
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 28 October 2022
Sec. Comparative and Clinical Medicine
Volume 9 - 2022 | https://doi.org/10.3389/fvets.2022.1010728
This article is part of the Research Topic Rising Stars in Comparative and Clinical Medicine: 2022 View all 29 articles
The need to supply quality food for the growing human population has led to the revolutionization of food production and processing in recent years. Meanwhile, food production sources are at risk of microbial attack, while the use of antibiotics to counter them is posing another threat to food safety and security. Vancomycin was used as the first line of defense against multiple drug-resistant bacteria salient of which is methicillin-resistant S. aureus. The emergence of the vancomycin resistance gene in bacteria impairs the efficacy of antibiotics on the one hand while its harmful residues impart food safety concerns on the other. Currently, a novel set of resistance genes “Van cluster” is circulating in a wider range of bacteria. Considerable economic losses in terms of low production and food safety are associated with this emerging resistance. The current review focuses on the emergence of vancomycin resistance and its impact on food safety. The review proposes the need for further research on the probable routes, mechanisms, and implications of vancomycin resistance from animals to humans and vice versa.
The use of antimicrobials poses a significant threat to animal production and profitability. Although there are Maximum Residue Limits (MRLs) of the approved veterinary drugs for their use to improve animal health, their implementation requires stern legislation and subsequent actions. On the other hand, there are a variety of guidelines in different countries but some of the countries still need to develop these guidelines (1). Implementing a withdrawal period of antibiotics in animal food products is an additional requirement to avoid food safety concerns. At the farm level, withdrawal periods are kept in line with the expected time of consumption of food. It is commonly required that drug residues must be below MRLs. Extra label utilization, miscalculated doses, non-specific use, and failure in observing the prescribed course of antibiotics may result in the spread of antimicrobial resistance.
Antimicrobial consumption through food or any other sources continuously leads to the suppression of intestinal microbiota that guards against pathogenic bacteria. Moreover, their presence in the bloodstream leads to the malfunctioning of various organs (2, 3). Extensive use of antimicrobials in animals may lead to the production of novel resistant bacteria in the environment. Some of the salient bacteria responsible for food safety and security include Salmonella, E. coli, and S. aureus isolated from various sources in humans and poultry (4–7), dairy milk (8–11), and farm animals' skin and nares (12, 13). The bacteria are developing resistance to multiple antibiotics and thus becoming a challenge to treating an infection with commonly used antibiotics. Among such antibiotics is vancomycin, a member of glycopeptides, and was the primary applied drug of choice against methicillin-resistant S. aureus (14). Recent studies showed S. aureus is a resistant pathogen against vancomycin (15, 16) thus increasing the burden of antimicrobial drug resistance.
Resistance against vancomycin is found to be due to a cluster of genes known as the “Van cluster” that is found in a number of bacteria such as S. aureus, E. fecalis, Actinomycetes spp., E. faceium, Clostridium difficile, and anaerobic bacteria. It is pertinent to note that Enterococcus spp. appeared to be a prominent reservoir of vancomycin resistance and its spread to other bacteria is horizontal using the conjugation method. It has been found that Enterococci contain Inc18 pSK41-like multiple drug-resistant conjugated plasmids while their presence in S. aureus has yet not been documented. The transfer to the latter is found to be by plasmids from E. faecalis to other bacteria. Worsening of this resistance is thought to be controlled by manipulation of Van cluster from the bacteria. Hence, it provides a better target for new drug development e.g., phosphinate, phosphonate, and hydroxyethylamines can inhibit Van clusters. These inhibitors are mostly used with vancomycin for the effective uptake of antibiotics inside the bacterial cell (17). Such discoveries are necessary for better control of vancomycin resistance. Vancomycin-resistant genes are extensively transferred from animal to human, animal to bacteria, human to bacteria, and bacteria to bacteria. There is a dire need to focus on novel genes in bacteria to address their resistance and impact on animal and public health. The current review thus focuses on vancomycin resistance in bacteria, its mode of spread, and its impact on food safety/security.
Vancomycin was isolated by E.C Kornfield in year 1957 from Streptomyces orientalis, a fungus, located in the deep jungles of Borneo (18). The obtained substance was named “compound 05865” having activity against gram-positive as well as anaerobic bacteria in initial findings. Vancomycin is a tricyclic glycopeptide that acts as a bactericidal against the polymerization of peptidoglycans in the bacterial cell wall. This antibiotic is equally effective against a wider range of pathogens and in surgical procedures to avoid secondary infection. The FDA has approved the use of this drug against Clostridium difficile, Staphylococcus enterocolitis, Pseudomembranous colitis, Enterococcal, Staphylococcal, and Streptococcal spp, which are responsible for diarrheas, endocarditis, sepsis, soft skin infection, bone infections, lower respiratory tract infections, and colitis (19). In other studies, there was an emergence of mecA or mecC gene in S. aureus that codes enzymes to crosslink the peptidoglycans in bacterial cell walls which is now considered a potential threat to control S. aureus. The enzymes had low affinity for β-lactams and hence there was a development of resistance. The resistance of Staphylococci against antibiotics was on the rise when this drug was developed therefore the FDA approved vancomycin with number 05865 in 1958 to control infection of Staphylococci. In such a scenario, there was a concern that this product would maintain its efficacy/activity or face resistance. A series of in-vitro and lab animal experiments were conducted to rule out this concern (20). The off-label uses of this drug consist of catheter-related infections, neonatal prophylaxis, bacterial meningitis, peritonitis, and many more (21). More than half of regularly used antibacterial medications are no longer effective against Gram-negative Pathogens (GNPs) including E. coli and K. pneumoniae, according to the WHO Global Report on Surveillance of Antimicrobial Resistance (22–24). It is important to note that drugs like colistin, which were used as a last resort, are no more effective against a GNP such as New Delhi metallo-lactamase-1.
Vancomycin resistance was first reported in Enterococcus faecium in 1986 (25) and the resistant genes were named VRE (Vancomycin-resistant Enterococci). Avoparcin was overused as a growth enhancer in animal production in European communities. In addition to Enterococci, there was also a development of novel strains of S. aureus against vancomycin (Figures 1, 2) that gave rise to three distinct categories (i) VSSA (S. aureus sensitive to vancomycin), (ii) VRSA (S. aureus resistant to vancomycin), and (iii) VISA (S. aureus intermediate susceptible to vancomycin).
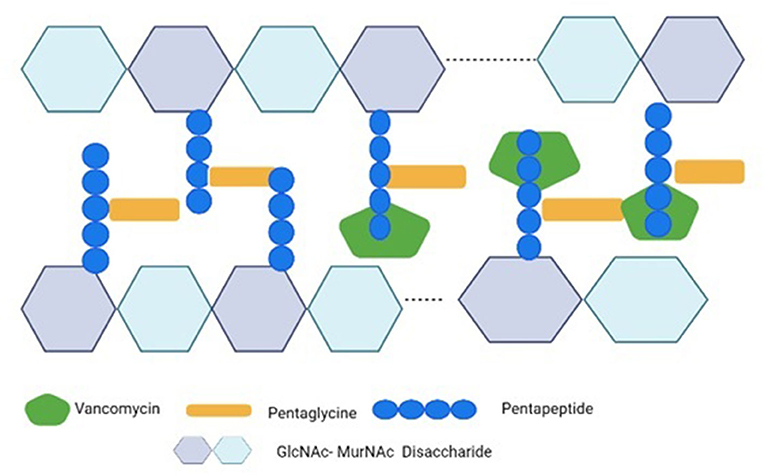
Figure 2. Vancomycin's mechanism of action against S. aureus. Vancomycin inhibits the cross-bridge formation between pentagen and pentaglycine by binding to the D-Ala-D-Ala residues of pentapeptide's C-terminal D-Ala-D-Ala. The GlcNAc stands for N-acetylglucosamine; the MurNAc stands for N-acetylmuramic acid.
This kind of resistance resulted in a synthesis of precursors of peptidoglycan which ends in D-Ala-D-Lac and D-Ala-D-Ser instead of D-Ala-D-Ala (Figure 3). These precursors are highly incompatible with vancomycin and thus give rise to a cluster of 11 genes found primarily in Enterococci to confer vancomycin resistance. The precursor ending with D-Ala-D-Lac is coded by vanA, B, D, F, I, and M clusters while that of D-Ala-DSer is coded by vanC, E, G, L, and N clusters. It was found that the vanA gene and vanB gene are frequently encountered genes when it comes to acquired resistance (26, 27). The ability of vancomycin to bind to targeted peptides is 1000 times less when these peptidoglycan precursors are produced with an association constant of 102 M-1 (28).
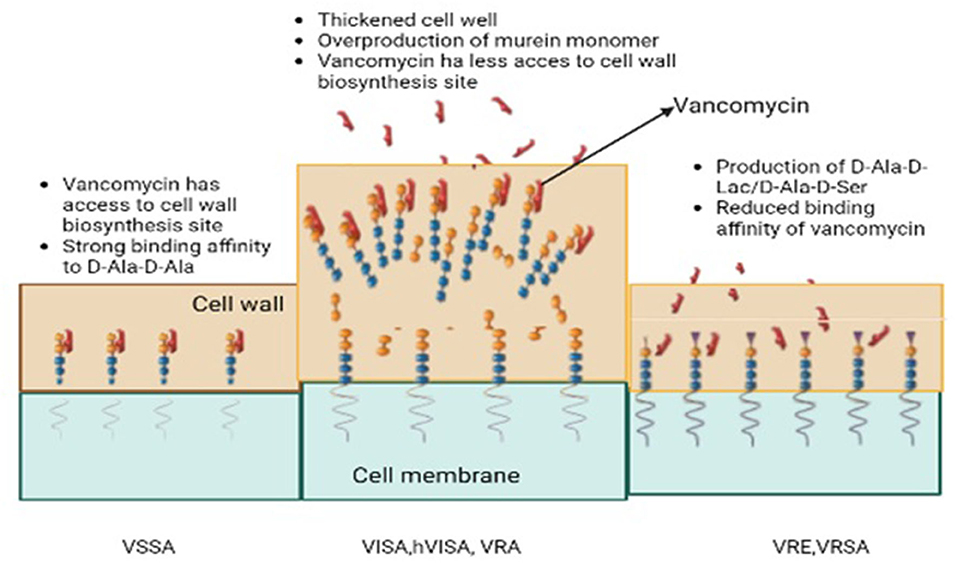
Figure 3. The development of acquired resistance; Vancomycin-resistant S. aureus (VRSA), Vancomycin-resistant Enterococci (VRE), Vancomycin susceptible S. aureus, Vancomycin intermediate S. aureus (VISA), Hetero-vancomycin intermediate S. aureus (hVISA).
The gene clusters vanA and vanB consist of vanX, vanH, vanA, and other related homologous variant genes (29). Activating transcription and regulating cytoplasmic responses is achieved by phosphorylating the vanR protein. Among them are vanR, which upregulates the expression of vanR & S genes, and vanHAX. They bind to the core of vanHAX and vanH which encodes a D-lactate dehydrogenase converting pyruvate into D-lactate and vanX. Finally, the products (D-lactate and vanX) encode a D, D-dipeptidase that further cleaves D-Ala–D-Ala (30). The vanA and B clusters also contain vanY that encodes a D, D-carboxypeptidase. A late membrane-bound peptidoglycan precursor is cleaved at the C-terminus by this enzyme. Vancomycin intermediate susceptible S. aureus (VISA) also contains mutations in the RNA polymerase gene rpoB which regulates transcription of genes in cell wall biosynthesis but mutated genes in the walkR system (walkR system, yvqF/vraSR system) seem to be directly or indirectly related to the biosynthesis or metabolism of those two-component sensory regulatory systems (31). In addition to this, lactamase-encoding genes have also exhibited contributions to the expression of VISA (32). VISA isolates demonstrate several common characteristics such as excessive quantities in the cell wall, abnormal daughter cells, post-cellular division, and reduced rates of autolysis. The altered phenomena behind these molecular mechanisms are yet to be understood due to abnormally high amounts of synthesized peptidoglycan. The activation of the arginine catabolism pathway (via an activated ADI pathway) in VISA strains (Mu50 lineage) has been suggested to provide ATP molecules and ammonia to compensate for increased cell wall biosynthesis. There is a need for further research to investigate this phenomenon globally in different strains of VISA. There was also a decrease in the cross-linking of peptidoglycan strands in the thickened wall, leading to an increase in non-cross-linked D-alanyl-D-alanine side chains. In addition to the binding of vancomycin to DAla-D-Ala, the bound vancomycin also serves as an obstacle to the free molecules found deeper within the structure (33).
The gram-negative bacteria had an additional outer membrane that acted as a barrier for the entry of glycopeptides-like hydrophilic molecules. The higher molecular weight and size of these molecules prevent them from entering through the porins of the outer membrane to reach the target site within the cell wall region. This mechanism results in resistance of gram-negative bacteria against glycopeptides and is called ‘intrinsic resistance'. The main contributor to this intrinsic resistance appears to be an extra outer membrane (OM) and numerous efflux pumps which successfully block the entry of several pharmacological compounds including glycopeptide antibiotics like vancomycin (34). According to Hancock and Brinkman (35) and Pagès et al. (36) hydrophobic medicines passively pass through the OM whereas hydrophilic antibiotics diffuse through porin channels. Glycopeptides have a complicated chemical structure, a large molecular weight, and are hydrophilic by nature (1,450–2,000 Da). They are hydrophilic but because of their enormous size and molecular weight, they cannot pass through the OM's porins to the cell wall region. GNPs inherently resist glycopeptides because they operate within cell walls.
Diseases were non-responsive to the antibiotics intended to be used as growth promotors for better FCR (feed conversion ratio) and reduced morbidity/mortality amid clinical/subclinical infections (37). Avoparcin was mainly used to treat animals including broilers, turkeys, pigs, and veal calves (38). Data from Denmark shows the breadth of avoparcin use where >24,000 kg was used for growth promotion whereas 24 kg of the drug was utilized to treat humans. In Australia, <600 kg of vancomycin and nearly 62,000 kg of avoparcin were purchased during 1992–996 in similar circumstances. Avoparcin provides cross-resistance to vancomycin, hence it is improperly used to treat VRE (38). The most clinically significant species is Enterococcus faecalis and the strains from animals may pose a risk to humans. For instance, it is believed that human VRE outbreaks in some countries were influenced by the rise of vancomycin-resistant enterococci (VRE). This factor is attributed to excessive usage of the vancomycin derivative avoparcin as a growth promoter in farm animals (39). Until it was forbidden in 1997, the widespread use of avoparcin as a growth enhancer in the agricultural sector was linked to the frequent isolation of VRE from farm animals (40). The first antibiotic feed additive used in food-producing animals banned in 1997 by the EU was avoparcin. This antibiotic was banned from use in food-producing animals because it belongs to the glycopeptide class, a critically important antibiotic used in human medicine. Resistant enterococci have also been isolated from the raw meat of animals fed avoparcin, creating a concern for the passage of resistant enterococci to people via the food chain (41). This is what led to the ban of avoparcin as an antibiotic feed additive in the EU.
During the 1990s, E. faecium with the vanA genotype, or VRE, was prevalent in the intestinal flora of farmed animals in Europe (42, 43). There are several explanations for why VRE still exists in livestock animals. Because the genes for both resistances were found on the same plasmid, a study in Denmark suggested the use of a combination of macrolide and tylosin as the solution for VRE (44). In another study, bacteria are forced to maintain resistance if plasmid addiction systems were present on the same plasmid as the vanA gene (45). The three most prevalent types are vanA, vanB, and vanC, and among these the vanA genotype is considered the most abundant (46, 47). A second variety (vanF) has also been discovered even though it has only been found in Paenibacillus popilliae thus far. P. popilliae has been proposed as a potential source of vancomycin resistance in enterococci due to a considerable degree of similarity between amino acid sequences of the vanF variant and vanA variant (48). Other potential origins include a variety of organisms that produce glycopeptides even if there is a more likely development of genetic variances as an earlier common source. The initial D-Ala-D-Ala pentapeptide found within the three-dimensional network of peptidoglycans that makes up the bacterial cell wall may be changed to either D-Ala-D-Lactate (D-Ala-D-Lac) or D-Ala-D-Serine (D-Ala-D-Ser) by all forms of vancomycin resistance in enterococci (49). Modifications to the cell wall's structure, brought on by several genes, are the root cause of all forms of vancomycin resistance no matter how many types, the number and arrangement of these genes remain remarkably stable. The sensor gene vanS activates the regulator gene vanR in the presence of a glycopeptide, vanH facilitates the conversion of pyruvate to lactate when the gene complex is active while vanA utilizes lactate to create the alternative D-Ala-D-Lac end of the pentapeptide. Resistance can only exist if the pentapeptide's typical D-Ala-D-Ala terminus is no longer produced. The vanX and vanY genes, respectively, hydrolyze and stop the formation of the pentapeptides and cleave any pentapeptides that might still grow and provide a solution to this problem (49). VanS starts the process of dephosphorylating vanR when a glycopeptide is absent making the gene inactive. Reportedly vanA-type resistance was the first kind among others that induced highly resistant transposon Tn1546 and the other closely linked factors. The vanA ligase catalyzes the ester bond synthesis between D-Ala and D-Lac while vanH dehydrogenase converts pyruvate to D-Lac (30). As a result, the D-Ala-D-Ala dipeptide is changed into the D-Ala-D-Lac dipeptide which significantly reduces the molecule's affinity for glycopeptides (26).
Lipopolysaccharide (LPS) helps Gram-negative bacteria maintain a strong permeability barrier in their outer membrane, blocking the introduction of toxic substances like antibiotics. The important gene cdsA, which is involved in the transformation of phosphatidic acid into CDP-diacylglycerol during phospholipid biosynthesis, was the target of all seven suppressors that were examined. These cdsA mutations lead to a partial functional impairment as phosphatidic acid accumulation. The fact that these cdsA mutations provide a general rise in vancomycin resistance, even in a wild-type cell, means that this suppression is not limited to mutations that lead to abnormalities in outer membrane biogenesis. Researchers demonstrated that the accumulation of phosphatidic acid by methods other than cdsA mutations also enhances resistance to vancomycin using genetics and quadrupole time of flight (Q-TOF) liquid chromatography-mass spectrometry (LC-MS) (50). The authors hypothesize that elevated amounts of phosphatidic acid alter the outer membrane's physical characteristics to prevent vancomycin from entering the periplasm and reaching its target, an intermediate needed for the production of the peptidoglycan cell wall (50). Like zoonotic bacteria, resistant bacteria from food animals' intestinal flora contaminate animal corpses after being killed and then pass down the food chain to infect humans' intestines (51). It is possible to determine the prevalence of resistance in various populations and to identify a potential transmission of resistant bacteria from animals to individuals, and vice versa, by examining the prevalence of resistance of specific indicator organisms like E. coli and enterococci in the intestinal tract of various populations of animals and humans.
The samples processed for isolation and identification of S. aureus may grow on a mannitol salt agar or, alternatively, an MRSA chromogenic agar. Whatever method of identification is applied, the obtained S. aureus is required to evaluate against vancomycin antibiotic, and, if minimum inhibitory concentration (MIC) is found to be ≥4 μg/mL, it ought to be reported to health departments as reportable isolates. The MIC of vancomycin against S. aureus of ≥8 μg/mL must be sent to CDC for confirmation of VRSA. All the S. aureus isolates showing MIC ≥16 μg/mL against vancomycin are requested by the CDC for further characterization of VRSA precursor organisms. Upon confirmation by the CDC, this is shared with public health partners (52).
The guidelines for declaring different strains of S. aureus with respect to vancomycin are as follows:
1. VSSA (S. aureus sensitive to vancomycin) presenting MIC ≤ 2 μg/mL.
2. VRSA (S. aureus resistant to vancomycin) presenting MIC ≥ 16μg/mL.
3. VISA (S. aureus intermediate susceptible to vancomycin) presenting MIC 4-8 μg/mL.
VRSA was identified in 2002 in a patient hospitalized for a catheter-related infection. The MIC was noted >32 μg/mL and was related to VRE while molecular analysis showed a vanA positive gene of the Van cluster (53). Vancomycin resistance mechanisms are discussed in the following section, including intrinsic resistance seen in Gram-negative bacteria. Although recently some antibiotic alternatives like bacteriophages are being investigated against potential pathogens (54).
According to Hospital Infection Control Practices Advisory Committee (HICPAC), hospitals should develop their institution-specific protocols while the focus should remain as follows (55).
• The clinicians ought to prescribe vancomycin keeping in view the circumstances and consequences.
• Hospital staff should be aware of the vancomycin resistance.
• Detection and prompt reporting of vancomycin resistance in microorganisms from laboratories and hospitals should be early and prompt.
• An appropriate implementation plan for the control of horizontal infection (person to person) of VRE ought to launch.
The CDC suggests decolonizing patients from VRSA to avoid the development of clinical signs and the further spread of resistance. The colonization of VRSA in persons is defined as the presence of VRSA in or on a person but has no development of any clinical signs. The persons may simultaneously colonize with VRSA infection e.g., wound infection, and colonization of VRSA through the colonization of nares. Here it is important to decolonize (reducing the burden of pathogen/organism by eradication strategy). There are approaches suggested for this and among such is decreasing the reservoir of vancomycin-resistant S. aureus (VRSA). Certain factors like the disease situation and status of the immune response, capacity of the person to tolerate dose regimen, and risk of transmission to others make the baseline for decolonization. Such a decision further relies on the consultation of the patient's physician and public authorities (52).
Methicillin-resistant S. aureus is not only a recognized source of infection in hospitals (56) but also outside healthcare setups. The excessive use of vancomycin to treat MRSA increases the selective pressure of vancomycin in the population, leading to higher VISAs and VRSA strains (57). The introduction of food animals into Vancomycin-resistant Enterococci production systems was mostly linked to the widespread use of sub-treatment avoparcin to promote the growth of animals in Europe and other countries since the mid-1990s. Avoparcin was prohibited as a growth promoter in the EU in 1997 as described in Commission Directive 97/6/EC. VRE-related human illnesses and outbreaks, as well as the frequency of VRE in asymptomatic human carriers, have grown throughout the EU since 1999. The prohibition of disinfectant growth promoters may have played an important role in reducing VRE carriers in animals and glycopeptide-resistant Enterococci species. Despite the greater frequency of VRE carriers in animals, fewer nosocomial VRE infections were noted in Europe than in the United States even though avoparcin was never prescribed to North American animals. A decade or two ago, people found it difficult to explain food-borne infections except in a few cases of similar VREs identified from humans and animals which were believed to be spread from animal contact rather than meat intake. Human infections with animal VRE stress resulted in temporary colonization. However, evidence of various molecular typing techniques brought forward a strong correlation between VREs or genetic determinants in animals and humans. These links between animals and human VRE carriers have not been proven to be causal but show that there are several reverse transmissions from wild animals and plants (57).
Antibiotics were initially licensed to be used as growth promoters, while the abrupt ban on their use has compromised health due to critical food animal diseases. On the other hand, this ban has resulted in a reduction of resistance to pathogens of animals and human origin thus enhancing animal health and safety. According to a recent report by Denmark's National Department of Poultry Production, the broiler industry has been “plagued by leg and skin problems” since the late 1990s which could have jeopardized the broiler industry's wellbeing (58).
Food safety concerns about antimicrobial resistance were increased after the ban was implemented. There was a rise in infections which were earlier controlled by the use of medicine in the feed. The utilization of therapeutics in food-producing animals was increased in various parts of Europe. Sales of therapeutic antimicrobials grew from 383 tons in 1999 to 437 tons in 2000 after the EU imposed a ban on growth promoters in 1999. This was due to an increase in tetracycline sales of 36 tons, trimethoprim/sulphonamides sales of 12 tons, and macrolides sales of 12 tons. The use of medicines in pigs increased by 7 tons and use in the poultry sector increased by 13 tons. More than one species of animals showed an increase in the use of medicine by 37 tons. The EU restriction in 1999, as well as the incidents of ailments such as porcine dermatitis, nephritis syndrome, and post-weaning multisystemic wasting syndrome, were factors in the pig business's expansion. Therapeutic antibiotic use in Denmark increased from 48 tons in 1996 to 94 tons in 2001. Tetracycline, which is primarily used in pigs, increased from 12.9 to 27.9 tons (a 116 percent increase) followed by macrolides and lincosamides (7.6 to 14.3 tons, an 88 percent increase), and aminoglycosides (7.1 to 11.9 tons, 68 percent). Experience in Sweden suggests this might be somewhat beneficial in the end, albeit at a greater financial cost. However, it is unknown whether this will apply to the rest of Europe where farming conditions differ from those in Scandinavia (59).
In Europe, 9.6 percent of nosocomial infections were caused by Enterococcus species while VRE nosocomial infections were very frequent in Intensive Care Units (ICU). Serious basic health issues including liver transplants, diabetes mellitus, or kidney failure are risk factors associated with VRE colonization and the subsequent development of nosocomial infections. Several investigations have shown that VRE bloodstream infections were common (BSI) and far more deadly than vancomycin-susceptible enterococci BSI (VSE) (60). VRE infections necessitate higher invasive procedures, increased use of antibiotics, and longer hospitalization that led to both increased hospital expenditure and disease risk. In the United States, hospital expenditure for BSI-like VRE infections ranged from $9,949 to $77,558, as per data gathered from a university-based teaching hospital. Another study conducted in the United States discovered that the expenditure associated with VRE-related surgical site infections (SSI) was approximately $12,766. Despite the expense associated with these preventive measures, VRE screening in elevated areas and VRE client isolation also are well-established in many institutions. A study reported the hospitalization expense of VRE-infected patients was more than VSE-infected individuals. This could be because of factors associated with VRE patients' expenses than VSE patients' expenses. The economic impact of VRE infections in hospitalized patients is now being investigated in the United States and other countries.
Vancomycin resistance has been found as an emerging issue that carries a wider range of resistant genes collectively called the “Van cluster”. These genes are found among a wider range of microbes that finally share these among each other. The transfer of resistance is extensive among animals, humans, and the environment with the full potential of zoonosis and reverse zoonosis. Significant economic damage is possible and food safety, food security, farm animal health, and public health are at risk. The adoption of a set of guidelines from the Centers for Diseases Controls and Prevention (CDC) ought to be implemented with an active reporting system. However, further protocols are required to control this resistance by developing coordinated but comprehensive therapeutic and preventive strategic plans.
AIA wrote the manuscript. AFA revised the manuscript. All authors contributed to the article and approved the submitted version.
The authors would like to thank the Deanship of Scientific Research, Qassim University, Saudi Arabia for funding the publication of this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Anonymous. APVMA. Veterinary drug residues in food commodities and overseas trade 2014. (2017). Availale online at: https://apvma.gov.au/node/669 (accessed May 05, 2017).
2. Reyes K, Zervos M, John J. “Enterococcal infections in adults,” in Antimicrobial Drug Resistance. Clinical and Epidemiological Aspects, eds Mayers DL. Sobel JD, Ouellette M, Kaye KS, Marchaim D. (Berlin: Springer International Publishing). (2017).
3. Alsaedi AA, El-Saed A, Althaqafi A, Bhutta MJ, Abukhzam B, Alshamrani M, et al. Antimicrobial therapy, resistance, and appropriateness in healthcare-associated and community-associated infections; a point prevalence survey. J Infect Chemother. (2022) 28:1358–63. doi: 10.1016/j.jiac.2022.06.003
4. Sulieman AME, Dafallah FE, Rahman EHA, Alshammari NI, Shommo SA, Ibrahim SE. Isolation, Identification and Characterization of Salmonella spp. from Chicken purchased at Wad Madani City, Gezira State, Sudan. Adv Life Sci. (2020) 8:98–102.
5. Elmossalamy DA Hamdy MM Aideia HAM Yassien NA and Zaki HMBA 2020. Incidence of Staphylococcus aureus and its enterotoxins in chicken meat and its products. Int J Vet Sci. 9:573–577. Available online at: http://www.ijvets.com/pdf-files/Volume-9-no-4-2020/573-577.pdf
6. Amer MM, Amer AM, Hassan ER, Ghetas AM. Salmonella Enteritidis in broiler chickens: isolation, antibiotic resistance phenotyping and efficacy of colistin on control of experimental infection. Int J Vet Sci. (2020) 9:267–72. doi: 10.37422/IJVS/20.027
7. Mannan MS, Sabuj AAM, Ray MK, Rabbi F, Bupasha ZB, Sarker MS. Antibiotic resistance of sorbitol non-fermenting Shiga toxin producing Escherichia coli isolated from buffaloes. Int J Vet Sci. (2021) 10:51–4. doi: 10.47278/journal.ijvs/2020.024
8. Elbayoumy MK, Allam AM, Omara ST, Elgabry EA, Abdelgayed SS. Role of artesunate in potentiation of β-lactam against methicillin resistant Staphylococcus aureus (MRSA) isolated from bovine mastitis and its histopathology impact in-vivo study. Int J Vet Sci. (2020) 9:337–42. doi: 10.37422/IJVS/030
9. Altaf M, Ijaz M, Iqbal MK, Rehman A, Avais M, Ghaffar A, et al. Molecularcharacterization of methicillin resistant Staphylococcus aureus (MRSA) and associated risk factors with the occurrence of goat mastitis. Pak Vet J. (2020) 40:1–6. doi: 10.29261/pakvetj/2019.079
10. Betelhem T, Shubisa AL, Bari FD. Isolation, identification and antimicrobial resistance of Staphylococcus aureus isolates from mastitis cases of lactating dairy cows found in Sululta and Holleta Towns, Oromia, Ethiopia. Agrobiological Records. (2022) 8:27–34. doi: 10.47278/journal.abr/2022.006
11. El Jalil MH, Khamar M, Maaninou S, Dahha M, Zinedine A, Ameur N. Antibiotic resistance of Escherichia coli strains isolated from broiler meat in Morocco. Int J Vet Sci. (2020) 9:305–308.
12. Shoaib M, Rahman SU, Aqib AI, Ashfaq K, Naveed A, Kulyar MFeA, et al. Diversified epidemiological pattern and antibiogram of mecA gene inStaphylococcus aureus isolates of pets, pet owners, and environment. Pak Vet J. (2020) 40:331–6. doi: 10.29261/pakvetj/2020.039
13. Sarwar I, Ashar A, Mahfooz A, Aqib AI, Saleem MI, Butt AA, et al. Evaluation of antibacterial potential of raw turmeric, nano-turmeric, and nsaids against multiple drug resistant Staphylococcus aureus and E. coli isolated from animal wounds. Pak Vet J. (2021) 41:209–14. doi: 10.29261/pakvetj/2021.014
14. Parveen S, Saqib S, Ahmed A, et al. Prevalence of mRSA colonization among healthcare-workers and effectiveness of decolonization regimen in ICU of a Tertiary care Hospital, Lahore, Pakistan. Adv Life Sci. (2020) 8:38–41.
15. Riaz S, Hussain A, Sohail M, et al. Isolation and characterization of Vancomycin resistant Staphylococcus aureus (VRSA) from Intensive Care Units (ICU) of different hospitals in Lahore, Pakistan. Adv Life Sci. (2021) 8:339–44.
16. Javed MU, Ijaz M, Fatima Z, Anjum AA, Aqib AI, Ali MM, et al. Frequency and antimicrobial susceptibility of methicillin and vancomycin-resistant Staphylococcus aureus from bovine milk. Pak Vet J. (2021) 41:463–468. doi: 10.29261/pakvetj/2021.060
17. Sova M, Cadez G, Turk S, Majce V, Polanc S, Batson S, et al. Design and synthesis of new hydroxyethylamines as inhibitors of D-alanyl-D-lactate ligase (VanA) and D-alanyl-D-alanine ligase (DdlB) Bioorg Med Chem Lett (2009) 19:1376–9. doi: 10.1016/j.bmcl.2009.01.034
18. Anderson RCGR, Higgins HMJr, Pettinga CD. Symposium: how a drug is born. Cincinnati J Med. (1961) 42:49–60.
19. Yablon SA, Krotenberg R, Fruhmann K. Diarrhea in hospitalized patients. Am J Phys Med Rehabil. (1992) 71:102–7. doi: 10.1097/00002060-199204000-00008
20. Moellering RC. Vancomycin: A 50-year reassessment. Clin Infect Dis. (2006) 42:1. doi: 10.1086/491708
21. Kampmeier S, Kossow A, Clausen LM, Knaack D, Ertmer C, Gottschalk A, et al. Hospital acquired vancomycin resistant enterococci in surgical intensive care patients - a prospective longitudinal study. Antimicrob Resist Infect Control. (2018) 7:103. doi: 10.1186/s13756-018-0394-1
23. Mehmood K, Bilal RM, Zhang H. Study on the genotypic and phenotypic resistance of tetracycline antibiotic in Escherichia coli strains isolated from free ranging chickens of Anhui Province, China. Agrobiological Records. (2020) 2:63–8. doi: 10.47278/journal.abr/2020.014
24. Pehlivanoglu F, Ozturk D, Turutoglu H. Carriage of plasmidic AmpC beta-lactamase producing Escherichia coli in cattle and sheep and characterisation of the isolates in terms of antibiogram profiles, phylogeny and virulence. Kafkas Univ Vet Fak Derg. (2020) 26:469–76.
26. Bugg TD, Wright GD, Dutka-Malen S, Arthur M, Courvalin P, Walsh CT, et al. Molecular basis for vancomycin resistance in Enterococcus faecium BM4147: biosynthesis of a depsipeptide peptidoglycan precursor by vancomycin resistance proteins VanH and VanA. Biochemistry. (1991) 30:10408–15. doi: 10.1021/bi00107a007
27. Evers S. Regulation of VanB-type vancomycin resistance gene expression by the VanSB-VanRB two-component regulatory system in Enterococcus faecalis V583. J Bacteriol. (1996) 178:1302–9. doi: 10.1128/jb.178.5.1302-1309.1996
28. McComas CC, Crowley BM, Boger DL. Partitioning the loss in vancomycin binding affinity for D-Ala-D-Lac into lost H-bond and repulsive lone pair contributions. J Am Chem. Soc. (2003) 125:9314–5. doi: 10.1021/ja035901x
29. Arthur MJ. Regulation of VanA- and VanB-type glycopeptide resistance in enterococci. Antimicrobial Agents Chemother. (2001) 45:375–81. doi: 10.1128/AAC.45.2.375-381.2001
30. Arthur M, Reynolds PE, Depardieu F, Evers S, Dutka-Malen S, Quintiliani R, et al. Mechanisms of glycopeptide resistance in enterococci. J Infect. (1996) 32:116. doi: 10.1016/s0163-4453(96)80003-x
31. Hafer C, Lin Y, Kornblum J, Lowy FD. Contribution of selected gene mutations to resistance in clinical isolates of vancomycin-intermediate staphylococcus aureus. Antimicrobial Agents Chemother. (2012) 56:5845–51. doi: 10.1128/AAC.01139-12
32. Rishishwar L, Petit RA, Kraft CS. Genome sequence-based discriminator for vancomycin-intermediate Staphylococcus aureus. J Bacteriol. (2014) 196:940–8. doi: 10.1128/JB.01410-13
33. Pereira PM, Filipe SR, Tomasz A. Fluorescence ratio imaging microscopy shows decreased access of vancomycin to cell wall synthetic sites in vancomycin-resistant Staphylococcus aureus. Antimicrobial Agents Chemother. (2007) 51:3627–33. doi: 10.1128/AAC.00431-07
34. Zgurskaya HI, López CA, Gnanakaran S. Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infect Dis. (2015) 1:512–522. doi: 10.1021/acsinfecdis.5b00097
35. Hancock REW, Brinkman FSL. Function of Pseudomonas porins in uptake and efflux. Annu Rev Microbiol. (2002) 56:17–38. doi: 10.1146/annurev.micro.56.012302.160310
36. Pagès JM, James CE, Winterhalter M. The porin and the permeating antibiotic: a selective diffusion barrier in Gram-negative bacteria. Nat Rev Microbiol. (2008) 6:893–903. doi: 10.1038/nrmicro1994
37. Butaye P, Devriese LA, Haesebrouck F. Antimicrobial growth promoters used in animal feed: effects of less well known antibiotics on gram-positive bacteria. Clin Microbiol Rev. (2003) 16:17588. doi: 10.1128/CMR.16.2.175-188.2003
38. Bager F, Madsen M, Christensen J, Aarestrup FM. Avoparcin used as a growth promoter is associated with the occurrence of vancomycin-resistant Enterococcus faecium on Danish poultry and pig farms. Prev Vet Med. (1997) 31:95112. doi: 10.1016/S0167-5877(96)01119-1
39. Nilsson O. Vancomycin resistant enterococci in farm animals—occurrence and importance. Infect Ecol Epidemiol. (2012) 2:16959. doi: 10.3402/iee.v2i0.16959
40. Bonten MJ, Willems R, Weinstein RA. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect Dis1. (2001) 314–25. doi: 10.1016/S1473-3099(01)00145-1
41. Chadwick PR, Woodford N, Kaczmarski EB, Gray S, Barrell RA, Oppenheim A. Glycopeptide-resistant enterococci isolated from uncooked meat. J Antimicrobial Chemother. (1996) 38:908–9. doi: 10.1093/jac/38.5.908
42. Aarestrup FM. Occurrence of glycopeptide resistance among Enterococcus faecium isolates from conventional and ecological poultry farms. Microb Drug Resist (Larchmont, NY). (1995) 1:2557. doi: 10.1089/mdr.1995.1.255
43. Klare I, Heier H, Claus H, Reissbrodt R, Witte W. vanA-mediated high-level glycopeptide resistance in Enterococcus faecium from animal husbandry. FEMS Microbiol lett. (1995) 125:16571. doi: 10.1111/j.1574-6968.1995.tb07353.x
44. Aarestrup FM. Characterization of glycopeptide-resistant enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J Clin Microbiol. (2000) 38:27747.
45. Johnsen PJ, Osterhus JI, Sletvold H, Sorum M, Kruse H, Nielsen K, et al. (2005). Persistence of animal and human glycopeptideresistant enterococci on two Norwegian poultry farms formerly exposed to avoparcin is associated with a widespread plasmid-mediated vanA element within a polyclonal Enterococcus faecium population. Appl Environ Microbiol. 71, 15968. doi: 10.1128/AEM.71.1.159-168.2005
46. Fisher K, Phillips C. The ecology, epidemiology and virulence of Enterococcus. Microbiology. (2009) 155:174957. doi: 10.1099/mic.0.026385-0
47. Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. (2008) 13. doi: 10.2807/ese.13.47.19046-en
48. Patel R, Piper K, Cockerill FR 3rd, Steckelberg JM, Yousten AA. The biopesticide Paenibacillus popilliae has a vancomycin resistance gene cluster homologous to the enterococcal VanA vancomycin resistance gene cluster. Antimicrob Agents Chemother. (2000) 44:705–9. doi: 10.1128/AAC.44.3.705-709.2000
49. Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. (2006) 42:S2534. doi: 10.1086/491711
50. Sutterlin HA, Zhang S. Accumulation of phosphatidic acid increases vancomycin resistance in Escherichia coli. J Bacteriol. (2014) 196:3214−20. doi: 10.1128/jb.01876-14
51. Murray BE. Problems and dilemmas of antimicrobial resistance. Pharmacotherapy. (1992) 12:86S−93S.
52. Walters M, Lonsway, D, Rasheed, K, Albrecht, V, McAllister, S, Limbago, B, . Investigation Control of Vancomycin-resistant Staphylococcus aureus: A Guide for Health Departments Infection Control Personnel. Atlanta, GA. (2015). Available online at: http://www.cdc.gov/hai/pdfs/VRSA-Investigation-Guide-05_12_2015.pdf
53. Dhanda G, Sarkar P, Samaddar S. Battle against vancomycin-resistant bacteria: recent developments in chemical strategies. J Med Chem. (2019) 62:3184–205. doi: 10.1021/acs.jmedchem.8b01093
54. Abdurahman MA, Tosun I, Durukan I, Khorshidtalab M, Kiliç AO. Four temperate bacteriophages from methicillin-resistant Staphylococcus aureus show broad bactericidal and biofilm removal activities. Kafkas Univ Vet Fak Derg. (2021) 27:29−36. doi: 10.9775/kvfd.2020.24680
55. Anonymous. Recommendations for Preventing the Spread of Vancomycin Resistance. Emerg Infect Dis. (1995) 1:66–67.
56. Qodrati M, SeyedAlinaghi S, Dehghan Manshadi SA, Abdollahi A, Dadras O. Antimicrobial susceptibility testing of Staphylococcus aureus isolates from patients at a tertiary hospital in Tehran, Iran, 2018-2019. Eur J Med Res. (2022) 27:152.
57. Samad MA, Sagor MS, Hossain MS, Karim MR, Mahmud MA, Sarker MS, et al. High prevalence of vancomycin non-susceptible and multi-drug resistant enterococci in farmed animals and fresh retail meats in Bangladesh. Vet Res Commun. (2022) 46:811–22.
58. Wierup M. The Swedish Experience of the 1986 year Ban of Antimicrobial Growth Promoters, with Special Reference to Animal Health, Disease Prevention, Productivity, and Usage of Antimicrobials. Microbial Drug Resistance. (2001) 7:183–90. Available online at: www.liebertpub.com
59. Hammerum AM, Heuer OE, Emborg HD, Bagger-Skjøt L, Jensen VF, Rogues AM, et al. (2007). Danish integrated antimicrobial resistance monitoring and research program. Emerg Infect Dis. 13:1632–9. doi: 10.3201/eid1311.070421
Keywords: food safety, vancomycin, resistance genes, bacteria, gram-positive, gram-negative
Citation: Aqib AI and Alsayeqh AF (2022) Vancomycin drug resistance, an emerging threat to animal and public health. Front. Vet. Sci. 9:1010728. doi: 10.3389/fvets.2022.1010728
Received: 01 September 2022; Accepted: 21 September 2022;
Published: 28 October 2022.
Edited by:
Isa Ozaydin, Kafkas University, TurkeyReviewed by:
Ahrar Khan, Shandong Vocational Animal Science and Veterinary College, ChinaCopyright © 2022 Aqib and Alsayeqh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abdullah F. Alsayeqh, YS5hbHNheWVxaEBxdS5lZHUuc2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.