- 1Bovine Tuberculosis and Brucellosis Programme, Department of Veterinary Tropical Diseases, Faculty of Veterinary Science, University of Pretoria, Pretoria, South Africa
- 2Department of Veterinary Medicine, School of Veterinary Medicine, University of Rwanda, Kigali, Rwanda
- 3Department of Animal Resources and Veterinary Services, Rwanda Agriculture and Animal Resources Board, Kigali, Rwanda
- 4National Reference Laboratory Division, Department of Biomedical Services, Rwanda Biomedical Centre, Kigali, Rwanda
Seroprevalence studies showed that brucellosis is prevalent in cattle in Rwanda with no recent study on the characterization of Brucella spp. Therefore, this study aimed to characterize Brucella spp. in seropositive herds of cattle farmed at the wildlife–livestock–human interface. Whole blood samples (n = 118), milk (n = 41), and vaginal swabs (n = 51) were collected from 64 seropositive herds. All samples (n = 210) were inoculated onto modified Centro de Investigacion y Tecnologia Agroalimentaria (CITA) selective medium. Cultures were analyzed to detect Brucella spp. using 16S−23S ribosomal DNA interspacer region (ITS) PCR, the Brucella cultures were speciated using AMOS and Bruce-ladder PCR assays. Brucella spp. were detected in 16.7% (35/210) of the samples established from the samples using ITS-PCR. The AMOS PCR assay identified mixed Brucella abortus and B. melitensis (n = 6), B. abortus (n = 7), and B. melitensis (n = 1) from cultures from blood samples; mixed B. abortus and B. melitensis (n = 1) and B. abortus (n = 4) from cultures from milk samples; mixed B. abortus and B. melitensis (n = 6), B. abortus (n = 8), and B. melitensis (n = 1) from cultures from vaginal swabs. Bruce-ladder PCR assay confirmed B. abortus and B. melitensis cultures. The isolation of Brucella spp. was significantly associated with districts, with the Nyagatare district having more isolates than other districts (p = 0.01). This study identified single or mixed B. abortus and B. melitensis infections in cattle samples in Rwanda, which emphasizes the need to improve brucellosis control at the wildlife–livestock–human interface and raise the awareness of cattle keepers, abattoir workers, laboratory personnel, and consumers of cattle products.
Introduction
Brucellosis is a widespread contagious bacterial disease in livestock, wildlife, marine animals, and humans (1). Brucellosis is caused by bacteria of the genus Brucella belonging to the family of alpha-2-Proteobacteriaceae (2, 3). The genus was initially subdivided into six classical species based on their intracellular colonization and host species preference (2). The six classical species include B. melitensis which affects primarily goats, Brucella abortus affecting cattle, Brucella ovis affecting sheep, B. suis affecting swine and rats, B. canis affecting dogs, and B. neotomae affecting wood rats (2, 4). Three classical Brucella species which are subdivided into biovars (bv.) include B. abortus with bv. 1, 2, 3, 4, 5, 6, and 9, B. melitensis with bv. 1, 2, and 3, and B. suis with bv. 1, 2, 3, 4, and 5 (1).
Brucella spp. are 96% genetically homologous (5) but can be distinguished based on their genetic polymorphisms (6–10). Two molecular markers (omp2a and omp2b) discovered within the outer membrane protein (omp25) were used in combination with restriction enzymes to differentiate Brucella spp. and some of their biovars (7, 11). Other Brucella spp. specific DNA sequences include repetitive extragenic palindromic (REP) (10), two repeated palindromic DNA sequences (BRU: RS1, Bru: RS2) (12), and the insertion sequence (IS) 711 (8). Insertion sequences are mobile genetic elements that code for proteins responsible for their transposition (13). The IS711 that was first discovered in 1993 from B. ovis (14) has 35 copies of the element and is different from that of B. abortus which has at least 6 copies (9). The IS711 is a unique sequence of Brucella spp. with multiple copies of which some occur at species and biovars-specific sites within the chromosomal locus, and this element is the basis of differentiation between B. abortus (bv. 1, 2, and 4), B. melitensis (bv. 1, 2, and 3), B. ovis, and B. suis bv.1 (AMOS PCR) (15). Furthermore, IS711 is the basis of discrimination between terrestrial Brucella spp. and biovars and vaccine strains using Bruce-ladder PCR assay (16, 17). These molecular PCR assays have reduced the long procedure of conventional phenotypic characterization of Brucella spp. in developed countries. However, serological methods are still prevailing in most developing countries with a lack of appropriate knowledge, and biosafety facilities (18, 19).
In Rwanda, the control of brucellosis falls under the animal health law which consists of regulations and procedures for reporting infectious diseases, guidelines for animal movement, and the prohibition of illegal slaughtering (20). Apart from this animal health law, there is no other published documentation about the brucellosis control scheme. However, routine serological testing of cattle and small ruminants is performed before important national ruminants trade for distribution to poor families by the government and other non-government organizations (NGOs), and during annual surveillance (once per year) in areas with high dairy production. Vaccination against brucellosis consists of administering RB51 to calves on demand by herders upon payment of US$ 0.5 per dose. Thus, vaccination is not systematic and coordinated at the national level. In Rwanda, the individual seroprevalence of brucellosis in cattle varies from 2.3 to 18.4% (21, 22), and ranged from 6.1 to 25.0% in women with a history of abortions (23, 24). However, apart from the study that characterized B. abortus bv. 3 from cattle in 1984 (25), there are no studies on Brucella spp. circulating in Rwandan cattle at the wildlife–livestock–human interface. The objective of this study was therefore to characterize the Brucella spp. that are circulating in Rwandan cattle farmed at the wildlife–livestock–human interface to document the updated control scheme for brucellosis in Rwanda.
Materials and methods
Description of the study area
The study area was selected at the wildlife–livestock–human interface in Rwanda. The interface comprised six districts including Gatsibo, Kayonza, and Nyagatare in the Eastern Province, Musanze in the Northern Province, Nyabihu in the Western Province, and a peri-urban district “Gasabo” of Kigali city. All the districts of Eastern Province border Akagera National Park, which is home to various wildlife animals. Musanze district borders Virunga National Park which accommodates buffaloes and primates, while Nyabihu hosts Gishwati-Mukura National Park which is home to primates and birds (Figure 1). The climate is warmer and drier in the Eastern Province, with annual average rainfalls ranging between 700 and 950 mm, and annual average temperatures ranging between 20 and 21°C. The vegetation is grassland with low inclined hills with an average altitude of 1513.5 m. The climate in the Northern and Western Provinces is the coolest and wettest. The annual rainfall ranges from 1,400 to 1,600 mm with annual average temperatures ranging from 15 to 17°C. The topology in the Northern Province is mountainous with the presence of volcanoes, and the average altitude ranges between 2,000 and 3,000 m (21).
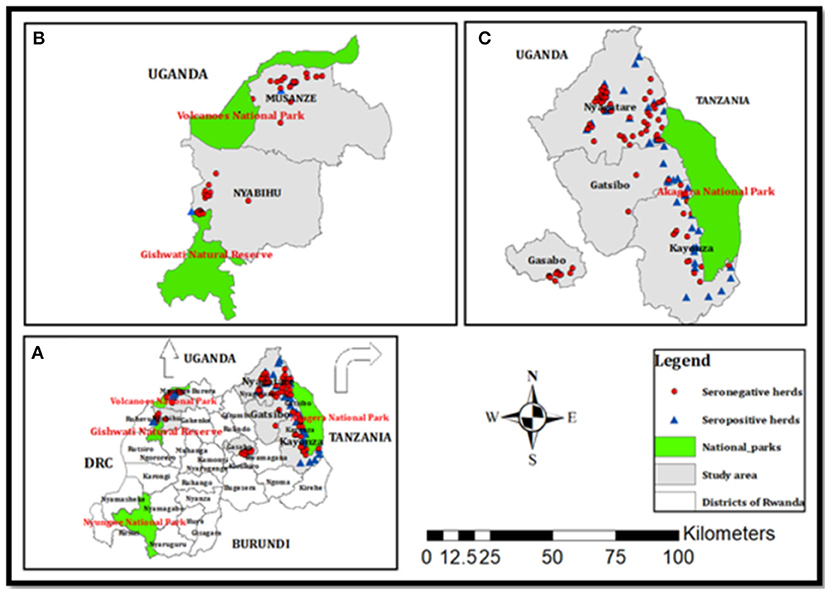
Figure 1. Maps of (A) Rwanda with different districts, (B) the Musanze and Nyabihu districts border the Virunga and Gishwati national parks, respectively, and (C) the Nyagatare, Gatsibo, and Kayonza districts border Akagera National Park, and Gasabo is an urban district with peri-urban areas. Red circles and blue triangles indicate seronegative and seropositive herds found in this study (21).
Study design and sample size
The target population was all cattle, which were brucellosis seropositive (RBT and i-ELISA) plus a few randomly selected cattle with seronegative status but belonging to brucellosis seropositive herds (Figure 1). In the cross-sectional brucellosis seroprevalence study that was previously described (21), the target population was all dairy herds present in the vicinity of national parks and those in the peri-urban areas of Kigali city (Figure 1). During the household visit, for each selected cow, we collected blood in clot activating vacutainers (seroprevalence) and milk from lactating cows or vaginal swabs from non-lactating cows. Due to logistic challenges encountered during sampling, no milk or vaginal swab was collected in the Kayonza district but instead whole blood in 4 ml heparin vacutainer tubes was collected from cattle from the Kayonza district and a few cattle in the Gatsibo district (Table 1). For this bacteriological study, 64 seropositive herds with seropositive cattle (n = 183) and seronegative cattle (n = 27) were recorded and the samples (n = 210) including milk (n = 41), vaginal swabs (n = 51), and whole blood (n = 118) were subjected to bacteriological culture (Table 1). Comparison between the types of samples was not analyzed in this study.
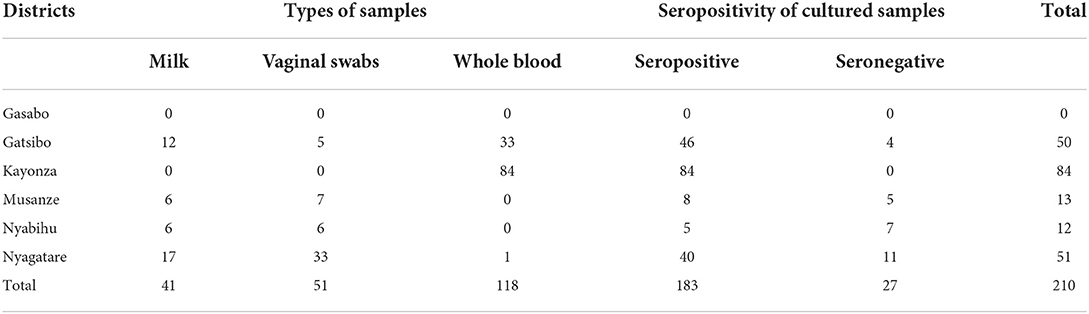
Table 1. Types of cultured samples, their brucellosis seropositivity, and their districts of origin.
Collection of whole blood, milk, and vaginal swabs
Animals were treated with humane care respecting their welfare. Whole blood was collected aseptically into a 4 ml vacutainer heparin tube from the jugular or tail veins of each animal. At least 20 ml of milk (5 ml from each teat) per cow was collected into the Falcon® 50 ml sterile conical centrifuge tubes (Thermo Fischer Scientific, Johannesburg, South Africa). For non-lactating cattle, a sterile transport swab (Aptaca, Canelli, Italy) was streaked on the walls of the vagina. Each sample was labeled with the corresponding animal identification and transported chilled to the nearest campus of the University of Rwanda. Milk was kept in –20°C while blood and vaginal swabs were kept in the fridge and cultured the following week. Culturing of samples and DNA extraction were done in the biosafety level 3 at National Reference Laboratory, Kigali Rwanda.
Isolation of Brucella spp. from whole blood, milk, and vaginal swabs
The pellet and supernatant of centrifuged milk (3,000 × g at 4°C for 15 min), whole blood, and vaginal swabs were inoculated onto modified CITA plates and incubated at 37°C with a 10% CO2 atmosphere as previously described (27). Plates were checked for bacterial growth every day for 3 weeks. For any suspect of Brucella spp. based on the morphology of colonies, streaking was performed to have pure colonies from which the DNA was extracted and screened for the presence of a 214 bp interspacer sequence (ITS) of the genus Brucella spp.
DNA extraction and identification of Brucella spp. using 16S−23S ribosomal DNA interspacer region (ITS) PCR assay
DNA was analyzed by PCR assays at Rwanda Agriculture Board, Department of Veterinary Services and visualization of PCR products at the Rwanda Biomedical Center, Entomology Laboratory. Genomic DNA was extracted from suspected cultures using the ReliaPrep gDNA tissue Miniprep system following the manufacturer's guidelines (Promega, Madison, USA). The identification of the genus Brucella was performed by amplification of the genomic DNA extracted from bacterial colonies using gene-specific primers (Table 2) and a protocol previously developed (28) with slight modifications. Brucella abortus strain 544 served as a positive control. The 15 μl PCR reaction mixture contained 1x of MyTaqTM Red PCR Mix (Bioline, South Africa), primers at 0.2 μM, and 2 μl of template DNA (Table 2). The PCR cycling condition was initial denaturation at 95°C for 3 min followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 2 min, extension at 72°C for 2 min, and a final extension step at 72°C for 5 min. The primers amplified a 214 bp fragment that was analyzed by electrophoresis using a 2% agarose gel stained with SYBR-safe DNA staining gel (Thermo Fischer, Johannesburg, South Africa) and visualized under UV light.
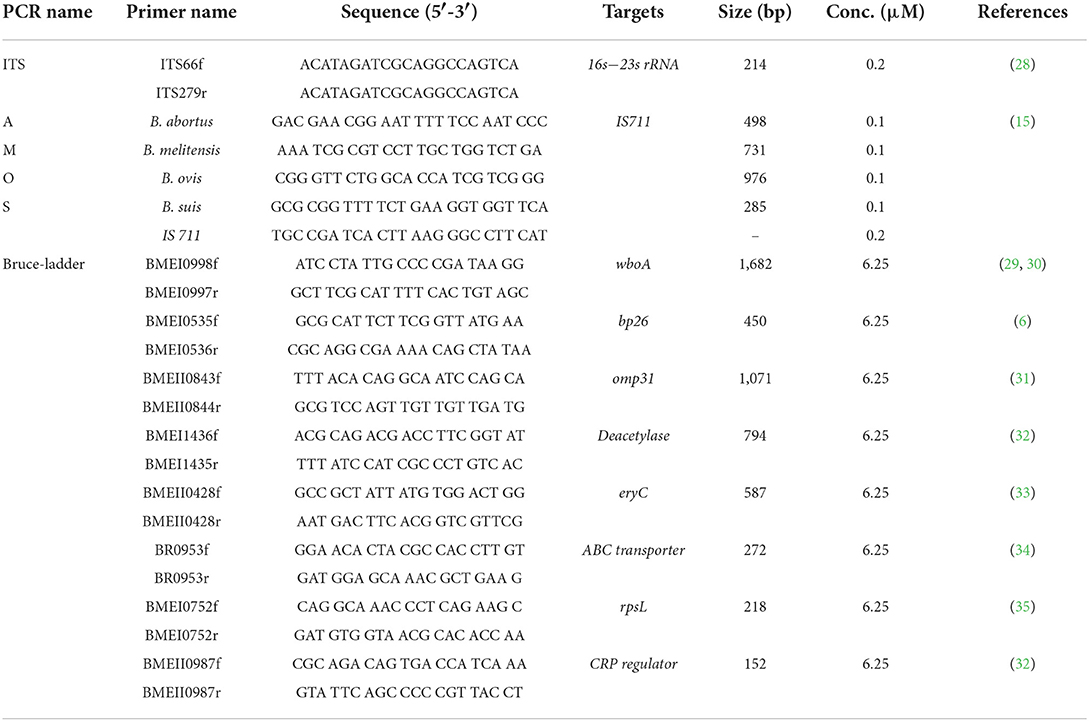
Table 2. Sequences of oligonucleotide primers used for the identification of the genus Brucella, the distinction of Brucella spp., and differentiation of terrestrial Brucella and vaccine strains using 16S−23S ribosomal DNA interspace region (ITS), AMOS, and Bruce-ladder PCR assays.
Identification of Brucella spp. using AMOS and Bruce-ladder PCR assays
Brucella abortus, B. melitensis, B. ovis, and B. suis were identified and differentiated using a multiplex AMOS PCR assay as previously described (15). A 25 μl reaction mixture contained 1x MyRaq Red PCR Mix (Bioline, South Africa), four species-specific forward primers, and reverse primer IS711 (Table 2) at a final concentration of 0.1 and 0.5 μM, respectively, and 2 μl of template DNA. Thermocycling conditions included initial denaturation at 95°C for 3 min followed by 35 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 2 min, an initial extension at 72°C for 2 min, and a final extension at 72°C for 5 min. PCR products were analyzed by gel electrophoresis using 2% agarose stained with SYBR safe DNA staining gel (Thermo Fischer, Johannesburg, South Africa) and visualized under UV light.
Vaccine strains and field isolates of Brucella spp. were identified and differentiated by a multiplex Bruce-ladder PCR assay developed as previously described (16, 17). A 25 μl PCR reaction contained 1x MyTaqTM Red Mix (Bioline, South Africa), eight species-specific forward and reverse primers at a final concentration of 6.25 μM (Table 2), and 2 μl of template DNA. The PCR cycling conditions included an initial denaturation at 95°C for 3 min followed by 25 cycles at 95°C for 30 s, at 64°C for 45 s, and at 72°C for 3 min, and a final extension step at 72°C for 10 min. PCR products were analyzed by gel electrophoresis using a 2% agarose stained with SYBR safe DNA staining gel (Thermo Fischer, Johannesburg, South Africa) and viewed under UV light.
Data analysis
Descriptive data were recorded and analyzed in excel spreadsheets. The districts of origin of samples were tested for significant associations with the culture prevalence confirmed by molecular detection of Brucella spp. using univariate logistic regression in the EpiInfo software version 7.2.4.0 at the significance level of 95% and p-value of 0.05.
Ethical considerations
This study was approved by the research screening and ethical clearance committee of the College of Agriculture, Animal Sciences and Veterinary Medicine, University of Rwanda (Ref:026/DRIPGS/2017). Ethical clearance was also obtained from the institutional review board of the College of Medicine and Health Sciences, University of Rwanda (N° 006/CMHS IRB/2018). Ethical clearance was also obtained from the Animal Ethics Committee of the Faculty of Veterinary Science, University of Pretoria, South Africa (V004/2018). Informed verbal consents were obtained from district officials and a consent form was signed by each participant before the commencement of this study.
Results
Identification of Brucella spp. using 16S−23S ribosomal DNA interspacer region (ITS) PCR assay
Of the 118 cultured blood samples, 14 amplified a 214 bp specific amplicon of the genus Brucella. Of the 41 cultured milk samples, 4 from seropositive cows and 2 from seronegative cows amplified a 214 bp sequence of the genus Brucella, respectively (Table 3, Figure 2). In total, 6 milk samples were positive for 16S−23S ribosomal DNA interspacer region (ITS) PCR assay. Of the 51 vaginal swabs, 13 from seropositive cows and 2 from seronegative cows resulted in amplification of 214 bp sequence of the genus Brucella, respectively (Table 3, Figure 2). Of the 183 samples from seropositive cows, 31 were identified as of Brucella spp. whereas, out of the 27 samples from seronegative cows, 4 were identified as of Brucella spp. In total, out of the 210 samples that were inoculated on a modified CITA medium, 35 were ITS-PCR positive (Table 3, Figure 2). Brucella spp. were detected in 11.9% (14/118), 9.8% (6/41), and 29.4% (15/51) of the samples of whole blood, milk, and vaginal swabs, respectively. Altogether, Brucella spp. were detected in 16.7% (35/210) of seropositive herds of cattle farmed at the wildlife–livestock–human interface.
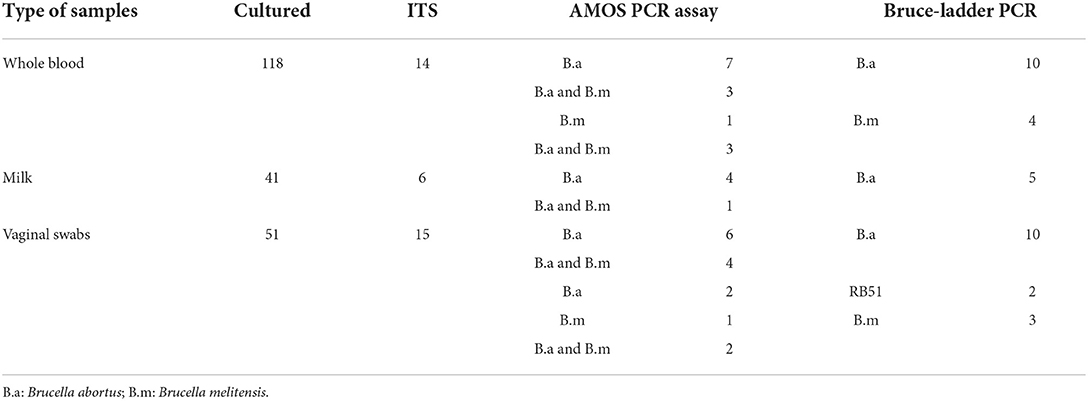
Table 3. Bacteriological, 16S−23S ribosomal DNA interspace region (ITS), AMOS, and Bruce-ladder PCR results of Brucella spp. isolated from seropositive herds of cattle farmed at the wildlife–livestock–human interface in Rwanda.
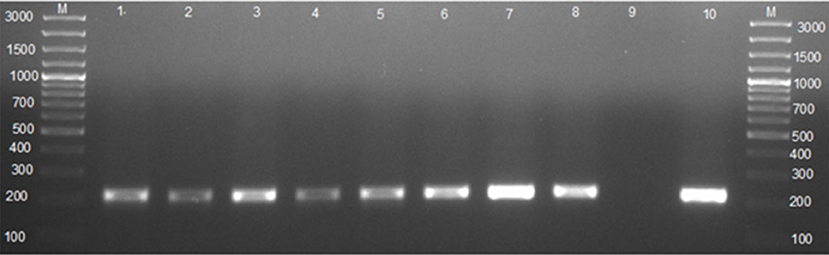
Figure 2. Agarose gel electrophoresis of the 16S−23S ribosomal DNA interspace region (ITS) PCR. Lane M: GeneRuler 100 bp (Thermo Fischer, Johannesburg, South Africa); lanes 1–3: Brucella spp. amplicon (214 bp) from whole blood; lanes 4–5: Brucella spp. amplicon (214 bp) from milk; lanes 6–8: Brucella spp. amplicon (214 bp) from vaginal swabs; lane 9: negative control containing sterile ultrapure water; lane 10: B. abortus 544.
Speciation of Brucella spp. using AMOS PCR assay
For whole blood, AMOS PCR assay identified mixed infections of B. abortus and B. melitensis (n = 6, simultaneous amplification of 731 and 496 bp), B. abortus (n = 7, amplification of 496 bp), and B. melitensis (n = 1, amplification of 731 bp) (Table 3, Figure 3). For milk samples, AMOS PCR assay identified one mixed infection of B. melitensis and B. abortus (n = 1) and B. abortus (n = 4) (Table 3, Figure 3). For vaginal swabs, AMOS PCR assay identified mixed infections of B. melitensis and B. abortus (n = 6), B. abortus (n = 8), and B. melitensis (n = 1) (Table 3, Figure 3).
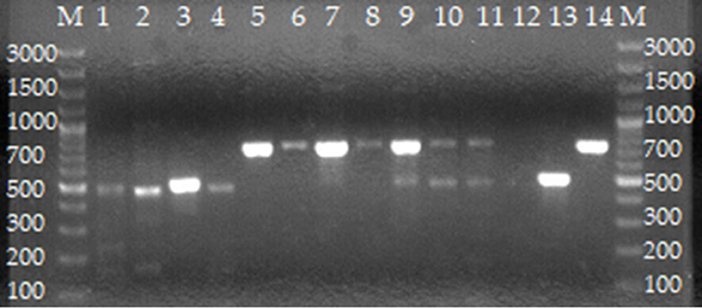
Figure 3. Agarose gel electrophoresis of AMOS PCR from cultures of isolates from cattle farmed at the wildlife–livestock–human interface. Lanes M: GeneRuler 100 bp (Thermo Fischer, Johannesburg, South Africa). Lanes 1–4: B. abortus; lanes 5–8: B. melitensis; lanes 9–11: mixed B. melitensis and B. abortus; lane 12: negative control containing sterile water; lane 13: B. abortus bv. 2 REF 544 strain; lane 14: B. melitensis bv. 1 16 M strain.
Distinction of terrestrial Brucella and vaccine strains using Bruce-ladder PCR assay
For whole blood, the Bruce-ladder PCR assay identified B. abortus (n = 10) and B. melitensis (n = 4) (Table 3, Figure 4). For individual milk samples, Bruce-ladder identified B. abortus (n = 5) (Table 3, Figure 4). For vaginal swabs, Bruce-ladder identified B. abortus (n = 10), B. abortus RB51 (n = 2), B. melitensis (n = 3) (Table 3, Figure 4).
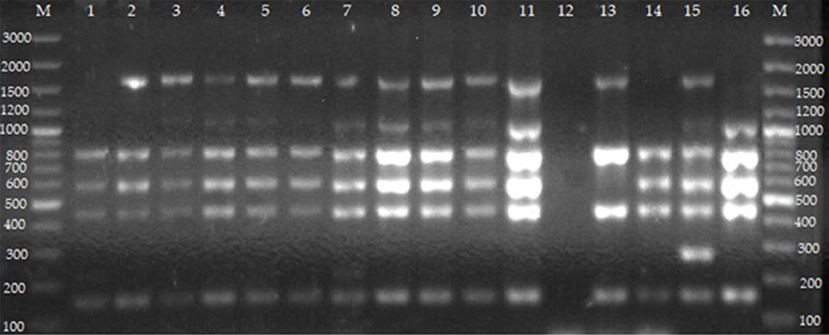
Figure 4. Agarose gel electrophoresis of Bruce-ladder PCR from cultures of isolates from cattle farmed at the wildlife–livestock–human interface. Lanes M: GeneRuler 100 bp (Thermo Fischer, Johannesburg, South Africa); lane 1: B. abortus RB51 from vaginal swabs; lane 2: B. abortus from whole blood; lanes 3–5: B. melitensis from whole blood; lane 6: B. abortus from milk; lanes 7–11: B. melitensis from vaginal swabs; lane 12: negative control containing sterile water, lane 13: B. abortus bv.1 S19; lane 14: B. abortus bv. 1 RB 51; lane 15: B. suis bv. 1 ZW 45; lane 16: B. melitensis bv. 1 16 M strain. Five PCR products (152, 450, 587, 794, and 1,682 bp) are expected for B. abortus (deletion of 25,061 bp in BMEII 0826–BMEII 0850). Five PCR products including 152, 450, 587, 794, and 2,524 bp are observed in B. abortus RB51 (insertion in BMEI 0998). Four PCR products including 152, 450, 794, and 1,682 bp are for B. abortus S19 (deletion of 702 bp in BMEII 0427–BMEII 0428). Six PCR products including 152, 450, 587, 794, 1,071, and 1,682 bp are expected in B. melitensis (insertion in BMEII 0843). Six PCR products including 152, 218, 450, 587, 794, and 1,682 bp are for B. melitensis rev 1 (point mutation in the BMEI 0752). Five PCR products including 152, 450, 587, 794, and 1,071 bp are for B. ovis (deletion of 1,507 bp in BMEI 0993–BMEI 1012). Six PCR products including 152, 272, 450, 587, 794, and 1,682 bp for B. suis. Five PCR products including 272, 450, 587, 1,071, and 1,682 bp are observed in B. canis (deletion of 976 bp in BME1435).
The isolation of Brucella spp. was significantly associated with district with Nyagatare having more isolates [36.6%, (15/41)] than Nyabihu [36.4%, (4/11)], Kayonza [24.5%, (13/53)], Gatsibo [7.4%, (3/27)], and Musanze [0.0%, (0/9)] (p = 0.01).
Discussion
Brucella spp. fall under category A pathogens and cause serious diseases in a wide range of animals and humans (1). Bovine brucellosis negatively affects national economies and public health worldwide (1, 36, 37). Seroprevalence studies showed that bovine brucellosis is prevalent in Rwanda (21, 22). However, serology does not provide a complete diagnosis and has drawbacks related to sensitivity and specificity (37, 38). Furthermore, Brucella spp. that are involved in the Brucella infections remain unknown in Rwanda. This study isolated Brucella spp. from blood, milk, and vaginal swabs of dairy cattle in Rwanda. The identified Brucella spp. included individual and mixed infections of B. melitensis and B. abortus. Two B. abortus RB51 were isolated from vaginal swabs using Bruce-ladder PCR assay.
The frequency of molecular detection of Brucella spp. (11.9%) from cultures of whole blood of cattle was higher than the finding (5.8%) of Caine et al. (39) and other previous studies which did not detect Brucella spp. from cultures of blood (40, 41). The present finding indicated that whole blood may be a good sample for isolation of Brucella spp. if processed immediately after collection (39, 42). The frequency of isolation of Brucella spp. in 12.2% of milk samples in this study is higher than the 6.5% recovered from raw milk informally marketed on streets in Uganda (43). The presence of Brucella spp. in milk is worrying since 21.7% of cattle keepers owning these seropositive cows reported drinking raw milk (21) which might be reflected in the human brucellosis cases detected in Rwanda (23, 24).
The isolation of Brucella spp. from seropositive and seronegative cows is consistent with earlier studies which also revealed the presence of Brucella spp. from seropositive and seronegative cows in Bangladesh (44), and China (45). The detection of Brucella spp. in seronegative animals may be because Brucella antibodies decrease in seronegative cows and chronically diseased cows while the organism remains intracellular in different tissues (44). The detection of Brucella spp. in seronegative cattle indicated that serological tests such as i-ELISA with cut-off points determined in Europe with no or low prevalence of brucellosis must be validated for Rwandan cattle. The isolation of Brucella spp. from the milk of seronegative cows is a problem of concern since serology is the only diagnostic method of brucellosis in Rwanda, and seronegative dairy cows continue shedding the pathogen in milk which is a valuable commodity, sometimes consumed unpasteurized and sold at the non-regulated market (21, 46). Furthermore, the traditional homemade cream milk known as “Ikivuguto” in the local language is frequently made of raw milk by several Rwandan families (47). Therefore, there is a need to investigate the presence of Brucella spp. in the homemade cream milk “ikivuguto” and to generate an awareness of this risk in Rwanda.
The detection of Brucella spp. in 29.4% of vaginal swabs was higher than the 12.6% previously reported in Pakistan (48), 1.5% in Mongolia (49), and 1.1% in Nigeria (50). This difference may be associated with the origin of samples and in this study, samples were collected from seropositive cows farmed in high-risk zones (21). In addition, the amount of Brucella isolation may also depend on the storage conditions and culture medium used (27, 51). The isolation of Brucella spp. from vaginal swabs confirms that Brucella organisms have the tropism for the reproductive organs of mature animals and massively multiply in the presence of reproductive hormones and erythritol (36, 52). Therefore, this finding support that vaginal swabs may be a good specimen for rapid molecular detection of brucellosis in animals (28).
Brucella spp. were more isolated from Nyagatare district compared to other districts. This difference may be due to the number of vaginal swabs (33/51) for the Nyagatare district compared to 5/50, 0/84, 7/13, and 6/12 for Gatsibo, Kayonza, Musanze, and Nyabihu districts, respectively. Brucella spp. colonize reproductive organs and were highly present and viable in vaginal swabs, which contained transport and storage medium (52). It was not surprising to detect B. abortus vaccine strain RB51 which is the vaccine used in the vaccination of cattle in high-risk zones in Rwanda. The identification of vaccine strains RB51 from cattle farmed at the interface in Rwanda indicates that RB51 is not safe for cattle and causes brucellosis in humans (53, 54) and that vaccinating pregnant animals should be done with caution.
It is of diagnostic importance that the 16S−23S ribosomal DNA interspace region (ITS) PCR detected Brucella DNA from seropositive cows as well as seronegative cows. The ITS PCR was able to detect as little as 3.8 fg of B. canis DNA mixed with 54 ng of template canine DNA extracted from vaginal swabs of non-infected bitches (28). The finding of this study confirms that ITS PCR can be used to detect Brucella spp. from vaginal swabs of animals that are seronegative, negative to blood culture, or blood PCR (28). However, there is a need to determine and validate the specificity and sensitivity of the ITS PCR in Rwanda since closely related Brucella pathogens that were not analyzed by Keid et al. (28) might be locally present and could generate false positives.
The recovery of B. abortus in the present study is consistent with earlier studies in the region (43, 55). This finding confirms that B. abortus is the main causal agent of brucellosis in dairy cattle. Although B. melitensis commonly cause the disease in goats, it was isolated in dairy cattle in the present study which could be due to the practice of co-rearing of animals (21). Hence there is a need to strengthen brucellosis control in cattle and avoid interspecies farming in Rwanda. AMOS-PCR detected a mixed infection of B. abortus and B. melitensis in the blood, milk, and vaginal swabs of cattle. Mixed infections of B. abortus and B. melitensis have been recently reported in aborted tissues of goats in Rwanda (26) and in herds where cattle graze together with small ruminants in South Africa and Kenya (56, 57). Keeping different animal species such as cattle and small ruminants on the same farm represents a risk of transmission of brucellosis to other animal species including humans. The purification of these cultures is recommended for future studies to isolate separately B. abortus and B. melitensis which primarily cause brucellosis in cattle and humans, respectively (36).
This is a problem of concern because diseased animals reduce production and Brucella spp. are present in the blood, milk, and vaginal secretions. This represents a great risk of contamination to handlers of live animals, carcasses, and consumers of raw milk and milk products.
Conclusion
This study identified mixed and single infections caused by B. abortus and B. melitensis from whole blood, vaginal swabs, and milk indicating the great risk of transmission to handlers of live cattle, carcasses, and consumers of unpasteurized milk and milk products. We, therefore, advise the revision of the vaccination program to include protection against B. melitensis in livestock. The study also isolated B. abortus RB51, a vaccine strain, in seropositive cattle. Education about the epidemiology of brucellosis and other zoonotic diseases is of paramount importance to all stakeholders in the animal sector and consumers of animal products.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by College of Agriculture, Animal Sciences and Veterinary Medicine, University of Rwanda (Ref: 026/DRIPGS/2017), Institutional Review Board of the College of Medicine and Health Sciences, University of Rwanda (No. 006/CMHS IRB/2018). Ethical clearance was also obtained from Animal Ethics Committee of the Faculty of Veterinary science, University of Pretoria, South Africa (V004/2018). Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
JN and HH: conceptualization and methodology. JN: formal analysis and writing—original draft preparation. JN, EU, VM, AI, RG, and LU: investigation and data collection. JN, FK, EM, and HH: writing—review and editing. HH and FK: supervision. HH: project administration, resources, and funding acquisition. All authors have read and approved the manuscript.
Funding
This study was supported by the Belgian Directorate-General for Development Cooperation, through its Framework Agreement with the Institute of Tropical Medicine (FA DGD-ITM 2017–2021). The funding body did not play a role in the design, analysis, and reporting of the study.
Acknowledgments
The authors would like to acknowledge the Belgian Directorate-General for Development Cooperation (DGD) through its framework agreement with the Institute of Tropical Medicine Antwerp and the Department of Veterinary Tropical Diseases for funding this research; the University of Rwanda, and the Department of Veterinary Services within Rwanda Agriculture and Animal Resources Board, National Reference Laboratory and Entomology laboratory within Rwanda Biomedical Centre for providing access the laboratories.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.1017851/full#supplementary-material
References
1. OIE. Brucellosis (Brucella abortus, Brucella melitensis, and Brucella suis). In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2021. Vols. 1 2 and 3. Terrestrial Manual 8th, ed., 2021 ed. Paris: World Organisation for Animal Health (2021). p. 355–98.
2. Meyer M. Host parasite relationships in Brucellosis. i. reservoirs of infection and interhost transmissibility of the parasite. Proc US Livestock Sanit Assoc. (1966) 70:129–34.
3. Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J Bacteriol. (1990) 172:3569–76. doi: 10.1128/jb.172.7.3569-3576.1990
4. Alton GG, Jones LM, Pietz DE, World Health Organization, Food and Agriculture Organization of the United Nations. Laboratory Techniques in Brucellosis. 2d ed. Geneva; Albany, NY: World Health Organization. (Available from Q Corp.) (1975).
5. Verger J-M, Grimont F, Grimont PA, Grayon M. Brucella, a monospecific genus as shown by deoxyribonucleic acid hybridization. Int J Syst Evol Microbiol. (1985) 35:292–5. doi: 10.1099/00207713-35-3-292
6. Cloeckaert A, Grayon M, Grepinet O. An IS711 element downstream of the bp26 gene is a specific marker of Brucella spp. isolated from marine mammals. Clin Diagn Lab Immunol. (2000) 7:835–9. doi: 10.1128/CDLI.7.5.835-839.2000
7. Cloeckaert A, Verger J-M, Grayon M, Grepinet O. Restriction site polymorphism of the genes encoding the major 25 kDa and 36 kDa outer-membrane proteins of Brucella. Microbiology. (1995) 141:2111–21. doi: 10.1099/13500872-141-9-2111
8. Halling SM, Tatum FM, Bricker BJ. Sequence and characterization of an insertion-sequence, IS711, from Brucella ovis. Gene. (1993) 133:123–7. doi: 10.1016/0378-1119(93)90236-V
9. Halling SM, Zehr ES. Polymorphism in Brucella spp. due to highly repeated DNA. J Bacteriol. (1990) 172:6637–40. doi: 10.1128/jb.172.12.6637-6640.1990
10. Mercier E, Jumas-Bilak E, Allardet-Servent A, O'Callaghan D, Ramuz M. Polymorphism in Brucella strains detected by studying distribution of two short repetitive DNA elements. J Clin Microbiol. (1996) 34:1299–302. doi: 10.1128/jcm.34.5.1299-1302.1996
11. Ficht T, Bearden S, Sowa B, Marquis H. Genetic variation at the omp2 porin locus of the brucellae: species-specific markers. Mol Microbiol. (1990) 4:1135–42. doi: 10.1111/j.1365-2958.1990.tb00688.x
12. Halling SM, Bricker BJ. Characterization and occurrence of 2 repeated palindromic DNA elements of Brucella spp. - Bru-Rs1 and Bru-Rs2. Mol Microbiol. (1994) 14:681–9. doi: 10.1111/j.1365-2958.1994.tb01306.x
13. Chandler MS. Insertion sequences and transposons. In: de Bruijn FJ, Lupski JR, Weinstock GM, editors. Bacterial Genomes. Boston, MA: Springer (1998) p. 30–7. doi: 10.1007/978-1-4615-6369-3_4
14. Mayfield JE, Bricker BJ, Godfrey H, Crosby RM, Knight DJ, Hailing SM, et al. The cloning, expression, and nucleotide sequence of a gene coding for an immunogenic Brucella abortus protein. Gene. (1988) 63:1–9. doi: 10.1016/0378-1119(88)90540-9
15. Bricker BJ, Halling SM. Differentiation of Brucella abortus bv. 1, 2, and 4, Brucella melitensis, Brucella ovis, and Brucella suis bv. 1 by PCR. J Clin Microbiol. (1994) 32:2660–6. doi: 10.1128/jcm.32.11.2660-2666.1994
16. Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, Vizmanos JL, Lopez-Goni I. Multiplex PCR assay for the identification and differentiation of all Brucella species and the vaccine strains Brucella abortus S19 and RB51 and Brucella melitensis Rev1. Clin Chem. (2006) 52:779–81. doi: 10.1373/clinchem.2005.062596
17. Lopez-Goni I, Garcia-Yoldi D, Marin CM, de Miguel MJ, Munoz PM, Blasco JM, et al. Evaluation of a multiplex PCR assay (Bruce-ladder) for molecular typing of all Brucella species, including the vaccine strains. J Clin Microbiol. (2008) 46:3484–7. doi: 10.1128/JCM.00837-08
18. Ducrotoy MJ, Bardosh KL. How do you get the Rose Bengal Test at the point-of-care to diagnose brucellosis in Africa? The importance of a systems approach. Acta Trop. (2017) 165:33–9. doi: 10.1016/j.actatropica.2016.10.004
19. Ducrotoy M, Bertu WJ, Matope G, Cadmus S, Conde-Alvarez R, Gusi AM, et al. Brucellosis in Sub-Saharan Africa: current challenges for management, diagnosis and control. Acta Trop. (2017) 165:179–93. doi: 10.1016/j.actatropica.2015.10.023
20. Minagri. Law Determining the Prevention and Fight Against Contagious Diseases for Domestic Animals in Rwanda Kigali. Ministry of Agriculture and Animal Resources (2009). Available online at: https://www.minagri.gov.rw/fileadmin/user_upload/documents/Law_and_Regulations/Official_Gazette_no_20_of_18.05.2009.pdf (cited August 16, 2020).
21. Ntivuguruzwa JB, Kolo FB, Gashururu RS, Umurerwa L, Byaruhanga C, van Heerden H. Seroprevalence and associated risk factors of bovine brucellosis at the wildlife-livestock-human interface in Rwanda. Microorganisms. (2020) 8:1553. doi: 10.3390/microorganisms8101553
22. Ndazigaruye G, Mushonga B, Kandiwa E, Samkange A, Segwagwe BE. Prevalence and risk factors for brucellosis seropositivity in cattle in Nyagatare District, Eastern Province, Rwanda. J South Afr Vet Assoc. (2018) 89:e1–8. doi: 10.4102/jsava.v89i0.1625
23. Rujeni N, Mbanzamihigo L. Prevalence of brucellosis among women presenting with abortion/stillbirth in Huye, Rwanda. J Trop Med. (2014) 2014:740479. doi: 10.1155/2014/740479
24. Gafirita J, Kiiza G, Murekatete A, Ndahayo LL, Tuyisenge J, Mashengesho V, et al. Seroprevalence of Brucellosis among patients attending a district hospital in Rwanda. Amer J Trop Med Hyg. (2017) 97:831–5. doi: 10.4269/ajtmh.16-0632
25. Verger JM, Grayon M. [Characteristics of 273 strains of Brucella abortus of African origin]. Dev Biol Stand. (1984) 56:63–71.
26. Ntivuguruzwa JB, Kolo FB, Mwikarago EI, van Heerden H. Characterization of Brucella spp. and other abortigenic pathogens from aborted tissues of cattle and goats in Rwanda. Vet Med Sci. (2022) 8:1655–63. doi: 10.1002/vms3.805
27. Ledwaba MB, Matle I, Van Heerden H, Ndumnego OC, Gelaw AK. Investigating selective media for optimal isolation of Brucella spp. in South Africa. Onders J Vet Res. (2020) 87:1–9. doi: 10.4102/ojvr.v87i1.1792
28. Keid LB, Soares RM, Vasconcellos SA, Chiebao DP, Salgado VR, Megid J, et al. A polymerase chain reaction for detection of Brucella canis in vaginal swabs of naturally infected bitches. Theriogenology. (2007) 68:1260–70. doi: 10.1016/j.theriogenology.2007.08.021
29. Garcia-Yoldi D, Marín C, Lopez-Goni I. Restriction site polymorphisms in the genes encoding new members of group 3 outer membrane protein family of Brucella spp. FEMS Microbiol Lett. (2005) 245:79–84. doi: 10.1016/j.femsle.2005.02.026
30. Vemulapalli R, McQuiston JR, Schurig GG, Sriranganathan N, Halling SM, Boyle SM. Identification of an IS711 element interrupting the wboA gene of Brucella abortus vaccine strain RB51 and a PCR assay to distinguish strain RB51 from other Brucella species and strains. Clin Diagn Lab Immunol. (1999) 6:760–4. doi: 10.1128/CDLI.6.5.760-764.1999
31. Vizcaino N, Verger J-M, Grayon M, Zygmunt MS, Cloeckaert A. DNA polymorphism at the omp-31 locus of Brucella spp.: evidence for a large deletion in Brucella abortus, and other species-specific markers. Microbiology. (1997) 143:2913–21. doi: 10.1099/00221287-143-9-2913
32. Rajashekara G, Glasner JD, Glover DA, Splitter GA. Comparative whole-genome hybridization reveals genomic islands in Brucella species. J Bacteriol. (2004) 186:5040–51. doi: 10.1128/JB.186.15.5040-5051.2004
33. Sangari FJ, García-Lobo JM, Agüero J. The Brucella abortus vaccine strain B19 carries a deletion in the erythritol catabolic genes. FEMS Microbiol Lett. (1994) 121:337–42. doi: 10.1111/j.1574-6968.1994.tb07123.x
34. Halling SM, Peterson-Burch BD, Bricker BJ, Zuerner RL, Qing Z, Li L-L, et al. Completion of the genome sequence of Brucella abortus and comparison to the highly similar genomes of Brucella melitensis and Brucella suis. J Bacteriol. (2005) 187:2715–26. doi: 10.1128/JB.187.8.2715-2726.2005
35. Cloeckaert A, Grayon M, Grépinet O. Identification of Brucella melitensis vaccine strain Rev.1 by PCR-RFLP based on a mutation in the rpsL gene. Vaccine. (2002) 20:2546–50. doi: 10.1016/S0264-410X(02)00159-7
36. Young EJ. An overview of human brucellosis. Clin Infect Dis. (1995) 21:283–9. doi: 10.1093/clinids/21.2.283
38. Nielsen K. Diagnosis of brucellosis by serology. Vet Microbiol. (2002) 90:447–59. doi: 10.1016/S0378-1135(02)00229-8
39. Caine L-A, Nwodo UU, Okoh AI, Green E. Molecular characterization of Brucella species in cattle, sheep and goats obtained from selected municipalities in the Eastern Cape, South Africa. Asian Pac J Trop Dis. (2017) 7:293–8. doi: 10.12980/apjtd.7.2017D6-431
40. Yagupsky P. Detection of Brucellae in blood cultures. J Clin Microbiol. (1999) 37:3437–42. doi: 10.1128/JCM.37.11.3437-3442.1999
41. O'Leary S, Sheahan M, Sweeney T. Brucella abortus detection by PCR assay in blood, milk and lymph tissue of serologically positive cows. Res Vet Sci. (2006) 81:170–6. doi: 10.1016/j.rvsc.2005.12.001
42. Gotuzzo E, Carrillo C, Guerra J, Llosa L. An evaluation of diagnostic methods for brucellosis—the value of bone marrow culture. J Infect Dis. (1986) 153:122–5. doi: 10.1093/infdis/153.1.122
43. Hoffman T, Rock K, Mugizi DR, Muradrasoli S, Lindahl-Rajala E, Erume J, et al. Molecular detection and characterization of Brucella species in raw informally marketed milk from Uganda. Infect Ecol Epidemiol. (2016) 6:32442. doi: 10.3402/iee.v6.32442
44. Islam M, Khatun M, Saha S, Basir M, Hasan M-M. Molecular detection of Brucella spp. from milk of seronegative cows from some selected area in Bangladesh. J Pathog. (2018) 2018:9378976. doi: 10.1155/2018/9378976
45. Ning P, Guo K, Xu L, Xu R, Zhang C, Cheng Y, et al. Evaluation of Brucella infection of cows by PCR detection of Brucella DNA in raw milk. J Dairy Sci. (2012) 95:4863–7. doi: 10.3168/jds.2012-5600
46. Kamana O, Jacxsens L, Kimonyo A, Uyttendaele M. A survey on hygienic practices and their impact on the microbiological quality and safety in the Rwandan milk and dairy chain. Int J Dairy Technol. (2017) 70:52–67. doi: 10.1111/1471-0307.12322
47. Karenzi E, Mashaku A, Nshimiyimana AM, Munyanganizi B, Thonart P. Kivuguto traditional fermented milk and the dairy industry in Rwanda. A review. Biotechnol Agron Soc Environ. (2013) 17:383–91.
48. Ali S, Ali Q, Melzer F, Khan I, Akhter S, Neubauer H, et al. Isolation and identification of bovine Brucella isolates from Pakistan by biochemical tests and PCR. Trop Anim Health Product. (2014) 46:73–8. doi: 10.1007/s11250-013-0448-6
49. Bayasgalan C, Chultemdorj T, Roth F, Zinsstag J, Hattendorf J, Badmaa B, et al. Risk factors of brucellosis seropositivity in Bactrian camels of Mongolia. BMC Vet Res. (2018) 14:342. doi: 10.1186/s12917-018-1664-0
50. Ocholi R, Kwaga J, Ajogi I, Bale J. Abortion due to Brucella abortus in sheep in Nigeria. Rev Scient Tech Off Int Épizoo. (2005) 24:973. doi: 10.20506/rst.24.3.1627
51. Falenski A, Mayer-Scholl A, Filter M, Göllner C, Appel B, Nöckler K. Survival of Brucella spp. in mineral water, milk and yogurt. Int J Food Microbiol. (2011) 145:326–30. doi: 10.1016/j.ijfoodmicro.2010.11.033
52. Keppie J, Williams A, Witt K, Smith H. The role of erythritol in the tissue localization of the brucellae. Brit J Exp Pathol. (1965) 46:104.
53. Yazdi HS, Kafi M, Haghkhah M, Tamadon A, Behroozikhah AM, Ghane M. Abortions in pregnant dairy cows after vaccination with Brucella abortus strain RB51. Vet Rec. (2009) 165:570–1. doi: 10.1136/vr.165.19.570
54. Dougherty AMF, Cornish TE, O'Toole D, Boerger-Fields AM, Henderson OL, Mills KW. Abortion and premature birth in cattle following vaccination with Brucella abortus strain RB51. J Vet Diagn Invest. (2013) 25:630–5. doi: 10.1177/1040638713499570
55. Mugizi DR, Muradrasoli S, Boqvist S, Erume J, Nasinyama GW, Waiswa C, et al. Isolation and molecular characterization of Brucella isolates in cattle milk in Uganda. Biomed Res Int. (2015) 2015:720413. doi: 10.1155/2015/720413
56. Muendo EN, Mbatha PM, Macharia J, Abdoel TH, Janszen PV, Pastoor R, et al. Infection of cattle in Kenya with Brucella abortus biovar 3 and Brucella melitensis biovar 1 genotypes. Trop Anim Health Prod. (2012) 44:17–20. doi: 10.1007/s11250-011-9899-9
Keywords: Brucella spp., dairy cattle, multiplex PCR assays, wildlife–livestock–human interface, Rwanda, seropositive herds
Citation: Ntivuguruzwa JB, Kolo FB, Gashururu R, Uwibambe E, Musanayire V, Ingabire A, Umurerwa L, Mwikarago EI and van Heerden H (2022) Molecular characterization of Brucella spp. from seropositive herds of cattle farmed at the wildlife–livestock–human interface in Rwanda. Front. Vet. Sci. 9:1017851. doi: 10.3389/fvets.2022.1017851
Received: 12 August 2022; Accepted: 20 September 2022;
Published: 11 October 2022.
Edited by:
Shengqing Yu, Shanghai Veterinary Research Institute (CAAS), ChinaReviewed by:
Aman Dev Moudgil, Lala Lajpat Rai University of Veterinary and Animal Sciences, IndiaRosa Estela Quiroz Castañeda, Instituto Nacional de Investigaciones Forestales, Agrícolas y Pecuarias (INIFAP), Mexico
Copyright © 2022 Ntivuguruzwa, Kolo, Gashururu, Uwibambe, Musanayire, Ingabire, Umurerwa, Mwikarago and van Heerden. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jean Bosco Ntivuguruzwa, Ym9zY3VzMkBnbWFpbC5jb20=
†Present address: Emil Ivan Mwikarago, Department of Human Medicine and Device Assessment and Registration, Division of Human Medicine, Assessment and Registration, Rwanda Food and Drug Administration, Kigali, Rwanda
 Jean Bosco Ntivuguruzwa
Jean Bosco Ntivuguruzwa Francis Babaman Kolo2
Francis Babaman Kolo2 Emil Ivan Mwikarago
Emil Ivan Mwikarago Henriette van Heerden
Henriette van Heerden