- Key Laboratory of Applied Technology on Green-Eco-Healthy Animal Husbandry of Zhejiang Province, College of Animal Science and Technology, College of Veterinary Medicine, Zhejiang A&F University, Hangzhou, China
Promyelocytic leukemia (PML) protein is a crucial component of PML-nuclear bodies (PML-NBs). PML and PML-NBs are involved in the regulation of various cellular functions, including the antiviral immune response. The human PML gene can generate several different isoforms through alternative splicing. However, little is known about the porcine PML alternative splicing isoforms and their expression profiles during Japanese encephalitis virus (JEV) infection. In the present study, we cloned seven mature transcripts of porcine PML, all of which contained the same N-terminal sequence but differed in the C-terminal sequences due to alternative splicing. These seven transcripts encoded five proteins all of which had the RBCC motif and sumoylation sites. Amino acid sequence homology analysis showed that porcine PML-1 had relatively high levels of identity with human, cattle, and goat homologs (76.21, 77.17, and 77.05%, respectively), and low identity with the mouse homolog (61.78%). Immunofluorescence analysis showed that the typical PML-NBs could be observed after overexpression of the five PML isoforms in PK15 cells. Quantitative reverse transcription PCR (RT-qPCR) analysis showed significant upregulation of PML isoforms and PML-NB-associated genes (Daxx and SP100) at 36 and 48 h post-infection (hpi). Western blotting analysis indicated that the PML isoforms were upregulated during the late stage of infection. Moreover, the number of PML-NBs was increased after JEV infection. These results suggest that porcine PML isoforms may play essential roles in JEV infection.
Introduction
Promyelocytic leukemia (PML) protein, also known as TRIM19, is a member of the tripartite motif (TRIM) protein family and was originally identified at the t(15;17) chromosomal translocation in acute promyelocytic leukemia (APL) (1). PML is critical for the formation of PML-nuclear bodies (PML-NBs) in the nuclei of normal cells (2). In APL cells, PML is fused with the retinoic acid receptor alpha (RARα) gene, resulting in the synthesis of chimeric PML-RARα protein (3). This chimeric protein disrupts PML-NBs leading to their microspeckled dispersion and the loss of their original functions (4). Arsenic trioxide (As2O3) is the most effective agent in treatment of APL and acts directly on PML. As2O3 promotes PML-RARα degradation through the SUMO (small ubiquitin-related modifier)-dependent polyubiquitination pathway (5, 6). PML-NBs are dynamic structures. PML maintains the structural stability of PML-NBs and recruits the permanently PML-NB-associated proteins Daxx (death domain-associated protein), SP100 (speckled protein 100), and SUMO, as well as many other proteins transiently residing in the PML-NBs depending on environmental and cellular conditions (2). PML and PML-NBs are involved in multiple biological processes, including tumorigenesis, apoptosis, DNA damage response, and antiviral defense (7). PML and PML-NBs play a vital role in the antiviral response in DNA and RNA virus infection. PML is an intrinsic restriction factor that counteracts viral infection by inhibiting viral replication. However, PML and PML-NBs are targeted and modified by viral proteins to overcome their antiviral activities during infection (8, 9).
Japanese encephalitis virus (JEV) belongs to the family Flaviviridae, and is the causative agent of Japanese encephalitis. JEV is a mosquito-borne virus transmitted by Culex species, which is prevalent in the eastern, southeastern, and southern regions of Asia (10, 11). Pigs are the main amplifying host for JEV. JEV can cause abortion, stillbirth, and weak piglets in pregnant swine, and neurological symptoms in piglets (12, 13). There is currently no effective therapeutic treatment for JEV, and outbreaks of JEV in pigs are difficult to control, with consequent catastrophic economic losses in the swine industry. PML belongs to the TRIM protein family, some members of which, such as TRIM52, have been shown to affect the replication of JEV. The interaction of JEV non-structural protein 2A (NS2A) and TRIM52 results in degradation of NS2A in a proteasome-dependent manner via the E3 ligase activity of TRIM52, which inhibits JEV replication (14). TRIM21 has roles in the regulation of the innate immune response to JEV infection (15). There is accumulating evidence that PML regulates the replication of flavivirus RNA genomes. For example, PML isoforms III and IV, but not the other PML isoforms, repress Dengue virus (DENV) replication. DENV non-structural protein NS5 polymerase complexes with PML III and IV to disrupt PML-NBs, contributing to suppression of the antiviral activity (16). Moreover, PML protein is a driver of anti-Zika virus (ZIKV) intrinsic immunity that inhibits ZIKV infection and replication (17). However, the expression patterns of PML isoforms and the potential roles of PML during JEV infection remain unclear.
Seven PML isoforms generated from a single PML gene via alternative splicing have been identified in humans. These isoforms share a conserved N-terminal domain but differ in the C-terminal domain. The C-terminal ends of the six PML isoforms (PMLI–PMLVI) contain nuclear localization signals (NLS), and are these isoforms are found in the nucleus, but PMLVII, which has a different C-terminus, is located in the cytoplasm (18). Therefore, the differences in the C-terminal sequences result in different biological functions of each PML isoform (19). Recently, based on the predicted cDNA sequences through analysis of the human and swine PML gene sequence, four porcine PML isoforms were cloned by PCR using the cDNAs generated from IFNα-stimulated PK15 cells as templates (20). However, there may be other porcine PML isoforms produced by alternative splicing.
In the present study, we performed molecular cloning and expression analysis of the porcine PML alternative splicing variants during JEV infection. We identified seven alternative splicing variants of porcine PML, all of which contained the same N-terminal sequence but differed in the C-terminal sequences due to alternative splicing. To investigate the role of porcine PML in JEV infection, we analyzed the expression of PML isoforms, Daxx, and SP100 after JEV infection in PK15 cells. The results showed that PML isoforms, Daxx, and SP100 were significantly upregulated during JEV infection. In addition, the number of PML-NBs was increased after JEV infection. These results suggest that porcine PML isoforms may be involved in JEV infection.
Materials and Methods
Cell Culture
PK15 cells obtained from the China Center for Type Culture Collection were cultured in Modified Eagle's Medium (MEM; Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS; HyClone) and 1% non-essential amino acids (Gibco-BRL Life Technologies, Grand Island, NY, USA). The cells were cultured in an incubator at 37°C and 5% CO2.
3′ and 5′ Race Assay
Reverse transcription of RNA to make cDNA for Rapid Amplification of cDNA Ends (RACE) assays was performed using a SMARTer® RACE 3′/5′ Kit (Takara, Dalian, China) according to the manufacturer's instructions. The reactions consisted of 1 μL RNA (1 μg/μL), 1 μL 3′-CDS Primer A (12 μM/L), and 10 μL sterile H2O (3′ RACE) or 1 μL RNA (1 μg/μL), 1 μL 5′-CDS Primer A (12 μM/L), and 9 μL sterile H2O (5′ RACE). The solution was mixed, centrifuged, and incubated at 72°C for 3 min. The tubes were cooled to 42°C for 2 min. For the 5′ RACE cDNA synthesis reaction, 1 μL SMARTer II A Oligonucleotide (24 μM/L) was added. Then, 4 μL 5 × First-Strand Buffer, 0.5 μL 100 μmol/L DTT, 20 U of RNase Inhibitor, and 200 U of SMARTScribe Reverse Transcriptase were added. The reaction was performed in a thermal cycler (T100; Bio-Rad, Hercules, CA, USA) for 90 min at 42°C, followed by heat inactivation for 10 min at 70°C. Samples were stored at −20°C for subsequent use.
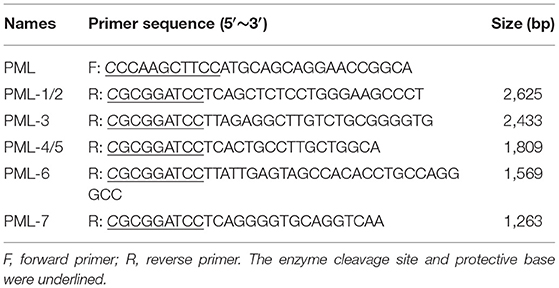
The 3′ and 5′ RACE gene-specific primers (GSPs) of the first-step PCR amplification were designed based on the conserved regions of porcine PML sequences in NCBI (GenBank Accession No: XM_001925572.5) (Table 1). The reaction consisted of 10 μL 3′ or 5′ RACE-Ready cDNA, 50 μL 2 × Vazyme LAmp Master Mix (Vazyme, Nanjing, China), 2.5 μL 10 × Universal Primer (UPM) long (10 μM/L), 2.5 μL 3′ Race-GSP (10 μM/L) or 5′ Race-GSP (10 μM/L), and 35 μL ddH2O. The PCR consisted of 5 min at 94°C followed by 35 cycles of 94°C for 30 s, 62°C for 30 s, and 72°C for 2 min, with a final step of 72°C for 5 min.
If the primary PCR failed to yield the distinct bands of interest, nested PCR was used. The reaction consisted of 30 μL the 3′ RACE first-step PCR product, 50 μL 2 × Vazyme LAmp Master Mix, 2.5 μL 10 × UPM short (10 μM/μL), 2.5 μL 3′ Race-NGSP (10 μM/μL), and 35 μL ddH2O.
Cloning and Sequencing
The products of 5′ RACE first-step PCR or 3′ RACE nested PCR were ligated with pMD19-T vector, and the recombinant vector was transformed into DH5α competent cells. We selected positive clones for sequence analysis. Sequencing results were assembled using DNAStar 8.0 software (DNAStar Inc., Madison, WI, USA). The sequences were compared to the published PML gene sequence, and the splicing and retention of exons and introns to obtain full-length cDNA sequences of alternative splicing variants were analyzed.
Sequence Analysis
DNAMAN 5.0 software (Lynnon Biosoft, Vandreuil, QC, Canada) was used for amino acid sequence alignment. The amino acid sequences of Homo sapiens (GenBank Accession No: NP_150241.2), Mus musculus (GenBank Accession No: NP_835188.2), Bos taurus (GenBank Accession No: XP_005221963.1), and Capra hircus (GenBank Accession No: XP_005695206.1) PML were downloaded from NCBI (https://www.ncbi.nlm.nih.gov). The open reading frames (ORFs) of the PML isoforms were analyzed using ORFfinder (https://www.ncbi.nlm.nih.gov/orffinder). Analyses of the conserved domains were performed using CD-Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de). The NLS was predicted using NucPred (https://nucpred.bioinfo.se/nucpred). The small ubiquitin-like modifier modification (sumoylation) sites were predicted using SUMOsp 2.0 software (http://sumosp.biocuckoo.org).
Plasmid Construction and Transfection
PML isoforms were amplified from cDNA of PK15 cells with the same forward primer and corresponding reverse primers (Table 2). The amplified fragments (PML-1/2, PML-3, PML-4/5, PML-6, and PML-7) were inserted into the pEGFP-C1 vector. PK15 cells were cultured in 12-well plates. The five PML recombinant eukaryotic vectors were transfected into the cells using Lipofectamine 3000 transfection reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. At 36 h post-transfection, cells were harvested for RT-qPCR and fluorescence analysis.
Immunofluorescence Analysis
PK15 cells were washed three times and fixed with 4% paraformaldehyde for 20 min at room temperature. Then the cells were permeabilized with 0.25% Triton X-100 for 15 min. After blocking with PBS containing 5% bovine serum albumin (BSA), the cells were stained with rabbit anti-PML polyclonal antibody (1:100; GeneCreate, Wuhan, China) at room temperature for 1 h. After washing three times with PBS, the cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit antibody (A-11008, 1:1,000; Invitrogen, Carlsbad, CA, USA). The nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI).
Viral Infection
PK15 cells were cultured in 12-well cell culture plates, and infected with JEV strain SA14–14–2 (MOI = 1) diluted in MEM (21). The infected cells were washed twice with PBS after 1 h of adsorption, and then maintained in MEM with 2% FBS. The infected cells were collected for RNA isolation at the indicated time points after infection.
RT-qPCR Assay
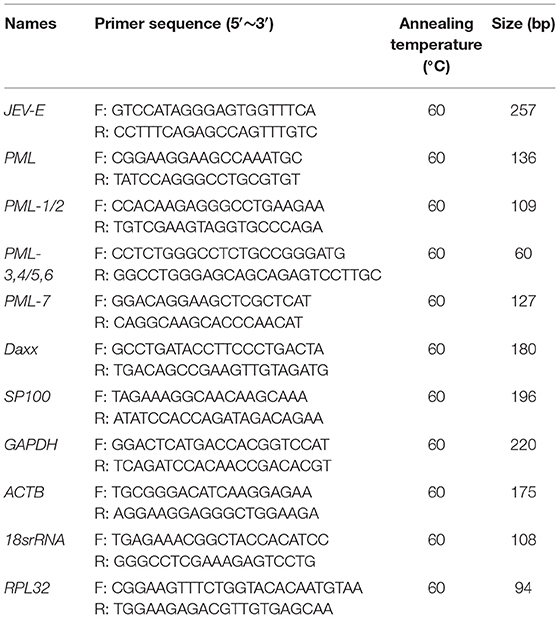
Total RNA was extracted using TRIzol reagent (Invitrogen). Total RNA (1 μg) was used to synthesize cDNA with an RT-PCR reagent kit (CWBiotech, Beijing, China). Quantitative analysis was performed using SYBR Premix Ex Taq II (Takara) on a CFX96 Touch instrument (Bio-Rad). The sequences of the qPCR primers used in the present study are listed in Table 3. The following PCR cycling conditions were used: denaturation at 95°C for 2 min, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s, and 72°C for 20 s, and then melting curve analysis. Relative expression levels were calculated using the 2−ΔΔCt method and GAPDH, ACTB, 18srRNA, and RPL32 as reference genes (22).
Western Blotting
Total proteins of JEV-infected and uninfected PK15 cells were extracted using RIPA lysis buffer with phosphatase and protease inhibitor (CWBiotech) respectively. Then the cell lysates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After blocking with 5% non-fat milk for 1 h, the membranes were incubated with rabbit anti-PML polyclonal antibody (1:1,000; GeneCreate) and rabbit anti-JEV (NS3) polyclonal antibody (GTX125868, 1:5,000; GeneTex, Irvine, CA, USA) overnight at 4°C. After washing three times, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (A21020; 1:10,000; Abbkine, Wuhan, China) for 1 h at room temperature. Then the membranes were imaged using a Tanon 5200 system (Biotanon, Shanghai, China) after treatment with HRP Substrate (Merck Millipore, Darmstadt, Germany). β-Actin was used as an internal control.
Statistical Analysis
All data are presented as the mean ± SEM of three independent experiments. Statistical significance was assessed using Student's t test. In all analyses, p < 0.05 was taken to indicate statistical significance.
Results
Identification and Cloning of 3′ and 5′ Porcine PML Gene cDNA Sequences
cDNA for RACE assay was amplified from PK15 cells by RT-PCR. The products of the first cycle (using 3′ Race-GSP and 5′ Race-GSP) and the second cycle of nested PCR (using 3′ Race-NGSP) were analyzed by agarose gel electrophoresis (Figure 1). As shown in Figure 1, specific bands were obtained from 3′ RACE nested PCR and 5′ RACE first-step PCR, and the PCR products were cloned into the pMD19-T vector. After transformation into DH5α cells, positive clones were selected and sequenced. From the sequencing and alignment results, we identified seven types of PML cDNA clone, which we designated, from longest to shortest, as PML-1, PML-2, PML-3, PML-4, PML-5, PML-6, and PML-7 (Figure 2A).
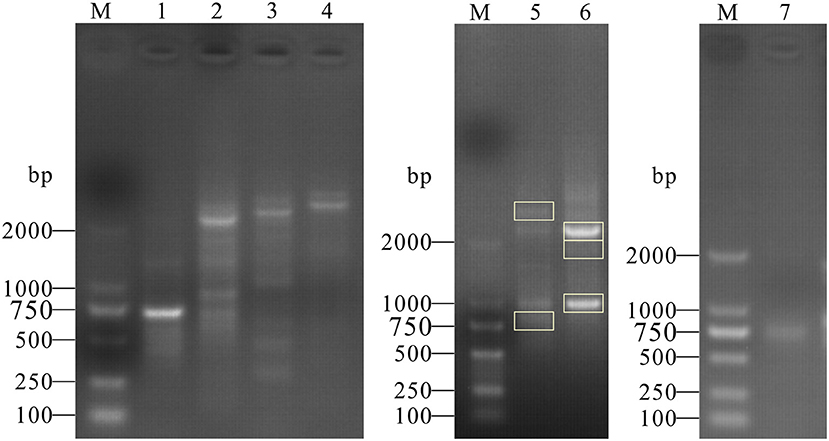
Figure 1. PCR products of 3′ and 5′ RACE of the porcine PML gene. M: DL2000 Marker; the marker fragment sizes were 100, 250, 500, 750, 1,000, and 2,000 bp. Lanes 1, 2, 3, 4: PCR products of the first 3′ RACE cycle. Labels on the lanes represent different groups of 3′ RACE primers (3′ Race-GSP1, GSP2, GSP3, and GSP4). Lanes 5, 6: PCR products of the second 3′ RACE cycle (3′ Race-GSP2 and 3′ Race-GSP3 first-step PCR products as 3′ RACE-Ready cDNA), and gel bands in the rectangular box were selected for cloning analysis. Lane 7: PCR products of the first 5′ RACE cycle.

Figure 2. Structure and sequence analysis of porcine PML isoforms. (A) The lengths of seven PML isoforms (PML-1 to PML-7) encoded by the different mRNA variants and their exon compositions from the translational start codons to the stop codons of the mRNA are shown. The full-length genomic sequence of PML includes nine exons (1–9). Alternative splicing of the PML gene leads to seven mRNA variants. Exon length: exon 1, 159 bp; exon 2, 467 bp; exon 3, 581 bp; exon 4, 71 bp; exon 5, 120 bp; exon 6, 256 bp; exon 7, 53 bp; exon 8, 151 bp; exon 9, 1,773 bp. (B) Conserved domain analysis of PML-1. (C) Sus scrofa, Homo sapiens, Bos taurus, Capra hircus, and Mus musculus PML-1 amino acid sequence alignment was performed with DNAMAN 5.0 software. Identical amino acids are indicated by a black background, and amino acids with substitutions are shown in light gray.
Structural Analysis of Seven Alternative Splicing Variants of Porcine PML Gene
Sequencing results were assembled using DNAStar 8.0 software (DNAStar Inc.), and the gene structures of the seven cloned different isoforms of the porcine PML gene are shown in Figure 2A. The full-length mRNA sequence of PML (PML-1) was 3,659 bp in length, and consisted of nine exons. The coding DNA sequence (CDS) of PML-1 was 2,625 bp and encoded 874 amino acids. The 3′ untranslated region (UTR) was 1,009 bp, and the 5′ UTR was much shorter at only 25 bp. PML-2 also encoded a product of 874 amino acids, but the 3′ UTR of PML-2 only retained 400 bp of the PML-1 3′ UTR. PML-3 contained exons 1–7 and retained part of the intron 7 sequence (7b: 641–1,788 bp), and encoded a product of 810 amino acids. PML-4 contained exons 1–7 and retained the partial sequence of intron 7 (7a: 1–1,785 bp), and encoded a product of 602 amino acids. PML-5 also contained exons 1–7 and retained part of the intron 7 sequence (7c: 1–434 bp, 7d: 1,245–1,788 bp). PML-6 contained exons 1–5 and retained a partial sequence of exon 6 (at 1–109 bp) and intron 7 (7e: 1,685–1,781 bp). PML-6 encoded a product of 522 amino acids. PML-7 contained exons 1–4 and retained a partial sequence of intron 4 (4a: 540–865 bp), and it encoded a product of only 420 amino acids. The PML-1 and PML-3 alternative splicing variants cloned here were the same as sPML-I and sPML-II cloned in a previous study (20).
All of the PML isoforms shared exons 1–4 and contained the same N-terminal domain but different C-terminal domains. The PML gene is a member of the TRIM gene family, and is also called TRIM19. Therefore, the first three exons of the PML isoforms contained the RING zinc finger, two B-box domains, and a coiled-coil motif (Figure 2B). Like other mammals, porcine PML-1 to PML-6 proteins included three sumoylation sites (at residues 65, 160, and 482), and PML-7 contained two sumoylation sites (at residues 65 and 160). Exon 6 encoded the NLS (Figure 2B). Therefore, six nuclear PML isoforms PML-1 to PML-6 were localized in the nucleus, while PML-7 was localized in the cytoplasm.
Sequence Alignment
To understand the evolutionary characteristics of the PML gene, we selected the porcine PML-1 protein and the longest PML isoform of human (H. sapiens), cattle (B. taurus), goat (C. hircus), and mouse (M. musculus) for amino acid sequence alignment using DNAMAN 5.0 software (Figure 2C). The amino acid sequence of porcine PML-1 showed 76.21, 77.17, 77.05, and 61.78% identity those of human, cattle, goat, and mouse, respectively. The identity of the PML gene varied highly at the N- and C-terminals. The RBCC motif and three sumoylation sites were very highly conserved. These results indicate that the PML protein was moderately conserved in these species and the functional domains were highly conserved.
Effects of Porcine PML Isoforms on PML-NB Distribution
PML is a critical component of PML-NB formation, and the PML-NBs exhibit a dot-like structure in immunofluorescence images (23). To determine the roles of the seven PML isoforms in the formation and distribution of PML-NBs, PML isoforms were overexpressed in PK15 cells by transfection with the recombinant eukaryotic vectors pEGFP-C1-PML1/2, PML3, PML4/5, PML6, and PML7, respectively. The transfection efficiency was determined by the expression of enhanced green fluorescent protein (EGFP). The transfection efficiency reached approximately 30%, as demonstrated by EGFP expression (Figure 3A). The expression of PML isoforms was examined by RT-qPCR. Compared to that of cells transfected with pEGFP-C1 as an empty vector control, PML was significantly overexpressed in pEGFP-C1-PML isoform-transfected cells (Figure 3B). Further, the typical PML-NBs could be observed after overexpression of the five PML isoforms in PK15 cells (Figure 3C). PML-1/2, PML-3, PML-4/5, and PML-6 formed typical PML-NBs in the nucleus, and the EGFP-PML fusion proteins were also expressed in the cytoplasm. However, PML-7 formed PML-NBs that were only observed in the cytoplasm.
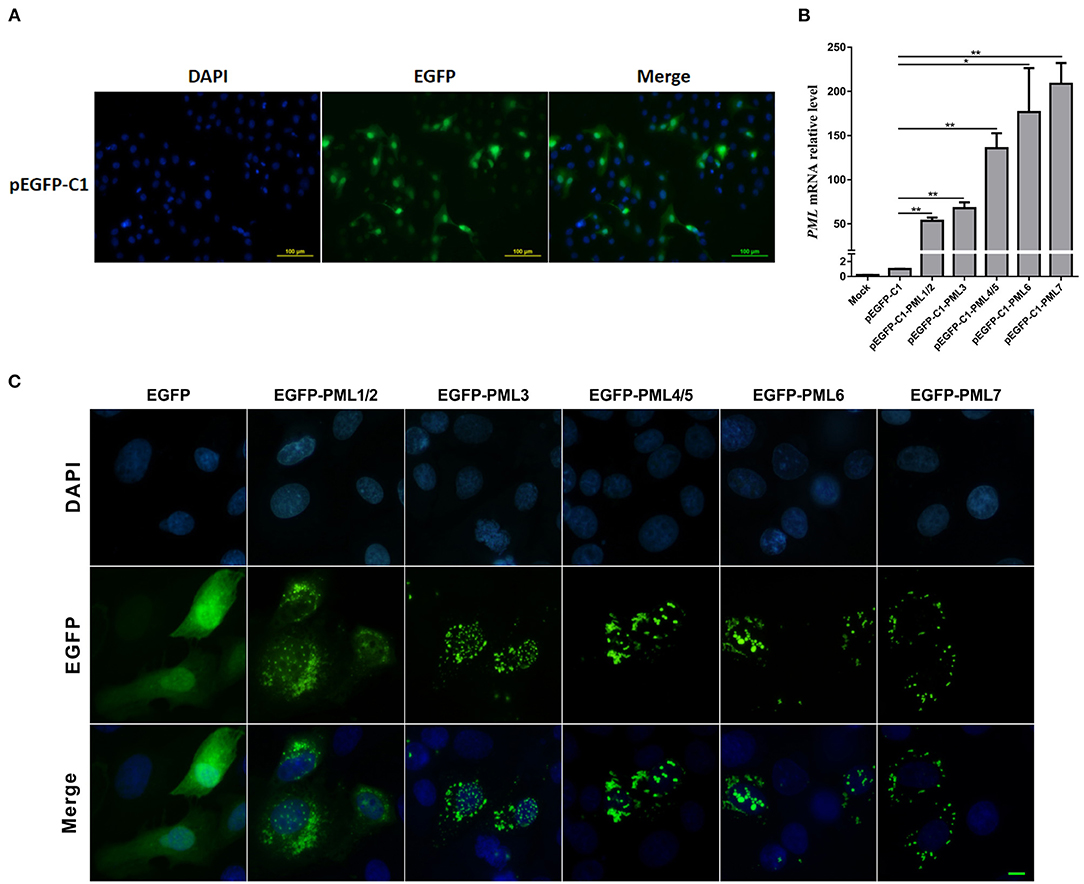
Figure 3. Subcellular distribution of PML-NBs in PK15 cells after transfection of pEGFP-C1-PML isoform recombinant plasmids. PK15 cells were transfected with pEGFP-C1-PML1/2, PML3, PML4/5, PML6, and PML7 plasmids for 36 h. (A) Fluorescence analysis of enhanced green fluorescent protein (EGFP) in PK15 cells transfected with the empty vector (pEGFP-C1). Nuclei were stained with DAPI (blue). (B) RT-qPCR analysis of the expression levels of PML isoforms after transfection with pEGFP-C1-PML1/2, PML3, PML4/5, PML6, and PML7. *p < 0.05, **p < 0.01. (C) The subcellular distribution of PML-NBs in PK15 cells overexpressing different PML isoforms. Scale bar: 10 μm.
Expression of PML Isoforms During JEV Infection
PML and PML-NBs play important roles in the innate immune response and antiviral defense (9). To investigate the porcine PML gene expression patterns after JEV infection, PK15 cells were infected with JEV, and the cells were harvested at the indicated time points. Viral replication was monitored by RT-qPCR, and the viral mRNA in PK15 cells peaked at 36 h post-infection (hpi) (Figure 4A). The expression of PML isoforms and PML-NB-associated genes (Daxx and SP100) were examined (Figure 4A). JEV infection significantly upregulated the mRNA expression of PML isoforms, Daxx and SP100 at 36 and 48 hpi. Among them, the PML-1/2 isoform expression was upregulated by 48-fold at 36 hpi. The PML protein levels were examined by Western blotting analysis using a polyclonal antibody against the N-terminal shared amino acids of the PML isoforms (Figure 4B). PML isoforms were upregulated at 36 and 48 hpi. To further determine the characteristics of the PML-NBs in JEV-infected PK15 cells, the cells were infected with JEV for 36 h and then we performed immunofluorescence analysis of the infected cells using anti-PML polyclonal antibody (Figure 4C). Compared to non-infected cells, the number of PML-NBs was significantly increased in JEV-infected cells, with approximately 20–30 PML-NBs in each cell. These results suggest that PML and PML-NBs may play important roles in JEV infection.
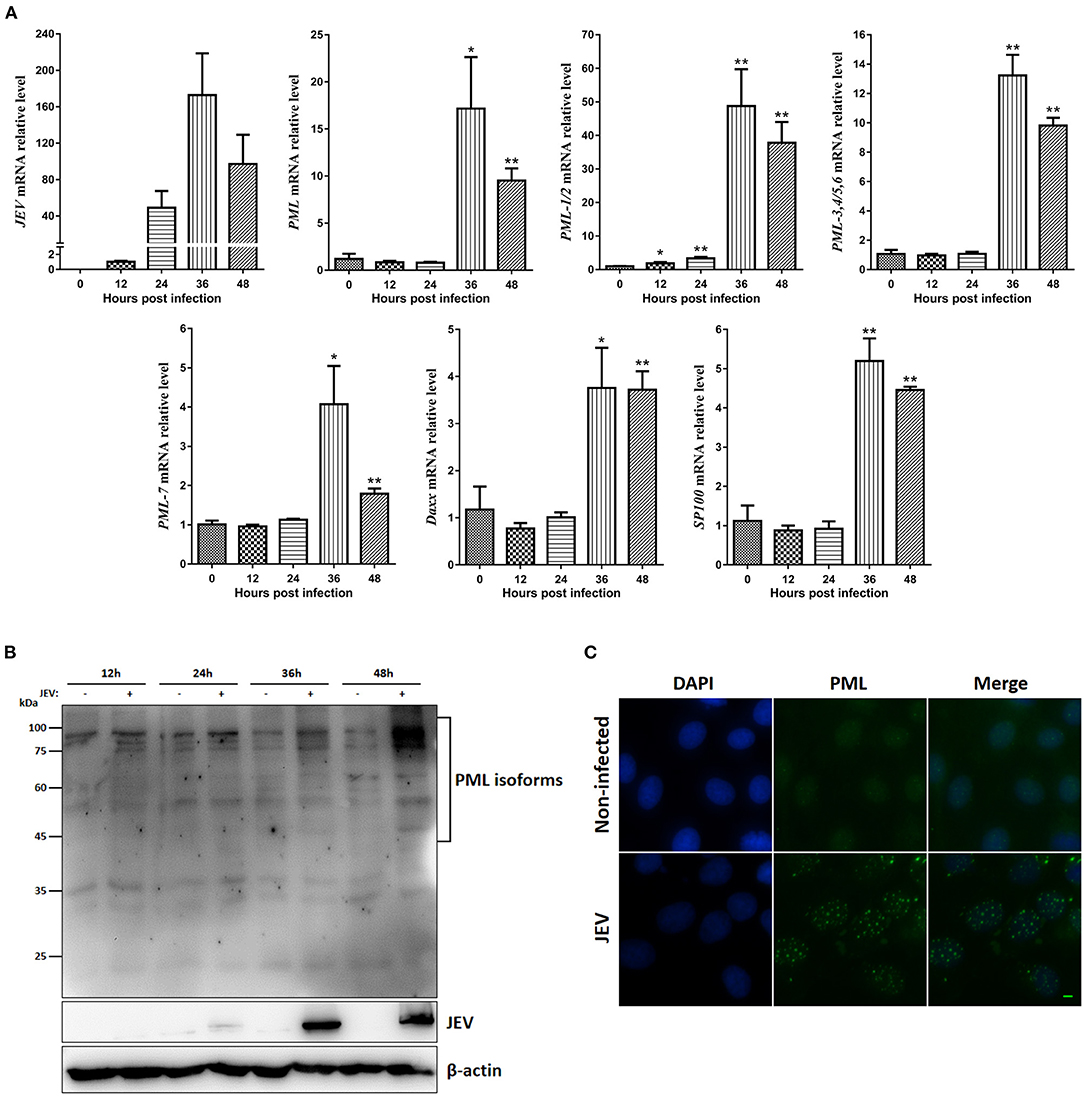
Figure 4. Expression of PML isoforms, Daxx, SP100, and PML-NBs during JEV infection. (A) JEV-E, PML isoform, Daxx, and SP100 mRNA levels were examined by RT-qPCR. GAPDH, ACTB, 18srRNA, and RPL32 levels were used as loading controls, and the relative expression levels were expressed as fold change relative to mock-infected cells. *p < 0.05, **p < 0.01. (B) Western blotting analysis of PML protein expression during JEV infection at the indicated time points. (C) PK15 cells were infected with JEV for 36 h, and then immunofluorescence staining of PML protein (green) was performed using anti-PML antibody. Nuclei were stained with DAPI (blue). Non-infected cells were used as controls. Scale bar: 10 μm.
Discussion
PML has multiple roles in biological processes, including antiviral defense (9). The human PML gene contains nine exons and expresses several mature transcripts via alternative splicing, yielding at least seven main PML isoforms (24). In the present study, we cloned the porcine PML gene and found seven PML mRNAs by 3′ and 5′ RACE assays. As alternative splicing of the PML gene primarily occurred at the C-terminus, PCR primers were designed to amplify the C-terminus of PML transcripts with a RACE kit according to the manufacturer's instructions. We obtained several fragments by gel electrophoresis, and five fragments were selected for sequence analysis. We ultimately obtained seven mature transcripts using sequence assembly with the 5-terminal amplified fragment. As we only selected five fragments with obviously amplified bands via gel electrophoresis for sequencing, additional PML isoforms may have been identified had we sequenced all of the amplified fragments.
We named the longest PML mRNA with nine exons PML-1. Notably, PML-2 contained a shorter exon 9 but encoded the same protein as PML-1. That is, PML-2 had a shorter 3′ UTR compared to PML-1. More miRNAs may regulate PML-1 via the 3′ UTR compared to PML-2. Alternative splicing of PML-3 to PML-6 was located in intron 7 and excluded exons 8 and 9. This feature of splicing patterns is similar to human PML. Human PML-II, PML-III, PML-V, and PML-VI also retain partial sequences of intron 7 (25). These results indicate that the splicing patterns of the PML gene were conserved in different species. Similar to PML-2, PML-4 and PML-5 encoded the same protein with a variable 3′ UTR. These two mRNAs may participate in different signal regulation processes. These PML isoforms all had the RBCC motif, and PML is a member of the TRIM family. PML-1 to PML-6 had the NLS, and they were nuclear proteins. PML-7 lacked the NLS, and was likely cytoplasmic. Similarly, PML-I to PML-VI in humans are present in the nucleus, while PML-VII is a cytoplasmic form (23, 25). PML sumoylation is crucial for the formation of PML-NBs (26). There were three sumoylation sites in PML-1 to PML-6, which were highly conserved in different species based on protein sequence alignment analysis. However, further studies are required to determine the functional roles of sumoylation of porcine PML.
PML protein was mainly localized in the PML-NBs in cells overexpressing each of the seven human isoforms (16, 27). This study identified the subcellular distribution of porcine PML isoforms by overexpressing different EGFP-fused PML isoforms. PML-1/2, PML-3, PML-4/5, and PML-6 contained the NLS, and the typical PML-NBs were found in the nucleus. These PML isoforms were also localized in the cytoplasm. The cellular localization of PML-NBs is dynamic due to the shuttling of PML-associated components, which is determined by specific PML isoforms (28, 29). We speculate that these isoforms are the critical shuttle proteins in the PML-NB reformation process. In addition, PML-7 was localized in the cytoplasmic PML-NBs due to its loss of the NLS (30). The NLS is essential for PML-NBs nuclear localization. The PML-NBs were only distributed in the cytoplasm when the NLS sequence was mutated (27).
There is accumulating evidence that the PML protein and PML-NBs have antiviral effects. For example, PML-NBs epigenetically modify the genomes of human herpes simplex virus type 1 (HSV-1) and human cytomegalovirus (HCMV), resulting in silencing of the viral genomes during the early stages of virus infection (9). PML-IV protein interacts with varicella-zoster virus (VZV) capsid protein and impedes the release of virus particles from the nuclei (31). Another study showed that only PML-III and PML-IV inhibited vesicular stomatitis virus (VSV), and PML-IV promoted the expression of IFN-β by enhancing IRF3 activity (32). The expression levels of PML isoforms, Daxx, and SP100 were significantly upregulated at 36 and 48 hpi. These expression trends were consistent with the viral growth curve, and the JEV viral particles peaked at 36 and 48 hpi (33). These results suggest that the PML-NB-associated proteins (PML isoforms, Daxx, and SP100) may play important roles in defense against JEV infection. In addition, the number of PML-NBs was significantly increased after JEV infection, suggesting that the components and morphology of PML-NBs vary in response to virus infection (9, 34). The results of Western blotting analyses showed that several PML isoforms were induced during JEV infection. The functions of the seven human PML isoforms are different, and further studies are required to determine the potential roles of these porcine PML isoforms and PML-NBs in JEV infection.
In summary, we cloned seven porcine PML alternative splicing variants, whose isoforms shared a common N-terminus and differed in the C-terminus. These seven alternative splicing variants encoded five proteins. Sequence alignment analysis showed that the RBCC and three sumoylation sites were highly conserved in different species. The expression levels of PML isoforms, Daxx, and SP100 were significantly upregulated, and the number of PML-NBs was increased during JEV infection. Further studies are required to determine the functions of these PML isoforms in JEV infection.
Data Availability Statement
The datasets generated and analyzed are available in the GenBank repository with the accession numbers PML-1 (MW490598), PML-2 (MW490599), PML-3 (MW490600), PML-4 (MW490601), PML-5 (MW490602), PML-6 (MW490603), and PML-7 (MW490604).
Author Contributions
SY conceived and designed the experiments. JZ, ZC, and ZD performed the experiments. XZ, HW, and XL analyzed the data. AZ and SY helped to write the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (LY19C170002, LY19C170003), the Zhejiang Provincial Key Research and Development Program (2021C04034), and the National Undergraduate Training Program for Innovation and Entrepreneurship (201910341043).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
As2O3, arsenic trioxide; Daxx, death domain-associated protein; DENV, Dengue virus; GSP, gene-specific primer; hpi, hours post-infection; JEV, Japanese encephalitis virus; NGSP, nested gene-specific primer; NLS, nuclear localization signal; PML, promyelocytic leukemia; PML-NBs, promyelocytic leukemia nuclear bodies; RARα, retinoic acid receptor alpha; RT-qPCR, quantitative reverse transcription PCR.; SP100, speckled protein 100; SUMO, small ubiquitin-related modifier; TRIM, tripartite motif; UTR, untranslated region; ZIKV, Zika virus.
References
1. Kakizuka A, Miller WH, Umesono K, Warrell RP, Frankel SR, Murty VV, et al. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. (1991) 66:663–674. doi: 10.1016/0092-8674(91)90112-C
2. Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. (2010) 6:51–67. doi: 10.7150/ijbs.6.51
3. Maimaitiyiming Y, Wang C, Xu S, Islam K, Chen YJ, Yang C. et al. Role of arsenic (+3 oxidation state) methyltransferase in arsenic mediated APL treatment: An in vitro investigation. Metallomics. (2018) 10:828–37. doi: 10.1039/C8MT00057C
4. Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. (2006) 24:331–9. doi: 10.1016/j.molcel.2006.09.013
5. Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB. et al. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. (2010) 328:240–3. doi: 10.1126/science.1183424
6. Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L. et al. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. (2008) 10:547–55. doi: 10.1038/ncb1717
7. Hsu KS, Kao HY. PML: Regulation and multifaceted function beyond tumor suppression. Cell Biosci. (2018) 8:5. doi: 10.1186/s13578-018-0213-7
8. Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. (2011) 31:145–58. doi: 10.1089/jir.2010.0111
9. Scherer M, Stamminger T. Emerging role of PML nuclear bodies in innate immune signaling. J Virol. (2016) 90:5850–4. doi: 10.1128/JVI.01979-15
10. Chai C, Palinski R, Xu Y, Wang Q, Cao S, Geng Y, et al. Aerosol and contact transmission following intranasal infection of mice with Japanese encephalitis virus. Viruses. (2019) 11. doi: 10.3390/v11010087
11. Solomon T. Control of Japanese encephalitis—within our grasp? N Engl J Med. (2006) 355:869–71. doi: 10.1056/NEJMp058263
12. Ruget AS, Beck C, Gabassi A, Trevennec K, Lecollinet S, Chevalier V. et al. Japanese encephalitis circulation pattern in swine of northern Vietnam and consequences for swine's vaccination recommendations. Transbound Emerg Dis. (2018) 65:1485–92. doi: 10.1111/tbed.12885
13. Chai C, Wang Q, Cao S, Zhao Q, Wen Y, Huang X, et al. Serological and molecular epidemiology of Japanese encephalitis virus infections in swine herds in China, 2006–2012. J Vet Sci. (2018) 19:151–5. doi: 10.4142/jvs.2018.19.1.151
14. Fan W, Wu M, Qian S, Zhou Y, Chen H, Li X. et al. TRIM52 inhibits Japanese encephalitis virus replication by degrading the viral NS2A. Sci Rep. (2016) 6:33698. doi: 10.1038/srep33698
15. Manocha GD, Mishra R, Sharma N, Kumawat KL, Basu A, Singh SK. Regulatory role of TRIM21 in the type-I interferon pathway in Japanese encephalitis virus-infected human microglial cells. J Neuroinflammation. (2014) 11:24. doi: 10.1186/1742-2094-11-24
16. Giovannoni F, Ladelfa MF, Monte M, Jans DA, Hemmerich P, García C. Dengue non-structural protein 5 polymerase complexes with promyelocytic leukemia protein (PML) isoforms III and IV to disrupt PML-nuclear bodies in infected cells. Front Cell Infect Microbiol. (2019) 9:284. doi: 10.3389/fcimb.2019.00284
17. Giovannoni F, Bosch I, Polonio CM, Torti MF, Wheeler MA, Li Z, et al. AHR is a Zika virus host factor and a candidate target for antiviral therapy. Nat Neurosci. (2020) 23:939–951. doi: 10.1038/s41593-020-0664-0
18. Lallemand-Breitenbach V, de Thé HPML. nuclear bodies: From architecture to function. Curr Opin Cell Biol. (2018) 52:154–61. doi: 10.1016/j.ceb.2018.03.011
19. Ha JS, Do YR Ki CS, Lee C, Kim DH, Lee W. et al. Identification of a novel PML-RARG fusion in acute promyelocytic leukemia. Leukemia. (2017) 31:1992–5. doi: 10.1038/leu.2017.167
20. Yu C, Xu A, Lang Y, Qin C, Wang M, Yuan X, et al. Swine promyelocytic leukemia isoform II inhibits pseudorabies virus infection by suppressing viral gene transcription in promyelocytic leukemia nuclear bodies. J Virol. (2020) 94:e01197-20. doi: 10.1128/JVI.01197-20
21. Yang S, Pei Y, Li X, Zhao S, Zhu M, Zhao A. miR-124 attenuates Japanese encephalitis virus replication by targeting DNM2. Virol J. (2016) 13:105. doi: 10.1186/s12985-016-0562-y
22. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
23. Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. (2007) 89:819–30. doi: 10.1016/j.biochi.2007.01.004
24. Nisole S, Maroui MA, Mascle XH, Aubry M, Chelbi-Alix MK. Differential roles of PML isoforms. Front Oncol. (2013) 3:125. doi: 10.3389/fonc.2013.00125
25. Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. (2007) 8:1006–16. doi: 10.1038/nrm2277
26. Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T. et al. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J Cell Biol. (1999) 147:221–34. doi: 10.1083/jcb.147.2.221
27. Jul-Larsen A, Grudic A, Bjerkvig R, Bøe SO. Subcellular distribution of nuclear import-defective isoforms of the promyelocytic leukemia protein. BMC Mol Biol. (2010) 11:89. doi: 10.1186/1471-2199-11-89
28. Luciani JJ, Depetris D, Usson Y, Metzler-Guillemain C, Mignon-Ravix C, Mitchell MJ. et al. PML nuclear bodies are highly organised DNA-protein structures with a function in heterochromatin remodelling at the G2 phase. J Cell Sci. (2006) 119:2518–31. doi: 10.1242/jcs.02965
29. Grobelny JV, Godwin AK, Broccoli D. ALT-associated PML bodies are present in viable cells and are enriched in cells in the G(2)/M phase of the cell cycle. J Cell Sci. (2000) 113 Pt 24:4577–85. doi: 10.1242/jcs.113.24.4577
30. Jensen K, Shiels C, Freemont PS. PML protein isoforms and the RBCC/TRIM motif. Oncogene. (2001) 20:7223–33. doi: 10.1038/sj.onc.1204765
31. Reichelt M, Wang L, Sommer M, Perrino J, Nour AM, Sen N. et al. Entrapment of viral capsids in nuclear PML cages is an intrinsic antiviral host defense against varicella-zoster virus. PLoS Pathog. (2011) 7:e1001266. doi: 10.1371/journal.ppat.1001266
32. El Asmi F, Maroui MA, Dutrieux J, Blondel D, Nisole S, Chelbi-Alix MK. Implication of PMLIV in both intrinsic and innate immunity. PLoS Pathog. (2014) 10:e1003975. doi: 10.1371/journal.ppat.1003975
33. Du C, Wang H, Yang S, Li X, Zhou X, Zhao A. Study on the role of lncRNA in the process of Japanese encephalitis virus infecting PK15 cells. China Anim Husb Vet Med. (2019) 46:2045–52. doi: 10.16431/j.cnki.1671-7236.2019.07.020
Keywords: porcine, PML gene, alternative splicing, clone, JEV
Citation: Zhu J, Chen Z, Dai Z, Zhou X, Wang H, Li X, Zhao A and Yang S (2021) Molecular Cloning of Alternative Splicing Variants of the Porcine PML Gene and Its Expression Patterns During Japanese Encephalitis Virus Infection. Front. Vet. Sci. 8:757978. doi: 10.3389/fvets.2021.757978
Received: 13 August 2021; Accepted: 21 October 2021;
Published: 23 November 2021.
Edited by:
Shao-Lun Zhai, Guangdong Academy of Agricultural Sciences, ChinaReviewed by:
Tao Lin, UCONN Health, United StatesLi Mao, Jiangsu Academy of Agricultural Sciences (JAAS), China
Copyright © 2021 Zhu, Chen, Dai, Zhou, Wang, Li, Zhao and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Songbai Yang, c2J5YW5nQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
 Jingjing Zhu
Jingjing Zhu Zhenyu Chen
Zhenyu Chen Zhenglie Dai
Zhenglie Dai Xiaolong Zhou
Xiaolong Zhou Xiangchen Li
Xiangchen Li Ayong Zhao
Ayong Zhao Songbai Yang
Songbai Yang