- 1Key Laboratory of Veterinary Etiological Biology, College of Veterinary Medicine, Lanzhou University, National Para-Reference Laboratory for Animal Echinococcosis, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2Department of Clinical Medicine and Surgery, University of Agriculture, Faisalabad, Pakistan
- 3Department of Biology, College of Science, University of Hafr Al Batin, Hafr Al Batin, Saudi Arabia
- 4Department of Parasitology, University of Agriculture, Faisalabad, Pakistan
- 5Faculty of Science, Al-Azhar University, Assuit, Egypt
- 6Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
Parasitic infestations are one of the major threats to the livestock industry in Pakistan. These have a negative impact on the production of domesticated livestock species. Paramphistomes belong to the superfamily Paramphistomoidea and are involved in infecting ruminants all over the world. To date, there was no information on mitochondrial DNA-based molecular characterization of Paramphistomum epiclitum from Pakistan. To close this research gap, this study was designed to provide insights into the epidemiology of Paramphistomum species. Paramphistomum epiclitum isolates were recovered from the rumen of small ruminants slaughtered at an abattoir located in Faisalabad city and animal demographics were recorded. DNA was extracted and mitochondrial cox1 was amplified and sequenced. Prevalence was calculated along with a 95% confidence interval in various groups. The chi-square test was applied to determine the association between different variables under investigation. A phylogenetic tree was constructed based on the Bayesian method. Population diversity indices were calculated using DnaSP 4.5 software. A total of 43 mutations were observed among 7 haplotypes. Negative values of Fu's Fs values, and Tajima's D indicated population expansion. Deworming, season, and grazing were the variables that significantly correlate (p < 0.05) with the prevalence of P. epiclitum. The high prevalence of P. epiclitum demonstrates that more studies are indeed needed to further understand the prevalence and distribution of P. epiclitum in definitive and all potential intermediate hosts in addition to intraspecies variation and relationship with populations from other locations.
Introduction
Paramphistomum infection is one of the neglected parasitic diseases affecting ruminants, which is widely distributed in tropical and subtropical areas of the world. Paramphistomum is derived from the Greek word amphistomes, which means paired mouth (1). The life cycle of Paramphistomum is completed between a snail (Planorbis planorbis, Bulinus spp., and Lymnaea bulimoides) as intermediate host and ruminants as definitive host (2). Miracdia are released from the eggs shed in the feces of infected animals. These miracidia invade snails. Parasitic larvae are developed in snails until cercaria is produced and these cercariae then encyst on hard surfaces, vegetation, or in water. Infective metacercariae are ingested by ruminants. Immature larvae present in the small intestine move to their predilection site such as fore stomach and liver where they are converted into adult flukes (3–5).
Parasites are capable of causing acute, chronic, and debilitating types of diseases leading to production losses in animals and researches have shown the positive role of alternative/complementary medicine to treat parasitic diseases (6–9). Generally, adult paramphistomes are considered non-pathogenic but anorexia, diarrhea, polydipsia, and mortality are caused by hemorrhagic enteritis due to migration of the larvae in duodenal mucosa. Paramphistomosis is caused by several species in different regions of the world but P. cervi and P. epiclitum are considered to be the most important species (10, 11). The prevalence of different Paramphistomum species has been reported in various species like sheep, goats, cattle, buffalo, and camels. The prevalence of Paramphistomum spp. in cattle was reported to be 36.9% in South-Eastern Iran (12). Raza et al. reported the prevalence of P. cervi to be 28.57, 23.80, 17.64, and 20% in sheep, goat, cattle, and buffalo, respectively (13). Prevalence of P. cervi was found to be 34% in camels reported from Maiduguri, Nigeria (14).
Paramphistomum epiclitum is the plug feeders (15) that burry themselves in the duodenal mucosa and feed on cells of Brunner's gland leading to diarrhea, anemia, hypoproteinemia, and weakness (16). The infection results in a poor feed conversion ratio, poor weight gain, and decreased milk production in affected animals (17). Recently the parasite has been found to cause significant losses in production (10, 18). Prevalence is high in tropical and subtropical areas, especially in Asia, Africa, and Eastern Europe (13, 19, 20). The prevalence in some areas of Asia such as Pakistan is recorded to be 30–60% (13). The prevalence of gastrointestinal helminths such as P. epiclitum in Pakistan has been previously recorded to be 25.1–92% in different regions at different times (13, 21–23).
Pakistan is endowed with a rich population of small ruminants that account for more than 100 million heads (24). One of the threatening issues faced by the farmers is a parasitic infection which results in decreased productivity of animals in terms of losses in weight gain, milk production, and reproduction of the infected animals. While addressing any problem one has to determine the magnitude of that problem to devise suitable and effective prevention and control strategies. Prevalence studies help to determine a true picture of diseases within the population of animals inhabiting that area. Currently, a few studies have been conducted on the prevalence of P. epiclitum in different areas of Pakistan but there is a lack of a comprehensive prevalence study on P. epiclitum in the Faisalabad region. Previously, numerous reports on molecular characterization based on the ITS gene are available from the country but the ITS gene is a hypervariable region and thus does not a candidate gene to molecularly characterize and identify the species. Similarly to date, no information on mitochondrial DNA-based molecular characterization is available from the region. To close these research gaps, the current study was designed.
Materials and methods
Study area
Pakistan is a South Asian country that shares its border with Iran in the southwest, China in the northeast, India in the east, and Afghanistan in the west. Faisalabad is the third most populous city in Pakistan and the second most populous city in Punjab Province. Geographically it is expanded from 730 to 740 in the east and 300 to 31.50 in the north over an area of 5,856 km2 and lies 186 meters above sea level. Annual rainfall in Faisalabad is about 350 millimeters. It is known for its well-developed canal system. The existence of a highly extensive irrigation system makes this land highly valuable for agriculture as well as livestock farming. The sampling of P. epiclitum was done from a central abattoir located in Faisalabad, Punjab Province. This slaughterhouse is a source of quality meat supply over the major area of the city which is why a huge number of small ruminants are pooled into this abattoir for slaughtering.
Parasite collection
From February 2021 till the end of January 2022, a total of 1,942 animals were examined at the abattoir post-slaughter. Sampling was done 5 days a week except for Tuesday and Wednesday due to weekly holidays in the slaughterhouse. One parasite per animal was collected from the animals found positive for P. epiclitum infection in Eppendorf tubes and data (species viz. sheep or goat, sex, age, breed, grazing status, flock size, and deworming status) of all the animals were recorded before slaughtering. Samples were labeled and transported to the Department of Clinical Medicine and Surgery, University of Agriculture, Faisalabad, Pakistan for further processing.
DNA extraction and PCR
DNA was extracted from a total of 40 P. epiclitum isolates using Qiagen® Blood and Tissue Kit strictly adhering to the manufacturer's guidelines. The final reaction mixture used for PCR contained a total volume of 50 μl comprising 10 pmol of each primer, 25 μl Premix Ex Taq™ version 2.0, 0.5 μl of sample DNA extract (≥20 ng), and volume made up to 50 μl by adding sufficient amount of DNase/RNase free distilled water (UltraPureTM, Introgen). PCR reaction mixture used as control negatives were added with nuclease-free water instead of DNA. Previously used primers JB4.5 (5′-TAAAGAAAGAACATAATGAAAATG-3′) and JB3 (5′-TTTTTTGGGCATCCTGAGGTTTAT-3′) were used to amplify the cox1 gene (25). PCR conditions used were as follows: initial denaturation at 94°C for 5 min followed by 35 cycles of 30 s at 94°C, 45 s at 50°C, and 35 s at 72°C, and a final extension at 72°C for 10 min (26). PCR products were run on 1.5% (w/v) agarose gel stained with GelRed™ and the products were viewed under the GelDoc system. A 500 bp ladder was used in each gel as a DNA marker for estimating the size of amplicons. Then PCR products were sent to Beijing Tsingke Biotechnology Co., Ltd., Beijing, China for sequencing.
Molecular analysis
DNA sequences were viewed and misread nucleotides were corrected as well as nucleotide sequences were aligned using Unipro UGENE v1.32.0 software while the identity and nucleotide sequence of each isolate was confirmed by using NCBI BLAST Program (27). By viewing the electropherogram using Chromas software, the low-quality parts at the beginning and end were trimmed. This was done to discard mismatching extremities. Population diversity indices that included haplotype diversity (Hd), no. of haplotypes (h), and nucleotide diversity (π) were calculated by using DnaSP 4.5 software (28). Neutrality indices of Tajima's D and Fu's Fs were determined using Arlequin 3.5.2.2 software (29–31). Nucleotide sequences of the cox1 gene of 7 haplotypes were used to construct a Bayesian phylogenetic tree using MrBayesv.3.1.1 software, taking Schistosoma japonicum as outgroup (32).
Statistical analysis
For analyzing data, chi-square values and Odds ratios were calculated using Statistix 10.0 and WinPepi software, respectively. The p < 0.05 was considered significant.
Results
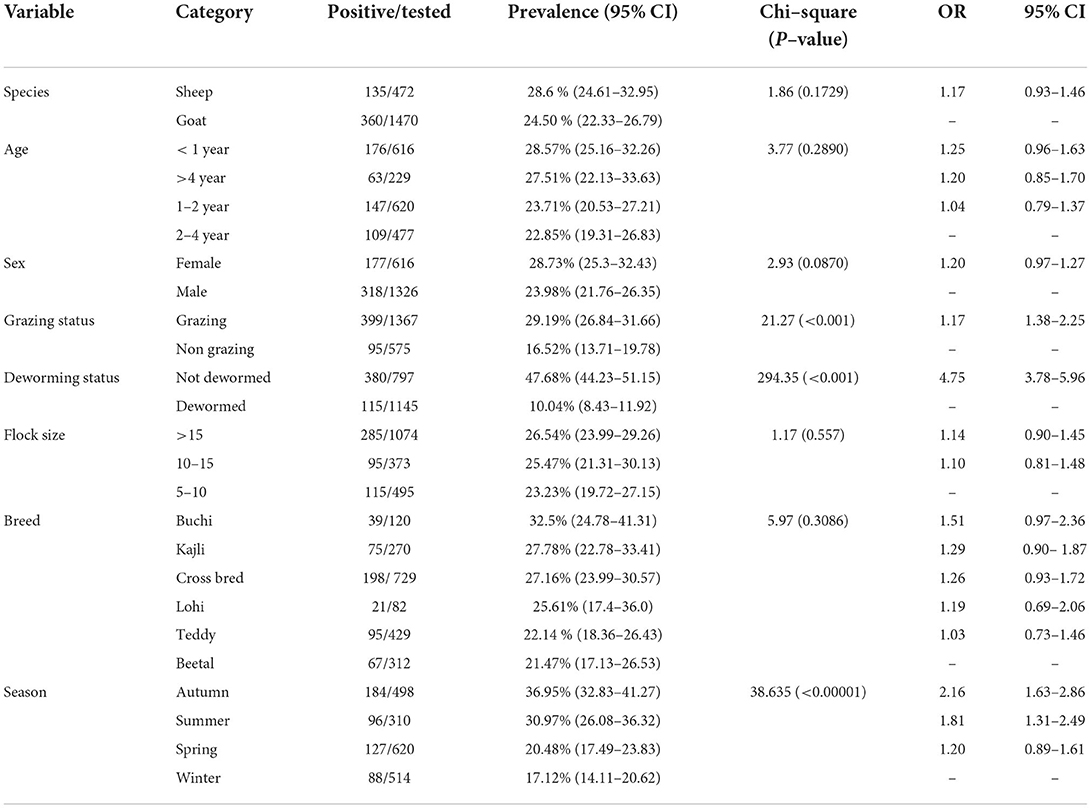
Higher prevalence was found in sheep (28.6%) as compared to goats (24.50%) but the difference in the prevalence between sheep and goats was statistically non-significant (p > 0.05) (Table 1). The highest prevalence was observed in animals with age < 1 year (28.57%) followed by age groups aging >4 years and 1–2 years having a prevalence of 27.51 and 23.71%, respectively. The lowest prevalence (22.85%) was found in animals aging between 2 and 4 years. There was no statistically significant difference (p > 0.05) in prevalence among various age groups. Concerning the sex of the animals, males showed a higher prevalence (23.98%) as compared to females (28.73%) and the difference was non-significant (p > 0.05).
The prevalence of P. epiclitum in grazing and stall-fed animals was 29.19 and 16.52%, respectively. Grazing animals had significantly (p < 0.05) higher rates of infection than stall-fed animals. Similarly, animals that were not treated with any anthelmintic showed a statistically significant difference (p < 0.05) from those which had a history of deworming medication. Seasonal calculation of prevalence showed a significant (p < 0.05) relationship between season and prevalence of P. epiclitum. Prevalence was found to be highest during the Autumn months (36.95%; September to November) followed by Summer (30.97%; June to August), and Winter (17.12%; December to February).
While comparing the prevalence of P. epiclitum among different breeds of sheep and goats, the prevalence was calculated to be highest in Buchi sheep and least in Beetal goats having a prevalence of 32.5 and 21.47%, respectively. No statistically significant difference in prevalence was found among various breeds of sheep and goat.
Amplification of the cox1 gene yielded PCR products of approximately 450 bp. Nucleotide sequences of all 40 isolates analyzed in this study were aligned with reference sequences of P. epiclitum, retrieved from GenBank. The newly generated sequences of P. epiclitum showed a total of 43 mutation sites. Among the 40 P. epiclitum isolates, 7 haplotypes were found for the cox1 gene. Table 2A shows the observed nucleotide polymorphism between haplotypes while the resulting amino acid changes are mentioned in Table 2B.
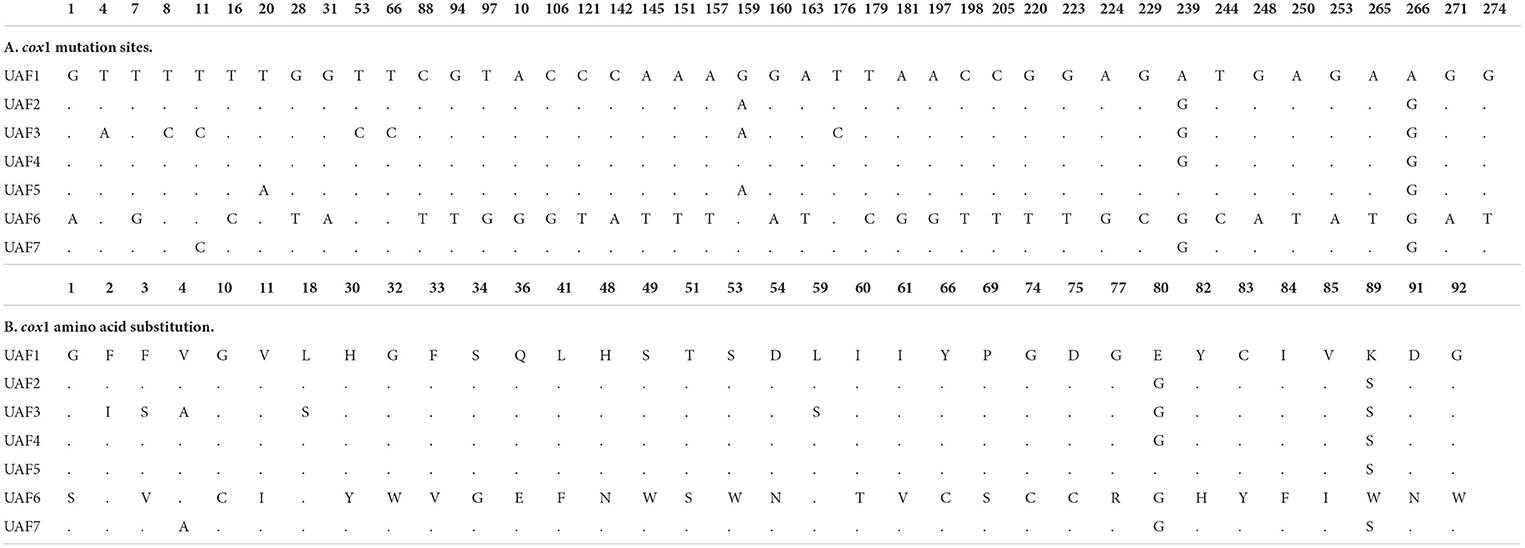
Table 2. Paramphistomum spp. partial cox1 gene nucleotide sequence polymorphism and corresponding amino acid changes.
Haplotype-1, -2, -3, -4, -5, -6, and -7 consisted of 10, 8, 7, 6, 4, 3, and 2 sequences, respectively.
The nucleotide diversity and neutrality indices for the entire P. epiclitum populations were calculated based on the sequences of the cox1 gene (Table 3). A low nucleotide (π) and high haplotype diversity (Hd) were observed for the cox1 gene. Tajima's D and Fu's Fs were negative and insignificant (p > 0.05).
The resulting sequences and those retrieved from GenBank were used to construct a phylogenic tree. The Bayesian phylogeny based on a dataset of the cox1 sequences placed most of the Pakistani P. epiclitum isolates in a distinct cluster (Figure 1). Only one sequence was found to be present within the cluster consisting of isolates from other countries [China (Accession No. KT198987), India (Accession No. JX678244), Laos (Accession No. LC113924), Saudi Arabia (Accession No. AB688990), and Thailand (Accession No. MZ602108)] of the globe.
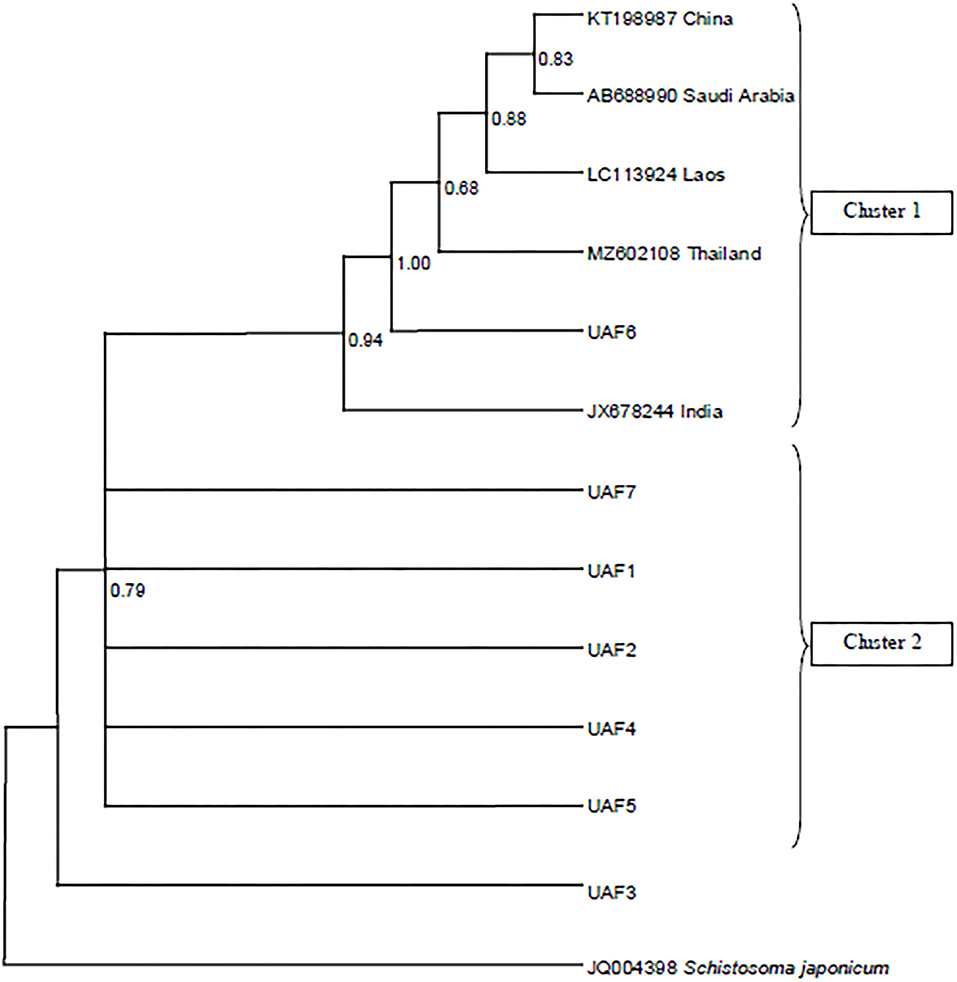
Figure 1. Bayesian phylogeny of Pakistani Paramphistomum epiclitum isolates inferred from the cox1 gene. Schistosoma japonicum is used as outgroup. Posterior probability values are depicted at the nodes.
Representative haplotype sequences from this study have been deposited in the GenBank database under the accession numbers ON899841–ON899847.
Discussion
Paramphistomum epiclitum is one of the most important parasites affecting almost every livestock species. This parasite poses a huge economic loss to the livestock industry in terms of mortality and morbidity along with the losses in the production of wool, meat, and milk in infected animals which leads to decreased profitability for livestock holders (17).
Keeping in view the economic importance of P. epiclitum scattered efforts have been made to estimate the burden of P. epiclitum infection among different livestock species in different areas of the country. This study is also aimed at estimating the burden of P. epiclitum infection in small ruminants in District Faisalabad and adjoining localities along with the analysis of various parameters that might positively or negatively correlate or affect the presence of Paramphistomum infection in small ruminants.
The prevalence of P. epiclitum in sheep was calculated to be 28.6% while that in goats was found to be 24.50% and the rate of infection did not differ significantly among the species (p > 0.05). Similar results were also reported in a study in which reported a prevalence of 28.57% in sheep and 23.80% in goats (13). These findings were close to our study. A higher prevalence of 32.51 and 42% was reported in sheep and goats, respectively, from Andhra Pradesh, India (33). While (34) published a study that showed a lower prevalence of P. epiclitum in sheep and goats 7.4 and 4.95%, respectively, in Uttarakhand, India. Another study from Jammu showed that the prevalence of P. epiclitum is 30.9% in goats and 36.2% in sheep (35). Like all the previous studies, the prevalence of P. epiclitum calculated in our study was also slightly higher in sheep than that in goats but the difference was statistically insignificant. This difference in prevalence can be attributed to the difference in the grazing habits of both species as well as other factors such as management, nutrition, and deworming status of the herd which was not observed in this study.
Age group-wise prevalence in sheep showed the prevalence of 32.97, 30.51, 28.97, and 22.03% among different age groups of animals (<1 year, >4 years, 1–2 year, and 2–4 years). Calculation of age-wise prevalence in goats showed the prevalence of 27.13, 26.95, 23.05, and 22.10% among different ages groups of animals (<1 year, >4 years, 2–4 year, and 1–2 years) showing a non-significant (p > 0.05) difference in prevalence. A study conducted by Patel et al. (36) also found that animals aged <1 year were highly susceptible to P. epiclitum infection than the aged animals and reported a prevalence of 61.9% in animals of < 1 year age and 49.36% in aged animals. Similar findings of higher infection rates in young animals were also reported previously (37).
The study also calculated breed-wise prevalence among three breeds of sheep and prevalence was calculated to be 32.5, 27.78, and 25.61% among Buchi, Kajli, and Lohi breeds, respectively, and no significant (p > 0.05) difference was found in the rate of infection among different breeds of sheep. Similar results have also been reported in three other breeds of sheep named Balochi, Harnai, and Babrik. Prevalence was reported to be 21.5% in both Balochi and Harnai sheep while it was 17.75% in Babrik breed of sheep (37). Breed-wise prevalence in cross-bred, Teddy, and Beetal goats also differed non-significantly (p > 0.05) being 27.16, 22.14, and 21.47%, respectively.
Prevalence among female and male sheep was found to be 29.55 and 28.24%, respectively. While prevalence recorded in male and female goats were 25 and 23.77%, respectively. Results showed that the rate of infection was higher among female animals than that in male animals in sheep while it was higher in male animals in goats but no statistically significant (p > 0.05) difference in rates of infection was found among both sexes in sheep as well as goats. The study conducted by Iqbal et al. (38) on the prevalence of Paramphistomum in cattle represented a higher prevalence among male cattle than that in females being 9.67 and 5.79%, respectively. Prevalence of Paramphistomum was reported to be slightly higher in female sheep than that of male sheep and reported prevalence of 22.33 and 17.83%, respectively, in female and male sheep (39). Similar results were also reported by Kifleyohannes et al. (40), and reported the prevalence of 29.49% in females while 15.79% in male sheep. Contrasting results were reported by Tariq et al. (37) who reported a higher prevalence in male sheep than that in female sheep.
The prevalence of P. epiclitum was calculated to be significantly (p < 0.05) higher in animals that were dewormed than in those which did not receive any anthelmintic treatment. Prevalence in dewormed animals was 10.04% as compared to 47.68% in animals that were not dewormed. A higher gastrointestinal parasitic prevalence of 63% in animals that were not dewormed was reported as compared to 57% in those animals that were dewormed (41). Prevalence in grazing and stall-fed animals was 29.19 and 16.52%, respectively. Grazing animals had significantly (p < 0.05) higher rates of infection than stall-fed animals. While conducting a cross-sectional study to investigate fasciolosis also the study reported a higher prevalence in grazing animals than in animals that were not grazed (42).
Seasonal prevalence showed a significant (p < 0.05) relationship between season and prevalence of P. epiclitum. Prevalence was found to be highest during the Autumn months (36.95%; September to November) followed by Summer (30.97%; June to August), and Winter (17.12%; December to February). A similar trend has also been reported by Tariq et al. (43) but the prevalence was lower than that of our study. The study reported a prevalence of 8.33, 5.18, 2.98, and 1.17% during Autumn, Summer, Spring, and Winter respectively, in sheep while reported prevalence of 14.10, 9.02, 7.83, and 6.61% during Autumn, Summer, Winter, and Spring respectively, in cattle. The highest prevalence of 72.44% in the rainy season followed by 61.82% in Summer and 56.72% in Winter (44). While (45) reported the highest prevalence of 3.3% in Summer followed by 2.7% in Autumn, 2.3% in Winter, and 1.4% in Spring.
Infectious diseases such as parasitic infestations are important health problems in both animals and humans (46–50), which cause economic losses and severe illness (51–56). In this study, a molecular description of P. epiclitum isolates from sheep and goats was reported for the first time in Pakistan based on the mt cox1 gene as mtDNA remains an important marker in exploring intraspecific variation due to maternal inheritance, conserved structure, higher evolution rate, high genetic divergence, and absence of recombination (57–59). Previously, numerous reports on molecular characterization based on the ITS gene are available from the country but the ITS gene is a hypervariable region and thus does not a candidate gene to molecularly characterize and identify the species.
The phylogenetic analysis of the cox1 sequences from these isolates revealed a separate cluster suggesting the existence of unique P. epiclitum haplotypes in Pakistan. All the sequences obtained from GenBank were found to be in one cluster irrespective of the country of origin. To the best of our knowledge, this is the first report about the cox1 gene-based phylogeny of P. epiclitum. The scarcity of mtcox1 sequences from different geographical regions in the GenBank repository limited a wide-range comparison and interpretation.
Keeping in view the preliminary nature of this study and the logistic constraints encountered, only a segment of the cox1 gene was amplified. However, the previous reports suggest that to have a better insight into the population genetics structure of parasites, full-length gene amplification is preferred (60). Thus, investigations employing full-length gene amplification are highly warranted in the future.
Conclusion
In this study, P. epiclitum was recovered from slaughtered sheep and goats in Faisalabad, Pakistan, and was characterized in what we believe is the first attempt to identify P. epiclitum in Pakistan based on mtDNA. The molecular analysis of the partial cox1 gene demonstrates a high degree of genetic variation with rare haplotypes among the Pakistani P. epiclitum population.
This study constitutes significant preliminary data for Pakistan as well as useful baseline information for future studies on the prevalence and population structure of P. epiclitum globally. The high prevalence of P. epiclitum demonstrates that more studies are indeed needed to further understand the prevalence and distribution of P. epiclitum in definitive and all potential intermediate hosts in addition to intraspecies variation and relationship with populations from other locations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical review and approval was not required for the study of animal participants in accordance with the local legislation and institutional requirements.
Author contributions
MA, LL, H-BY, and W-ZJ conceptualized the study while methodology was designed by MA, W-ZJ, MS, and RA. Validation and formal analysis was carried out by MA, W-ZJ, LL, H-BY, MK, MI, and MS. Investigations were made by MA, FA, RA, AA, and WQ. The original draft was prepared by MA and FA. AA, HA, and NA revised the manuscript. Writing—review and editing of the manuscript was performed by W-ZJ, H-BY, and B-QF. Supervision, project administration, and funding acquisition were obtained by W-ZJ. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge funding received from the National Key Research and Development Program (2021YFE0191600), Cultivation of Achievements of State Key Laboratory of Veterinary Etiological Biology (SKLVEB2020CGPY01), and Central Public-Interest Scientific Institution Basal Research Fund (1610312020016).
Acknowledgments
We are thankful to Dr. Khurram Ashfaq, Dr. Imaad Rasheed, and other supporting staff of Department of Clinical Medicine and Surgery, University of Agriculture, Faisalabad for logistic support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1018854/full#supplementary-material
References
1. Murphy TM, Power EP, Sanchez-Miguel C, Casey MJ, Toolan DP, Fagan JG. Paramphistomosis in Irish cattle. Vet Rec. (2008) 162:831. doi: 10.1136/vr.162.25.831-a
2. González-Warleta M, Lladosa S, Castro-Hermida JA, Martínez-Ibeas AM, Conesa D, Munoz F, et al. Bovine paramphistomosis in Galicia (Spain): prevalence, intensity, aetiology and geospatial distribution of the infection. Vet Parasitol. (2013) 191:252–63. doi: 10.1016/j.vetpar.2012.09.006
3. Prasad A, Singh NK. Evaluation of antibody response to various developmental stage specific somatic antigens of Paramphistomum epiclitum in goats. BioMed Res Int. (2014) 2014:505484. doi: 10.1155/2014/505484
4. Ali Q, Rashid I, Shabbir MZ, Akbar H, Shahzad K, Ashraf K, et al. First genetic evidence for the presence of the rumen fluke Paramphistomum epiclitum in Pakistan. Parasitol Int. (2018) 67:533–7. doi: 10.1016/j.parint.2018.05.005
5. Khan I, Afshan K, Ullah R, Komal M, Khan MA, Firasat S. Serological and immuno-histopathological detection of Paramphistomum epiclitum infection in large ruminant population in Punjab, Pakistan. J Hell Vet Medical Soc. (2021) 72:3203–12. doi: 10.12681/jhvms.28515
6. Strbac F, Bosco A, Amadesi A, Rinaldi L, Stojanović D, Simin N, et al. Ovicidal potential of five different essential oils to control gastrointestinal nematodes of sheep. Pak Vet J. (2021) 41:353–8. doi: 10.29261/pakvetj/2021.026
7. Zaman MA, Qamar W, Yousaf S, Mehreen U, Shahid Z, Khan MK, et al. In vitro experiments revealed the anthelmintic potential of herbal complex against Haemonchus contortus. Pak Vet J. (2020) 40:271–3. doi: 10.29261/pakvetj/2019.128
8. Jalil PJ, Shnawa BH, Hammad SM. Silver nanoparticles: Green synthesis, characterization, blood compatibility and protoscolicidal efficacy against Echinococcus granulosus. Pak Vet J. (2021) 41:393–99. doi: 10.29261/pakvetj/2021.039
9. Wajiha, Qureshi NA. I vitro anticoccidial, antioxidant activities and biochemical screening of methanolic and aqueous leaves extracts of selected plants. Pak Vet J. (2021) 41:57–63. doi: 10.29261/pakvetj/2020.071
10. Anuracpreeda P, Wanichanon C, Sobhon P. Paramphistomum cervi: antigenic profile of adults as recognized by infected cattle sera. Exp Parasitol. (2008) 118:203–7. doi: 10.1016/j.exppara.2007.08.005
11. Javed Khan U, Tanveer A, Maqbool A, Masood S. Epidemiological studies of paramphistomosis in cattle. Vet Arh. (2008) 78:243–51.
12. Khedri J, Radfar MH, Borji H, Mirzaei M. Prevalence and intensity of Paramphistomum spp. in cattle from South-Eastern. Iran Iran J Parasitol. (2015) 10:268.
13. Raza MA, Murtaza S, Bachaya HA, Hussain A. Prevalence of Paramphistomum cervi in ruminants slaughtered in district Muzaffar Garh. Pak Vet J. (2009) 28:34–6.
14. Biu AA, Abbagana A. Prevalence of paramphistomes in camels slaughtered at Maiduguri, Nigeria. Niger J Parasitol. (2007) 28:44–6. doi: 10.4314/njpar.v28i1.37858
15. Sintayehu M, Mekonnen A. Prevalence and intensity of Paramphistomum in ruminants slaughtered at Debre Zeit industrial abattoir, Ethiopia. Glob Vet. (2012) 8:315–9.
16. Dube S, Aisien MS. Descriptive studies on Paramphistomes of small domestic ruminants in Southern Nigeria. (2010). Available online at: https://www.semanticscholar.org/paper/Descriptive-studies-on-Paramphistomes-of-small-in-Dube/bc830d8351a8ae3504fbdbebf4db2353cf96fccc
17. Rafiq N, Niaz S, Zeb I, Ayaz S, da Silva Vaz I Jr, Ali A. Molecular characterization of Paramphistomum cervi in buffaloes. Acta Sci Vet. (2020) 48:107107. doi: 10.22456/1679-9216.107107
18. Gordon DK, Roberts LC, Lean N, Zadoks RN, Sargison ND, Skuce PJ. Identification of the rumen fluke, Calicophoron daubneyi, in GB livestock: possible implications for liver fluke diagnosis. Vet Parasitol. (2013) 195:65–71. doi: 10.1016/j.vetpar.2013.01.014
19. Pfukenyi DM, Mukaratirwa S, Willingham AL, Monrad J. Epidemiological studies of amphistome infections in cattle in the highveld and lowveld communal grazing areas of Zimbabwe. Onderstepoort J Vet Res. (2005) 72:67–86. doi: 10.4102/ojvr.v72i1.224
20. Arias M, Lomba C, Dacal V, Vázquez L, Pedreira J, Francisco I, et al. Prevalence of mixed trematode infections in an abattoir receiving cattle from northern Portugal and north-west Spain. Vet Rec. (2011) 168:408. doi: 10.1136/vr.d85
21. Sajid A, Khan MQ, Qayyum M, Khan MF. Prevalence of gastrointestinal parasites in sheep and goats maintained at NARC, Islamabad. Pak Vet J. (2000) 20:157–8.
22. Al-Shaibani IR, Phulan MS, Arijo A, Qureshi TA. Epidemiology of ovine gastrointestinal nematodes in Hyderabad district, Pakistan. Pak Vet J. (2008) 1:28.
23. Ijaz M, Khan MS, Avais M, Ashraf K, Ali MM. Infection rate and chemotherapy of various helminths in goats in and around Lahore. Pak Vet J. (2008) 28:167–70.
24. Anonymous. Pakistan Economic Survey, Finance Division, Government of Pakistan (2021). Available online at: https://www.finance.gov.pk/survey/chapter_22/PES02-AGRICULTURE.pdf
25. Bowles J, Blair D, McManus DP. Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Biochem Parasitol. (1992) 54:165–73. doi: 10.1016/0166-6851(92)90109-W
26. Sharbatkhori M, Fasihi Harandi M, Mirhendi H, Hajialilo E, Kia EB. Sequence analysis of cox1 and nad1 genes in Echinococcus granulosus G3 genotype in camels (Camelus dromedarius) from central Iran. Parasitol Res. (2011) 108:521–7. doi: 10.1007/s00436-010-2092-7
27. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. (1990) 215:403–10. doi: 10.1016/S0022-2836(05)80360-2
28. Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol Biol Evol. (2017) 34:3299–302. doi: 10.1093/molbev/msx248
29. Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. (1989) 123:585–95. doi: 10.1093/genetics/123.3.585
30. Fu YX. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. (1997) 147:915–25. doi: 10.1093/genetics/147.2.915
31. Excoffier L, Laval G. Schneider S. Arlequin (version 30): an integrated software package for population genetics data analysis. Evol Bioinform. (2005) 1:117693430500100003. doi: 10.1177/117693430500100003
32. Huelsenbeck JP, Ronquist F, MRBAYES. Bayesian inference of phylogenetic trees. Bioinformatics. (2001) 17:754–5. doi: 10.1093/bioinformatics/17.8.754
33. Preethi M, Venu R, Srilatha C, Rao KS, Rao PV. Prevalence of paramphistomosis in domestic ruminants in Chittoor District of Andhra Pradesh, India. Agric Sci Digest. (2020) 40:61–8. doi: 10.18805/ag.D-5014
34. Maitra A, Yadav CL, Sanjukta RK. Seasonal prevalence of paramphistomosis in domestic ruminants in different agro-climatic zones of Uttarakhand, India. Asian Pac j trop med. (2014) 4:S748–53. doi: 10.1016/S2222-1808(14)60720-9
35. Godara R, Katoch R, Yadav A, Rastogi A. Epidemiology of paramphistomosis in sheep and goats in Jammu, India. J Parasit Dis. (2014) 38:423–8. doi: 10.1007/s12639-013-0264-y
36. Patel MD, Nauriyal DS, Hasnani JJ, Gupta RS. Prevalence of gastrointestinal parasitism in goats maintained under semi-intensive and field management systems. Indian Vet J. (2001) 21:99–101.
37. Tariq KA, Chishti MZ, Ahmad F, Shawl AS. The epidemiology of paramphistomosis of sheep (Ovis aries L.) in the north west temperate Himalayan region of India. Vet Res Commun. (2008) 32:383–91. doi: 10.1007/s11259-008-9046-x
38. Iqbal MN, Shahzad KA, Muhammad A. Identification and prevalence of Paraphistomum cervi in naturally infected water buffaloes of central Punjab, Pakistan. Veterinaria. (2013) 1:9–12.
39. Tehmina S, Shahina R, Razzaq A, Marghazani IB, Khosa AN. Prevalence of Paramphistomum cervi in different sheep breeds of Balochistan (Pakistan). Rev Vet. (2014) 25:12–5. doi: 10.30972/vet.251542
40. Kifleyohannes T, Kebede E, Hagos Y, Weldu K, Michael MG. Prevalence of paramphistomosis in ruminants in Ashenge, Tigray Ethiopia. Acta Parasitol Glob. (2015) 6:83–6.
41. Amran MA, Yadav SK, Akter F, Sarkar S, Hossain MA, Joy SM, et al. Prevalence of gastrointestinal parasitic infections in different existing goat breeds in different districts of Bangladesh. J. Adv. Parasitol. (2018) 5:11–21.
42. Tesfay MM, Tadele BA, Kebede AT, Asfaw YT, Woldie BM, Tsegay AK, et al. Investigation of an Acute Fasciolosis Complicated by Clostridia Infection Outbreak in Korem Town, Southern Tigray of Ethiopia. (2021). doi: 10.21203/rs.3.rs-557724/v1
43. Ozdal N, Gul A, Ilhan FA, Deger S. Prevalence of Paramphistomum infection in cattle and sheep in Van Province, Turkey. Helminthologia. (2010) 47:20–4. doi: 10.2478/s11687-010-0003-1
44. Rahman MA, Labony SS, Dey AR, Alam MZ. An epidemiological investigation of gastrointestinal parasites of small ruminants in Tangail, Bangladesh. J Bangladesh Agric Univ. (2017) 15:255–9. doi: 10.3329/jbau.v15i2.35071
45. Tehrani A, Javanbakht J, Khani F, Hassan MA, Khadivar F, Dadashi F, et al. Prevalence and pathological study of Paramphistomum infection in the small intestine of slaughtered ovine. J Parasit Dis. (2015) 39:100–6. doi: 10.1007/s12639-013-0287-4
46. Ismael SMM, Salem SA, Elshahidy MS. Isolation and molecular characterization of circulating foot and mouth disease virus in Egypt during 2018-2020. Int J Vet Sci. (2021) 10:162–71. doi: 10.47278/journal.ijvs/2021.046
47. Nasr EA, Fawzy RE, Marian GS, Abbas AM, Khalifa E. Using of gamma interferon γIFN and multiplex PCR (m-PCR) for detection of bovine tuberculosis in dairy herds in Egypt. Int J Vet Sci. (2021) 10:229–33. doi: 10.47278/journal.ijvs/2021.035
48. Osman SA, Tharwat M, Saeed EMA. An outbreak of ovine listeriosis in Qassim region, Saudi Arabia: Epidemiological, clinical and treatment outcomes. Int J Vet Sci. 2021 10:312–16. doi: 10.47278/journal.ijvs/2021.060
49. Ali S, Ijaz M, Ahmed A, Aziz MU, Naveed M, Javed MU, et al. Prevalence and associated risk factors of bovine babesiosis in Lahore, Pakistan. Agrobiological Rec. (2020) 2:17–23. doi: 10.47278/journal.abr/2020.007
50. Zaman MA, Mehreen U, Qamar W, Qamar MF, Kashif M, Shahid Z, Abbas RZ. Brief account of bovine theileriosis prevalence in some South Asian countries. Agrobiological Rec. (2020) 2:38–48. doi: 10.47278/journal.abr/2020.010
51. Du XX, Sherein SA, Liu P, Haque MA, Khan A. Bovine mastitis: behavioral changes, treatment and control. Continental Vet J. (2022) 2:15–23.
52. Sharif M, Tunio SA, Bano S. Synergistic effects of Zinc oxide nanoparticles and conventional antibiotics against methicillin resistant Staphylococcus aureus. Adv Life Sci. (2021) 8:167–71.
53. Reshetnikova TI, Zenkin AS, Krylova TG. Experimental use of the triazavirin antiviral medication in conditions of group administration at the pig-breeding unit. Adv Life Sci. (2021) 8:381–86.
54. Özcan U, Sezener MG, Sayilkan BU, Ergüden VE, Küllük E, Yaman S, et al. New Aspect in Neonatal Calf Diarrhea: Presence of Escherichia coli CS31A at Unexpected Ratio. Kafkas Univ Vet Fak Derg. (2021) 27:133–4. doi: 10.9775/kvfd.2020.25280
55. Özdemir O, Ortatatli M, Terzi F, Hatipoglu FH, Çiftçi MK, Ateş MB. The Usability of Cytological and Immunocytological Methods for Rapid Diagnosis of Encephalitic Listeriosis in Ruminants. Kafkas Univ Vet Fak Derg. (2021) 27:225–33.
56. Aksel EG, Akçay A, Arslan K, Sohel MH, Güngör G, Akyüz B. The Effects of MBL1 Gene Polymorphism on Subclinical Mastitis in Holstein Cows. Kafkas Univ Vet Fak Derg. (2021) 27:389–95.
57. Mueller RL, Macey JR, Jaekel M, Wake DB, Boore JL. Morphological homoplasy, life history evolution, and historical biogeography of plethodontid salamanders inferred from complete mitochondrial genomes. Proc Natl Acad Sci. (2004) 101:13820–5. doi: 10.1073/pnas.0405785101
58. Shen X, Wang H, Ren J, Tian M, Wang M. The mitochondrial genome of Euphausia superba (Prydz Bay) (Crustacea: Malacostraca: Euphausiacea) reveals a novel gene arrangement and potential molecular markers. Mol Biol Rep. (2010) 37:771–84. doi: 10.1007/s11033-009-9602-7
59. Wei SJ, Tang P, Zheng LH, Shi M, Chen XX. The complete mitochondrial genome of Evania appendigaster (Hymenoptera: Evaniidae) has low A+ T content and a long intergenic spacer between atp8 and atp6. Mol Biol Rep. (2010) 37:1931–42. doi: 10.1007/s11033-009-9640-1
Keywords: prevalence, risk factors, genetic diversity, cox1, sheep and goat, Pakistan
Citation: Alvi MA, Alshammari A, Asghar F, Ali RMA, Li L, Saqib M, Khan MK, Imran M, Qamar W, Askar H, Abdelsater N, Fu B-Q, Yan H-B and Jia W-Z (2022) Prevalence, risk factors and first record of mitochondrial cox1 gene-based molecular characterization of Paramphistomum epiclitum from Pakistan. Front. Vet. Sci. 9:1018854. doi: 10.3389/fvets.2022.1018854
Received: 14 August 2022; Accepted: 29 August 2022;
Published: 21 November 2022.
Edited by:
Khalid Mehmood, Islamia University of Bahawalpur, PakistanReviewed by:
Eric Tzyy Jiann Chong, Universiti Malaysia Sabah, MalaysiaHafiz Ishfaq Ahmad, University of Veterinary and Animal Sciences, Pakistan
Copyright © 2022 Alvi, Alshammari, Asghar, Ali, Li, Saqib, Khan, Imran, Qamar, Askar, Abdelsater, Fu, Yan and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-Bin Yan, eWFuaG9uZ2JpbkBjYWFzLmNu; Wan-Zhong Jia, amlhd2FuemhvbmdAY2Fhcy5jbg==
†These authors have contributed equally to this work
 Mughees Aizaz Alvi
Mughees Aizaz Alvi Ayed Alshammari3†
Ayed Alshammari3† Rana Muhammad Athar Ali
Rana Muhammad Athar Ali Muhammad Saqib
Muhammad Saqib Bao-Quan Fu
Bao-Quan Fu Hong-Bin Yan
Hong-Bin Yan Wan-Zhong Jia
Wan-Zhong Jia