, and the toxin gene
A+B+CDT+ toxic strain ST1 (RT027) was identified. The strain belonged to MLST clade 2, with a single nucleotide deletion at position 117 and 18 nucleotide deletions in the tcdC gene from positions 330–348 (Figure 3). This strain is highly virulent and has caused many large-scale outbreaks globally (38).
ST42 (RT106), ST2 (RT014), ST2 (LW01), ST133 (LW04), ST-UN (LW04), and ST3 (RT001) were all found in 15 A+B+CDT-strains and belonged to the MLST clade 1 and were closely related (Figures 4, 5). ST3 and ST42 had a strong relationship. ST133 and ST-UN were similar and shared the same RT LW04 (Figure 4). Furthermore, the 355th adenine of ST133's housekeeping gene atpA was mutated to guanine. It led to the change in the atpA gene's allele from 1–4, thus forming a new mutant strain ST-UN.
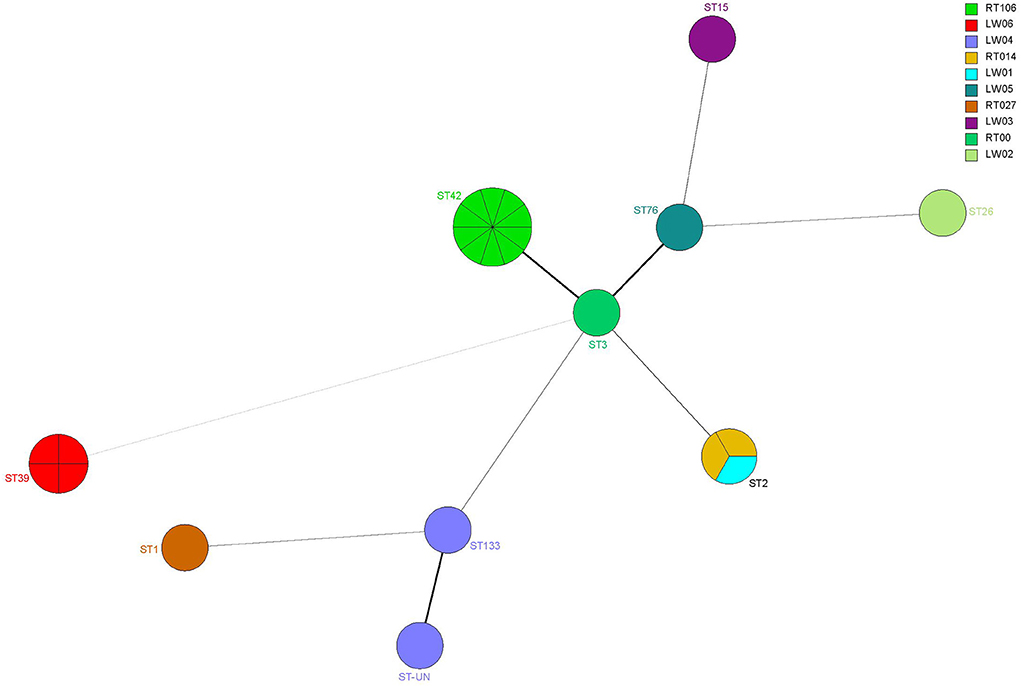
Figure 4. Analysis of the MLST minimum spanning tree for 24 C. difficile strains. Each circle represents a ST type, while the number next to the circle represents the ST type. The size of the circle represents the number of isolated strains; The colors represent different ribotypes; Kinship is shown by the lines connecting the circles.
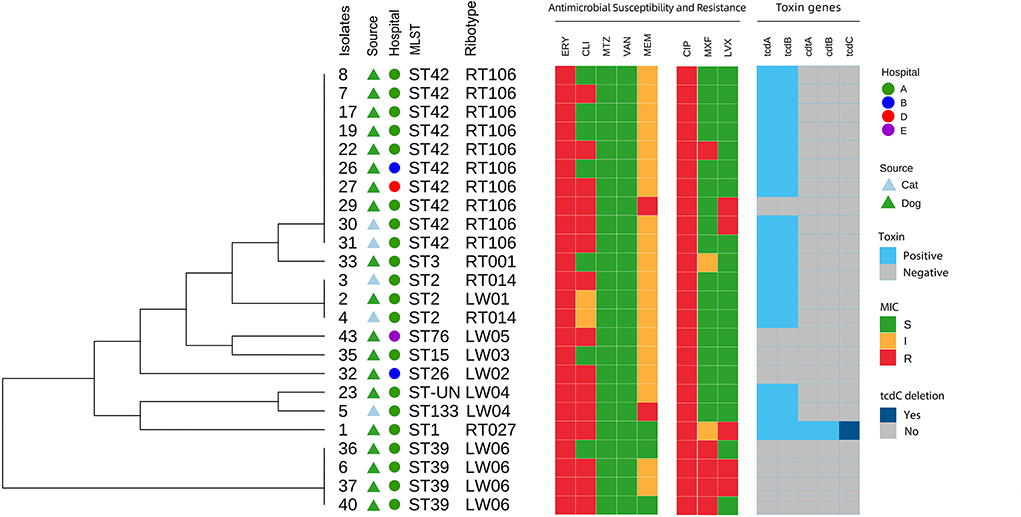
Figure 5. Core genome-based phylogenetic tree and distribution of C. difficile source, hospitals, toxin genes, MLST, ribotypes, antimicrobial susceptibility and resistance among isolates from dogs and cats.
Eight A-B-CDT- strains included ST39 (LW06), ST26 (LW06), ST76 (LW05), ST15 (LW03) and ST42 (RT106). ST42, ST76, ST15, and ST26 were members of MLST clade 1 and were closely related (Figures 4, 5). ST39 belonged to MLST clade 4. These five STs also had different RTs. In addition, nine strains of ST42 (RT106) were toxin-producing, while one was non-toxin-producing (Table 4 and Figure 5).
Antimicrobial susceptibility analysis
All 24 C. difficile strains were susceptible to vancomycin and metronidazole but were resistant to erythromycin and ciprofloxacin. Clindamycin resistance was also high (62.5%). The resistance rates for levofloxacin, moxifloxacin, and meropenem were 20.8, 16.7, and 8.3%, respectively. The 50% minimum inhibitory concentrations (MIC50) of vancomycin and metronidazole were 0.5 and 4 μg/mL, respectively (Table 5).
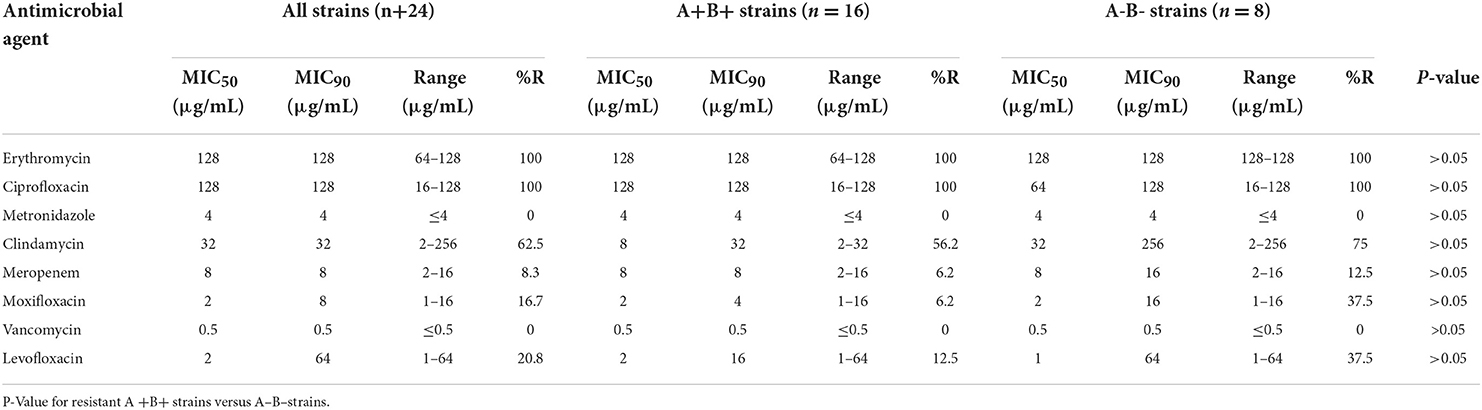
Table 5. Antibiotic resistance rates and minimum inhibitory concentration (MIC) ranges for the 24 C. diffificile clinical isolates.
The resistance rates of 16 toxin-producing strains to clindamycin, levofloxacin, moxifloxacin, and meropenem were 56.2, 12.5, 6.2, and 6.2%, respectively. For eight non-toxin-producing strains, the resistance rates of clindamycin, moxifloxacin and levofloxacin and meropenem were 75, 37.5, 37.5, and 12.5%, respectively. The resistance of non-toxic strains was slightly higher than that of the toxic strains. The resistance rates of 10 ST42 (RT106) strains to clindamycin, meropenem, levofloxacin, and moxifloxacin were 60, 10, 30, and 20%, respectively. The resistance rates of the above antibiotics for four ST39 (LW06) strains were 75, 25, 25, and 75%, respectively. Moreover, two ST2 (RT014) strains exhibited 50.0% resistance to clindamycin but were susceptive to meropenem, moxifloxacin, and levofloxacin. Among above three common genotypes, ST42 (RT106) exhibited the highest resistance rate to levofloxacin and ST39 (LW06) were resistant to clindamycin and moxifloxacin (Figure 6).
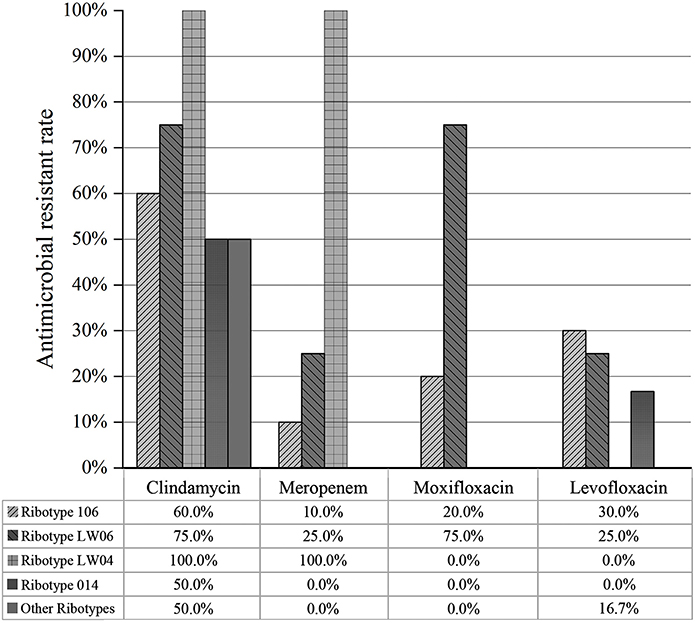
Figure 6. Drug resistance of different PCR ribotypes of 24 C. difficile isolates to antibiotics (clindamycin, meropenem, moxifloxacin and levofloxacin).
Discussion
C. difficile is an obligate anaerobe and needs specific sampling and isolation methods. In this study, samples were transported in the Amies transport medium to reduce the possibility of bacterial death due to oxygen exposure. Cefoxitin and cycloserine in the CCFA selective medium effectively inhibited the growth of other bacteria but did not affect C. difficile. In addition, sodium taurocholate in CCFA promoted the germination of C. difficile spores (39). As a result, the CCFA medium outperformed the traditional medium for C. difficile isolation. Currently, the principal method for identifying bacteria is 16S rRNA sequencing, which is labor-intensive and time-consuming. Therefore, the MALDI-TOF-MS identification method was applied in this study, which is rapid, high-throughput, and accurate (40).
C. difficile isolation rates in companion dogs and cats in Lanzhou city of China were 10.3% and 8.1%, respectively. The isolation rates were higher than those reported recently in China by Wei et al. (dogs 0.7% and cats 7%) (41). The CDI positive rates of dogs and cats from hospital A were significantly higher than other four hospitals (P < 0.01). The possible reasons for this for this difference were not determined as this was not the focus of the study. However, it possible that hospital A served as a potential source of C. difficile for patients or samples. This result was consistent to the molecular typing result that the prevalent strain in hospital A was RT106 (ST42). It is also possible higher CDI positivity was found in hospital A due to external conditions such as the geographic location, community socioeconomic conditions, animal population differences, or types of problems treated. On the other hand, the isolation rate of CDI from pets was less relative to pet status including age, diarrhea, and health condition (P > 0.05). The similar results were also observed from CDI epidemiologic study in dogs and cats in Madrid in Spain (20).
C. difficile is classified into different toxin types according to the type of toxin gene it carries. A+B+CDT- is the most commonly reported toxin type among toxic C. difficile strains, accounting for 70–90% of the total (42, 43). The toxin type with the highest toxin-producing capacity and prevalence is A+B+CDT+, such as RT027 and RT078, which caused CDI outbreaks worldwide (44). Furthermore, the clinical A-B+CDT- type is common in Chinese inpatients (45). The presence of only the tcdB gene may increase virulence due to its strong regulatory capacity (46). Therefore, the toxicity of A-B+CDT- toxin type cannot be ignored. There was also an identified C. difficile strain that only expressed the binary toxin CDT and CDT could alone cause CDI recurrence (27). However, there is currently no information on A+B-CDT- type of C. difficile.
In this study, bacterial toxin genes were amplified using five-fold PCR technology. There were 16 toxin-producing and 8 non-toxin-producing strains identified with toxin production rates of 57.9% in dogs (11/19) and 100% in cats (5/5). In North America, 50–73% and 7.1–34.8% of the toxin-producing C. difficile strains were isolated from dogs and cats, respectively (47–50). In Europe, toxic type isolation rates for dogs and cats were 5.5–58.0% and 5.6–80%, respectively (51–54). The isolation rate of toxin-producing strains in dogs in this study was similar to those in Europe and the United States, but the isolation rate in cats was higher. We isolated a C. difficile strain carrying the tcdA, tcdB, and CDT genes. C. difficile strains carrying the binary toxin usually leads to a high risk of morbidity and mortality and were of worldwide concern (8, 55).
A total of 10 STs and 10 RTs were identified in this study. Among them, ST42 (RT106) and ST2 (RT014/LW01) were the most common isolates, accounting for 41.7 and 12.5% of all isolates, respectively. ST39 (LW06), ST1 (RT027), and the remaining eight STs were located in MLST clade4, clade2 and clade1, respectively. STs from the same MLST clade were more closely related. The percentages of ST42 (RT106) and ST2 (RT014/LW01) among 16 toxin-producing strains were the highest. However, ST3 (RT001), ST1 (RT027), ST133 (LW04), and ST-UN (LW04) all had only one strain. ST42 (RT106) was the most common genotype. Recent reports showed that the most common RTs were RT014 and RT010 in dogs and RT014 in cats (56–58). RT106 was mainly found in some studies conducted in Brazil and Germany, and the isolation rate was relatively low (56). In Europe, RT014 is the most common cause of CDI-related diarrhea in human (13, 59). Although RT014 rarely causes outbreaks of CDI, it is highly adaptive and widespread in animals (60). In Berlin of Germany, the isolation rate of RT014 C. difficile in companion animals is 22.2% (4/18) (56). Two strains of RT014 were identified among 24 C. difficile strains in this study, with a relatively low isolation rate of 8.3%. Moreover, a RT027 strain of A+B+CDT+ was isolated, which possessed high toxin-producing capacity due to the binary toxin genes cdtA and cdtB. This strain had a single nucleotide deletion at site 117 and a nucleotide sequence deletion at sites 330–347 of the tcdC gene. The tcdC gene plays a negative regulatory role, and its deletion can increase the expression of toxin A and toxin B, thus enhancing the strain's virulence (61). Isolation of RT027 strain from companion animals was rarely reported worldwide. In Canada, a RT027 strain from a healthy dog in an animal hospital was firstly reported and then another report from dog in 2018 (56, 62). This study is the first report of the isolation of the RT027 strain in China on a healthy dog from an animal hospital. Hence, some highly virulent strains with high prevalence and pathogenicity in human may exist in animal hospitals, which increases the risk of cross-transmission between pets and human beings. In the future, enhancing inspection, prevention, and control are needed to avoid outbreaks of highly virulent C. difficile strains.
Eight non-toxin-producing C. difficile strains were isolated in this study and their STs were ST39, ST15, ST26, ST42 and ST76. Among them, ST39 (n = 4) was the most abundant, whereas the rest had only one strain. Moreover, we identified one non-toxin-producing ST42 strain among 10 strains. This same ST strain with both toxin-producing and non-toxin-producing properties exists in the PUBMLST database, but such a case is relatively rare.
The drug susceptibility analysis suggested that 24 C. difficile strains isolated in this study were susceptible to metronidazole and vancomycin with MIC90 of 4 and 0.5 μg/mL, respectively. Almost all C. difficile were susceptible to metronidazole and vancomycin. Therefore, they are currently the preferred antibiotics for the clinical treatment of CDI in human. Metronidazole is more commonly used in dogs than vancomycin for treating acute diarrhea and chronic enteritis (63, 64). Twenty-four C. difficile strains were resistant to erythromycin (100%) and ciprofloxacin (100%), which were similar to erythromycin (13–100%), ciprofloxacin (99%), clindamycin (55%), and moxifloxacin (34%) (65). C. difficile isolated from animals in China had a resistance rate of 93.7% to ciprofloxacin (66). The resistance rate of C. difficile to ciprofloxacin ranged between 80 and 100% in Shanghai of China (16). C. difficile also produced complete resistance to erythromycin. Although erythromycin was used less frequently in Spanish companion animals, resistance is relatively high (67). The resistance rate of 345 toxin-producing C. difficile strains to erythromycin was 69.5% in China (66). The resistance rates of toxin-producing C. difficile isolates to clindamycin and levofloxacin were 62.5 and 20.8%, respectively. The main MLST types of resistant isolates were ST42 (RT106), ST39 (LW06) and ST1 (RT027). All four strains of ST39 (LW06) developed antibiotic resistance. Therefore, ST39 (LW06) had high resistance to other antibiotics, which may be attributed to the different resistance rates among various regions and prevalent strains.
Conclusion
The positive rates of C. difficile for dogs and cats in Lanzhou city of China were 10.3% (19/185) and 8.1% (5/62), respectively. The main pandemic strains were ST42 (RT106) and ST2 (RT014/LW01), which were also frequently reported in human-related studies. All isolated C. difficile strains were susceptible to metronidazole and vancomycin, while the resistant rates to erythromycin, ciprofloxacin, clindamycin, levofloxacin, moxifloxacin, and meropenem were 100, 100, 62.5, 20.8, 16.7, and 8.3%, respectively. The CDI of companion animals in China should be paid more attention to ensure animal welfare and public health security.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository and accession numbers can be found below: NCBI; OP503451-OP503473.
Ethics statement
The animal study was reviewed and approved by the Lanzhou Institute of Husbandry and Pharmaceutical Sciences of CAAS.
Author contributions
X-WL and J-YL conceived of, proposed the idea, and designed the study. G-LW, S-HL, ZQ, L-XB, and W-BG conducted the experiments. J-YL and G-LW performed the acquisition and analysis of data. J-YL, G-LW, and X-WL wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by grants from the National Key R&D Program of China (2021YFD1800900), Natural Science Foundation of Gansu Province (22JR5RA042) and Science-technology innovation engineering of CAAS (25-LZIHPS-02).
Acknowledgments
We would like to thank Wu Qing-feng from Institute of Modern Physics of the Chinese Academy of Sciences for MALDI-TOF MS determination.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1032945/full#supplementary-material
References
1. Hall JC, O'Toole E. Intestinal flora in new-born infants with a description of a new pathogenic anaerobe, bacillus difficilis. Am J Dis Child. (1935) 49:390–402. doi: 10.1001/archpedi.1935.01970020105010
2. Bartlett JG, Moon N, Chang TW, Taylor N, Onderdonk AB. Role of clostridium difficile in antibiotic-associated pseudomembranous colitis. Gastroenterology. (1978) 75:778–82. doi: 10.1016/0016-5085(78)90457-2
3. Kim KH, Fekety R, Batts DH, Brown MD, Cudmore Silva JJ, et al. Isolation of clostridium difficile from the environment and contacts of patients with antibiotic-associated colitis. J Infect Dis. (1981) 143:42–50. doi: 10.1093/infdis/143.1.42
4. Riegler M, Sedivy R, Pothoulakis C, Hamilton G, Zacherl J, Bischof G, et al. Clostridium difficile toxin b is more potent than toxin a in damaging human colonic epithelium in vitro. J Clin Invest. (1995) 95:2004–11. doi: 10.1172/JCI117885
5. Thelestam M, Chaves-Olarte E. Cytotoxic effects of the clostridium difficile toxins. Curr Top Microbiol Immunol. (2000) 250:85–96. doi: 10.1007/978-3-662-06272-2_4
6. Geric B, Carman RJ, Rupnik M, Genheimer CW, Sambol SP, Lyerly DM, et al. Binary toxin-producing, large clostridial toxin-negative clostridium difficile strains are enterotoxic but do not cause disease in hamsters. J Infect Dis. (2006) 193:1143–50. doi: 10.1086/501368
7. Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. (2009) 7:526–36. doi: 10.1038/nrmicro2164
8. Kelly CP, LaMont JT. Clostridium difficile–more difficult than ever. N Engl J Med. (2008) 359:1932–40. doi: 10.1056/NEJMra0707500
9. Loo VG, Poirier L, Miller MA, Oughton M, Libman MD, Michaud S, et al. A predominantly clonal multi-institutional outbreak of clostridium difficile-associated diarrhea with high morbidity and mortality. N Engl J Med. (2005) 353:2442–9. doi: 10.1056/NEJMoa051639
10. Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (shea) and the infectious diseases society of America (IDSA). Infect Control Hosp Epidemiol. (2010) 31:431–55. doi: 10.1086/651706
11. Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, et al. The changing epidemiology of clostridium difficile infections. Clin Microbiol Rev. (2010) 23:529–49. doi: 10.1128/CMR.00082-09
12. Gonzales-Luna AJ, Carlson TJ, Dotson KM, Poblete K, Costa G, Miranda J, et al. Pcr ribotypes of clostridioides difficile across texas from 2011 to 2018 including emergence of ribotype 255. Emerg Microbes Infect. (2020) 9:341–7. doi: 10.1080/22221751.2020.1721335
13. Bauer MP, Notermans DW, van Benthem BH, Brazier JS, Wilcox MH, Rupnik M, et al. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. (2011) 377:63–73. doi: 10.1016/S0140-6736(10)61266-4
14. Tang C, Cui L, Xu Y, Xie L, Sun P, Liu C, et al. The incidence and drug resistance of clostridium difficile infection in mainland China: a systematic review and meta-analysis. Sci Rep. (2016) 6:37865. doi: 10.1038/srep37865
15. Chen YB, Gu SL, Wei ZQ, Shen P, Kong HS, Yang Q, et al. Molecular epidemiology of clostridium difficile in a tertiary hospital of China. J Med Microbiol. (2014) 63:562–9. doi: 10.1099/jmm.0.068668-0
16. Huang H, Weintraub A, Fang H, Nord CE. Antimicrobial resistance in clostridium difficile. Int J Antimicrob Agents. (2009) 34:516–22. doi: 10.1016/j.ijantimicag.2009.09.012
17. Cheng JW, Xiao M, Kudinha T, Kong F, Xu ZP, Sun LY, et al. Molecular epidemiology and antimicrobial susceptibility of clostridium difficile isolates from a university teaching hospital in China. Front Microbiol. (2016) 7:1621. doi: 10.3389/fmicb.2016.01621
18. Yan QR, Cheng Y, Lu JX. molecular typing methods on clostridium difficile and their application. Zhonghua Liu Xing Bing Xue Za Zhi. (2011) 32:1046–9. doi: 10.1038/cdd.2010.68
19. Stamper PD, Alcabasa R, Aird D, Babiker W, Wehrlin J, Ikpeama I, et al. Comparison of a commercial real-time PCR assay for tcdB detection to a cell culture cytotoxicity assay and toxigenic culture for direct detection of toxin-producing clostridium difficile in clinical samples. J Clin Microbiol. (2009) 47:373–8. doi: 10.1128/JCM.01613-08
20. Álvarez-Pérez S, Blanco JL, Harmanus C, Kuijper EJ, García ME. Prevalence and characteristics of clostridium perfringens and clostridium difficile in dogs and cats attended in diverse veterinary clinics from the madrid region. Anaerobe. (2017) 48:47–55. doi: 10.1016/j.anaerobe.2017.06.023
21. Lefebvre SL, Arroyo LG, Weese JS. Epidemic clostridium difficile strain in hospital visitation dog. Emerg Infect Dis. (2006) 12:1036–7. doi: 10.3201/eid1206.060115
22. Schneeberg A, Neubauer H, Schmoock G, Grossmann E, Seyboldt C. Presence of clostridium difficile PCR ribotype clusters related to 033, 078 and 045 in diarrhoeic calves in Germany. J Med Microbiol. (2013) 62:1190–8. doi: 10.1099/jmm.0.056473-0
23. Weese JS, Finley R, Reid-Smith RR, Janecko N, Rousseau J. Evaluation of clostridium difficile in dogs and the household environment. Epidemiol Infect. (2010) 138:1100–4. doi: 10.1017/S0950268809991312
24. Loo VG, Brassard P, Miller MA. Household transmission of clostridium difficile to family members and domestic pets. Infect Control Hosp Epidemiol. (2016) 37:1342–8. doi: 10.1017/ice.2016.178
25. Allen CA, Babakhani F, Sears P, Nguyen L, Sorg JA. Both fidaxomicin and vancomycin inhibit outgrowth of clostridium difficile spores. Antimicrob Agents Chemother. (2013) 57:664–7. doi: 10.1128/AAC.01611-12
26. Karaaslan A, Soysal A, Yakut N, Akkoc G, Demir SO, Atici S, et al. Hospital acquired clostridium difficile infection in pediatric wards: a retrospective case-control study. Springerplus. (2016) 5:1329. doi: 10.1186/s40064-016-3013-8
27. Legenza L, Barnett S, Rose W, Safdar N, Emmerling T, Peh KH, et al. Clostridium difficile infection perceptions and practices: a multicenter qualitative study in south Africa. Antimicrob Resist Infect Control. (2018) 7:125. doi: 10.1186/s13756-018-0425-y
28. Bjöersdorff OG, Lindberg S, Kiil K, Persson S, Guardabassi L, Damborg P. Dogs are carriers of Clostridioides difficile lineages associated with human community-acquired infections. Anaerobe. (2021) 67:102317. doi: 10.1016/j.anaerobe.2020.102317
29. Griffiths D, Fawley W, Kachrimanidou M, Bowden R, Crook DW, Fung R, et al. Multilocus sequence typing of clostridium difficile. J Clin Microbiol. (2010) 48:770–8. doi: 10.1128/JCM.01796-09
30. Cheng JW, Liu C, Kudinha T, Xiao M, Yu SY, Yang CX, et al. Use of matrix-assisted laser desorption ionization-time of flight mass spectrometry to identify MLST clade 4 clostridium difficile isolates. Diagn Microbiol Infect Dis. (2018) 92:19–24. doi: 10.1016/j.diagmicrobio.2018.04.011
31. Cheng JW, Xiao M, Kudinha T, Xu ZP, Sun LY, Hou X, et al. The role of glutamate dehydrogenase (GDH) testing assay in the diagnosis of clostridium difficile infections: a high sensitive screening test and an essential step in the proposed laboratory diagnosis workflow for developing countries like China. PLoS ONE. (2015) 10:e144604. doi: 10.1371/journal.pone.0144604
32. Tang YJ, Gumerlock PH, Weiss JB, Silva JJ. Specific detection of Clostridium difficile toxin a gene sequences in clinical isolates. Mol Cell Probes. (1994) 8:463–7. doi: 10.1006/mcpr.1994.1066
33. Persson S, Torpdahl M, Olsen KE. New multiplex pcr method for the detection of Clostridium difficile toxin a (tcda) and toxin b (tcdb) and the binary toxin (cdta/cdtb) genes applied to a danish strain collection. Clin Microbiol Infect. (2008) 14:1057–64. doi: 10.1111/j.1469-0691.2008.02092.x
34. Curry SR, Marsh JW, Muto CA, O'Leary MM, Pasculle AW, Harrison LH. Tcdc genotypes associated with severe tcdc truncation in an epidemic clone and other strains of clostridium difficile. J Clin Microbiol. (2007) 45:215–21. doi: 10.1128/JCM.01599-06
35. Indra A, Schmid D, Huhulescu S, Hell M, Gattringer R, Hasenberger P, et al. Characterization of clinical clostridium difficile isolates by PCR ribotyping and detection of toxin genes in austria, 2006-2007. J Med Microbiol. (2008) 57:702–8. doi: 10.1099/jmm.0.47476-0
36. CLSI. Methods for antimicrobial susceptibility testing of anaerobic bacteria; approved standard eighth edition. DocumentM11-A8. Clinical and laboratory Standards Institute, Wayne, PA. (2012). Available online at: https://community.clsi.org/media/1468/m11a8
37. CLSI. Performance standards for antimicrobial susceptibility testing;27th ed. CLSI supplement M100. Wayne, PA (2017). Available online at: https://ansi.cachefly.net/preview-pages/CLSI/preview_CLSI+M100-S27
38. Jia H, Du P, Yang H, Zhang Y, Wang J, Zhang W, et al. Nosocomial transmission of clostridium difficile Ribotype 027 in a Chinese hospital, 2012-2014, traced by whole genome sequencing. BMC Genomics. (2016) 17:405. doi: 10.1186/s12864-016-2708-0
39. Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for clostridium difficile spores. J Bacteriol. (2008) 190:2505–12. doi: 10.1128/JB.01765-07
40. Jamal WY, Shahin M, Rotimi VO. Comparison of two matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry methods and api 20an for identification of clinically relevant anaerobic bacteria. J Med Microbiol. (2013) 62:540–4. doi: 10.1099/jmm.0.053256-0
41. Wei Y, Sun M, Zhang Y, Gao J, Kong F, Liu D, et al. Prevalence, genotype and antimicrobial resistance of clostridium difficile isolates from healthy pets in eastern China. BMC Infect Dis. (2019) 19:46. doi: 10.1186/s12879-019-3678-z
42. Jin D, Luo Y, Huang C, Cai J, Ye J, Zheng Y, et al. Molecular epidemiology of clostridium difficile infection in hospitalized patients in eastern China. J Clin Microbiol. (2017) 55:801–10. doi: 10.1128/JCM.01898-16
43. Bacci S, St-Martin G, Olesen B, Bruun B, Olsen KE, Nielsen EM, et al. Outbreak of clostridium difficile 027 in north Zealand, Denmark, 2008-2009. Euro Surveill. (2009) 14:19183. doi: 10.2807/ese.14.16.19183-en
44. McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, et al. Clinical practice guidelines for clostridium difficile infection in adults and children: 2017 update by the infectious diseases society of America (IDSA) and society for healthcare epidemiology of America (shea). Clin Infect Dis. (2018) 66:e1–48. doi: 10.1093/cid/ciy149
45. Cheng JW, Liu C, Kudinha T, Xiao M, Fan X, Yang CX, et al. The TcdA-negative and tcdB-positive clostridium difficile st81 clone exhibits a high level of resistance to fluoroquinolones: a multi-Centre study in Beijing, China. Int J Antimicrob Agents. (2020) 56:105981. doi: 10.1016/j.ijantimicag.2020.105981
46. Lanis JM, Barua S, Ballard JD. Variations in Tcdb activity and the hypervirulence of emerging strains of clostridium difficile. PLoS Pathog. (2010) 6:e1001061. doi: 10.1371/journal.ppat.1001061
47. Marks SL, Kather EJ, Kass PH, Melli AC. Genotypic and phenotypic characterization of clostridium perfringens and clostridium difficile in diarrheic and healthy dogs. J Vet Intern Med. (2002) 16:533–40. doi: 10.1111/j.1939-1676.2002.tb02383.x
48. Clooten J, Kruth S, Arroyo L, Weese JS. Prevalence and risk factors for clostridium difficile colonization in dogs and cats hospitalized in an intensive care unit. Vet Microbiol. (2008) 129:209–14. doi: 10.1016/j.vetmic.2007.11.013
49. Perrin J, Cosmetatos I, Gallusser A, Lobsiger L, Straub R, Nicolet J. Clostridium difficile associated with typhlocolitis in an adult horse. J Vet Diagn Invest. (1993) 5:99–101. doi: 10.1177/104063879300500122
50. Weese JS, Mshelbwala PP, Lohr F. Clostridium difficile shedding by healthy dogs in Nigeria and Malawi. Zoonoses Public Health. (2019) 66:618–21. doi: 10.1111/zph.12613
51. Schneeberg A, Rupnik M, Neubauer H, Seyboldt C. Prevalence and distribution of Clostridium difficile PCR ribotypes in cats and dogs from animal shelters in Thuringia, Germany. Anaerobe. (2012) 18:484–8. doi: 10.1016/j.anaerobe.2012.08.002
52. Hensgens MP, Keessen EC, Squire MM, Riley TV, Koene MG, de Boer E, et al. Clostridium difficile infection in the community: a zoonotic disease? Clin Microbiol Infect. (2012) 18:635–45. doi: 10.1111/j.1469-0691.2012.03853.x
53. Wetterwik KJ, Trowald-Wigh G, Fernstrom LL, Krovacek K. Clostridium difficile in Faeces from healthy dogs and dogs with diarrhea. Acta Vet Scand. (2013) 55:23. doi: 10.1186/1751-0147-55-23
54. Hussain I, Sharma RK, Borah P, Rajkhowa S, Hussain I, Barkalita LM, et al. Isolation and characterization of clostridium difficile from pet dogs in Assam, India. Anaerobe. (2015) 36:9–13. doi: 10.1016/j.anaerobe.2015.09.006
55. Huang J, Kelly CP, Bakirtzi K, Villafuerte GJ, Lyras D, Mileto SJ, et al. Clostridium difficile toxins induce vegf-a and vascular permeability to promote disease pathogenesis. Nat Microbiol. (2019) 4:269–79. doi: 10.1038/s41564-018-0300-x
56. Rabold D, Espelage W, Abu SM, Eckmanns T, Schneeberg A, Neubauer H, et al. The zoonotic potential of Clostridium difficile from small companion animals and their owners. PLoS ONE. (2018) 13:e193411. doi: 10.1371/journal.pone.0193411
57. Orden C, Neila C, Blanco JL, Alvarez-Perez S, Harmanus C, Kuijper EJ, et al. Recreational sandboxes for children and dogs can be a source of epidemic ribotypes of Clostridium difficile. Zoonoses Public Health. (2018) 65:88–95. doi: 10.1111/zph.12374
58. Andres-Lasheras S, Martin-Burriel I, Mainar-Jaime RC, Morales M, Kuijper E, Blanco JL, et al. Preliminary studies on isolates of Clostridium difficile from dogs and exotic pets. BMC Vet Res. (2018) 14:77. doi: 10.1186/s12917-018-1402-7
59. Janezic S, Ocepek M, Zidaric V, Rupnik M. Clostridium difficile genotypes other than ribotype 078 that are prevalent among human, animal and environmental isolates. BMC Microbiol. (2012) 12:48. doi: 10.1186/1471-2180-12-48
60. Janezic S, Zidaric V, Pardon B, Indra A, Kokotovic B, Blanco JL, et al. International Clostridium difficile animal strain collection and large diversity of animal associated strains. BMC Microbiol. (2014) 14:173. doi: 10.1186/1471-2180-14-173
61. Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, et al. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet. (2005) 366:1079–84. doi: 10.1016/S0140-6736(05)67420-X
62. Lefebvre SL, Reid-Smith RJ, Waltner-Toews D, Weese JS. Incidence of acquisition of methicillin-resistant staphylococcus aureus, Clostridium difficile, and other health-care-associated pathogens by dogs that participate in animal-assisted interventions. J Am Vet Med Assoc. (2009) 234:1404–17. doi: 10.2460/javma.234.11.1404
63. Surawicz CM, Brandt LJ, Binion DG, Ananthakrishnan AN, Curry SR, Gilligan PH, et al. Guidelines for diagnosis, treatment, and prevention of Clostridium difficile infections. Am J Gastroenterol. (2013) 108:478–98. doi: 10.1038/ajg.2013.4
64. Bauer MP, van Dissel JT, Kuijper EJ. Clostridium difficile: controversies and approaches to management. Curr Opin Infect Dis. (2009) 22:517–24. doi: 10.1097/QCO.0b013e32833229ce
65. Spigaglia P, Barbanti F, Morandi M, Moro ML, Mastrantonio P. Diagnostic testing for Clostridium difficile in Italian microbiological laboratories. Anaerobe. (2016) 37:29–33. doi: 10.1016/j.anaerobe.2015.11.002
66. Zhang LJ, Yang L, Gu XX, Chen PX, Fu JL, Jiang HX. The first isolation of Clostridium difficile rt078/st11 from pigs in China. PLoS ONE. (2019) 14:e212965. doi: 10.1371/journal.pone.0212965
Keywords: Clostridium difficile, dog, cat, molecular typing, antimicrobial resistance
Citation: Wen G-L, Li S-H, Qin Z, Yang Y-J, Bai L-X, Ge W-B, Liu X-W and Li J-Y (2022) Isolation, molecular typing and antimicrobial resistance of Clostridium difficile in dogs and cats in Lanzhou city of Northwest China. Front. Vet. Sci. 9:1032945. doi: 10.3389/fvets.2022.1032945
Received: 31 August 2022; Accepted: 31 October 2022;
Published: 18 November 2022.
Edited by:
Valentina Virginia Ebani, University of Pisa, ItalyReviewed by:
Hazim Khalifa, Kafrelsheikh University, EgyptTimothy Frana, Boehringer Ingelheim, United States
Copyright © 2022 Wen, Li, Qin, Yang, Bai, Ge, Liu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xi-Wang Liu, eGl3YW5nbGl1QDEyNi5jb20=; Jian-Yong Li, bGlqeTE5NzFAMTYzLmNvbQ==
 Gui-Lin Wen
Gui-Lin Wen Shi-Hong Li
Shi-Hong Li Zhe Qin
Zhe Qin Ya-Jun Yang
Ya-Jun Yang Li-Xia Bai
Li-Xia Bai Wen-Bo Ge
Wen-Bo Ge Xi-Wang Liu
Xi-Wang Liu