
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Vet. Sci. , 10 September 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.744276
Porcine epidemic diarrhea virus (PEDV) is a causative agent of a highly contagious enteric disease in swine of all ages, leading to severe economic losses for the swine industry in many countries. One of the most effective approaches in controlling PEDV infection is vaccination. The ORF3 accessory protein has been proposed as a crucial viral virulence factor in a natural host. However, due to the lack of an extensive comparative study of ORF3, exactly how the ORF3 takes part in virus replication and pathogenesis as well as its role in host-virus interaction is unclear. In this review, we aim to discuss the current knowledge of ORF3 concerning its dispensability for viral replication in vitro, ability to modulate host responses, contribution to virus pathogenicity, and research gaps among ORF3 functional studies. These will be beneficial for further studies to a better understanding of PEDV biology and PEDV vaccine development.
Porcine epidemic diarrhea virus (PEDV) is an enteric pathogen that has spread in the swine population. PEDV infection causes severe watery diarrhea, dehydration, vomiting, and death, particularly in neonatal piglets, resulting in massive economic losses in pig industries worldwide, particularly in the United States, China, South Korea, and Thailand (1–4). According to the phylogenetic analysis, PEDV has been classified into two major groups, namely genotype 1 (G1) and genotype 2 (G2) (5). The G1 group could be divided into two sub-genotypes: G1a and G1b. The classical PEDV strains CV777 and DR13 are the G1 representatives (6). During the 1980-the 2000s, the G1a PEDV had caused outbreaks in Asia (7). Despite its moderate virulence, vaccines against this diarrheal disease had been developed and used in many countries, including Japan, China, and South Korea (8). The highly virulent G2 strain has emerged in China in 2010 and spread to many countries worldwide (2, 3), with the mortality rate in nursing piglets almost 100% (9). Although various G1a-based vaccines have been used to control the outbreaks, their efficacy against these highly virulent strains is minimal (10). Vaccines specifically designed for the G2 genotype are thus necessary to effectively control the ongoing PEDV epidemics.
Belonging to the genus Alphacoronavirus, PEDV is an enveloped virus bearing positive-sense single-stranded RNA of approximately 28 kb in length. The viral genome comprises at least seven open reading frames (Figure 1A) encoding two polyproteins, pp1a and pp1ab, which can be processed into 16 non-structural proteins (nsps), four structural proteins (spike, S; envelope, E; membrane, M and nucleocapsid, N), and only one accessory protein, the ORF3 (12). For decades, numerous studies have been carried out to gain more insights into various aspects of PEDV, including basic virology, pathogenesis, immune responses and vaccine design. Despite the rapidly accumulating data of the coronavirus structural proteins, those related to the accessory proteins are relatively limited. It has been demonstrated that PEDV ORF3 participates in increased virus infection and lesion in the swine intestinal tract (11, 13). The defective PEDV ORF3 with deletion at a C-terminus acquired after virus adaptation in cell culture appeared to reduce virus virulence (13, 14). However, the evidence so far could not delineate how the ORF3 governs virus pathogenesis and virus replication in vitro and in vivo. Many questions remain unanswered and need comprehensive studies to understand the functions of this protein.
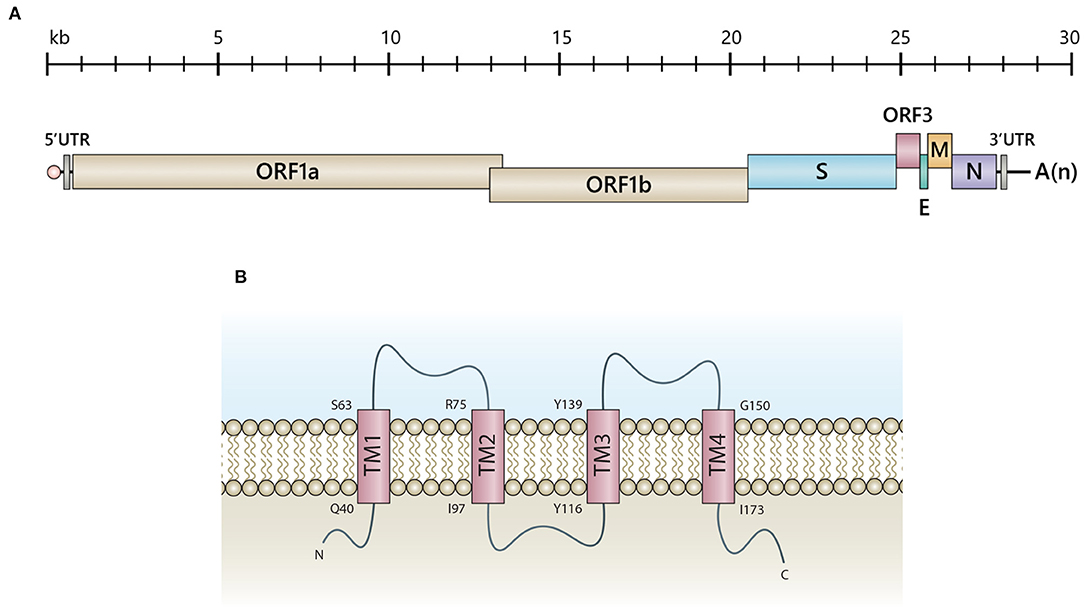
Figure 1. Schematic representation of PEDV genomic RNA and the predicted ORF3 structure. (A) The PEDV genome consists of 5'-capped UTR (5'UTR), ORF1a, and ORF1b followed by the genes encoding the spike (S), accessory protein (ORF3), envelope (E), membrane protein (M) and nucleoprotein (N), and 3'polyadenylated UTR (3'UTR). (B) Illustration of a predicted ORF3 structure proposed by computational modeling and transmembrane prediction programs (11). Transmembrane (TM) domains include TM1 (aa positions 40-63), TM2 (aa positions 75-97), TM3 (aa positions 116-139) and TM4 (aa positions 150-173). Q, Glutamine; S, Serine; R, Arginine; I, Isoleucine; Y, Tyrosine; G, Glycine.
The PEDV ORF3 [675 nucleotides; 224 amino acids (aa)] is a viroporin, consisting of four transmembrane domains possessing a potassium ion channel activity (11) (Figure 1B). Viral proteins bearing ion channel activity, including E proteins of MHV (15), SARS-CoV (16), and IBV (17), have been reported to play a role in pathogenesis, virus assembly, and release. Studies performed in Xenopus laevis oocytes and yeast cells expressing transmembrane-truncated PEDV ORF3 demonstrated that the constructs bearing deleted amino acid at positions 82-98 and 151-172 completely lost the channel activity (11), which might be associated with the reduced pathogenicity of PEDV strain DR13 (aa82-99 deletion) in piglets (18). However, the evidence showing that the ion channel activity of ORF3 is necessary for PEDV replication is still lacking. Most of the available data are based on studies in ORF3a of SARS-CoV. The ion channel activity of SARS-CoV ORF3a has been shown to play a role in vial pro-apoptotic property (19), virus replication, and pathogenesis in an animal model (20). Nevertheless, the SARS-CoV ORF3a's ion activity is not related to the induction of IL-1β, virus replication and virus virulence in cell culture (20, 21). The study also demonstrated a functional compensation between ORF3a and other viroporins on virus viability and disease severity, impacting the rational vaccine design (20). However, it should be noted that PEDV ORF3 and SARS-CoV ORF3a are quite remarkably distinct in terms of protein structure and function. PEDV ORF3 likely may not utilize its ion channel activity in the same manner as observed in SARS-CoV or other coronaviruses.
Wild-type PEDV and cell culture adapted PEDV (ca-PEDV) have been shown to exert different degrees of disease severity in the infected hosts. Bernasconi et al. (22) showed that only 5 out of 21 newborn piglets inoculated with ca-PEDV had mild watery diarrhea, and no other clinical signs were observed. Likewise, piglets orally inoculated with KPEDV-9, a highly adapted variant, did not have signs of diarrhea or other clinical symptoms (23). PEDV DR13 strain that was serially passaged in Vero cells also lost virus pathogenicity (24). When the genetic constellation of wild-type and attenuated PEDVs were analyzed and compared, it is evident that PEDV ORF3 gene derived from attenuated PEDVs, but not the virulent variant, displays shortened or truncated ORF3 with 51 nucleotide deletions (18), suggesting that the PEDV ORF3 might involve in disease severity. A recent study by Wang et al. (13) demonstrated that ORF3 might play a crucial role in viral virulence. By swapping the S gene derived from the virulent strain into the genome of the avirulent strain, the authors showed that the chimeric virus did not alter the infectivity and invasiveness of attenuated strain, suggesting that S alone is not the only satisfactory factor obligated for virus virulence. The authors later showed that the inclusion of other structural genes, especially the full-length ORF3 derived from the virulent strain, could markedly enhance the virulence of attenuated strain resulting in severe diarrhea in infected pigs (13). To clarify this point, further investigation is required to confirm whether the ORF3 could indeed function synergistically with the S protein to determine the pathogenicity of the virus. Furthermore, using a reverse genetics system to generate the infectious-clone-derived PEDV (icPEDV), Beall et al. (25) demonstrated that pigs infected with icPEDVΔORF3 had a substantially lower diarrheic score than those infected with the wild-type PEDV strain PC22A or icPEDV carrying the intact ORF3. It is also notable that the icPEDVΔORF3 is also efficiently transmitted from infected pigs to uninfected pigs via indirect contact leading to disease outcomes (25). These findings altogether suggest the role of PEDV ORF3 as a virulence factor in the in vivo model. While the function of ORF3 remains to be determined, mounting evidence points to its strong association with viral virulence. As PEDV targets intestinal epithelial cells for productive infection in vivo, this process may require the functional ORF3, possibly via its ion channel activity. However, most in vitro studies are based on cultured cells such as Vero, MARC-145, and IPEC-J2, which may not be biologically relevant to what happens in vivo. As organoid systems, especially 3D intestinal organoids, could be highly relevant models for assessing differences between PEDV variants, future studies using this platform are needed to gain more insights into the functional role of ORF3.
Despite its critical role in viral pathogenesis, ORF3 is dispensable for virus growth in vitro (25–27). Given that most of the field PEDV strains carry the full-length ORF3 gene in the genome except for only a few isolates harboring a truncation of ORF3 (28, 29), it is likely that the ORF3 may be beneficial for its growth in the natural host. Nonetheless, several studies have reported the inhibitory effects of the ORF3 against the viral growth in vitro, especially the recovery of PEDV via reverse genetics (Table 1). It is also important to note that the effect of ORF3 might largely depend on the PEDV variant used in each study. While many studies demonstrated that the intact ORF3 could suppress cell-adapted PEDV replication in cultured cells (27, 31, 32), a few studies showed that the ORF3 could promote viral replication of specific PEDV variants (11, 14) or no effect of the ORF3 at all (25).
Thus far, the mechanism by which the ORF3 regulates PEDV replication in vitro has been suggested but not yet clearly defined. Wongthida et al. (32) demonstrated that phenylalanine at position 81 (F81) and methionine at position 167 (M167) are possibly responsible for suppressing PEDV growth in the cell culture. A single substitution from phenylalanine to leucine at position 81 (F81L) was sufficient for a productive replication of PEDV carrying the full-length ORF3 in VeroE6-APN cells (32). Likewise, a recent study reported that PEDV strains PC22A, AH2012/12, and CH7 bearing L81 in the ORF3 could be successfully rescued by reverse genetics, underscoring the importance of the amino acid residue 81 in ORF3 in virus adaptation in VeroE6 cells. It is also important to note that the inhibitory effect of ORF3 was also demonstrated in another porcine nidovirus, porcine reproductive and respiratory syndrome virus (PRRSV) (32). This finding leads to the possibility that the PEDV ORF3 might exert its inhibitory activity by affecting the replication step(s) common among nidoviruses.
Despite its inhibitory effect against PEDV reverse genetics rescue, the ORF3 has been shown to enhance PEDV growth in some studies. For example, attenuated PEDV strains, AH-M (bearing truncated ORF3) and CV777 (bearing the full-length ORF3), were reported to grow more efficiently in Vero cells stably expressing ORF3 than those propagated in parental Vero cells (14). Moreover, Si et al. (33) compared the growth kinetics of cell-adapted PEDV (DR13) carrying either intact or truncated ORF3. They found that PEDVs with intact/naturally truncated ORF3 (91 residues) appeared to grow to higher titer than the PEDV lacking the ORF3. While it is still not determined how ORF3 could be regulated to result in different outcomes regarding the PEDV replication, we speculate that this might be partially due to different PEDV strains, variations of PEDV ORF3 length, and cell lines used.
Besides its direct role in PEDV replication, accumulating evidence has suggested that ORF3 might play a role in several cellular processes. It has been shown that ORF3 could regulate the cell cycle progression by prolonging the S phase (14), inhibiting early cell apoptosis (33), and promoting autophagy (34), consequently provide a proper environment for viral propagation. Furthermore, it is also demonstrated that the ORF3 could specifically interact with PEDV S during virus replication (31). This interaction between PEDV proteins suggests that ORF3 might play a pivotal part in the virus life cycle, especially in the natural host (31).
In addition, deletion or truncation of the ORF3 acquired during propagation in cell culture has been used as a genetic marker to differentiate between the field and cell-adapted strains. However, the recombination of a highly pathogenic PEDV and a low pathogenic vaccine strain (30), and a natural PEDV co-infection (35) can result in pathogenic PEDV variants carrying truncated ORF3. These data thus indicate that ORF3 deletion or truncation alone may not be sufficient for safe vaccine candidates. This point underscores the necessity of precise genetic modification on ORF3 combined with other viral genes for vaccine design, which will benefit the surveillance study and disease monitoring in pig farms.
Even though the underlying mechanisms of ORF3 functions related to pathogenicity, virus adaptation, and virus proliferation have not been elucidated, several lines of evidence have emphasized the impact of the interplay between ORF3 and the host's machinery. Microscopic inspection of cells over-expressing the ORF3 showed that the protein was typically localized in the cytoplasm (14, 34, 36), specifically in the perinuclear area and the vesicles (31), the ER, and the Golgi (31, 34, 37). These findings point to the possibility that ORF3 may be associated with distinct cellular pathways that might more or less modulate viral replication and pathogenesis.
A panel of intracellular sorting motif mutations located on the C-terminus of PEDV ORF3 revealed that the YLAI motif (residues 170–173; 170YLAI173) is essential for the translocation of ORF3 from the ER to the plasma membrane (37). Despite no clear association of 170YLAI173 with the viral growth and pathogenesis, the finding indicates that the disruption of the 170YLAI173 signal could affect ORF3 cellular localization and transport, consequently influencing the intracellular interplay with host proteins or the virus itself. By this means, we postulate that the ORF3 with reported C-terminus deletions [91aa (33), 144aa (30), and 155aa (13) length] loss the functional motifs or proper protein folding, leading to disruption of protein trafficking and the interaction with cellular components. Furthermore, an interactome study has demonstrated that ORF3 interacting cellular proteins are enriched in the endo-lysosomal components (endosome, lysosome, and vacuole); many of them are associated with ubiquitination and immune signaling pathways (JAK/STAT and interleukin signaling) (36). In particular, the binding of ORF3 and vacuolar protein-sorting-associated protein 36 (VPS36), a component of the endosomal sorting complexes required for transport II (ESCRT-II), was found to trigger ORF3 degradation, which, in turn, suppressed PEDV replication in VeroE6 cells (36). In addition, ORF3 was shown to modulate the NF-κB signaling pathway to downregulate proinflammatory cytokines (interleukin-8; IL-8 and TNF-α). Interestingly, ORF3 could also interact with the IκB kinase β (IKBKB), resulting in the up-regulation of the IKBKB-meditated NF-κB promoter activity but down-regulation of the IKBKB-meditated IFN-β promoter and IFN-β mRNA expression (38). Consistently, Wu et al. (39) also demonstrated that ORF3 could inhibit the NF-κB activation by hampering nuclear factor p65 phosphorylation and nuclear translocation as well as down-regulating p65 expression, resulting in a reduction of proinflammatory cytokines IL-6 and IL-8 production. Therefore, these findings highlight the role of ORF3 in governing host immune responses that account for virus growth and disease severity.
PEDV has been reported to induce apoptosis by activating mitochondrial apoptosis-inducing factors (40) and accelerating autophagy associated with inflammatory cytokine expression (41). Notably, ORF3 was also shown to localize in ER and trigger ER stress by increasing the expression level of GRP78 and activating the PERK-eIF2α signaling pathway and inducing autophagy in vitro (34). These findings point to a possible link between PEDV infection/ORF3 and mitochondria. Even though the contribution of ORF3 in PEDV-induced apoptosis has not been well-defined, Si et al. (33) demonstrated that ORF3 and its naturally truncated form could inhibit apoptosis through the suppression of procaspase-3 activation to facilitate virus replication. However, conflicting results of ORF3-induced apoptosis have been reported in studies using ORF3-expressing cells and cells infected with the PEDV bearing ORF3 (33, 34).
The current data thus suggest that, besides being an ion channel or viroporin, ORF3 could interact with a large number of host's proteins as a means to manipulate cellular machinery to regulate virus replication and pathogenesis. Thus, characterization of specific amino acids or functional domains would be beneficial to precisely modify the PEDV ORF3 gene as a principle for vaccine design concerning virus cultivation and attenuation. A summary of the interaction of ORF3 and cellular proteins, based on current published data, is depicted in Figure 2.
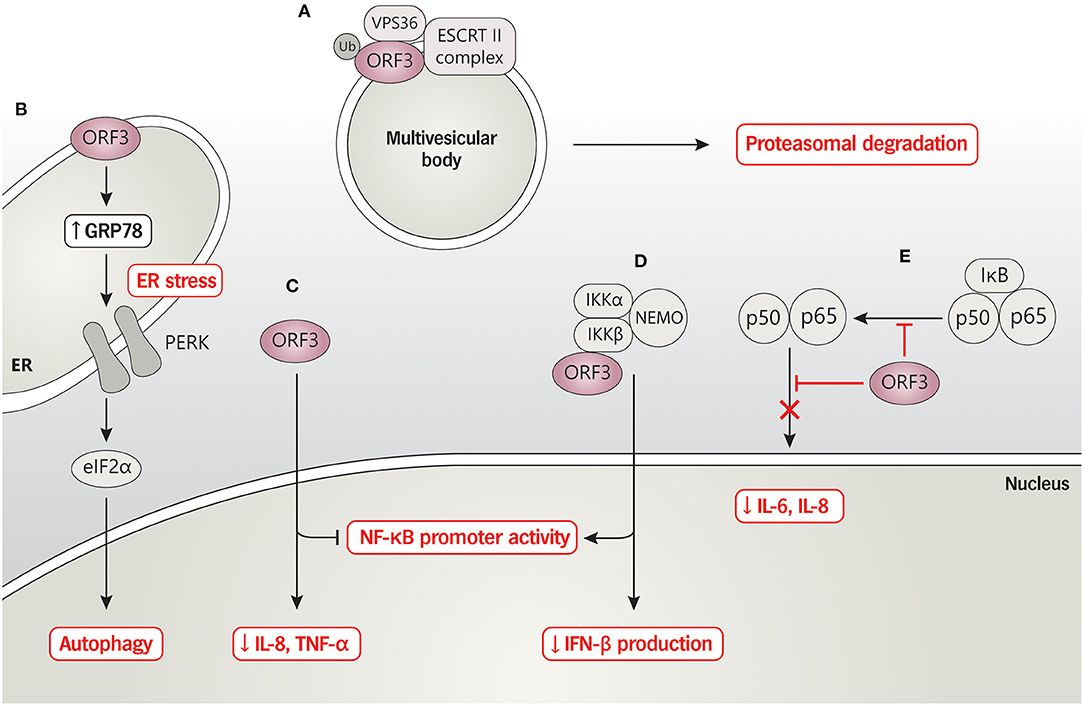
Figure 2. PEDV ORF3 and host protein interaction. (A) ORF3 protein interacts with VPS36 leading to ORF3 degradation via proteasomal degradation pathway. (B) PEDV ORF3 induces the expression level of GRP78 and activates the PERK-eIF2α signaling pathway resulting in enhancing cell autophagy. (C) ORF3 inhibits NF-κB promoter activity and down-regulates IL-8 and TNF-α mRNA expression while (D) induces NF-κB promoter activity and reduces IFN-β production in the binding with an overexpressed-IKBKB. (E) ORF3 suppresses phosphorylation and nuclear translocation of NF-κB, resulting in reduced proinflammatory cytokines IL-6 and IL-8.
Serial passages in host cells to obtain PEDV vaccine candidates often result in the loss or truncation of the ORF3 gene. For example, Chen et al. (30) showed that when the virulent PEDV strain YN was subjected to multiple passages in Vero cells, a high-growth PEDV with shortened ORF3 (144 residues) emerged during viral propagation as early as passage 15, designated YN15. However, the YN15 was still highly pathogenic in piglets, suggesting that shortening of ORF3 is not sufficient for virus attenuation (30). Interestingly, when YN15 was further propagated in Vero cells up to 144 passages, the virus, termed YN144, showed a markedly attenuated phenotype with no further mutations in the ORF3 gene (30). Likely, modification of ORF3 and mutations of critical residues especially of the spike protein, are crucial for developing a PEDV-based live virus vaccine. Comparing the effect of PEDV ORF3 variants (full-length vs. various truncated forms) in distributing viral pathogenesis in the identical viral genetic background would give rise to solid evidence of its role.
A reverse genetic system is a powerful tool for molecular studies and vaccine development. Given that ORF3 has been shown to have inhibitory effects on virus rescue, understanding its mechanism and region(s) of amino acid sequence responsible for this particular aspect can help overcome difficulties in PEDV isolation in vitro. As mentioned earlier, the presence of ORF3 in different forms can markedly influence PEDV virus rescue by reverse genetics and virus pathogenicity in the natural host. Extensive deletion of ORF3 might likely support efficient virus rescue and propagation in Vero cells. However, a high-growth virus with deleted ORF3 could poorly infect intestinal cells in vivo, affecting the virus's property to be used as an oral vaccine. Therefore, the strategy of ORF3 gene engineering for virus vaccine production should be considered to enable the virus to grow well in cell culture and, at the same time, to become less virulent but still maintaining its infectivity in intestinal cells.
ORF3 is a multi-functional protein that plays essential roles in modulating cellular mechanisms, particularly the host immune system and apoptosis. These roles of ORF3 are likely associated with virus replication and pathogenicity. Due to the lack of comprehensive studies of ORF3 and inconsistent findings of ORF3 among available literature, its impacts on virus adaptation in cell culture, virus replication, and pathogenesis are still not well-elucidated. Future studies utilizing various approaches are needed to gain more precise insights into our understanding of this protein. For example, a series of PEDV ORF3 mutations (variants) should be constructed together with the S protein to generate recombinant PEDVs to show the effect of each mutation in the virus phenotype. The impact of ORF3 variants on PEDV growth kinetics should be investigated in detail in vitro and in vivo. The mechanistic role of ORF3 in interferon signaling pathway/inflammasome activation could be further explored. Knowledge obtained from these studies would be a groundwork for viral gene mutagenesis to improve a better live-attenuated PEDV vaccine accomplishing high yield in cell culture with a minimum passaging, induction of specific immune response, and no reversion to virulence in the vaccinated animal.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This work was supported by National Vaccine Institute (NVI), Thailand, under grant number P2150344.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Temeeyasen G, Srijangwad A, Tripipat T, Tipsombatboon P, Piriyapongsa J, Phoolcharoen W, et al. Genetic diversity of ORF3 and spike genes of porcine epidemic diarrhea virus in Thailand. Infect Genet Evol. (2014) 21:205–13. doi: 10.1016/j.meegid.2013.11.001
2. Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, et al. Emergence of Porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest. (2013) 25:649–54. doi: 10.1177/1040638713501675
3. Sun R-Q, Cai R-J, Chen Y-Q, Liang P-S, Chen D-K, Song C-X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg Infect Dis. (2012) 18:161–3. doi: 10.3201/eid1801.111259
4. Park S-J, Kim H-K, Song D-S, Moon H-J, Park B-K. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field isolates in Korea. Arch Virol. (2011) 156:577–85. doi: 10.1007/s00705-010-0892-9
5. Lee C. Porcine epidemic diarrhea virus: an emerging and re-emerging epizootic swine virus. Virol J. (2015) 12:193. doi: 10.1186/s12985-015-0421-2
6. Lin C-M, Saif LJ, Marthaler D, Wang Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. (2016) 226:20–39. doi: 10.1016/j.virusres.2016.05.023
7. Pensaert MB, Martelli P. Porcine epidemic diarrhea: a retrospect from Europe and matters of debate. Virus Res. (2016) 226:1–6. doi: 10.1016/j.virusres.2016.05.030
8. Gerdts V, Zakhartchouk A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet Microbiol. (2017) 206:45–51. doi: 10.1016/j.vetmic.2016.11.029
9. Guo J, Fang L, Ye X, Chen J, Xu S, Zhu X, et al. Evolutionary and genotypic analyses of global porcine epidemic diarrhea virus strains. Transbound Emerging Dis. (2019) 66:111–8. doi: 10.1111/tbed.12991
10. Wang D, Fang L, Xiao S. Porcine epidemic diarrhea in China. Virus Res. (2016) 226:7–13. doi: 10.1016/j.virusres.2016.05.026
11. Wang K, Lu W, Chen J, Xie S, Shi H, Hsu H, et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. (2012) 586:384–91. doi: 10.1016/j.febslet.2012.01.005
12. Kocherhans R, Bridgen A, Ackermann M, Tobler K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes. (2001) 23:137–44. doi: 10.1023/A:1011831902219
13. Wang D, Ge X, Chen D, Li J, Cai Y, Deng J, et al. The S gene is necessary but not sufficient for the virulence of porcine epidemic diarrhea virus novel variant strain BJ2011C. J Virol. (2018) 92:e00603–18. doi: 10.1128/JVI.00603-18
14. Ye S, Li Z, Chen F, Li W, Guo X, Hu H, et al. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. (2015) 51:385–92. doi: 10.1007/s11262-015-1257-y
15. Madan V, García M de J, Sanz MA, Carrasco L. Viroporin activity of murine hepatitis virus E protein. FEBS Lett. (2005) 579:3607–12. doi: 10.1016/j.febslet.2005.05.046
16. Liao Y, Yuan Q, Torres J, Tam JP, Liu DX. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology. (2006) 349:264–75. doi: 10.1016/j.virol.2006.01.028
17. Ye Y, Hogue BG. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J Virol. (2007) 81:3597–607. doi: 10.1128/JVI.01472-06
18. Park S-J, Moon H-J, Luo Y, Kim H-K, Kim E-M, Yang J-S, et al. Cloning and further sequence analysis of the ORF3 gene of wild- and attenuated-type porcine epidemic diarrhea viruses. Virus Genes. (2008) 36:95–104. doi: 10.1007/s11262-007-0164-2
19. Chan C-M, Tsoi H, Chan W-M, Zhai S, Wong C-O, Yao X, et al. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int J Biochem Cell Biol. (2009) 41:2232–9. doi: 10.1016/j.biocel.2009.04.019
20. Castaño-Rodriguez C, Honrubia JM, Gutiérrez-Álvarez J, DeDiego ML, Nieto-Torres JL, Jimenez-Guardeño JM, et al. Role of severe acute respiratory syndrome coronavirus viroporins E, 3a, and 8a in replication and pathogenesis. mBio. (2018) 9:e02325–17. doi: 10.1128/mBio.02325-17
21. Siu K-L, Yuen K-S, Castaño-Rodriguez C, Ye Z-W, Yeung M-L, Fung S-Y, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. (2019) 33:8865–77. doi: 10.1096/fj.201802418R
22. Bernasconi C, Guscetti F, Utiger A, Reeth KV, Ackermann M, Pospischil A. Experimental Infection of Gnotobiotic Piglets With a Cell Culture Adapted Porcine Epidemic Diarrhoea Virus: Clinical, Histopathological and Immunohistochemical Findings. Lyon: Foundation Marcel Merieux Lyon France (1994).
23. Kweon CH, Kwon BJ, Lee JG, Kwon GO, Kang YB. Derivation of attenuated porcine epidemic diarrhea virus (PEDV) as vaccine candidate. Vaccine. (1999) 17:2546–53. doi: 10.1016/S0264-410X(99)00059-6
24. Song DS, Yang JS, Oh JS, Han JH, Park BK. Differentiation of a Vero cell adapted porcine epidemic diarrhea virus from Korean field strains by restriction fragment length polymorphism analysis of ORF 3. Vaccine. (2003) 21:1833–42. doi: 10.1016/S0264-410X(03)00027-6
25. Beall A, Yount B, Lin C-M, Hou Y, Wang Q, Saif L, et al. Characterization of a pathogenic full-length cDNA clone and transmission model for porcine epidemic diarrhea virus strain PC22A. mBio. (2016) 7:e01451–15. doi: 10.1128/mBio.01451-15
26. Fan B, Yu Z, Pang F, Xu X, Zhang B, Guo R, et al. Characterization of a pathogenic full-length cDNA clone of a virulent porcine epidemic diarrhea virus strain AH2012/12 in China. Virology. (2017) 500:50–61. doi: 10.1016/j.virol.2016.10.011
27. Jengarn J, Wongthida P, Wanasen N, Frantz PN, Wanitchang A, Jongkaewwattana A. Genetic manipulation of porcine epidemic diarrhoea virus recovered from a full-length infectious cDNA clone. J Gen Virol. (2015) 96:2206–18. doi: 10.1099/vir.0.000184
28. Chen X, Zeng L, Yang J, Yu F, Ge J, Guo Q, et al. Sequence heterogeneity of the ORF3 gene of porcine epidemic diarrhea viruses field samples in Fujian, China, 2010–2012. Viruses. (2013) 5:2375–83. doi: 10.3390/v5102375
29. Lu Y, Su X, Du C, Mo L, Ke P, Wang R, et al. Genetic diversity of porcine epidemic diarrhea virus with a naturally occurring truncated ORF3 gene found in Guangxi, China. Front Vet Sci. (2020) 7:435. doi: 10.3389/fvets.2020.00435
30. Chen F, Zhu Y, Wu M, Ku X, Ye S, Li Z, et al. Comparative genomic analysis of classical and variant virulent parental/attenuated strains of porcine epidemic diarrhea virus. Viruses. (2015) 7:5525–5538. doi: 10.3390/v7102891
31. Kaewborisuth C, He Q, Jongkaewwattana A. The accessory protein ORF3 contributes to porcine epidemic diarrhea virus replication by direct binding to the spike protein. Viruses. (2018) 10:399. doi: 10.3390/v10080399
32. Wongthida P, Liwnaree B, Wanasen N, Narkpuk J, Jongkaewwattana A. The role of ORF3 accessory protein in replication of cell-adapted porcine epidemic diarrhea virus (PEDV). Arch Virol. (2017) 162:2553–2563. doi: 10.1007/s00705-017-3390-5
33. Si F, Hu X, Wang C, Chen B, Wang R, Dong S, et al. Porcine epidemic diarrhea virus (PEDV) ORF3 enhances viral proliferation by inhibiting apoptosis of infected cells. Viruses. (2020) 12:214. doi: 10.3390/v12020214
34. Zou D, Xu J, Duan X, Xu X, Li P, Cheng L, et al. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet Microbiol. (2019) 235:209–9. doi: 10.1016/j.vetmic.2019.07.005
35. Lu Y, Huang W, Zhong L, Qin Y, Liu X, Yang C, et al. Comparative characterization and pathogenicity of a novel porcine epidemic diarrhea virus (PEDV) with a naturally occurring truncated ORF3 gene coinfected with PEDVs possessing an intact ORF3 gene in piglets. Viruses. (2021) 13:1562. doi: 10.3390/v13081562
36. Kaewborisuth C, Yingchutrakul Y, Roytrakul S, Jongkaewwattana A. Porcine epidemic diarrhea virus (PEDV) ORF3 interactome reveals inhibition of virus replication by cellular VPS36 protein. Viruses. (2019) 11:382. doi: 10.3390/v11040382
37. Si F, Chen B, Hu X, Yu R, Dong S, Wang R, et al. Porcine Epidemic Diarrhea Virus ORF3 Protein Is Transported through the Exocytic Pathway. J Virol. (2020) 94:e00808–20. doi: 10.1128/JVI.00808-20
38. Kaewborisuth C, Koonpaew S, Srisutthisamphan K, Viriyakitkosol R, Jaru-Ampornpan P, Jongkaewwattana A. PEDV ORF3 independently regulates IκB kinase β-mediated NF-κB and IFN-β promoter activities. Pathogens. (2020) 9:376. doi: 10.3390/pathogens9050376
39. Wu Z, Cheng L, Xu J, Li P, Li X, Zou D, et al. The accessory protein ORF3 of porcine epidemic diarrhea virus inhibits cellular interleukin-6 and interleukin-8 productions by blocking the nuclear factor-κB p65 activation. Vet Microbiol. (2020) 251:108892. doi: 10.1016/j.vetmic.2020.108892
40. Kim Y, Lee C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology. (2014) 460–1:180–93. doi: 10.1016/j.virol.2014.04.040
Keywords: PEDV, ORF3, virus replication, pathogenesis, virus and host interaction, vaccine development
Citation: Jantraphakorn Y, Viriyakitkosol R, Jongkaewwattana A and Kaewborisuth C (2021) Interaction Between PEDV and Its Hosts: A Closer Look at the ORF3 Accessory Protein. Front. Vet. Sci. 8:744276. doi: 10.3389/fvets.2021.744276
Received: 20 July 2021; Accepted: 19 August 2021;
Published: 10 September 2021.
Edited by:
Levon Abrahamyan, Université de Montréal, CanadaReviewed by:
Bin Li, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaCopyright © 2021 Jantraphakorn, Viriyakitkosol, Jongkaewwattana and Kaewborisuth. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Challika Kaewborisuth, Y2hhbGxpa2Eua2FlQGJpb3RlYy5vci50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.