
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 11 February 2022
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.756438
This article is part of the Research Topic Ruminant Mastitis: A 360° View View all 21 articles
Helcococcus ovis (H. ovis) was first reported in ovine subclinical mastitis milk and post-mortem examination organs in Spain and the United Kingdom in 1999; subsequently, it appeared in cattle, horse, goat, and human. However, isolation and characterization of the pathogen from clinical bovine mastitis is unknown. The objective of this study was to identify the pathogen in clinical bovine mastitis. A total of four strains were isolated from bovine mastitis milk samples from a Chinese dairy farm, and they were identified as H. ovis by microscopic examination and 16S rRNA gene sequencing. Phylogenetic tree was constructed using 16S rRNA gene, and the isolates were closely related to other China strains and strains from Japan. The growth speed of the H. ovis isolated was relatively slower than Streptococcus agalactiae, and the phenotypic characteristics were similar to H. ovis CCUG37441 and CCUG39041 except to lactose. The isolates were sensitive to most of the common used antimicrobials. The H. ovis isolates could lead to mild murine mastitis alone and induce severe mastitis when co-infected with Trueperella pyogenes in the murine mammary infection model constructed.
Bovine mastitis is one of the most costly diseases in dairy industry due to milk discarding and related treatment costs, as well as culling of cows (1–3).
In China, the most frequently isolated mastitis pathogens were Escherichia coli (14.4%), Klebsiella spp. (13.0%), coagulase-negative staphylococci (11.3%), Streptococcus dysgalactiae (10.5%), and Staphylococcus aureus (10.2%) (4). Once these pathogens enter the mammary gland and detected by epithelial cells together with resident macrophages, the cells will release inflammatory cytokines such as TNF-α (tumor necrosis factor–alpha), IL-1β (interleukin-1 beta), humoral components (alexin, antibodies, antimicrobial peptides) are also participate in the process, and then, more leukocytes (neutrophils and lymphocytes) will be attracted under the chemotaxis, and more cytokines be synthesized and secreted; in the end, bovine mastitis is induced till the mastitis pathogens are eliminated (5).
Traditional diagnosis for bovine mastitis includes visual examination, California Mastitis Test, and Somatic Cells Counting. Pathogens isolation and identification are help for mastitis diagnosis and treatment; whereas, due to characteristics of microorganism, some of them are hard to isolate, which limits the research of such mastitis pathogens. Because of the limits of farm diagnostic tools in China, certain uncommon mastitis pathogens cannot be accurately identified on site. This might result in false negative results for mastitis diagnosis.
Helcococcus ovis (H. ovis) was first isolated from colonies mixed with Trueperella pyogenes (T. pyogenes) and Staphylococcus spp. from the lung, liver, spleen, and mastitis milk of two sheep in the United Kingdom and Spain (6), respectively. Subsequently, studies indicated that it can induce many other diseases in species of animals, even in human being, which suggest that it may be a zoonotic pathogen. H. ovis was also isolated from cows with abortions (7), puerperal metritis (8, 9), valvular endocarditis (10), horses with pulmonary abscess (11), and sheep with pleuritis and bronchopneumonia (12, 13).
Bacteriology methods and biochemistry tests are usually used to identify the biology characteristics of H. ovis; results indicated that this bacterium is a catalase-negative, facultatively anaerobic, and gram-positive cocci; and it often co-infected with other bacteria such as Staphylococci and T. pyogenes.
To the best of our knowledge, only four isolates of H. ovis were genome sequenced by Cunha et al. (8), whose results indicated that the guanine cytosine (GC) content of H. ovis was about 27.5%, the number of 33 tRNAs was identified in H. ovis, and all four isolates of H. ovis contained a ribosomal protection gene (tetB) and major facilitator superfamily (MFS) efflux gene (tetA), which confer resistance to tetracyclines. In addition, one of the isolates contained AcrEF-TolC, which can confer resistance to fluoroquinolones, cephalosporins, cephamycins, and penams (8).
Recently, H. ovis was found in human with pyogenic disease. The patient was reported to have a contact history with wool and cowhide leather before the disease and recovered after treatments of cefotaxime and ornidazole. The H. ovis was isolated from the samples collected from the skin around the eyes of the patient. Further antimicrobial resistance test indicated that the H. ovis strain is susceptible to penicillin, ampicillin, teicoplanin, ceftriaxone, vancomycin, and linezolid (14). Schwaiger et al. (15) reported coexistence of H. ovis with T. pyogenes detected by PCR; however, they failed to isolate the bacteria from the samples.
The objective of this study is to describe the H. ovis isolates first found in clinical mastitis cases in China, to determine the phylogeny relation to H. ovis strains isolated from other species, and investigate the antimicrobial resistance profiles and pathogenicity.
All experiments followed the Regulations of Experimental Animals (2008) promulgated by China Ministry of Science and Technology. All animal procedures were approved by the Institutional Animal Care and Use Committee of Yunnan Agricultural University (Approval No: IACUC-20132030301).
During the daily etiology examination of clinical mastitis milk samples in a dairy farm in Hebei province, four blood agar plates with tiny and transparent colonies were classified as gram-positive cocci but could not be further classified into Staphylococcus spp. nor Streptococcus spp. by a Streptococcal grouping kit (Lancefield's classification kit, Hopebio, Qingdao, China) test.
To identify these isolates, the four blood agar plates were delivered to the Mastitis Diagnostic Laboratory in China Agriculture University (Beijing). The pin-point colonies were transplanted onto blood agar plates and were incubated in 37°C until 36 h. Heavy growth of two types of colonies was observed, one was tiny and transparent, whereas the other type was white. All the two types of colonies were exposed to gram-staining and microscope examination.
Partial 16S rRNA gene (1,300 bp) amplification (forward primer: 5′-TACCTTGTTACGACTT-3′; reverse primer: 5′-AGAGTTTGATCCTGGCTCAG-3′) (16) and sequencing were conducted, and these sequences were BLAST with the available sequences in GenBank. The phylogenetic tree of these H. ovis isolates was constructed using the clustal V method (DNASTAR Lasergene-Megalign, version 7.1).
The growth curves of H. ovis and S. agalactiae were assessed simultaneously as S. agalactiae is one of the typical G+ cocci pathogens of bovine mastitis. The volume of 1 ml of culture media of each isolate was incubated into 100-ml brain heart infusion broth with 5% fetal bovine serum and placed on a constant temperature shaker (37°C, 220 rpm). The volume of 3 ml of bacterial suspension was collected at 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22, and 24 h, respectively, for optical density (OD) examination at 600 nm in a UV spectrophotometer (Jingke Scientific Instrument Co., Ltd., Shanghai, China).
Biochemical tests were performed on the four isolates using an HBI biochemical kit according to the manufacturer's instructions (Hopebio, Qingdao, China). Isolates were plated onto tryptic soy blood agar, and then, single colony was suspended into saline equivalent to a turbidity of 0.5 McFarland standard. The volume of 200 μl of saline suspension was added into each micro biochemical pool and then incubated at 37°C for 24 h. The culture medium will turn yellow when the test is positive to ribose, mannitol, sorbitol, lactose, raffinose, maltose, melibiose, and sucrose; red to urease; modena to hippurate hydrolysis; and blue to esterase.
Antimicrobial resistance tests were performed using the broth microdilution method with Streptococcus pneumoniae ATCC 49619 as quality control strain according to the Clinical and Laboratory Standards Institute (17). Common used antimicrobials for bovine mastitis treatment and in human medicine was selected for antimicrobial resistance testing, which includes penicillin, cefalexin, ceftiofur, oxacillin, clindamycin, tetracycline, enrofloxacin, daptomycin, erythromycin, and vancomycin (18). The dosage was based on previous research (14). Plates were incubated at 37°C in a humidified atmosphere for 24 h. The minimum inhibitory concentration (MIC) of each strain was defined as the lowest concentration of an antimicrobial that completely inhibited growth in broth (no growth) during the 24-h incubation. The MIC was determined as per the CLSI's guidelines. The concentrations of the antimicrobials agents range from 0.015 to 16 μg/ml. Isolates were classified following the clinical breakpoints described in CLSI (17).
Pregnant (20 days of gestation) 6- to 8-week-old SPF BALB/c mice (SiPeiFu Laboratory Animal Technology, Beijing, China) were used to determine the pathogenic role of H. ovis during intramammary infection, as described before (19). On the third day after parturition, mice were anesthetized with intramuscular injection of Zoletil 50 (50 mg/kg; Virbac, Carros, France). Four groups (n = 5 per group) of mice were allocated with three challenge groups (12, 24, and 36 h) and one negative control group [sterile phosphate buffered saline (PBS)]. Teat ducts of both the L4 (left) and R4 (right) abdominal mammary glands were exposed under a binocular stereoscopic microscope, and 100 μl of bacterial suspension (105 CFU/ml) was injected using a syringe with a 34G blunt needle (19). The clinical signs and pathological changes of each mouse were observed and recorded. Then, at 12, 24, and 36 h after challenge (five mice for each time point), the murine mammary gland was separated, part of mammary gland tissue (0.1 g) was separated into sterile tube under a germ-free environment to measure the bacterial burden, and the other part of mammary gland tissue was separated and fixed in 5% of paraformaldehyde to conduct histological evaluation.
To illustrate the pathogenicity of the co-infection of H. ovis and T. pyogenes, four groups (n = 5 per group) of the abovementioned mice were allocated with three challenge groups (H. ovis, T. pyogenes, and H. ovis and T. pyogenes) and one negative control group (sterile PBS). The challenge method and dosage is same (105 CFU/ml) with the single infection model above. The clinical signs and pathological changes of each mouse were observed and recorded. Murine blood was collected for serum separation to conduct ELISA at 24 h after challenge. Then, at 24 h after challenge (five mice for each time point), histological evaluation and immunohistochemical assay were conducted, as in the single challenge model above.
The IL-16 in blood was measured by ELISA kits (MULTI SCIENCES, Hangzhou, Jiangsu, China) according to the manufacturer's instructions. The absorbance was read at 450 nm by Multiskan MK3 (Thermo-Fisher Scientific, Waltham, MA, USA). All absorbance results were normalized by standard curves.
After embedded in paraffin wax, and sectioned and stained with hematoxylin-eosin, histological evaluation was performed to assess tissue necrosis, polymorphonuclear neutrophilic granulocyte inflammation (i.e., neutrophilic inflammation), and lymphocytic inflammation. For immunohistochemical assays, the sections were incubated with rabbit immunoglobulin G (IgG) against mouse IL-1β or TNF-α and then with a HRP-conjugated secondary antibody. A microscope (Olympus, Tokyo, Japan) was used for the examination.
The prevalence of H. ovis and its 95% confidence interval (95% CI) was calculated by using online tool VassarStats (http://www.vassarstats.net/).
All experiments (growth curve determination, bacterial burden, and ELISA) consisted of three independent repeats, and results were analyzed using SPSS 20 and GraphPad Prism 8. 0. 2. Data were assessed using one-way analysis of variance (one- way ANOVA). The data were expressed as the mean ± SD. P-values < 0.05 were significant.
The small and transparent colonies were single or in pair gram-positive cocci, whereas the white colonies were gram-positive single irregular rod-shaped bacteria (Figure 1). The gram-positive cocci were identified as H. ovis, whereas the rod-shaped bacteria were identified as T. pyogenes [information of the four H. ovis isolates (prevalence: 11.76%, 95% CI: 4.67–26.62%) was shown in Table 1]. The sequences of the H. ovis isolates were submitted to GenBank (https://www.ncbi.nlm.nih.gov/genbank/) with the accession numbers MT758192.1, MT758194.1, MT758195.1, and MT758196.1. All the four isolates (bold fonts) were closely related to the strains isolated from goat (H. ovis–YYQ1403) and bovine case (H. ovis–XJDY-N1-3) in China; meanwhile, they were also closely related to strains isolated from swine and bovine cases in Japan (e.g., H. ovis–Ymagata-080813 and H. ovis–Ymagata-160927) (Figure 2).
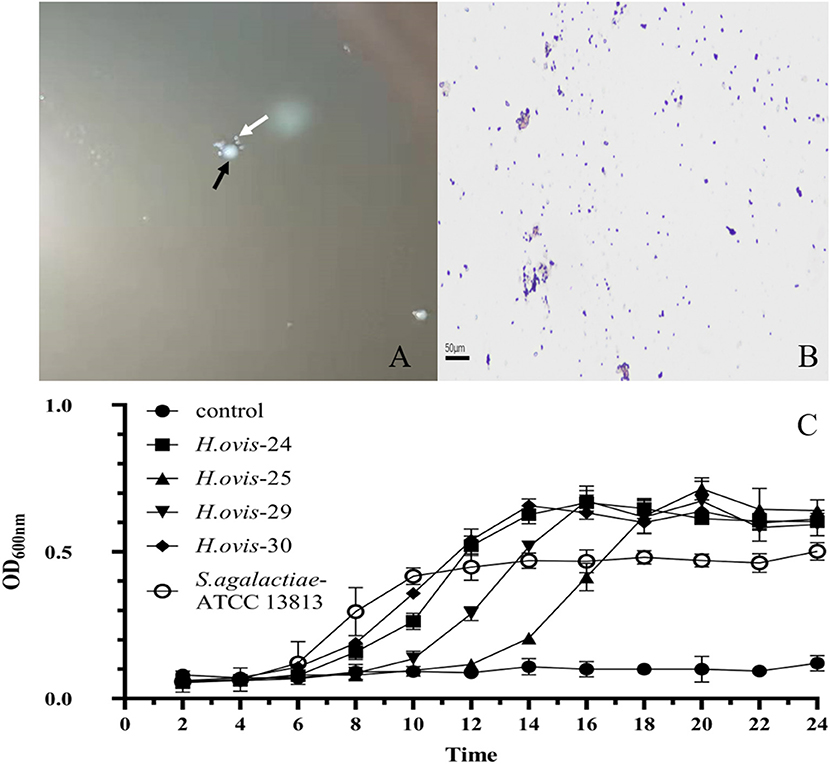
Figure 1. Morphological characteristics and growth curves of Helcococcus ovis. (A) White arrow shows the tiny, transparent colony; the black arrow indicates white colony. (B) Gram-positive cocci (400×). (C) Gram-positive irregularity rod-shaped bacteria (400×).
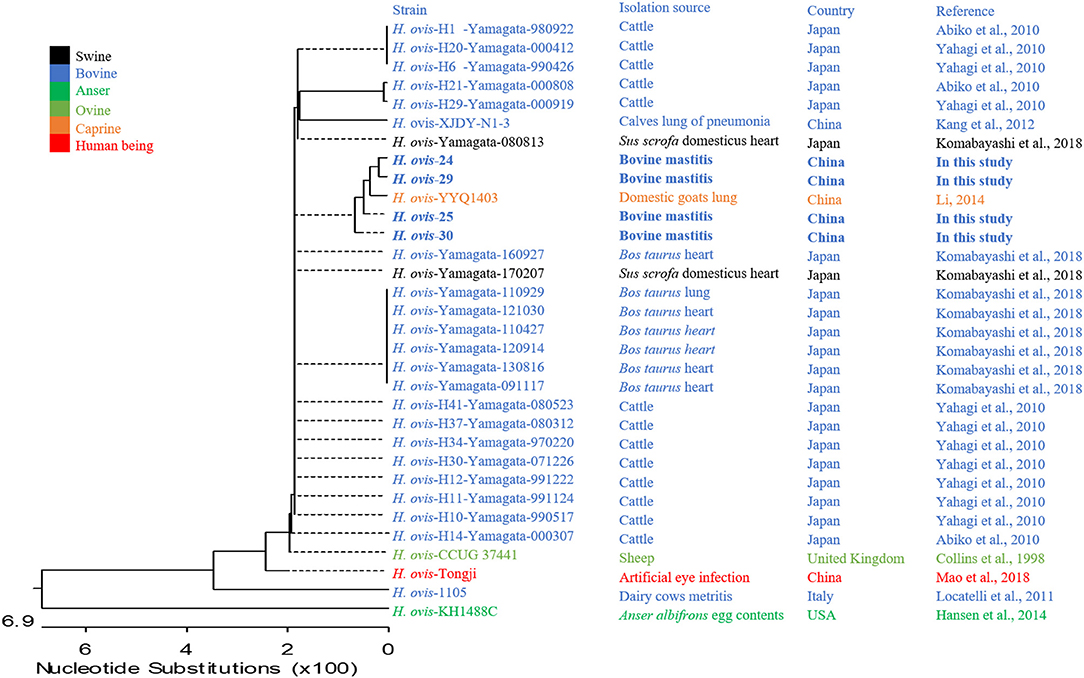
Figure 2. Phylogenetic analysis of four strains of Helcococcus ovis. Bold fonts indicate Helcococcus ovis isolates in this study.
The growth curve of the 4 H. ovis isolates consisted of a lag phase (~6 h), a log phase (~10 h), and a stationary phase. The H. ovis isolates have a long lag phase and relatively higher OD600nm value comparing with Streptococcus agalactiae (Figure 1D).
The ribose, mannitol, sorbitol, raffinose, melibiose, sucrose, β-mannosidase, urease, hippurate, hydrolysis, and esterase reactions were negative for the four isolates while positive to lactose and maltose (Table 2). The biochemistry test characterization of the H. ovis isolates was same to the standard strain of H. ovis CCUG 37441 and CCUG 39041 except for lactose and esterase.
As shown in Table 3, results showed that all isolates were susceptible to penicillin, cefalexin, ceftiofur, oxacillin, clindamycin, erythromycin, and vancomycin; except, one isolate (Hebei-24) was intermediate to tetracycline and enrofloxacin.
Swollen and hyperemia mammary glands were observed 12 h after challenge with H. ovis, and more profound pathological changes observed after 24 and 36 h (Figure 3A) after challenge. For histological characteristics of murine mammary glands (see Figure 3B). Slight infiltrations of inflammatory cells and progressive mammary alveolar damage were observed, stromal hyperplasia appeared in the infected mammary gland 12 and 24 h after challenge, large quantity of lymphocytes formed a tumor-like structure after 36 h, and degeneration and necrosis of mammary epithelial cells were observed. The bacterial load of the H. ovis isolates in the murine mammary gland tissue was 4.6 × 108 CFU/g after 12-h challenge and gradually increased gradually to 6.8 × 108 CFU/g and 1.8 × 109 CFU/g at 24 and 36 h after challenge (Figure 3C).
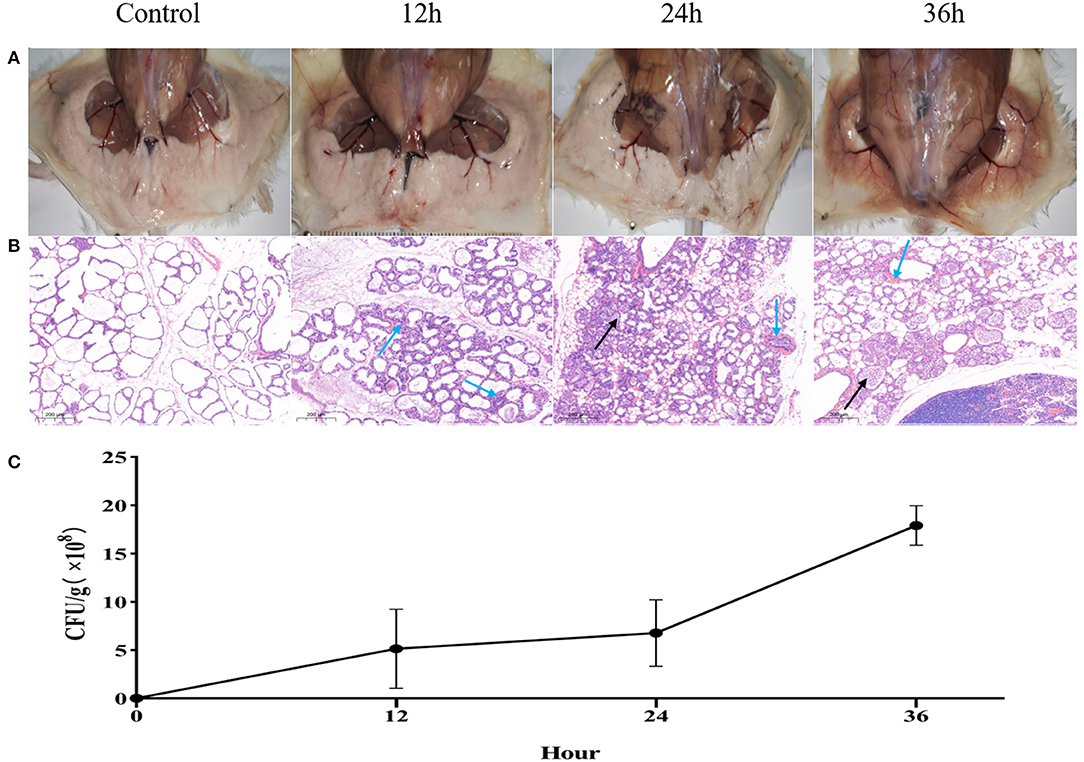
Figure 3. Inflammation and histological evaluation and bacterial burden of murine mammary gland after intramammary inoculation with Helcococcus ovis. (A) Pathological changes in murine mammary glands challenged with Helcococcus ovis. (B) Helcococcus ovis infection induced slight murine mastitis: infiltration of inflammatory cells (mainly neutrophils) into gland alveoli (black arrow) and stromal hyperplasia and progressive mammary alveolar damage (blue arrow). (C) Bacterial burden in mammary glands of mice (up to 36 h after inoculation). Each time point represents five mice in each of the four groups. Data are mean ± SD of three independent experiments.
At 24-h after challenge, the bacterial load was even among the three infected groups (Figure 4C). Swollen, hyperemia, and edema mammary gland were observed after individual challenge with H. ovis and T. pyogenes separately; severer pathological changes exhibited in the group co-infected by H. ovis and T. pyogenes (Figure 4A). Histological examination revealed severer inflammation in the co-infection group than the individual challenge group: stromal hyperplasia, slight infiltrations of inflammatory cells, and mammary alveolar damage exhibited in H. ovis infection group; numerous inflammatory cells infiltrations and mammary alveolar damage were observed in T. pyogenes infection group; whereas massive infiltrations of inflammatory cells and stromal hyperplasia together with mammary alveolar damage exhibited in the H. ovis and T. pyogenes co-infection group (Figure 4B).
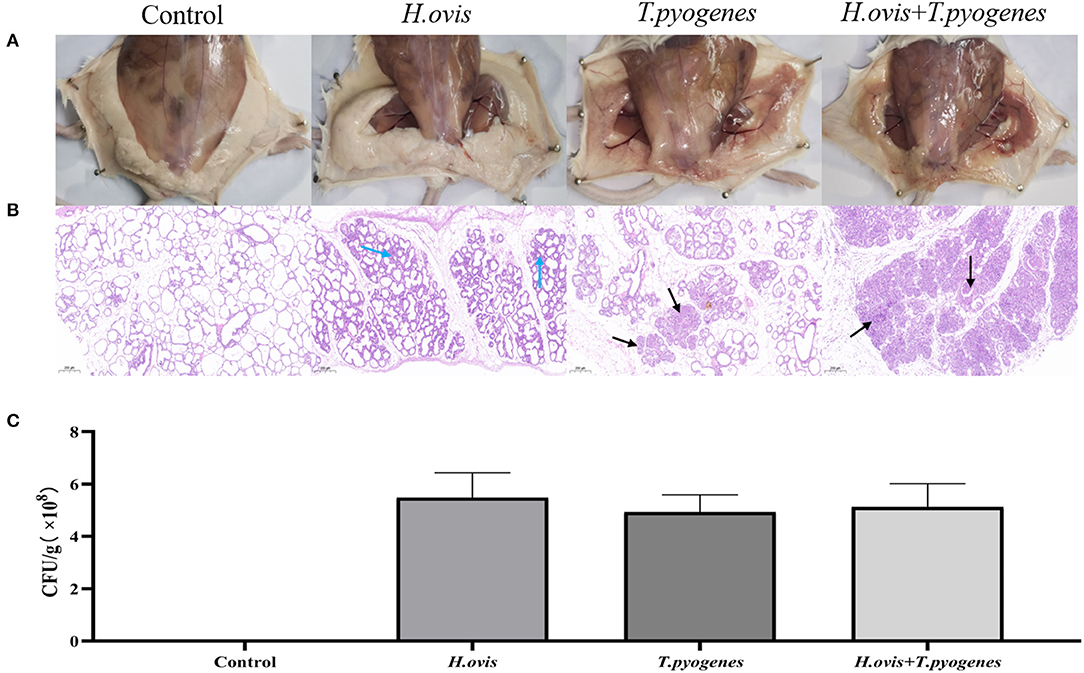
Figure 4. Inflammation and histological evaluation and bacterial burden of murine mammary gland after intramammary inoculation with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. (A) Pathological changes in murine mammary glands challenged with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. (B) Helcococcus ovis induced murine mastitis, with stromal hyperplasia and mammary alveolar damage (blue arrow) after infection. Trueperella pyogenes induced murine mastitis, with an increased infiltration of inflammatory cells (mainly neutrophils) into the gland alveoli (black arrow) and progressive mammary alveolar damage after infection. Helcococcus ovis and Trueperella pyogenes co-infection induced murine mastitis, with an increasing number of inflammatory cells (mainly neutrophils) infiltrated into the gland alveoli (black arrow), stromal hyperplasia, and progressive mammary alveolar damage after infection. (C) Bacterial burden in mammary glands of mice (24 h after inoculation). Each group represents five mice in each of the four groups. Data are mean ± SD of three independent experiments.
The intensity of brown color revealed that TNF-α and IL-1β in murine mammary gland was upregulated in the co-infection group than the H. ovis and T. pyogenes individual infectiongroups in immunohistochemical analysis. The expression of IL-16 in murine serum was increased significantly in the co-infection group than in the individual infection group by the ELISA test results (Figure 5).
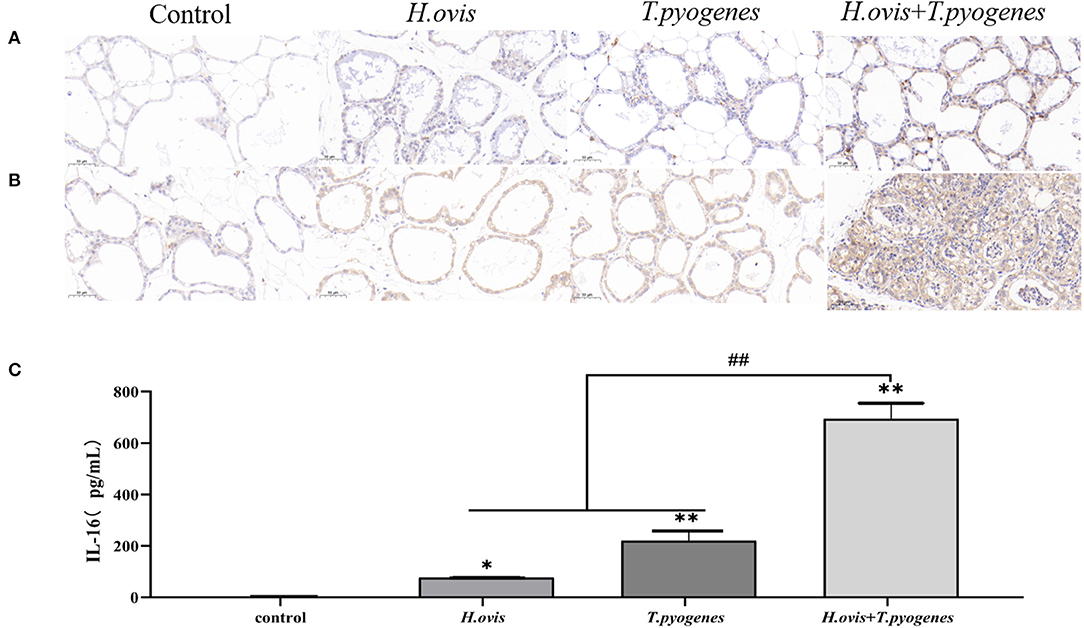
Figure 5. Immunohistochemical (IHC) detection of IL-1β and TNF-α in murine mammary glands and enzyme linked immunosorbent assay (ELISA) detection of IL-16 in murine serum after intramammary inoculation with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. (A) IHC detection of IL-1β in murine mammary glands challenged with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. (B) IHC detection of TNF-α in murine mammary glands challenged with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. (C) ELISA detection of IL-16 in murine serum challenged with Helcococcus ovis, Trueperella pyogenes, Helcococcus ovis, and Trueperella pyogenes. Data are mean ± SD of three independent experiments. *P < 0.05, ** and ##P < 0.01.
To the best of our knowledge, this study is the first report of H. ovis isolated from bovine mastitis in China, although the prevalence was relatively low. In China, there were only four publications of H. ovis, which includes three cases in goat and bovine pneumonia (18, 21, 22) and one in human artificial eye, which raised concern over the zoonotic property of H. ovis (14). More mastitis milk samples should be invesgated and etiological research need to be conducted to confirm the pathogenicity of H. ovis in bovine mastitis and other infection diseases according to Koch's postulates. Rothschild (11) assumed that this bacteria is one of the skin microbiota of cows, which also required further study. The colony of H. ovis is so tiny that it may be ignored by laboratory technicians when mixes with other major bacteria or milk droplets during routine milk sample examination. Similar with mycoplasma, it is a critical mastitis pathogen but difficult to isolate due to slow growth and tiny colonies, which makes it hard to diagnose and research (23). Molecular diagnostics can overcome this and, at the same time, etiology investigation of an emerging pathogen is also pivotal.
Bovine mastitis is predominantly caused by bacteria; therefore, antimicrobials are extensively applied to mastitis prevention and treatment (19), which raised the concern of AMR that threatens human health (18). In this study, the resistance of the four isolates to daptomycin was observed, and one of the isolates was intermediary resistant to tetracycline and enrofloxacin, which were common used in treatments against bovine mastitis. Genome sequencing of H. ovis isolated from bovine puerperal metritis in previous research has indicated that the isolates in that study contained tetracycline-resistant gene: a ribosomal protection gene (tetB) and MFS efflux gene (tetA), and one of the isolates contained AcrEF-TolC, an inner membrane proton that confers resistance to fluoroquinolones, cephalosporins, cephamycins, and penams (8). However, in our study, only slight resistance to antimicrobials was demonstrated, which may indicated that the isolates were rarely exposed to antimicrobial treatments and their emergence in mastitis milk was recent.
The phenotypic characteristics difference of the four isolates with CCUG 37441 and CCUG 39041 may be due to the fact that isolates were isolated from mastitis milk and that lactose was the main carbohydrate source. Because of the unreliability of biochemical methods, they can only be used as auxiliary inspection method, and molecular identification is required to confirm it (14).
The murine model of intramammary challenge with bovine mastitis pathogens has been successfully used to assess bacterial infection and tissue damage (19). The murine model of H. ovis may improve our understanding on the correlation between this bacteria and bovine mastitis, the treatment efficacy applied in clinical trials, as well as the relationship among the bacteria, bovine immune response, and lactation (24). The mammary epithelial cells and the resident macrophages are the first line to interact with pathogens at the onset of mastitis, and then, cytophagocytosis is activated, and pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-8 are released. Consequently, more immunocytes such as neutrophils and lymphocytes are gathered to the inflammation site, and then, more inflammatory cytokines secreted and inflammatory worsened (5).
In this study, the pathogenicity of H. ovis to mice was examnied at different periods after challenge. The slight infiltrations of inflammatory cells, stromal hyperplasia, mild mammary alveolar damage, and bacteria burden in tissue indicated that H. ovis cannot cause severe mastitis. Disease caused by the co-infection of H. ovis and other pathogens have been reported before (6, 9, 10), whereas the pathogenicity of the co-infection H. ovis infection has not been researched. To discover the pathogenicity of the co-infection of H. ovis and T. pyogenes, an advanced study was conducted. Individual infection by T. pyogenes leads to typical bovine mastitis (such as the symptoms of redness, swelling, heat, pain, and dysfunction of mammary glands, as well as massive infiltration of inflammatory cells, and mammary alveolar damage) in this study, which was consistent with previous study that T. pyogenes was distributed in variaty of animals and can lead to bovine mastitis (25). Although the co-infection of H. ovis and T. pyogenes induced severer mastitis than individual infections by each of the two bacteria, the murine mammary gland alveolar was filled with rupt epithelial cells and neutrophils. Leukocytes are always circulating in the blood around mammary gland; when the barriers of mammary gland are broke through by bacteria, the epithelial cells are damaged, cytokines such as IL-1β and TNF-α are released, and chemotaxis moves neutrophils into alveolar to eliminate the invading pathogen. When the infection occurs and neutrophils apoptosis being delayed, it helps limiting the extent of infection (5). The infection of H. ovis and T. pyogenes also increased the IL-16 concentration in murine serum. As IL-16 is secreted by T cells and is chemoattractant to CD4+T cells, the two bacteria may be able to activate cellular immunity. The interaction among microbiota plays a crucial role in biofilm forming, metabolism, and pathogenicity (26–29). Stipkovits also demonstrated the synergy between Mycoplasma arginini (M. arginini) and Streptococcus dysgalactiae (Strep. dysgalactiae); due to the ability of M. arginini to inhibit the T cells growth and the cytotoxic T cells activity, M. arginini infection does not produce clinical signs of mastitis but it can induce severe mastitis together with Strep. dysgalactiae (30). In this study, H. ovis was isolated from samples that T. pyogenes exist, and the co-infection of H. ovis and T. pyogenes caused much serious murine mastitis than individual infection by T. pyogenes; meanwhile, the challenge with H. ovis alone only caused mild mastitis, which means that H. ovis may be able to induce immunosuppression in cows or improve the inflammation.
The H. ovis isolates in our research were closely related to other strains isolated from China and the strains from Japan, the growth speed of the isolates was relatively slower than S. agalactiae, and the phenotypic characteristics were similar to CCUG37441 and CCUG39041 except to lactose; isolates were sensitive to most of antimicrobials; H. ovis could lead to mild murine mastitis and could induce severe mastitis when co-infected with T. pyogenes. As an emerging pathogen, it draws importance when designing mastitis control plans against H. ovis.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
The animal study was reviewed and approved by Institutional Animal Care and Use Committee of Yunnan Agricultural University (Approval No: IACUC-20132030301).
The study was designed by KL, LZ, and XG. The experiments were performed by YL and PC. The data were analyzed by JG, WQ, and BH, who also drafted the manuscript, which was reviewed and revised by ZD and GL. The final version was read and approved by all authors.
This study was supported by the National Natural Science Foundation of China (no. 31660730), Yunnan Provincial Science and Technology Department-Yunnan Expert Workstation (no. 202005AF150041), and Science Research Foundation of Yunnan Education Bureau (grant no. 2020y134).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Shaheen M, Tantary HA, Nabi SU. A treatise on bovine mastitis: disease and disease economics, etiological basis, risk factors, impact on human health, therapeutic management, prevention and control strategy. J Adv Dairy Res. (2016) 1:1000150. doi: 10.4172/2329-888X.1000150
2. Krömker V, Leimbach S. Mastitis treatment-reduction in antibiotic usage in dairy cows. Reprod Domest Anim. (2017) 3:21–9. doi: 10.1111/rda.13032
3. Gussmann M, Denwood M, Kirkeby C, Farre M, Halasa T. Associations between udder health and culling in dairy cows. Prev Vet Med. (2019) 171:104751. doi: 10.1016/j.prevetmed.2019.104751
4. Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. (2017) 100:4797–806. doi: 10.3168/jds.2016-12334
5. Alhussien MN, Dang AK. Potential roles of neutrophils in maintaining the health and productivity of dairy cows during various physiological and physiopathological conditions: a review. Immunol Res. (2019) 67:21–38. doi: 10.1007/s12026-019-9064-5
6. Collins MD, Falsen E, Foster G, Monasterio LR, Dominguez L, Fernandez-Garazabal JF. Helcococcus ovis sp. nov, a gram-positive organism from sheep. Int J Syst Bacteriol. (1999) 4:1429–32. doi: 10.1099/00207713-49-4-1429
7. AHVLA's disease surveillance report. Bovine abortions caused by Helcococcus ovis. Vet Rec. (2014) 175:38–41. doi: 10.1136/vr.g4376
8. Cunha F, Jeon SJ, Kutzer P, Jeong KC, Galvão KN. Draft genome sequences of Helcococcus ovis strains isolated at time of metritis diagnosis from the uterus of holstein dairy cows. Microbiol Resour Announc. (2019) 8:e00402–19. doi: 10.1128/MRA.00402-19
9. Locatelli C, Scaccabarozzi L, Pisoni G, Bronzo V, Casula A, Testa F, et al. Helcococcus kunzii and Helcococcus ovis isolated in dairy cows with puerperal metritis. J Gen Appl Microbiol. (2013) 59:371–4. doi: 10.2323/jgam.59.371
10. Kutzer P, Schulze C, Engelhardt A, Wieler LH, Nordhoff M. Helcococcus ovis, an emerging pathogen in bovine valvular endocarditis. J Clin Microbiol. (2008) 46:3291–5. doi: 10.1128/JCM.00867-08
11. Rothschild CM, Oaks JL, Schaupp JK, Rurangirwa FR, Sellon DC, Hines MT. Helcococcus ovis isolated from a pulmonary abscess in a horse. J Clin Microbiol. (2004) 42:2224–6. doi: 10.1128/JCM.42.5.2224-2226.2004
12. Zhang Y, Cui J, Parkinson A, Hayes J, Ott K, Byrum B. Isolation of Helcococcus ovis from sheep with pleuritis and bronchopneumonia. J Vet Diagn Invest. (2009) 21:164–6. doi: 10.1177/104063870902100130
13. Li R, Yang Y, Tang Q. Diagnosis and treatment of mixed infection of Pasteurella and Helcococcus ovis in Nubian goats (in Chinese). Chin J Vet Med. (2015) 52:48–9.
14. Mao L, Chen Z, Lu Y, Yu J, Zhou Y, Lin Q, et al. Helcococcus ovis in a patient with an artificial eye: a case report and literature review. BMC Infect Dis. (2018) 18:401. doi: 10.1186/s12879-018-3310-7
15. Schwaiger K, Wimmer M, Huber-Schlenstedt R, Fehlings K, Hölzel CS, Bauer J. Hot topic: Bovine milk samples yielding negative or nonspecific results in bacterial culturing–the possible role of PCR-single strand conformation polymorphism in mastitis diagnosis. J Dairy Sci. (2012) 95:98–101. doi: 10.3168/jds.2011-4700
16. Frank JA, Reich CI, Sharma S, Weisbaum JS, Wilson BA, Olsen GJ. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl Environ Microbiol. (2008) 74:2461–70. doi: 10.1128/AEM.02272-07
17. Clinical and Laboratory Standard Institute. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI (2019). p. M100–S124.
18. Cheng J, Qu W, Barkema HW, Nobrega DB, Gao J, Liu G, et al. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J. Dairy Sci. (2019) 102:2416–26. doi: 10.3168/jds.2018-15135
19. Chen P, Qiu Y, Liu G, Li X, Cheng J, Liu K, et al. Characterization of Streptococcus lutetiensis isolated from clinical mastitis of dairy cows. J Dairy Sci. (2021) 104:702–14. doi: 10.3168/jds.2020-18347
20. Adkins PR, Middleton JR. Laboratory Handbook on Bovine Mastitis. Madison, WI: National Mastitis Council Incorporated (2017).
21. Zhou W, Li N, Wei X, Hu J, Xia T, Wang Z, et al. Isolation and identification of Helcococcus ovis from bovine (in Chinese). Chin J Prev Vet Med. (2012) 34:584–6.
22. Guo H. Diagnosis and treatment of a case of mixed infected of Mycoplasma bovine and Helcococcus ovis pneumonia (in Chinese). China Cattle Sci. (2015) 41:91–2.
23. Liu Y, Zhou M, Xu S, Khan MA, Shi Y, Qu W, et al. Mycoplasma bovis-generated reactive oxygen species and induced apoptosis in bovine mammary epithelial cell cultures. J Dairy Sci. (2020) 103:10429–45. doi: 10.3168/jds.2020-18599
24. Ingman WV, Glynn DJ, Hutchinson MR. Mouse models of mastitis - how physiological are they? Int Breastfeed J. (2015) 10:12. doi: 10.1186/s13006-015-0038-5
25. Rzewuska M, Kwiecień E, Chrobak-Chmiel D, Kizerwetter-Swida M, Stefańska I, Gieryńska M. Pathogenicity and virulence of Trueperella pyogenes: a review. Int J Mol Sci. (2019) 20:2737. doi: 10.3390/ijms20112737
26. Bäumler AJ, Sperandio V. Interactions between the microbiota and pathogenic bacteria in the gut. Nature. (2016) 535:85–93. doi: 10.1038/nature18849
27. Peleg AY, Hogan DA, Mylonakis E. Medically important bacterial-fungal interactions. Nat Rev Microbiol. (2010) 8:340–9. doi: 10.1038/nrmicro2313
28. Røder HL, Sørensen SJ, Burmølle M. Studying bacterial multispecies biofilms: where to start? Trends Microbiol. (2016) 24:503–13. doi: 10.1016/j.tim.2016.02.019
29. Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. (2014) 24:40–9. doi: 10.1016/j.cub.2013.10.077
Keywords: pathogenicity, phylogeny, antimicrobial resistance profile, Helcococcus ovis, clinical bovine mastitis
Citation: Liu K, Deng Z, Zhang L, Gu X, Liu G, Liu Y, Chen P, Gao J, Han B and Qu W (2022) Biological Characteristics and Pathogenicity of Helcococcus ovis Isolated From Clinical Bovine Mastitis in a Chinese Dairy Herd. Front. Vet. Sci. 8:756438. doi: 10.3389/fvets.2021.756438
Received: 10 August 2021; Accepted: 29 December 2021;
Published: 11 February 2022.
Edited by:
Paolo Moroni, Cornell University, United StatesReviewed by:
Mohanned Naif Alhussien, Technical University of Munich, GermanyCopyright © 2022 Liu, Deng, Zhang, Gu, Liu, Liu, Chen, Gao, Han and Qu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Weijie Qu, MTQwMTA3NTAxMEBxcS5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.