- 1The International Joint Laboratory for Cooperation in Agriculture and Agricultural Product Safety, Ministry of Education, Yangzhou University, Yangzhou, China
- 2Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
- 3Jiangsu Key Laboratory of Zoonosis, Yangzhou, China
African swine fever virus (ASFV) can infect domestic pigs and wild boars and causes huge economic losses in global swine industry. Therefore, early diagnosis of ASFV is important for the control and eradication of African swine fever (ASF). In this study, a SYBR Green-based real-time polymerase chain reaction (PCR) assay targeting the viral encoded A137R gene was established for the detection of ASFV infection. For the evaluation of the established real-time PCR, 34 clinical samples were assessed by both the A137R gene-based real-time PCR and OIE-recommended TaqMan PCR. The results showed that 85.29% (29/34) were detected by A137R gene-based real-time PCR, but only 79.41% (27/34) positive using OIE-recommended TaqMan PCR. Moreover, no cross-reaction with other common swine pathogens was found in the A137R gene-based real-time PCR. These results demonstrated that the established real-time PCR assay in this study showed better performance than the OIE-recommended method in detecting ASFV from clinical samples, which could be applied for control and eradication programs of ASF.
Introduction
African swine fever (ASF) is a contagious viral disease characterized by fever, cutaneous congestion, multiple hemorrhages in the internal organs, ataxia, severe depression, and high mortality in acute cases. The virus affects pigs of all species, breeds, and ages (1). In August 2018, ASF was reported for the first time in China, which heralded a new transmission era. Subsequently, ASF spreads to the entire Southeast Asia in the next year (2–4). Currently, the outbreaks of ASF are still ongoing in Africa, the trans-Caucasus region, Eastern Europe, Russian Federation and Asia, which caused a huge challenge to the swine industry in these regions (4).
The improvement of African swine fever virus (ASFV) diagnostic methods is one of the essential steps to control and prevent the ASF. Up to date, there is no commercial vaccine for ASF. Moreover, considering the similarities in clinical symptoms between ASF and other swine diseases, such as classical swine fever (CSF), and the potential appearance of non-specific clinical symptoms, the rapid and reliable diagnosis is essential for timely implementation of control measures to block the spread of this devastating disease (5, 6). In World Organization for Animal Health (OIE) handbook, the common laboratory diagnostic methods of ASFV include virus isolation (VI), the haemadsorption (HAD) test, polymerase chain reaction (PCR), loop-mediated isothermal amplification (LAMP), recombinase polymerase amplification (RPA), enzyme-linked immunosorbent assay (ELISA), immunoblotting test and indirect immunofluorescent assay (IFA) (7–9). Virus isolation and HAD using porcine macrophages cells are prestigious methods for ASFV diagnosis. But both methods require excellent laboratory skills and more than 6 days to get the result (10, 11). The conventional PCR recommended by OIE showed lower sensitivity when detecting field and experimental samples infected with vp72 genotype II strains, which may be due to a nucleotide mismatch between primer and viral target gene (10, 12). Recent studies of real-time PCR amplification have shown that low numbers of mismatches in the primer binding regions resulted in a quantification error up to 63.12% (13). Mainly, a conflicting result between real-time PCR and the reference method, direct immuno-fluorescence assay, was reported in which the mismatches in primer and probe binding region have affected real-time PCR accuracy (14). The real-time PCR assays developed in recent years have been commonly used due to their efficiency, high sensitivity and specificity. These assays have been adopted for routine diagnosis in national and reference laboratories (5, 10, 15). Most of them have been designed depend on the VP72-coding region, a highly conserved gene coding the major viral structure protein (8, 10). However, there are relatively few detection methods that target other genes. According to the analysis of genetic variation of the virus, we have recently found several mismatches between the OIE-recommended primers and multiple ASFV isolates (16). Due to new viral isolates displaying new genetic patterns, it is necessary to update the current real-time PCR assay to be able to detect all genotypes of the currently prevalent ASFV.
The A137R gene of ASFV encodes p11.5 protein, which is an important structural protein of ASFV. The high abundance expression of p11.5 protein in the three susceptible cell lines (WSL-HP, HEK 293, and Vero cells) infected with ASFV indicates that it plays an important role in the virus replication cycle (17). A137R gene is also highly expressed in Vero cells infected with ASFV according to transcriptome analysis reported by Cackett et al. (18). Considering the high expression level of the A137R gene, it is likely important throughout infection, which makes it an interesting candidate detection target. In this study, we developed and evaluated a SYBR Green-based real-time PCR assay for detection of ASFV which targeted the A137R gene. A comparative analysis between the A137R gene-based real-time PCR assay and OIE-recommended TaqMan PCR assay was performed. The results indicated that the A137R gene-based real-time PCR had higher sensitivity and advantages of saving time, labor and cost, which is a valuable feature for diagnosis and surveillance of ASFV infections among the pigs. This method will be beneficial for the prevention and control of ASF.
Materials and Methods
Virus and Nucleic Acid Extraction
The full length of A137R gene (414 bp) of ASFV genome (Pig/HLJ/2018, GenBank Accession Number MK333180) were synthetized by Sangon Biotech (Shanghai, China). Classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), porcine epidemic diarrhea virus (PEDV), porcine transmissible gastroenteritis virus (TGEV) and porcine rotavirus (PoRV) were obtained from our laboratory. Porcine circovirus 2 (PCV2) genome was a kind gift from professor Shaobin Shang at the College of Veterinary Medicine, Yangzhou University (Yangzhou, Jiangsu, China). The ASFV genome (GenBank accession number MH766894) was a kind gift from professor Rongliang Hu (Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, China). DNA was extracted by a FastPure Cell/Tissue DNA Isolation Mini Kit (Vazyme Biotech Co., Ltd., Nanjing, China). Total RNA was extracted with a FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme Biotech Co., Ltd., Nanjing, China). Reverse transcription of RNA into cDNA was conducted using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China) following the manufacturer's instruction. To avoid template degradation, eluted nucleic acid was appropriately diluted, aliquoted and stored in −70°C freezer.
Design of the Primers
The forward primer A137R-F (5′-GGACATCGAGTGGTATTAAAAGG-3′, nt 250–272) and the reverse primer A137R-R (5′-TGGCCTGAAAGTCAACATTGA-3′, nt 326–346) were designed based on the alignment of 100 complete coding sequences (CDS) of the ASFV A137R gene available in the GenBank database (The Genbank accession numbers are shown in Figure 1). The sequences were analyzed using MegAlign and Protean software, version 7.1.0 (DNAstar). The primers were chosen in highly conserved regions of the targeted sequences, and the expected size of the real-time PCR product was 97 bp. The primers A137R-F/A137R-R corresponded to sites that were more conserved than those used in the OIE-recommended TaqMan PCR method (19) (Figure 1). To genotype ASFV in the samples that were positive in the developed assay, the forward primer B646L-F (5′-ATGGCATCAGGAGGAGCTTTTTG-3′, nt 1–23) and the reverse primer B646L-R (5′-TTAGGTACTGTAACGCAGCACAGC-3′, nt 1918–1941) were designed and used for automated dideoxynucleotide cycle sequencing of the B646L (p72) gene. All primers were synthesized by Sangon Biotech (Shanghai, China).

Figure 1. Alignment of sequences of the ASFV A137R gene and B646L gene in GenBank. (A) Locations of the target sequences of the primers A137R-F/A137R-R designed in the present study. (B) Locations of the target sites of the primers OIE-recommended TaqMan PCR method (19). Dots (.) indicate identical bases. The primer-binding sequences are boxed.
Construction of Recombinant Standard Plasmid
The A137R gene was cloned into the TA vector pGEM-T easy (TaKaRa, Dalian, China) using common molecular biology techniques. The recombinant vector were transformed into E. coli DH5α competent cells (Vazyme Biotech Co., Ltd., Nanjing, China). Plasmid constructs were confirmed by Sanger sequencing (Sangon Biotech, Shanghai, China), and quantified by a NanoDrop ND-1000 Spectrophotometer (NanoDrop, Wilmington, Delaware, USA). Moreover, the standard plasmid copy number (N) was calculated by the following formula: N (copies/ml) = [6.02 × 1023 (copies/mol) × C (g/ml)]/MW (g/mol), where C is the concentration, and MW is the average molecular weight (double-stranded DNA) calculated as the number of bases (B) ×660 (g/mol) bases. B was determined by adding the number of bases in the plasmid and in the insert.
Optimization of Real-Time PCR
The real-time PCR assay was developed and validated using the LightCycler (Roche Diagnostics, Mannheim, Germany) and TaKaRa TB Green Premix Ex Taq II (TaKaRa, Dalian, China). With other factors constant, the concentration of primers and annealing temperature were optimized to make the method more specific and sensitive (20). Four concentrations of the primer: 0.1, 0.2, 0.4, 0.6 μM and four different annealing temperatures: 56, 58, 60, 62°C were used in the experiment. According to the results, the optimum conditions were determined.
Standard Curve
The standard plasmid was diluted 10-fold serially and amplified with the optimized real-time PCR system at the concentration of 1.0 × 108−1.0 × 101 copies/μL. The final standard curve is generated based on the CT value and the logarithm of standard copy number. Meanwhile, nuclease free water was used as negative control.
Sensitivity
To determine the sensitivity of the assay, the standard plasmid was diluted 10-fold serially with concentrations ranging from 1.0 × 108 to 1.0 × 101 copies/μL. Prepared standards were amplified with the optimized real-time PCR system to investigate the sensitivity of the method. In addition, one of ASFV DNA samples with 10-fold dilution was tested using both A137R gene-based real-time PCR assay and the OIE-recommended TaqMan PCR assay developed by King et al. to compare the detection limit of the assays (19).
Specificity
The real-time PCR assay was performed with other templates from PCV2, CSFV, PRRSV, PEDV, TGEV, and PoRV to evaluate the specificity of the reaction.
Repeatability
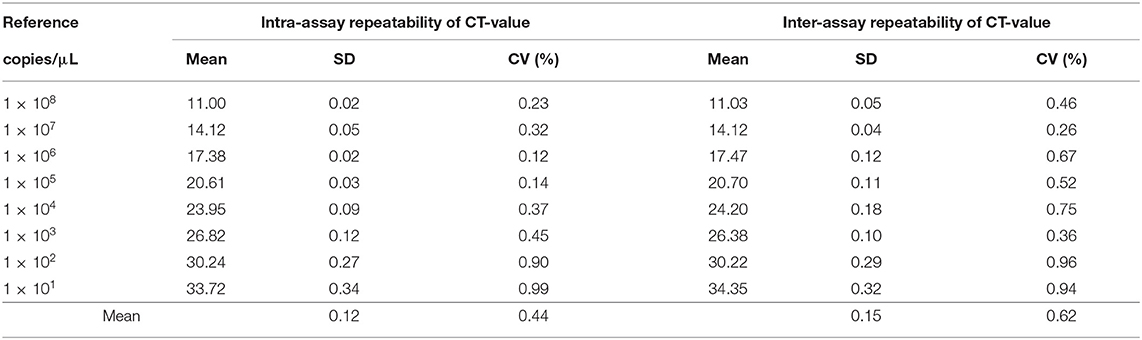
The standard plasmids with 10-fold serial dilution from 1.0 × 108 to 1.0 × 101 copies/μL were used for the repeatability test. The diluted standard plasmids were stored in −70°C freezer immediately after each dilution was made. For the intra-assay test, three replicate samples from each dilution were tested in the same run. For the inter-assay test, each dilution of standard plasmids was tested in three independent runs to measure the test reliability or reproducibility regarding the mean CT-values with standard deviation (SD) and coefficient variation (CV).
Clinical Sample Detection
A total of 34 specimens, including 10 swine anticoagulant blood, 10 tissue samples, 10 cloacal swabs and 4 samples of pig livers were processed for DNA extraction. Four samples of pig livers were collected from different supermarkets in Yangzhou, China. Other specimens were a kind gift from professor Rongliang Hu (Institute of Military Veterinary Medicine, Academy of Military Medical Sciences, Changchun, China). All the samples were stored at −70°C for subsequent use. DNA extracted from each of specimens was detected by A137R gene-based real-time PCR assay. The results were compared with those obtained by OIE-recommended TaqMan PCR that was performed in parallel. The samples with different results were sequenced and analyzed using primers B646L-F/B646L-R. Subsequently, 7 selected DNA of ASFV positive specimens, which included 2 swine anticoagulant blood, 3 tissue samples and 2 cloacal swabs were diluted with serial 10-fold dilutions. These dilution series DNA were tested both by the developed method and the OIE-recommended TaqMan PCR.
Results
Optimization of Real-Time PCR System
According to the results of Orthogonal test, the concentration of primers at 0.4 μM was the most efficient. The optimized real-time PCR reaction volume was 20 μL containing 10 μL TB Green Premix Ex Taq II, 0.4 μM forward primer, 0.4 μM reverse primer, 6.4 μL RNase Free ddH2O and 2.0 μL DNA template. The optimal program was as follows: one cycle of 30 s at 95°C, 45 cycles of 5 s at 95°C and 30 s at 62°C. Fluorescence signals for each sample were harvested at the end of each step at 62°C. Finally, the dissolution curve was added: one cycle at 95°C for 15 s, 60°C for 30 s and 95°C for 15 s.
Establishment of Standard Curve
To construct a standard curve with the logarithm of plasmid copy numbers and the measured CT value (Figure 2A), serial plasmid dilutions at the concentration of 1.0 × 108-1.0 × 101 copies/μL were prepared. Three replicates were tested for each dilution. The optimal curve was selected as the standard curve. Establishment of ASFV standard curve with abscissa as logarithm of copy number and ordinate as CT-value, the correlation coefficient (R2) was 0.9992, the slope was −3.252, and the intercept was 36.26. The standard formula is y = −3.252x + 36.26, and R2 = 0.9992. Moreover, negative or extraction blank controls should have a CT > 38.0.

Figure 2. Standard curve and sensitivity tests for A137R gene-based real-time PCR assay of ASFV. (A) Establishment of standard curve for A137R gene-based real-time PCR. The 10-fold serial dilutions ranging from 1.0 × 108 to 1.0 × 101 copies/μL of DNA plasmid were tested in the real-time PCR. Each point corresponds to the mean value of three replicates. The optimal standard formula is y = −3.252x + 36.26, and the correlation coefficient is 0.9992. (B) Sensitivity tests for A137R gene-based real-time PCR assay of ASFV. Ten-fold serial dilutions of the DNA plasmid were used to perform the real-time PCR to obtain the expanded curve of the assay.
Sensitivity of Real-Time PCR Targeting the A137R Gene
Sensitivity tests were performed using plasmids ranging from 1.0 × 108 to 1.0 × 101 copies/μL. The 10-fold serial dilutions of the standard plasmids were simultaneously detected by the established real-time PCR. The minimum detection template concentration of method was 10 copies/μL (Figure 2B). As shown in Figure 3, the A137R gene-based real-time PCR had 10 times lower detection limit when compared with that of the OIE-recommended TaqMan PCR (19), indicating that the established real-time PCR was more sensitive than OIE-recommended TaqMan PCR.
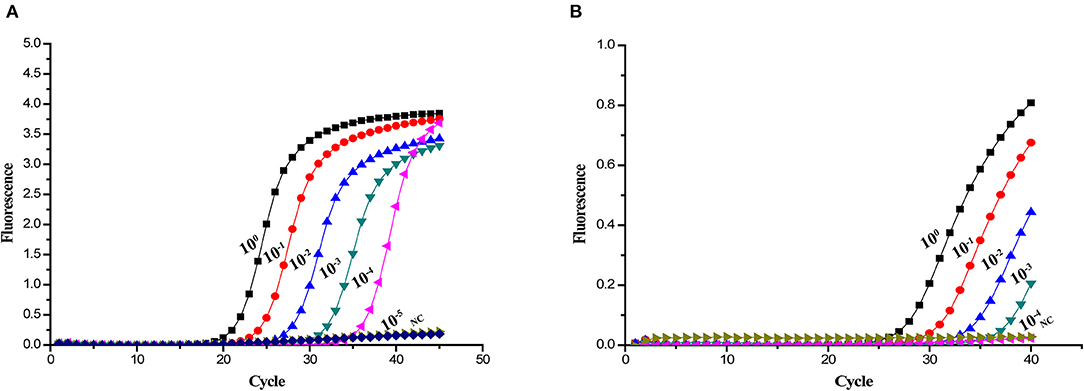
Figure 3. Sensitivity of different real-time PCR assays for detection of ASFV DNA. Ten-fold serial dilutions of the ASFV DNA were used to perform the real-time PCR to obtain the expanded curve of the assays. (A) Detection limit of the A137R gene-based real-time PCR assay. (B) Detection limit of the OIE-recommended TaqMan PCR assay.
Specificity of Real-Time PCR Targeting the A137R Gene
To investigate the specificity of the real-time PCR, other templates from common swine viruses were tested with the assay. The ASFV DNA samples got strong fluorescent signals. However, the negative control and other swine viruses such as PCV2, CSFV, PRRSV, PEDV, TGEV, and PoRV showed no signal amplification (Figure 4). Therefore, there is a significant distinction between ASFV and other swine viruses by comparing the signal strength at different levels.
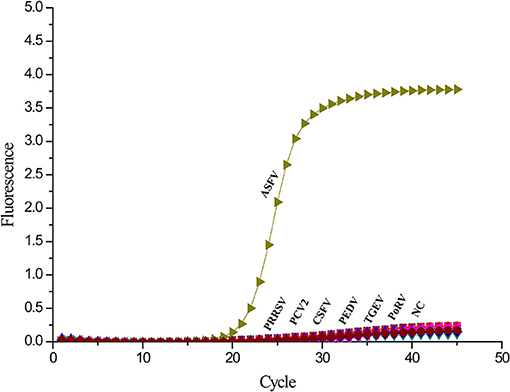
Figure 4. Specificity analysis of the A137R gene-based real-time PCR. Only ASFV showed a positive fluorescence signal, and no positive signal was observed with other swine viruses.
Repeatability of Real-Time PCR Targeting the A137R Gene
The intra-assay repeatability was assessed by testing 10-fold serial dilutions of the standard plasmids ranging from 1.0 × 108 to 1.0 × 101 copies/μL in three replicates. To determine the intre-assay repeatability, the same standard plasmids were analyzed in triplicate on 3 different days. In result, intra-assay standard deviations (SD) ranged from 0.02 to 0.34, and inter-assay standard deviations (SD) ranged from 0.04 to 0.32. The coefficient of variation (CV) value of intra-assay and inter-assay were 0.12–0.99% and 0.26–0.96%, respectively (Table 1). The results showed that the real-time PCR targeting the A137R gene developed in this study is repeatable.
Clinical Sample Application
To determine the practical application of this method, we used 34 specimens to compare the real-time PCR targeting the A137R gene and recommended TaqMan PCR. The positivity rates of ASFV in swine anticoagulant blood, tissue samples and cloacal swabs were determined to be 90% (9/10), 100% (10/10), and 80% (8/10) by OIE-recommended TaqMan PCR. While, they were 100% (10/10) of swine anticoagulant blood, 100% of tissue samples (10/10) and 90% (9/10) of cloacal swabs in the A137R gene-based real-time PCR assay. Four samples of pig livers from supermarket were detected negative by both methods. As shown in Table 2, the positivity rate of the A137R gene-based real-time PCR was 85.29% (29/34) and only 79.41% (27/34) in the OIE-recommended TaqMan PCR. The positive samples in this method but negative in the OIE-recommended method were sequenced. Sequencing results showed that these sequences are identical to the genotype II viruses circulating in China, without any mutations. The positive detection rate of these clinical samples tested by our assay was higher than that of OIE-recommended TaqMan PCR, which may correlate with different expression level of target genes in early infection. The results of 7 selected DNA of ASFV positive specimens in Table 3 showed that A137R gene-based real-time PCR had an equal or 10 times higher sensitivity when compared with the OIE-recommended TaqMan PCR. Therefore, the method developed in this study showed better performance than OIE-recommended TaqMan PCR assay in detecting ASFV gene from clinical samples.

Table 2. Comparison of detecting clinical samples by A137R gene-based real-time PCR and OIE-recommended TaqMan PCR.
Discussion
African swine fever is a highly contagious disease with 100% morbidity and mortality. China, the largest pork producer and consumer, had attracted global concerns because of the prevalence of ASFV (21). Various ASFV vaccine strategies focusing on DNA-, antigen- and virus vector-based have been investigated. However, there is still no successful vaccine until now (22). Many control measures were conducted by Chinese government to inhibit the rapid spread of ASFV which emphasize the need for early, rapid, high rates of sensitive and specific diagnosis in ASFV. For this purpose, PCR assays have been recommended as the methods for detecting ASFV.
Real-time PCR and conventional PCR assay are the most widely used assay and also recommended by OIE for virological and molecular diagnosis of ASF. However, one report showed that the OIE-recommended conventional PCR for ASFV had low sensitivity, most likely due to an imperfect match of the primers with the target sequences of some ASFV genotypes (5, 10). Although OIE-recommended TaqMan PCR has shown excellent sensitivity and specificity rates, the high fidelity of the method is slightly decreased when the samples showed low or weak ASFV-positive (23, 24). Therefore, it is necessary to find more effective detection targets to improve the detection of ASFV. Some scholars have analyzed ASFV protein expression in three susceptible mammalian cell lines representing a susceptible host (wild boar) and two non-susceptible species (human and green monkey) by mass spectrometry. The results showed that p11.5 protein encoded by A137R gene was highly expressed in three susceptible cell lines infected with ASFV (17). The results are consistent with transcriptome analysis (18). It is speculated that p11.5 protein may play an important role in the viral replication cycle. Due to SYBR Green-based real-time PCR have similar sensitivity and specificity as TaqMan-based PCR (25), we developed and evaluated a SYBR Green-based real-time PCR assay which target the A137R gene for detection of ASFV.
In present study, a pair of primers was designed based on the alignment of A137R gene sequences targeting conserved areas of the gene and making these primers more universally applicable than those used in OIE-recommended TaqMan PCR. BLAST searches of the new ASFV primers confirmed that the primer sequences target highly conserved regions of the ASFV A137R gene sequences. Only one ASFV isolate was found to have mismatches with the primers used in the new real-time PCR, while several mismatches were found between the OIE-recommended primers and multiple ASFV isolates. These indicated that the novel assay could be used for universal detection of ASFV. After optimization, the A137R gene-based real-time PCR assay was established. A correlation coefficient higher than 0.98 indicates that the linear correlation of the standard curve was good (26). The results showed that the correlation standard curve equation was y = −3.252x + 36.26, and the correlation coefficient was 0.9992. Therefore, the standard curve showed a good linear correlation. Based on the data, the detection limit of A137R gene-based real-time PCR was as low as 10 copies/μL. No positive signal was observed with other swine viruses, indicating favorable specificity of the assay. Repeatability analysis revealed the intra-assay and inter-assay CVs to be lower than 1%, which indicated the repeatable of the method. DNA of ASFV specimens was tested after 10-fold serial dilution. The results showed that A137R gene-based real-time PCR was more sensitive than OIE-recommended TaqMan PCR in most ASFV specimens. When testing swine anticoagulant blood and cloacal swabs with lower virus levels, the positivity rates of the A137R gene-based real-time PCR was higher than OIE-recommended TaqMan PCR. Recently, genotype I ASFV has been reported in domestic pigs in China (27). The A137R primers used in the developed SYBR Green-based real-time PCR targeting the A137R gene of ASFV are highly conserved across all known ASFV genotypes, including genotype I, making it possible to detect both genotype I and II ASFV co-circulating in China at the moment. The real-time PCR assay developed in this study enables the detection of virus in the early stage of infection or low virus concentration, providing a good method for the early diagnosis of the disease.
In conclusion, our data indicated that SYBR Green-based real-time PCR targeting the A137R gene is a sensitive, specific, repeatable and reliable method to detect ASFV. In addition, SYBR Green-based method has advantages of saving time, labor, and cost. It could therefore be a promising tool for clinical diagnosis and epidemiological investigation of ASFV as well as for future research on the virus.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
Material preparation, data collection, and analysis were performed by DY, RG, HL, CB, KQ, and AQ. The first draft of the manuscript was written by DY and revised by AQ and KQ. All authors contributed to the study conception and design, commented on previous versions of the manuscript, and read and approved the final manuscript.
Funding
This work was supported by National Science Foundation of China (31941016), the National Key Research and Development Program (Grant No. 2021YFD180001), the Priority Academic Program Development of Jiangsu Higher Education Institutions, and the Jiangsu Co-innovation Center for the Prevention and Control of Important Animal Infectious Diseases and Zoonoses. The funding bodies did not play direct roles in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gallardo MC, Reoyo AT, Fernandez-Pinero J, Iglesias I, Munoz MJ, Arias ML. African swine fever: a global view of the current challenge. Porcine Health Manag. (2015) 1:21. doi: 10.1186/s40813-015-0013-y
2. Zhou X, Li N, Luo Y, Liu Y, Miao F, Chen T, et al. Emergence of African Swine fever in China, 2018. Transbound Emerg Dis. (2018) 65:1482–4. doi: 10.1111/tbed.12989
3. Bao J, Wang Q, Lin P, Liu C, Li L, Wu X, et al. Genome comparison of African swine fever virus China/2018/AnhuiXCGQ strain and related European p72 Genotype II strains. Transbound Emerg Dis. (2019) 66:1167–76. doi: 10.1111/tbed.13124
4. Danzetta ML, Marenzoni ML, Iannetti S, Tizzani P, Calistri P, Feliziani F. African swine fever: lessons to learn from past eradication experiences. A systematic review. Front Vet Sci. (2020) 7:296. doi: 10.3389/fvets.2020.00296
5. Galindo I, Alonso C. African swine fever virus: a review. Viruses. (2017) 9:103. doi: 10.3390/v9050103
6. Luo Y, Atim SA, Shao L, Ayebazibwe C, Sun Y, Liu Y, et al. Development of an updated PCR assay for detection of African swine fever virus. Arch Virol. (2017) 162:191–9. doi: 10.1007/s00705-016-3069-3
7. Cubillos C, Gomez-Sebastian S, Moreno N, Nunez MC, Mulumba-Mfumu LK, Quembo CJ, et al. African swine fever virus serodiagnosis: a general review with a focus on the analyses of African serum samples. Virus Res. (2013) 173:159–67. doi: 10.1016/j.virusres.2012.10.021
8. Oura CA, Edwards L, Batten CA. Virological diagnosis of African swine fever–comparative study of available tests. Virus Res. (2013) 173:150–8. doi: 10.1016/j.virusres.2012.10.022
9. Sanchez-Vizcaino JM, Mur L, Gomez-Villamandos JC, Carrasco L. An update on the epidemiology and pathology of African swine fever. J Comp Pathol. (2015) 152:9–21. doi: 10.1016/j.jcpa.2014.09.003
10. Gallardo C, Nieto R, Soler A, Pelayo V, Fernandez-Pinero J, Markowska-Daniel I, et al. Assessment of African swine fever diagnostic techniques as a response to the epidemic outbreaks in eastern European union countries: how to improve surveillance and control programs. J Clin Microbiol. (2015) 53:2555–65. doi: 10.1128/JCM.00857-15
11. Malmquist WA, Hay D. Hemadsorption and cytopathic effect produced by African Swine fever virus in swine bone marrow and buffy coat cultures. Am J Vet Res. (1960) 21:104–8.
12. Aguero M, Fernandez J, Romero L, Sanchez Mascaraque C, Arias M, Sanchez-Vizcaino JM. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J Clin Microbiol. (2003) 41:4431–4. doi: 10.1128/JCM.41.9.4431-4434.2003
13. Suss B, Flekna G, Wagner M, Hein I. Studying the effect of single mismatches in primer and probe binding regions on amplification curves and quantification in real-time PCR. J Microbiol Methods. (2009) 76:316–9. doi: 10.1016/j.mimet.2008.12.003
14. Kamau E, Agoti CN, Lewa CS, Oketch J, Owor BE, Otieno GP, et al. Recent sequence variation in probe binding site affected detection of respiratory syncytial virus group B by real-time RT-PCR. J Clin Virol. (2017) 88:21–5. doi: 10.1016/j.jcv.2016.12.011
15. Fernandez-Pinero J, Gallardo C, Elizalde M, Robles A, Gomez C, Bishop R, et al. Molecular diagnosis of African Swine Fever by a new real-time PCR using universal probe library. Transbound Emerg Dis. (2013) 60:48–58. doi: 10.1111/j.1865-1682.2012.01317.x
16. Tran HTT, Dang AK, Ly DV, Vu HT, Hoang TV, Nguyen CT, et al. An improvement of real-time polymerase chain reaction system based on probe modification is required for accurate detection of African swine fever virus in clinical samples in Vietnam. Asian-Aust J Anim Sci. (2020) 33:1683–90. doi: 10.5713/ajas.19.0525
17. Kessler C, Forth JH, Keil GM, Mettenleiter TC, Blome S, Karger A. The intracellular proteome of African swine fever virus. Sci Rep. (2018) 8:14714. doi: 10.1038/s41598-018-32985-z
18. Cackett G, Matelska D, Sykora M, Portugal R, Malecki M, Bahler J, et al. The African swine fever virus transcriptome. J Virol. (2020) 94:e00119–20. doi: 10.1128/JVI.00119-20
19. King DP, Reid SM, Hutchings GH, Grierson SS, Wilkinson PJ, Dixon LK, et al. Development of a TaqMan PCR assay with internal amplification control for the detection of African swine fever virus. J Virol Methods. (2003) 107:53–61. doi: 10.1016/S0166-0934(02)00189-1
20. Bora DP, Venkatesan G, Bhanuprakash V, Balamurugan V, Prabhu M, Siva Sankar MS, et al. TaqMan real-time PCR assay based on DNA polymerase gene for rapid detection of Orf infection. J Virol Methods. (2011) 178:249–52. doi: 10.1016/j.jviromet.2011.09.005
21. Dixon LK, Sun H, Roberts H. African swine fever. Antiviral Res. (2019) 165:34–41. doi: 10.1016/j.antiviral.2019.02.018
22. Gaudreault NN, Richt JA. Subunit vaccine approaches for African swine fever virus. Vaccines. (2019) 7:56. doi: 10.3390/vaccines7020056
23. McKillen J, McMenamy M, Hjertner B, McNeilly F, Uttenthal A, Gallardo C, et al. Sensitive detection of African swine fever virus using real-time PCR with a 5' conjugated minor groove binder probe. J Virol Methods. (2010) 168:141–6. doi: 10.1016/j.jviromet.2010.05.005
24. Tignon M, Gallardo C, Iscaro C, Hutet E, Van der Stede Y, Kolbasov D, et al. Development and inter-laboratory validation study of an improved new real-time PCR assay with internal control for detection and laboratory diagnosis of African swine fever virus. J Virol Methods. (2011) 178:161–70. doi: 10.1016/j.jviromet.2011.09.007
25. Wang Y, Fu Z, Guo X, Zhang D, Bai C, Li W, et al. Development of SYBR Green I-based real-time reverse transcription polymerase chain reaction for the detection of feline astrovirus. J Virol Methods. (2021) 288:114012. doi: 10.1016/j.jviromet.2020.114012
26. Nieto D, Aramouni M, Grau-Roma L, Segales J, Kekarainen T. Dynamics of Torque teno sus virus 1 (TTSuV1) and 2 (TTSuV2) DNA loads in serum of healthy and postweaning multisystemic wasting syndrome (PMWS) affected pigs. Vet Microbiol. (2011) 152:284–90. doi: 10.1016/j.vetmic.2011.05.020
Keywords: African swine fever virus, A137R gene, SYBR Green, real-time PCR, detection
Citation: Yin D, Geng R, Lv H, Bao C, Shao H, Ye J, Qian K and Qin A (2021) Development of Real-Time PCR Based on A137R Gene for the Detection of African Swine Fever Virus. Front. Vet. Sci. 8:753967. doi: 10.3389/fvets.2021.753967
Received: 05 August 2021; Accepted: 04 November 2021;
Published: 29 November 2021.
Edited by:
Mary-Louise Penrith, University of Pretoria, South AfricaReviewed by:
Gerald Misinzo, Sokoine University of Agriculture, TanzaniaJuanita Van Heerden, Agricultural Research Council of South Africa (ARC-SA), South Africa
Copyright © 2021 Yin, Geng, Lv, Bao, Shao, Ye, Qian and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kun Qian, cWlhbmt1biYjeDAwMDQwO3l6dS5lZHUuY24=; Aijian Qin, YWlqaWFuJiN4MDAwNDA7eXp1LmVkdS5jbg==
 Dan Yin1,2,3
Dan Yin1,2,3 Hongxia Shao
Hongxia Shao Jianqiang Ye
Jianqiang Ye Kun Qian
Kun Qian Aijian Qin
Aijian Qin