
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 26 November 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.756764
This article is part of the Research Topic Recent Advances in Porcine Respiratory Diseases View all 10 articles
 Kunli Zhang1,2
Kunli Zhang1,2 Pinpin Chu1,2
Pinpin Chu1,2 Shuai Song1,2
Shuai Song1,2 Dongxia Yang1,2
Dongxia Yang1,2 Zhibiao Bian1,2
Zhibiao Bian1,2 Yan Li1,2
Yan Li1,2 Hongchao Gou1,2
Hongchao Gou1,2 Zhiyong Jiang1,2
Zhiyong Jiang1,2 Rujian Cai1,2
Rujian Cai1,2 Chunling Li1,2*
Chunling Li1,2*Haemophilus parasuis has emerged as an important bacterial pathogen in pig husbandry, as H. parasuis can coinfect pigs with a variety of pathogenic microorganisms and further cause an aggravation of the disease. It is crucial to investigate its pathogenetic mechanism. Gram-negative bacteria naturally secrete outer membrane vesicles (OMVs), and their potent virulence factors play prominent roles that affect the interaction between bacteria and host. Still, the pathogenesis that is associated with the bacterial OMVs has not been well-elucidated. In this study, we investigated the secretion of OMVs from a clinical H. parasuis isolate strain (H45). In addition, we further analyzed the characterization, the comprehensive proteome, and the virulence potential of OMVs. Our data demonstrated that H. parasuis could secrete OMVs into the extracellular milieu during infection. Using liquid chromatography with tandem mass spectrometry (MS/MS) identification and bio-information analysis, we identified 588 different proteins associated with OMVs. Also, we also analyzed the subcellular location and biological function of those proteins. These proteins are mainly involved in immune and iron metabolism. Moreover, we confirmed the pathogenicity of H. parasuis OMVs by observing a strong inflammatory response in J774A.1 and porcine alveolar macrophages. Taken together, our findings suggested that OMVs from H. parasuis were involved in the pathogenesis of this bacterium during infection.
Haemophilus parasuis is a Gram-negative bacterium of the Pasteurellaceae family. Haemophilus parasuis is an early colonizer of the upper respiratory tract and also is a part of the normal microbiota of healthy pigs (1). Haemophilus parasuis is the causative agent of Glässer's disease, which is characterized by fibrinous polyserositis, polyarthritis, meningitis, and arthritis and causes significant economic losses. Glässer's disease prevention and control continue to be challenging, as the pathogenicity of H. parasuis is not fully characterized. Fifteen standard H. parasuis serovars have been recognized in addition to non-typeable isolates. Among the standard serovars, 1, 5, 10, 12, 13, and 14 are considered highly virulent, 2, 4, 15, and 8 are mildly virulent, and serovars 3, 6, 7, 9, and 11 are considered avirulent (2, 3). Serovars 4 and 5 are the most prevalent in China, followed by serovars 7, 12, 13, and 14 (4–6). Virulence factors play a primary role during the bacterial pathogenic process. So far, various virulence factors of H. parasuis have been identified, including 6-phosphogluconate dehydrogenase, lipooligosaccharide, cytolethal distending toxin protein (CDT, CdtA, CdtB, and CdtC), immunoglobulin A protease, capsule, porin family proteins (OmpP2 and OmpP5), monomeric autotransporters (bmaA), virulence-associated trimeric autotransporters (VtaA2, VtaA3, vtaA8, and vtaA9), and GalU and GalE (7–13). These virulence factors are involved in different pathogenesis processes, including invasion of the host, evasion of host's defenses, and the assistance of bacterial multiplication.
It has been found that most Gram-negative bacteria can secrete outer membrane vesicles (OMVs) during both in vitro growth and in vivo infection (14). Studies show that OMVs are 20–250 nm in diameter, containing lipopolysaccharide (LPS), outer membrane proteins (OMPs), outer membrane lipids, periplasmic proteins (OPs), cytoplasmic proteins (CPs), and DNA or RNA (15). Several kinds of pathogenic Gram-negative bacteria secrete OMVs, including Escherichia coli (16), Neisseria meningitidis (17), Shigella flexneri (18), Helicobacter pylori (19), and Pseudomonas aeruginosa (20). OMVs contain diverse virulence factors of pathogenic bacteria, such as heat-labile toxin and cytolysin A of E. coli, b-lactamase, hemolytic phospholipase C of P. aeruginosa, and VacA of H. pylori, and their roles in bacterial pathogenesis have been relatively well-characterized (21). OMVs also play important roles in microbial community interactions, including quorum sensing, biofilm formation, nutrient acquisition, antibiotic resistance, and competition against other microbes. For example, the hydrophobic quorum senses molecules Pseudomonas quinolone signal of P. aeruginosa, C16-HSL of Paracoccus denitrificans, and CAI-1 of Vibrio harveyi are packaged into OMVs, which allows for stabilization, concentration, and dispersal through the environment to mediate the generation of virulence factors, iron acquisition, and host immune responses (22–24). In the case of Haemophilus influenzae, higher OMV levels correlated with increased serum resistance (25). Recent research found that outer membrane vesiculation facilitates surface exchange and in vivo adaptation of vibrio cholera (26). However, the secretion of OMVs and their contribution to pathogenesis have been rarely reported in H. parasuis. In the present study, we have purified the OMVs from a clinical H. parasuis and analyzed the proteome of the H. parasuis-derived OMVs. We demonstrated that H. parasuis could secrete OMVs during in vitro growth. The liquid chromatography with MS/MS analysis showed that several virulence-associated proteins and immune modulators from H. parasuis-derived OMVs were identified. Moreover, we found that OMVs of H. parasuis could induce interleukin (IL)-1β production in mouse mononuclear macrophages (J774A.1) and porcine alveolar macrophages (PAMs). These results suggest that OMVs can be vehicles for the transport of effector molecules into host cells.
To investigate whether H. parasuis can secrete OMVs during in vitro growth, a clinical isolate of H. parasuis H45 was grown in tryptic soy broth (TSB) and observed with electron microscopy. The H45 strain had the typical morphology of HPS, which has various forms, from a single short bacillus to a long rod (Figure 1A). Further observation revealed that some vesicle-like structures were found in and around the bacterium (Figures 1B,C). As displayed in Figure 1C, the spherical vesicles budded from bacterial outer membranes. These results suggested that H. parasuis H45 could secrete OMVs during in vitro growth.
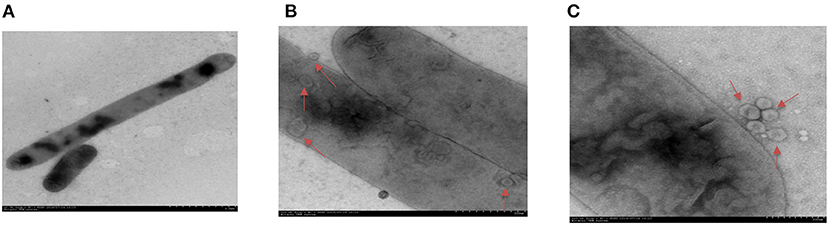
Figure 1. Transmission electron micrographs image of H. parasuis and nascent OMVs. (A) Image of H. parasuis H45 strain which stained with uranyl acetate. (B) Transmission electron micrographs image of OMVs (red arrow) located at intracellular of H. parasuis H45 strain. (C) Transmission electron micrographs image of OMVs (red arrow) located at extracellular of H. parasuis H45 strain.
The cell-free OMV fractions isolated from H. parasuis broth cultures were repurified by density gradient ultracentrifugation. The electron microscopy results showed that OMVs' fraction exhibited bilayered spherical vesicles (Figures 2A,B). The vesicle diameters ranged from 20 to 160 nm, with an average 50–70 nm diameter. Dynamic light scattering resulted that the Rh of the vesicles is ~69.8 nm; the size of purified OMVs ranged in diameter from ~50 to 100 nm, which was consistent with other reports (Figure 2C). To further determine whether the vesicles carried bacterial proteins, OMPs, OPs, and CPs were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) along with OMV fraction (Figure 2D). High similarity of protein bands among OMP, OP, CP, and the OMV fraction was observed, suggesting that the vesicles were mainly responsible for carrying the extracellular proteome of Haemophilus parasuis.
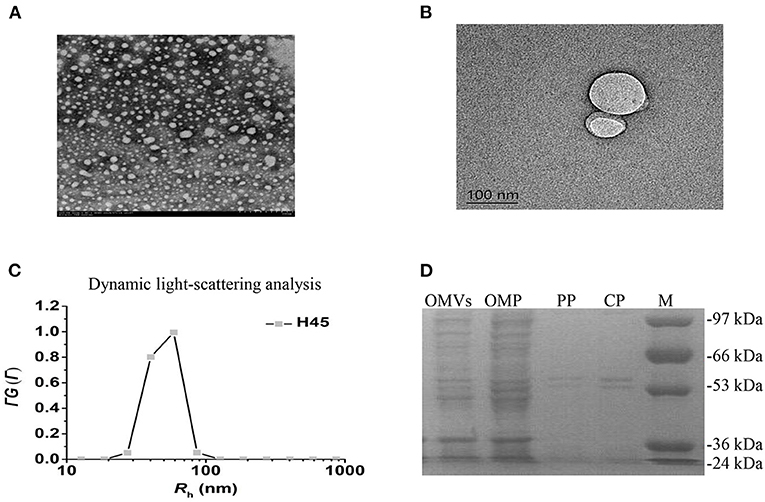
Figure 2. Characteristics of OMVs isolated from H. parasuis H45 strain. (A) Negatively stained preparation of isolated H. parasuis OMVs showing size of OMVs. Scale bar: 200 nm. (B) Transmission electron microscopy image of generation of one OMV from H45. Scale bar: 100 nm. (C) CONTIN analysis of dynamic light scattering data. (D) Comparison of protein components in different parts of HPS by SDS-PAGE. OMV, Outer membrane vesicles; OMP, Outer membrane protein; PP, Periplasmic protein; CP, Cytoplasmic protein.
To identify proteins associated with H. parasuis-derived OMVs, proteome analysis was performed by liquid chromatography with MS/MS. Firstly, the purified OMVs were digested with trypsin enzyme to obtain the freeze-dried peptide liquid without salt. Then, the dried peptide samples were separated by Shimadzu LC-20AD model nanoliter liquid chromatography. The liquid phase chromatography-separated peptides were detected by electrospray ionization (ESI) MS/MS (Triple TOF 5600). Subsequently, aligning the experimental MS/MS data with theoretical MS/MS data from UniProt and National Center for Biotechnology Information database is used to identify the proteins of OMVs. In all, 195,271 spectra were obtained. After identification by the search engine, 1,614 spectra were matched; 915 peptides and 588 proteins were identified in H. parasuis OMVs (Figure 3A). The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD029451. The identified proteins were classified into six groups according to the subcellular location of the proteins in the bacteria: cytoplasmic (n = 356), cytoplasmic membrane (n = 78), periplasmic (n = 27), outer membrane (n = 20), extracellular (n = 3), and unknown (n = 104) (Figure 3B). OMVs are enriched in membrane proteins (n = 98), including CPs and OMPs. In contrast, 5% of the total bacterial proteins are OPs. Those subcellular location results indicated that the OMVs were successfully purified from Haemophilus parasuis. Notably, proteomic results indicated that a large number of CPs were highly enriched in H. parasuis OMVs. Furthermore, we carried out a Gene Ontology (GO) functional annotation analysis for all identified proteins. The molecular function and biological processes genes are shown in Figures 3C,D. Similar to other reports, the H. parasuis OMVs were rich in proteins that have catalytic activity (n = 344) and binding function (n = 290) (Figure 3C). The classification of biological processes showed that H. parasuis-derived OMVs also contain numbers of proteins associated with cellular processes (n = 282) and metabolic processes (n = 257) (Figure 3D). These results suggested that OMVs play important roles in the pathogenesis of H. parasuis during infection.
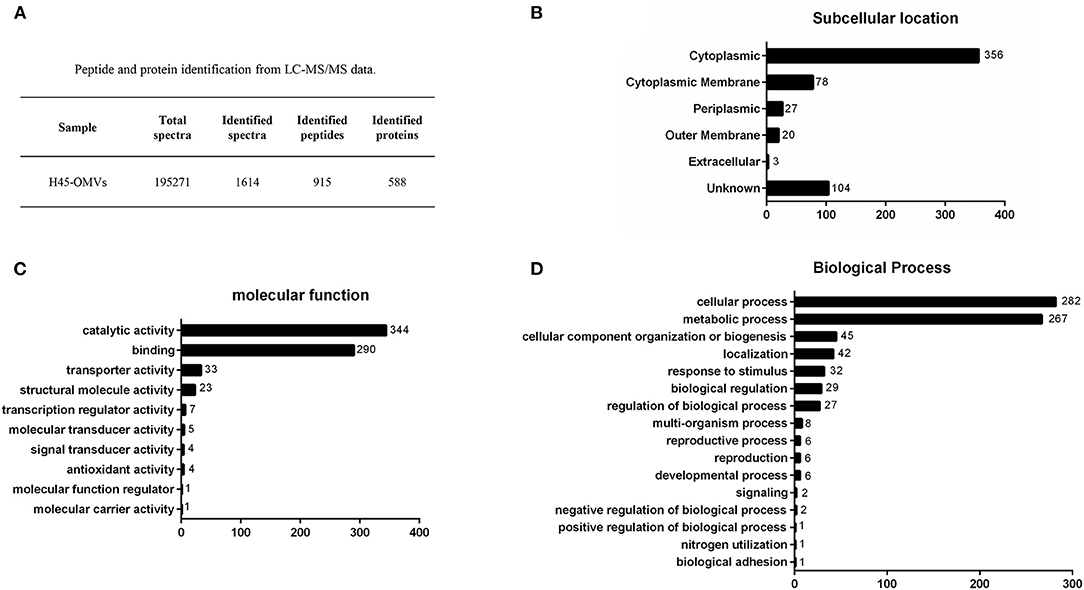
Figure 3. Overview of proteome identification of H45-derived OMVs. (A) Peptide and protein of OMVs from liquid chromatography with tandem mass spectrometry data. (B–D) All identified proteins of H45-OMVs were analyzed by GO functional annotation (http://www.geneontology.org.), subcellular location (B), molecular function (C), and biological process of genes (D).
OMVs are considered to represent a specialized bacterial secretion pathway. Several studies showed that OMVs could deliver bacterial bioactive proteins, toxins, and virulence factors (15). According to the proteome result of OMVs, we found that H. parasuis (H45)-derived OMVs are rich in immune modulators and virulence-associated proteins. According to the protein mass score, the detected proteins related to immune modulators and virulence-associated proteins are listed in Table 1. Lots of immunogenic proteins are identified, such as ompP2 (ACL31860.1), oppA (ACL32119.1), ompD15 (ACL31912.1), palA (ACL31779.1), AfuA (ACL31862.1), LppC (ACL32393.1), espP2 (ACL32961.1), and so on. These results indicated that OMV might induce a degree of protective immunity and could be selected as a new type subunit vaccine to prevent H. parasuis infection. Iron is an essential nutrient for almost all bacteria. To acquire enough free iron from the host, pathogenic bacteria have developed different mechanisms to obtain iron from the host (siderophores, hemophores, or host-molecule-binding proteins), which are involved in the expression of surface-exposed proteins (27). The iron metabolism-related proteins yfeA (ACL32714.1), dsbC (ACL31879.1), hxuA (ACL33616.1), fbpA (ACL32742.1), and HAPS_0679 (ACL32327.1) were found in OMV proteome data. In addition, several virulence-associated proteins also were identified: they are vtaA9 (ADZ54070.1), tbpA (ACL33656.1), pgi (ACL32784.1), manB (ACL32480.1), ompP5 (ACL33726.1), TolC (ACL33604.1), and yfeA (ACL32714.1). Serum resistance is potential pathogenesis of H. parasuis, which is associated with systemic diseases in swine. Several serum-resistance associated proteins of H45-derived OMVs were identified, including ctdA (ACL32011.1), ctdB (ACL32010.1), ctdC (ACL32009.1), lsgB (ACL31747.1), galE (ACL32657.1), HAPS_1019 (ACL32638.1), and oppA (ACL32119.1). Among them, ctdA, ctdB, ctdC are considered to contribute to cell cycle arrest, apoptosis, adherence, and invasion. Haemophilus parasuis (H45)-derived OMVs can carry such numbers of proteins that associated with virulence and/or pathogenic mechanism, which suggested OMVs might have an important role in H. parasuis infection.
Severe serous inflammation in multi-tissues is a typical feature of H. parasuis infection. To evaluate the ability of H45 infection for inducing IL-1β transcription and secretion, mouse mononuclear macrophage cells (J774A.1) were infected with H45 at different MOIs of infection. As shown in Figures 4A,B, the IL-1β messenger RNA (mRNA) expression and secretion were significantly increased in a dose-dependent manner during H45 infection. To further investigate whether OMVs take part in IL-1β secretion during H. parasuis H45 infection, J774A.1 were treated with OMVs at different doses for 24 h. Then, the IL-1β mRNA expression and secretion were detected by quantitative polymerase chain reaction and enzyme-linked immunosorbent assay, respectively. We found that the IL-1β expression and secretion induced by H45-derived OMVs are also in a dose-dependent manner (Figures 4C,D). This result suggested that OMV is an essential component in the inflammatory response caused by H45 infection. At last, PAMs were used to confirm the results mentioned earlier. As expected, the expressed and secreted IL-1β levels were both significantly increased in PAMs after H45 infection at 10 MOI of infection or H45 OMVs stimulation at 10 μg/ml for 24 and 48 h (Figures 4E,F). Taken together, these results indicate that H. parasuis H45 infection induces IL-1β expression and secretion, and OMVs were required for IL-1β secretion.
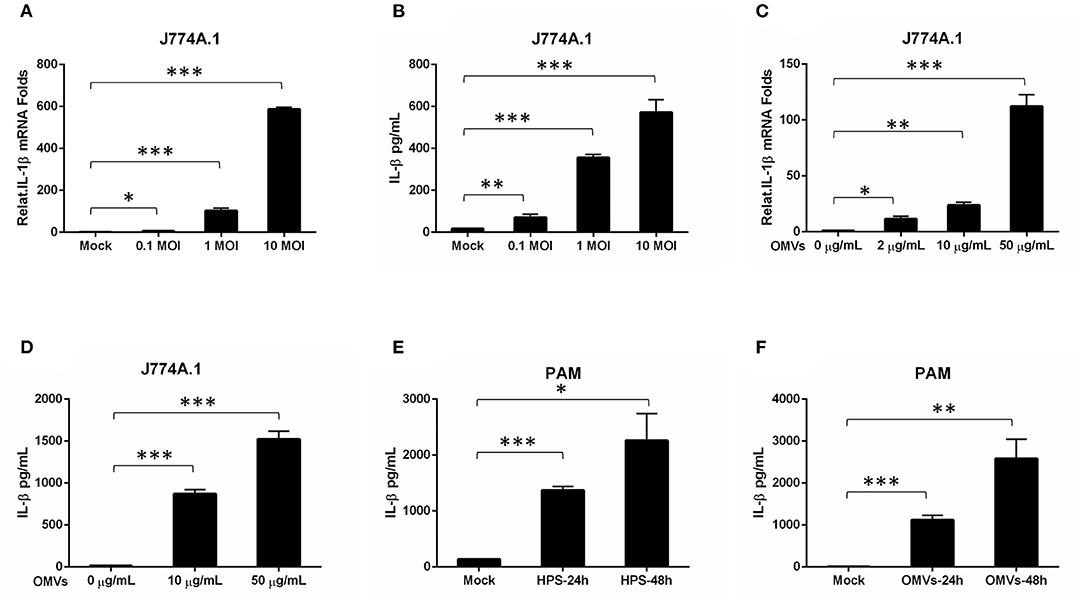
Figure 4. OMV is sufficient to stimulate an inflammatory response in macrophage cells. (A,B) Mouse Mononuclear macrophage cells (J774A.1) were either mock-infected or infected with H. parasuis at different doses (0.1, 1, 10 MOI) for 24 h. The mRNA levels of IL-1β in the H. parasuis infected cells were analyzed by qRT-PCR (A). The IL-1β levels in the cell supernatants were analyzed by ELISA (B). (C) J774A.1 were either mock-stimulated or stimulated with OMVs at 2 μg/mL, 10 μg/mL, 50 μg/mL for 24 h. The mRNA levels of IL-1β in the cell supernatants were analyzed by qRT-PCR. (D) J774A.1 were either mock-stimulated or stimulated with OMVs at 10 μg/mL, 50 μg/mL for 24 h. The IL-1β levels in the cell supernatants were analyzed by ELISA. (E) PAMs were either mock-infected or infected with H. parasuis at 10 MOI for different times (24 h, 48 h). The I L-1β levels in the cell supernatants were analyzed by ELISA. (F) PAMs were either mock-stimulated or stimulated with 10 μg/mL OMVs at different times (24 h, 48 h). The IL-1β levels in the cell supernatants were analyzed by ELISA. *p < 0.05, **p < 0.01, ***p < 0.001, Student t test, in comparison groups during statistical analysis.
Haemophilus parasuis is the causative agent of Glässer's disease and also is the main bacteria in porcine mixed infection. Haemophilus parasuis can cause severe system inflammation alone or induce secondary infection after porcine reproductive and respiratory syndrome virus, pseudorabies virus, porcine circovirus, or another virus infection in the infected pigs (28–30). Haemophilus parasuis infection is increasingly harmful to the global swine industry, and the prevention and control of H. parasuis have always been a significant challenge, as the pathogenicity of H. parasuis is not fully characterized. Production of OMVs is a particular function of Gram-negative bacteria, which plays a prominent role in establishing connections between bacteria and hosts. We confirmed that H. parasuis secreted OMVs into the extracellular environment during in vitro culture and many Gram-negative bacteria. Therefore, identifying the component of H. parasuis-derived OMVs is a pivotal factor in ascertaining such infections' pathogenesis.
In our study, the size of extracellular vesicles of H. parasuis clinical isolate strain (H45) is in the range of 20–160 nm. Further proteomic analysis revealed that H. parasuis OMVs contain 588 proteins and are rich in cytoplasmic and membrane proteins. According to GO functional annotation analysis, a number of proteins of OMVs are involved in catalytic or binding activity functions. OMVs also carry those proteins related to transporter activity, structural molecule activity, transcription activity, molecular transducer activity, signal transducer activity, and antioxidant activity. Classification of biological processes analysis showed that the proteins of OMVs are also involved in several biological processes such as metabolic, biogenesis, ion transport and localization, and biological regulation. The characteristics mentioned earlier of OMVs indicated that OMVs might be involved in biological processes of H. parasuis proliferation, infection, and pathogenicity.
The trivalent inactivated vaccine of H. parasuis developed by our team has obtained the approval number of three new veterinary drug certificates in China. The H45 strain of H. parasuis is the primary strain of the vaccine. The immune protection rate of the vaccine in piglets reached 80%. Many studies have proven that native OMVs isolated from pathogenic bacteria showed strong protective efficacy against pathogen infection (31–33). In this study, we found that H. parasuis H45 strain-derived OMVs contain numerous immunogenic antigens, including ompP2, oppA, ompD15, palA, AfuA, LppC, espP2, and vtaA1 (Table 1). These results indicate that the OMVs of H45 have the potential to replace the whole inactivated vaccine of H. parasuis to protect piglets from H. parasuis infection. However, the immune protection efficiency of OMV in piglets still needs to be further studied.
We also confirmed the pathogenicity of H. parasuis OMVs by observing strong inflammatory responses in J774A.1 cells and PAMs. Furthermore, H45-derived OMVs stimulation also can induce the IL-1β mRNA expression and secretion in a dose- and time-dependent manner. Our data confirmed the inflammation-inducing effect of the OMVs on target cells. The characteristic of the inflammatory response induced by OMVs may be related to LPS, which is one of the most abundant host immunogenic components of OMVs. It has been demonstrated that the OMVs of enterohemorrhagic E. coli acted as cargo to deliver LPS into the cytosol, which trigger caspase-11-dependent effector responses in vitro and in vivo (34). OMPs and porin are major structural components of both the outer membrane and secreted vesicles involved in the formation and secretion of OMVs. The previous study has shown that loss of porin inhibited macrophage inflammatory responses to OMVs of Klebsiella pneumoniae, inducing extremely low levels of pro-inflammatory cytokine (35). In this study, we found that OMVs contain ompP2, which affects the adherence capacity of H. parasuis in PAM (11). Moreover, many virulence factors related to iron metabolism, serum resistance, adherence, and invasion were also identified. For example, vtaA9 was involved in the phagocytosis resistance during H. parasuis infection (8, 9); CDTs (cdtA, cdtB, and cdtC) contribute to serum resistance, adherence, and invasion in PK-15 and PUVEC (10). The hosts' inflammation response induced by OMVs might partly relate to the adherence and invasion proteins. These OMV proteins can promote the effect of pathogenic internalization. Numerous studies have clearly shown that the delivery of toxins and other virulence factors via OMVs essentially influences the interactions between pathogen and host cells. Hence, the functions of these virulence factors of OMVs during the H. parasuis infection would be further investigated.
In conclusion, this study describes the typical morphology and the high-throughput proteomic characterization of OMVs derived from H. parasuis clinically isolated strain H45. We have identified 588 proteins in H. parasuis-derived OMVs and discovered several proteins associated with immunogenicity, iron metabolism, serum resistance, adherence, and invasion. Our further study shows that H. parasuis-derived OMVs can induce the production of pro-inflammatory cytokine IL-1β. According to these results, we speculated OMVs from H. parasuis were involved in the pathogenesis of this bacterium during infection.
Haemophilus parasuis H45 strain was a clinically isolated strain in China. H45 belongs to serovar 5, which is high-level virulent and is the most popular strain in China. Bacteria were cultured in TSB (Difco Laboratories, Detroit, MI, USA) supplemented with 1 mg/ml nicotinamide adenine dinucleotide and 10% bovine serum. After incubation at 37°C overnight with circular agitation (200 rpm), bacteria were harvested at an optical density at 600 nm 0.7–0.9. A volume of each strain was mixed with 20% glycerol and stored at −80°C in cryogenic storage tubes for further use.
Mouse mononuclear macrophage cells (J774A.1) were purchased and cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C with 5% CO2. Four-week-old piglets were used to isolate PAMs as previously described (36) and maintained in RPMI-1640 medium containing 10% heat-inactivated fetal bovine serum (Hyclone), 100 U/ml penicillin, 100 μg/ml streptomycin, and non-essential amino acid (Gibco) at 37°C in a humidified atmosphere of 5% CO2.
The OMVs of H. parasuis were collected from the supernatant of bacterial culture (1,000 ml). In brief, H. parasuis were inoculated overnight in 5 ml of TSB. The next day, 1,000 ml of TSB with the 1 mL of starter was cultured at 37°C with shaking until the optical density at 600 nm reached 1.0. Bacterial cells were removed with centrifugation at 5,000 × g for 20 min at 4°C. The supernatants were concentrated with a 100-kDa hollow-fiber membrane, and then, the concentrate was vacuum filtered through a 0.22-μm membrane to remove any remaining bacteria. The OMVs were collected by ultracentrifugation at 150,000 × g for 4.5 h at 4°C (rotor Type 50.2 Ti, Beckman-Coulter). The pelleted OMVs were suspended with sterile phosphate-buffered saline (PBS). The resuspended OMVs were purified by density gradient centrifugation with OptiPrep [60% iodixanol (w/v)]. The OptiPrep solution was diluted with 0.85% (w/v) NaCl containing 10-mM 4-(2-hydroxyethyl) piperazine-1-ethanesulfonic acid–sodium hydroxide pH 7.4 into 45, 40, 35, 30, 25, and 20% (v/v) OptiPrep. The 45% OptiPrep solution was prepared with suspended OMVs and layered 2 ml at the bottom of an Ultra-Clear centrifuge tube (13.2 ml, Beckman Coulter). The discontinuous gradients were loaded in 2-ml increments in degressive concentrations on top of the prior layer [36]. Prepared tubes were centrifuged at 150,000 × g for 16 h at 4°C (rotor SW 41 Ti, BeckmanCoulter). Each 1-ml fraction was carefully collected by pipet from top to bottom. The OMV content of each fraction was analyzed by 12% SDS-PAGE. The fractions containing OMVs were collected and pooled into an ultracentrifugation tube with 27-ml sterile PBS (at least 10-fold of the sample volume) and then ultracentrifuged at 200,500 × g for 2 h at 4°C (rotor Type 45 Ti, Beckman Coulter) to remove the OptiPrep solution. The purified OMVs were resuspended with sterile PBS, and the aliquots were stored at −80°C for future use.
Transmission electron microscopy was used to investigate OMV morphology. A drop of OMV suspension was placed on Formvar/carbon-coated grids and adsorbed for 5 min. Grids were washed with distilled water and blotted with filter paper. For negative staining, grids were treated with 1.5% (wt/vol) uranyl acetate for 1 min, air-dried, and viewed with a Hitachi HT7700 electron microscope operating at 80 kV.
OMV proteins were quantified by BCA Protein Assay Kit (Thermo fisher scientific, Cat# 23225). Vesicles proteins from H. parasuis H45 (Serum type 5) were analyzed by 12% SDS-PAGE. The gel was stained with Coomassie brilliant blue. Protein molecular weight standard (size range 35–180 KDa, Thermo Fisher Scientific) was used.
The vesicle proteins were separated by SDS-PAGE and digested with the trypsin enzyme. The digested peptide liquid was desalted and freeze-dried. The dried peptide samples were reconstituted with mobile phase A (2% ACN, 0.1% FA), and the supernatant after centrifugation at 20,000 × g for 10 min was taken for high-performance liquid chromatography–ESI-MS/MS analysis. The peptide samples were subjected to Shimadzu LC-20AD model nanoliter liquid chromatography (75 μm × 3 μm × 15 cm, C18 column) at a flow rate of 300 nl/min. The liquid phase chromatography-separated peptides were passed to an ESI-MS/MS, which was carried out using a TripleTOF 5600 (SCIEX, Framingham, MA, USA) mass spectrometer equipped with a Nanospray III source, and coupled with an emitter which drawn from quartz material (New Objectives, Woburn, MA, USA). For data acquisition, the mass spectrometer parameters were set as follows: ion source spray voltage 2,300 V, nitrogen pressure 35 psi, spray gas 15, and spray interface temperature 150°C. Scanning in high sensitivity mode, the MS1 scan cumulative time was 250 ms, and the scan quality range was 350–1,500 Da. Based on the MS1 scanning information, according to the ionic strength in the MS1 spectrum from high to low, the first 30 ions exceeding 150 cp were selected for fragmentation, and the MS2 information was scanned. The scan accumulation time of the MS2 mass spectrum was 50 ms. The collision energy was set to “rolling collision energy.”
The protein identification uses experimental MS/MS data and aligns them with theoretical MS/MS data from the database to obtain results. The process starts from converting raw MS data into a peak list and then searching matches in the Mascot search engine (Mascot 2.3.02). The UniProt protein database and National Center for Biotechnology Information database were selected. We used e-value ≤ 0.05 from Mascot as a filtering condition in this pipeline (the PSM FDR column is the Mascot value). Then, based on the parsimony principle, we performed protein inference on peptides and generated a series of protein groups. Finally, functional annotation analyses such as GO, Clusters of Orthologous Groups of protein/EuKaryotic Orthologous Groups, and Pathway analysis are performed from the final protein identification list.
J774A.1 or PAMs cells were stimulated by H. parasuis or OMVs as indicated. After 24 or 48 h, supernatants from cell culture were harvested and assayed with the production of murine IL-1β (R&D Systems, Cat# MLB00C) and swine IL-1β (RayBio, Cat# ELP-IL1b-1) by enzyme-linked immunosorbent assay, in accordance with the manufacturer's instructions. The absorbance was quantified at 450 nm by a microplate spectrophotometer (Synergy H1, BioTek, USA).
The original contributions of mass spectrometry proteomics data presented in the study are publicly available. This data can be found here: http://proteomecentral.proteomexchange.org. Further inquiries can be directed to the corresponding author.
KZ and CL designed and supervised the research. KZ wrote the paper and revised the manuscript. KZ, PC, ZB, DY, SS, YL, ZJ, HG, PC, and RC performed and analyzed the experiments. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (32002298), Independent Research and Development Projects of Maoming Laboratory (2021ZZ003), the Science and Technology Planning Project of Guangzhou (202102020385), and the Special Fund for Scientific Innovation Strategy—Construction of High-Level Academy of Agriculture Science (R2019YJ-YB2005, R2020QD-047) and the Special fund for scientific innovation strategy-construction of high level Academy of Agriculture Science-Prominent Talents (R2020PYJC001).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Moller K, Kilian M. V factor-dependent members of the family Pasteurellaceae in the porcine upper respiratory tract. J Clin Microbiol. (1990) 28:2711–6. doi: 10.1128/jcm.28.12.2711-2716.1990
2. Rafiee M, Blackall PJ. Establishment, validation and use of the Kielstein-Rapp-Gabrielson serotyping scheme for Haemophilus parasuis. Aust Vet J. (2000) 78:172–4. doi: 10.1111/j.1751-0813.2000.tb10586.x
3. Del Rio ML, Gutierrez CB, Rodriguez Ferri EF. Value of indirect hemagglutination and coagglutination tests for serotyping Haemophilus parasuis. J Clin Microbiol. (2003) 41:880–2. doi: 10.1128/JCM.41.2.880-882.2003
4. Cai X, Chen H, Blackall PJ, Yin Z, Wang L, Liu Z. Serological characterization of Haemophilus parasuis isolates from China. Vet Microbiol. (2005) 111:231–6. doi: 10.1016/j.vetmic.2005.07.007
5. Zhang J, Xu C, Guo L, Ke B, Ke C, Zhang B. A rapid pulsed-field gel electrophoresis method of genotyping Haemophilus parasuis isolates. Lett Appl Microbiol. (2011) 52:589–95. doi: 10.1111/j.1472-765X.2011.03048.x
6. Zhao Y, Wang Q, Li J, Lin X, Huang X, Fang B. Epidemiology of Haemophilus parasuis isolates from pigs in China using serotyping, antimicrobial susceptibility, biofilm formation and ERIC-PCR genotyping. PeerJ. (2018) 6:e5040. doi: 10.7717/peerj.5040
7. Bouchet B, Vanier G, Jacques M, Auger E, Gottschalk M. Studies on the interactions of Haemophilus parasuis with porcine epithelial tracheal cells: limited role of LOS in apoptosis and pro-inflammatory cytokine release. Microb Pathog. (2009) 46:108–13. doi: 10.1016/j.micpath.2008.10.008
8. Olvera A, Ballester M, Nofrarias M, Sibila M, Aragon V. Differences in phagocytosis susceptibility in Haemophilus parasuis strains. Vet Res. (2009) 40:24. doi: 10.1051/vetres/2009007
9. Costa-Hurtado M, Ballester M, Galofre-Mila N, Darji A, Aragon V. VtaA8 and VtaA9 from Haemophilus parasuis delay phagocytosis by alveolar macrophages. Vet Res. (2012) 43:57. doi: 10.1186/1297-9716-43-57
10. Zhang B, He Y, Xu C, Xu L, Feng S, Liao M. Cytolethal distending toxin (CDT) of the Haemophilus parasuis SC096 strain contributes to serum resistance and adherence to and invasion of PK-15 and PUVEC cells. Vet Microbiol. (2012) 157:237–42. doi: 10.1016/j.vetmic.2011.12.002
11. Zhang B, Xu C, Liao M. Outer membrane protein P2 of the Haemophilus parasuis SC096 strain contributes to adherence to porcine alveolar macrophages cells. Vet Microbiol. (2012) 158:226–7. doi: 10.1016/j.vetmic.2012.01.023
12. Xu C, Zhang L, Zhang B, Feng S, Zhou S, Li J. Involvement of lipooligosaccharide heptose residues of Haemophilus parasuis SC096 strain in serum resistance, adhesion and invasion. Vet J. (2013) 195:200–4. doi: 10.1016/j.tvjl.2012.06.017
13. Costa-Hurtado M, Garcia-Rodriguez L, Lopez-Serrano S, Aragon V. Haemophilus parasuis VtaA2 is involved in adhesion to extracellular proteins. Vet Res. (2019) 50:69. doi: 10.1186/s13567-019-0687-2
14. Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. (1999) 181:4725–33. doi: 10.1128/JB.181.16.4725-4733.1999
15. Jan AT. Outer membrane vesicles (OMVs) of gram-negative bacteria: a perspective update. Front Microbiol. (2017) 8:1053. doi: 10.3389/fmicb.2017.01053
16. McBroom AJ, Kuehn MJ. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol Microbiol. (2007) 63:545–58. doi: 10.1111/j.1365-2006.05522.x
17. Devoe IW, Gilchrist JE. Release of endotoxin in the form of cell wall blebs during in vitro growth of Neisseria meningitidis. J Exp Med. (1973) 138:1156–67. doi: 10.1084/jem.138.5.1156
18. Kadurugamuwa JL, Beveridge TJ. Delivery of the non-membrane-permeative antibiotic gentamicin into mammalian cells by using Shigella flexneri membrane vesicles. Antimicrob Agents Chemother. (1998) 42:1476–83. doi: 10.1128/AAC.42.6.1476
19. Keenan J, Day T, Neal S, Cook B, Perez-Perez G, Allardyce R. A role for the bacterial outer membrane in the pathogenesis of Helicobacter pylori infection. FEMS Microbiol Lett. (2000) 182:259–64. doi: 10.1111/j.1574-6968.2000.tb08905.x
20. Kadurugamuwa JL, Beveridge TJ. Virulence factors are released from Pseudomonas aeruginosa in association with membrane vesicles during normal growth and exposure to gentamicin: a novel mechanism of enzyme secretion. J Bacteriol. (1995) 177:3998–4008. doi: 10.1128/jb.177.14.3998-4008.1995
21. Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. (2010) 74:81–94. doi: 10.1128/MMBR.00031-09
22. Mashburn LM, Whiteley M. Membrane vesicles traffic signals and facilitate group activities in a prokaryote. Nature. (2005) 437:422–5. doi: 10.1038/nature03925
23. Toyofuku M, Morinaga K, Hashimoto Y, Uhl J, Shimamura H, Inaba H. Membrane vesicle-mediated bacterial communication. ISME J. (2017) 11:1504–9. doi: 10.1038/ismej.2017.13
24. Brameyer S, Plener L, Muller A, Klingl A, Wanner G, Jung K. Outer membrane vesicles facilitate trafficking of the hydrophobic signaling molecule CAI-1 between Vibrio harveyi cells. J Bacteriol. (2018) 200:e00740–17. doi: 10.1128/JB.00740-17
25. Roier S, Zingl FG, Cakar F, Durakovic S, Kohl P, Eichmann TO. A novel mechanism for the biogenesis of outer membrane vesicles in gram-negative bacteria. Nat Commun. (2016) 7:10515. doi: 10.1038/ncomms10515
26. Zingl FG, Kohl P, Cakar F, Leitner DR, Mitterer F, Bonnington KE. Outer membrane vesiculation facilitates surface exchange and in vivo adaptation of vibrio cholerae. Cell Host Microbe. (2020) 27:225–37.e228. doi: 10.1016/j.chom.2019.12.002
27. Parrow NL, Fleming RE, Minnick MF. Sequestration and scavenging of iron in infection. Infect Immun. (2013) 81:3503–14. doi: 10.1128/IAI.00602-13
28. Palzer A, Ritzmann M, Wolf G, Heinritzi K. Associations between pathogens in healthy pigs and pigs with pneumonia. Vet Rec. (2008) 162:267–71. doi: 10.1136/vr.162.9.267
29. Zhao Z, Qin Y, Lai Z, Peng L, Cai X, Wang L. Microbial ecology of swine farms and PRRS vaccine vaccination strategies. Vet Microbiol. (2012) 155:247–56. doi: 10.1016/j.vetmic.2011.09.028
30. Palzer A, Eddicks M, Zoels S, Stark J, Reese S, Strutzberg-Minder K. Field evaluation of the efficacy, compatibility and serologic profiling of a combined vaccine against porcine reproductive and respiratory syndrome and Haemophilus parasuis in nursery pigs. Prev Vet Med. (2015) 119:134–40. doi: 10.1016/j.prevetmed.2015.03.005
31. Liu Q, Yi J, Liang K, Zhang X, Liu Q. Salmonella choleraesuis outer membrane vesicles: proteomics and immunogenicity. J Basic Microbiol. (2017) 57:852–61. doi: 10.1002/jobm.201700153
32. Hays MP, Houben D, Yang Y, Luirink J, Hardwidge PR. Immunization with skp delivered on outer membrane vesicles protects mice against enterotoxigenic escherichia coli challenge. Front Cell Infect Microbiol. (2018) 8:132. doi: 10.3389/fcimb.2018.00132
33. Solanki KS, Varshney R, Qureshi S, Thomas P, Singh R, Agrawal A. Non-infectious outer membrane vesicles derived from Brucella abortus S19Deltaper as an alternative acellular vaccine protects mice against virulent challenge. Int Immunopharmacol. (2021) 90:107148. doi: 10.1016/j.intimp.2020.107148
34. Vanaja SK, Russo AJ, Behl B, Banerjee I, Yankova M, Deshmukh SD. Bacterial outer membrane vesicles mediate cytosolic localization of LPS and caspase-11 activation. Cell. (2016) 165:1106–19. doi: 10.1016/j.cell.2016.04.015
35. Turner KL, Cahill BK, Dilello SK, Gutel D, Brunson DN, Alberti S. Porin Loss Impacts the Host Inflammatory Response to Outer Membrane Vesicles of Klebsiella pneumoniae. Antimicrob Agents Chemother. (2015) 60:1360–9. doi: 10.1128/AAC.01627-15
Keywords: Haemophilus parasuis, outer membrane vesicles, purification, proteome analysis, inflammatory response
Citation: Zhang K, Chu P, Song S, Yang D, Bian Z, Li Y, Gou H, Jiang Z, Cai R and Li C (2021) Proteome Analysis of Outer Membrane Vesicles From a Highly Virulent Strain of Haemophilus parasuis. Front. Vet. Sci. 8:756764. doi: 10.3389/fvets.2021.756764
Received: 11 August 2021; Accepted: 25 October 2021;
Published: 26 November 2021.
Edited by:
Arkadiusz Dors, National Veterinary Research Institute (NVRI), PolandReviewed by:
Xiaoli Liu, Ludong University, ChinaCopyright © 2021 Zhang, Chu, Song, Yang, Bian, Li, Gou, Jiang, Cai and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunling Li, bGNsY2xhcmVAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.