
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 18 November 2021
Sec. Livestock Genomics
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.732440
This article is part of the Research Topic Sheep and Goat Gene Exploration View all 19 articles
 Yufang Liu1,2
Yufang Liu1,2 Yulin Chen1,2
Yulin Chen1,2 Zuyang Zhou1,2
Zuyang Zhou1,2 Xiaoyun He1
Xiaoyun He1 Lin Tao1
Lin Tao1 Yanting Jiang3
Yanting Jiang3 Rong Lan3
Rong Lan3 Qionghua Hong3*
Qionghua Hong3* Mingxing Chu1*
Mingxing Chu1*Granulosa cell (GC) proliferation provides essential conditions for ovulation in animals. A previous study showed that DENND1A plays a significant role in polycystic ovary syndrome. However, the modulation of DENND1A in GCs remains unclear. Our previous integrated analysis of miRNA–mRNA revealed that the 3'-untranslated region of DENND1A could be a target of chi-miR-324-3p. In this study, we used quantitative reverse transcription polymerase chain reaction (RT-qPCR) to investigate DENND1A expression in ovarian tissues of high- and low-yielding goats. Furthermore, dual-fluorescent reporter vector experiments, Cell Counting Kit-8 (CCK-8) assay, and RT-qPCR were used to elucidate the regulatory pathway of chi-miR-324-3p-DENND1A in GCs. The results revealed an opposite tendency between the expressions of chi-miR-324-3p and DENND1A in the ovaries of high- and low-yielding goats. The CCK-8 assay indicated that chi-miR-324-3p overexpression significantly suppressed GC proliferation, whereas chi-miR-324-3p inhibition promoted GC proliferation. In addition, the expressions of GC proliferation markers LHR, Cylin D2, and CDK4 showed the same tendency. The dual-fluorescent reporter assay revealed that chi-miR-324-3p directly targeted DENND1A, and the RT-qPCR results revealed that DENND1A expression was inhibited by chi-miR-324-3p. In summary, chi-miR-324-3p inhibited the proliferation of GCs by targeting DENND1A.
Ovaries, which have two functions, the cyclical production of fertilizable ova and steroid hormones, are important organs for economic animals (1). Follicles are the fundamental unit of mammalian ovaries, and the complex regulation of follicular dynamics dictates the degree of prolificacy in different species (2). The process of physical contact between granulosa cells (GCs) and oocytes can aid in oocyte growth, maturation, and, ultimately, ovulation. The proliferation of GCs is elementary for follicle and oocyte development, ovulation, and luteinization (3). Many factors can promote the proliferation of GCs, including genes, transcription factors, non-coding RNAs, and hormones. BMPR-IB, a gene that is well-known for its mutation in prolific sheep, was knocked out using the CRISPR-Cas technology in goat GCs, which resulted in the higher expressions of R-Smads, LHR, and FSHR in the BMPR-IB knocked out GCs compared with those in the GCs in which BMPR-IB was not knocked out (4). Other genes were also found to play an important role in GC development, such as GDF9, BMP15, and YAP1 (5–7).
Studies have shown that miRNA as a regulator influences the development of GCs. miR-224 is a promising marker of polycystic ovary syndrome (PCOS), and the overexpression of miR-224 reduces cell expansion and expression of oocyte development-associated genes (8). miR-101-3p mimics and negative control were transfected into goat GCs, and RNA-seq was used to construct the cDNA libraries. Studies have suggested that miR-101-3p targeted to STC1 regulates the expression of STAR, CYP19A1, CYP11A1, and 3β-HSD steroid hormone synthesis-associated gene, thereby promoting the secretion of E2 and P4 (3). Additionally, miR-383 and miR-320 regulate steroid hormone secretion in GCs, whereas miR-21-5p, miR-503, miR-424, and miR-503 are involved in ovulation, follicular-luteal transformation, and luteinization (9–11). miR-324-3p is involved in various cancer development processes, which promote tumor growth by targeting DACT1 and activating the Wnt/β-catenin pathway in hepatocellular carcinoma (12). Recently, a new study demonstrated that lncRNA H1FX-AS1 targeted DACT1 and inhibited cervical cancer by sponging miR-324-3p (13). Fang et al. revealed that lncRNA SNHG22 facilitated malignant phenotypes in triple-negative breast cancer by sponging miR-324-3p and upregulating SUDS3 (14). However, the function of miR-324-3p in regulating GC proliferation remains unknown.
DENND1A, which encodes for a clathrin-binding protein that is a member of the connecdenn family and a guanine nucleotide-exchange factor, is located at 9q22.32 (15). A previous study revealed a DENND1A variant, DENND1A.V2, which was upregulated in the theca cells of women with PCOS, and reduced CYP17A1 expression and androgen secretion (16). Several studies have found that polymorphisms of DENND1A were related to an increased risk of PCOS (17, 18). Therefore, the previous studies showed that DENND1A plays a crucial role in PCOS. However, the function of DENND1A in GCs is still unknown. In our previous study, integrated analysis of miRNA–mRNA results revealed that DENND1A is a target gene of chi-miR-324-3p. Hence, we deduced that chi-miR-324-3p inhibits GC proliferation in PCOS by suppressing DENND1A expression.
In this study, we aimed to identify the roles of chi-miR-324-3p in the proliferation of goat GCs and the relationship of chi-miR-324-3p with DENND1A. The findings demonstrated that chi-miR-324-3p could regulate GC proliferation by targeting DENND1A. Overall, this study provides new evidence supporting the potential application of chi-miR-324-3p to prolific goats.
This study and all the experimental procedures were approved by the Science Research Department (in charge of animal welfare issue) of the Institute of Animal Sciences and Chinese Academy of Agricultural Sciences (IAS-CAAS) (Beijing, China). Ethical approval was also provided by the animal ethics committee of IAS-CAAS (No. IAS2019-63).
Female native domestic goats, known as Yunshang black goats, were used in this study. According to their yield records (at least two-born litter size), 277 female goats (152 high yielding goat and 125 low yielding goat) with no significant differences in weight and height were grouped into either the high-yielding group (average litter size 3.00 ± 0.38) or low-yielding group (average litter size 1.32 ± 0.19) (p < 0.05). All goats were bred under the same conditions, with free access to water and feed in a goat farm located in Yunnan Province. In the follicular phase, three high-yielding (high litter size group, HF) and three low-yielding goats (low litter size group, LF) were selected from the batch of goats according to their litter size. The ovarian tissues were collected from goats, frozen immediately in liquid nitrogen, and subsequently stored at −80°C until RNA extraction.
Following previously described methods, primary goat GCs were isolated from the follicles of ovarian tissues in the follicular phase (19). The diameter of the GCs in the follicular phase was over 3.5 mm. The isolated cells were seeded in 6-mm plates and maintained in a complete medium [DMEM/F12 (1:1), 10% FBS, and 1% penicillin/streptomycin] as described by Yang et al. (20). When the cell confluence was >90%, the cells were transferred to 10-mm plates for the next experiment.
According to the manufacturer's protocol, total RNA was isolated from ground ovarian tissue powder and goat primary GCs using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The purity and concentration of the RNA samples were evaluated using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Standard denaturing agarose gel electrophoresis was used to determine the presence of contamination and degradation.
The 3′ untranslated region (UTR) of DENND1A containing the predicted target site, including wild type (WT) and mutant type (MUT), were cloned into pmiR-RB-Report vector [named pmiR-RB-DENND1A-WT and pmiR-RB-DENND1A-MUT; isolated using XhoI and NotI (Takara, Dalian, China)], respectively. The insert sequence was synthesized by RiboBio company, including the wild/mutant sequences, miRNA mimic, mimic-NC, inhibitor, and inhibitor NC (RiboBio, Guangzhou, China).
HEK293T, a human cell line of renal epithelial cells, was used to validate the miRNA target. Cells were seeded into 24-well plates. Cotransfection with 200 ng of target mRNA-3'-UTR-WT or target mRNA-3'-UTR-MUT and 10 μl of miRNA mimic or mimic-NC was performed using Lipofectamine 2000 (Invitrogen, USA). Subsequently, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, WI, USA) at 48 h post-Transfection. The assays were performed in triplicate.
According to the manufacturer's instructions, reverse transcription was performed using a PrimeScript™ RT reagent kit (TaKaRa, Dalian, China), for mRNA and miRcute Plus miRNA First-Strand cDNA kit (TIANGEN, Beijing, China) for miRNA. RT-qPCR was performed using SYBR Green qPCR Mix (TaKaRa, Dalian, China) for mRNAs and miRcute Plus miRNA qPCR kit (TIANGEN, Beijing, China) for miRNAs using RocheLight Cycler®480 II system (Roche Applied Science, Mannheim, Germany). The qPCR for mRNAs was performed as follows: initial denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 5 s, and finally annealing at 60°C for 30 s. In contrast, the qPCR for miRNAs was performed as follows: initial denaturation at 95°C for 15 min, followed by 40 cycles of denaturation at 94°C for 20 s, and finally annealing at 60°C for 34 s. The data were analyzed using the 2−ΔΔCt method. LHR, Cylin D2, and CDK4 were selected as GC proliferation markers (21). Goat RPL19 gene and U6 gene were used as a reference gene for normalizing target gene data. The sequences of RT-qPCR primers are listed in Table 1.
GC proliferation was quantified using an ethynyldeoxyuridine (EdU) kit according to the manufacturer's protocol (Beyotime). Cells were seeded into 96-well plates as described above. One hundred microliters of 50 μM EdU was aliquoted to each well and cells were incubated an additional 3 h. Cells were then washed with PBS and fixed with 4% paraformaldehyde for 30 min. To neutralize excess aldehyde groups, 50 μl of 2 mg/ml of glycine was aliquoted per well and incubated with cells for 15 min. Subsequently, 100 μl of 0.5% Triton X-100 in PBS was aliquoted per well and incubated with cells for 15 min. After rinsing, 100 μl of Apollo reagent was added, and the cells were incubated in the dark for 30 min at room temperature. Cells were washed with PBS and then nuclei were stained with Hoechst 33,342 reaction solution for 30 min in the dark. EdU-stained cells were visualized and quantified using a fluorescence microscope. Three fields were randomly selected for quantification and statistical analysis.
GCs were inoculated into 96-well plates with approximately 100 μl of cell suspension per well in three replicates. Cells were then incubated for 2–4 h at 37°C in an incubator, and used for subsequent experiments after cell apposition/adequate confluency was reached more than 90%. Cell proliferation assay was conducted using the Cell Counting Kit-8 (CCK-8) method (Beyotime, Beijing, China). Goat GCs were cultured in 96-well plates and measured by adding 10 μl of CCK-8 solution per well as per protocol. After the addition of CCK-8 solution, absorbance at 450 nm was measured at 0, 6, 12, 24, 48, and 72 h.
Statistical analyses of RT-qPCR results and graphs were conducted using GraphPad Prism (v.5.0) (San, Diego, CA, USA). Statistical significance of the data was tested using paired t-tests. The results were presented as means ± SEM of three replicates, and the statistical significance was represented by p-values < 0.05 (*p < 0.05) and p-values < 0.01 (**p < 0.01).
It has been predicted in a previous study that as a DENND1A regulator, chi-miR-324-3p is expressed in goat ovarian tissues in the follicular phase. To illustrate the correlation of the predicted miRNA–mRNA pair, the expression patterns of chi-miR-324-3p and its putative target DENND1A were investigated in HF (high-yielding group) and LF (low-yielding group). As shown in Figure 1, opposite expression patterns of chi-miR-324-3p and DENND1A were observed in the high-yielding group (Figure 1). This indicated a negative correlation between the expressions of chi-miR-324-3p and DENND1A.
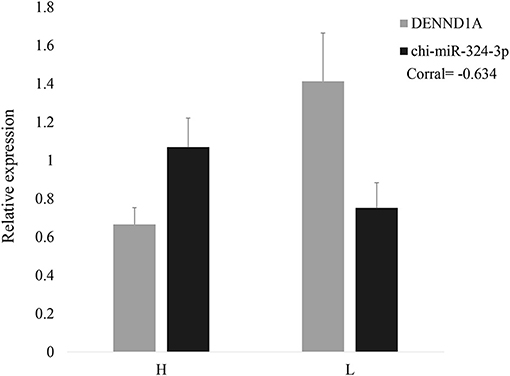
Figure 1. Relative expressions of DENND1A and chi-miR-324-3p in goat ovarian tissues in high- and low-yielding groups. H represents the high-yielding group; L represents the low-yielding group.
To investigate the regulatory function of chi-miR-324-3p on GC proliferation, an overexpression experiment was performed by transfecting a chi-miR-324-3p mimic into goat GCs (Figure 2A). The EdU, CCK-8 assay and analysis of the expressions of GC proliferation factors were used for validating the functions of chi-miR-324-3p in GCs. The results revealed that the GCs proliferation was significantly lower in the mimic group than that in the mimic-NC group (Figures 2B,C) (p < 0.05). The expressions of LHR, Cylin D2, and CDK4 were also significantly lower in the mimic group than in the mimic-NC group (Figure 2D) (p < 0.05). These results demonstrated that the overexpression of chi-miR-324-3p inhibited the proliferation of goat GCs.
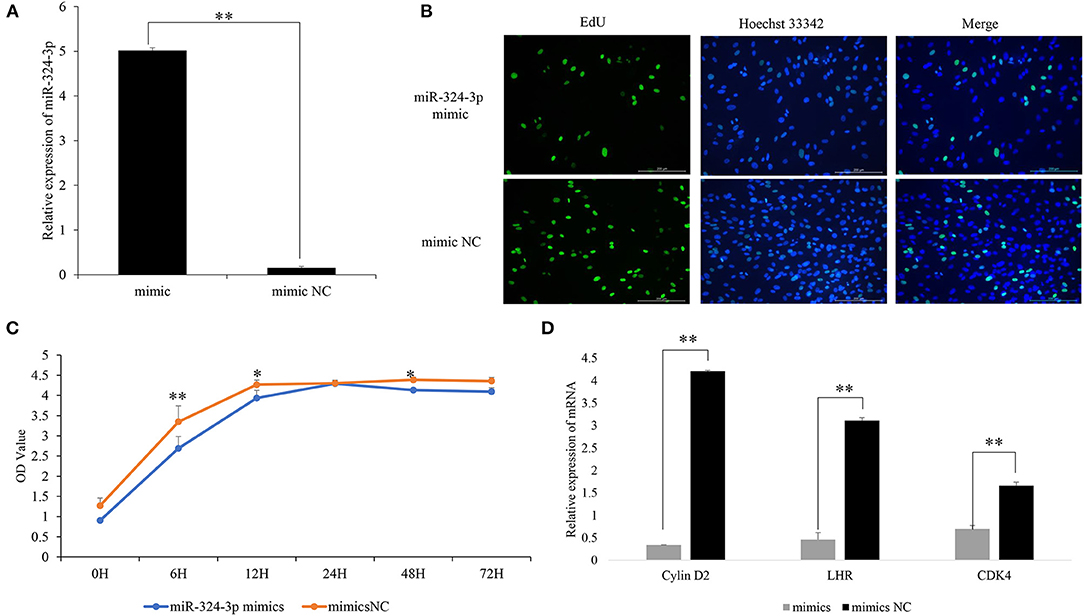
Figure 2. Overexpression of chi-miR-324-3p in goat GCs. (A) Relative expression of miR-324-3p. (B) The result of EdU assay. (C) CCK-8 assay of goat GCs with miR-324-3p mimic and mimic NC transfection at 0–72 h. (D) Relative expressions of LHR, CDK4, and Cylin D2 in transfected chi-miR-324-3p mimic and mimic NC at 48 h. *p < 0.05; **p < 0.01. CCK-8, Cell Counting Kit-8; GC, granulosa cell.
To further explore the role of chi-miR-324-3p in goat GCs, chi-miR-324-3p inhibitor was transfected into goat GCs (Figure 3A). The EdU and CCK-8 assay revealed that the GC proliferation in the inhibitor group was significantly higher than that of the inhibitor-NC group (Figures 3B,C) (p < 0.05). The mRNA expression of GC proliferation markers was significantly upregulated after inhibitor transfection (Figure 3D) (p < 0.05). These results demonstrated that the inhibition of chi-miR-324-3p could promote the proliferation of goat GCs.
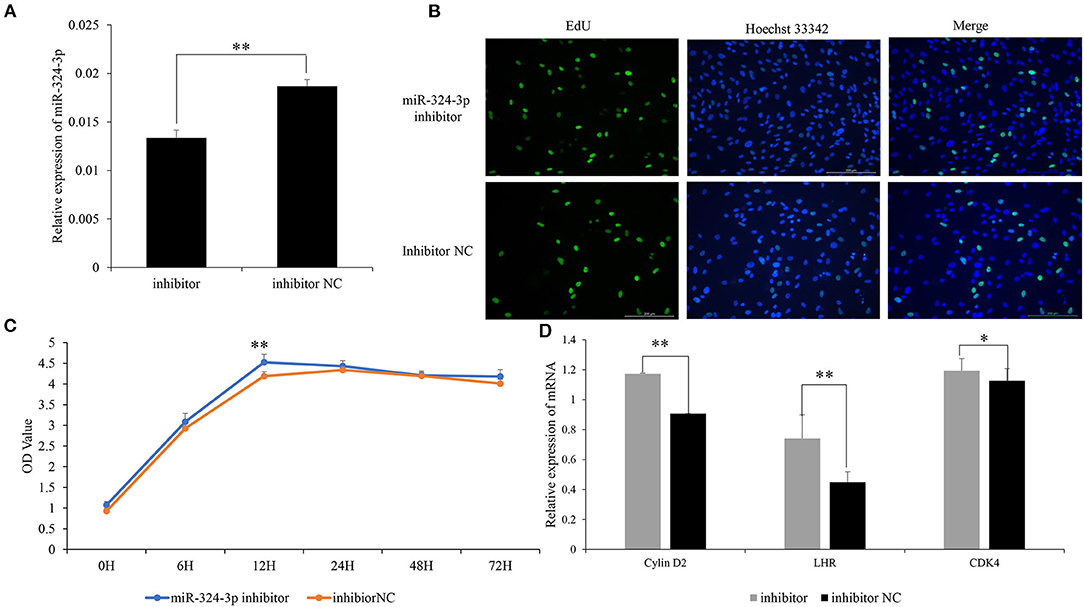
Figure 3. Inhibited chi-miR-324-3p expression in goat GCs. (A) Relative expression of miR-324-3p. (B) The result of EdU assay. (C) CCK-8 assay of goat GCs with miR-324-3p mimic and mimic NC transfection at 0–72 h. (D) Relative expressions of LHR, CDK4, and Cylin D2 in transfected chi-miR-324-3p mimic and mimic NC at 48 h. *p < 0.05; **p < 0.01. CCK-8, Cell Counting Kit-8; GC, granulosa cell.
In a previous study, it was revealed that DENND1A is a potential target of chi-miR-324-3p. To verify whether DENND1A is the direct target gene of chi-miR-324-3p, two plasmids containing either the WT or MUT 3'-UTR of DENND1A were constructed (Figure 4A). Subsequently, cotransfection of chi-miR-324-3p mimic or mimic-NC into 293T cells was performed. Notably, chi-miR-324-3p markedly decreased the activity of WT DENND1A compared with the negative control, but no significant changes were noted for the MUT (Figure 4B). These findings indicated that chi-miR-324-3p can directly target the 3'-UTR of DENND1A.
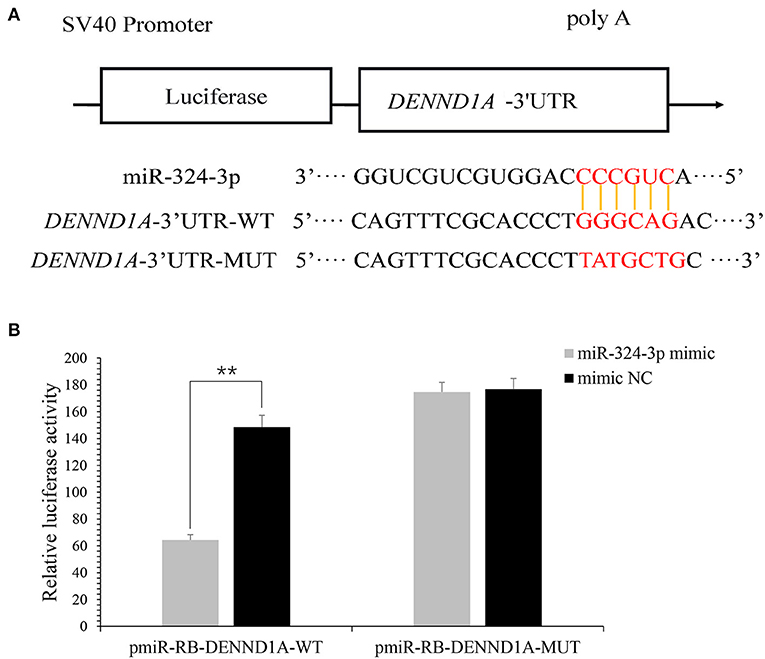
Figure 4. DENND1A is directly targeted by chi-miR-324-3p. (A) Predicted binding sites of chi-miR-324-3p in the 3'-UTR of DENND1A. The red section is the seed region of chi-miR-324-3p. (B) Relative luciferase activities of 293T cells cotransfected with the DENND1A 3'-UTR wild-type and mutant dual-luciferase reporter and chi-miR-324-3p mimic and mimic NC at 48 h. UTR, untranslated region. **P < 0.01.
To examine the validity of the putative target, chi-miR-324-3p mimic or mimic-NC was transfected into goat GCs. RT-qPCR data revealed that the transcriptional levels of DENND1A were different between the mimic and mimic-NC groups (Figure 5). Thus, we concluded that chi-miR-324-3p directly targets the 3'-UTR of DENND1A to suppress its mRNA translation in goat GCs.
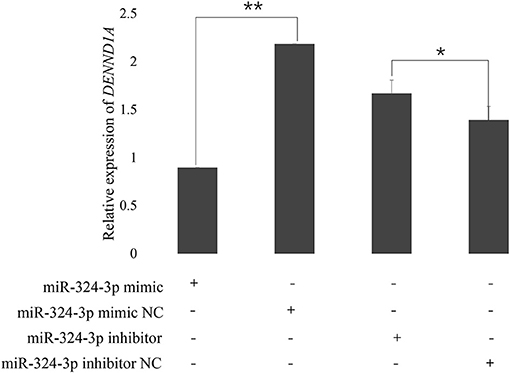
Figure 5. Expression of DENND1A in goat GCs transfected with chi-miR-324-3p mimic or mimic NC, and inhibitor or inhibitor-NC at 48 h. RPL19 was used as the reference gene. Results are representative of the mean (±standard deviation) of three independent analyses. *p < 0.05; **p < 0.01.
The key candidate gene associated with prolific traits in goats remains unknown. In our study of RNA-seq, chi-miR-324-3p and DENND1A showed differential expression levels in high- and low-yielding goats and chi-miR-324-3p was predicted to target DENND1A (data unpublished). In this study, we found and validated the function of chi-miR-324-3p in goat GCs. Our results indicated that chi-miR-324-3p regulated GC proliferation by targeting DENND1A.
The DENN domain is a common, evolutionarily ancient, and conserved protein module; there are eight families and DENND1A is a member of these. DENND1A was detected by proteomic analysis of clathrin-coated vesicles (22). Its variant is associated with hyperandrogenism and irregular menses and is a risk variant for PCOS as documented by the NIH criteria (23). DENND1A is expressed in the theca cells and testes; therefore, it may be associated with hyperandrogenism by increasing androgen levels (24). In this study, the high expression of DENND1A was associated with prolific traits in goats; chi-miR-324-3p directly acted on the 3'-UTR of DENND1A in the 293T cell line. Meanwhile, our overexpression and inhibition experiments on chi-miR-324-3p further verified that the expressions of DENND1A and chi-miR-324-3p showed opposite trends in goat GCs. Some studies have suggested that DENND1A regulates Rab GTPases, which are important for calcium regulated exocytosis in pituitary cells and for basal and GnRH-induced gonadotropin release (25, 26). Moreover, DENND1A has been reported to participate in endocytosis and receptor trafficking in the cellular membrane. Furthermore, high methylation existed in the DENND1A intron in aging oocytes (16, 27). Regardless, the effects of DENND1A on goat GC proliferation remain experimentally unclarified. Here, we provide new insights for understanding that DENND1A might be a key gene involved in goat GC proliferation that is regulated by chi-miR-324-3p. Taken together, these experimental results support the conclusion that DENND1A is involved in GC proliferation.
A previous study reported that miR-324-3p can promote the dysregulation of the expression of genes involved in cell death and apoptosis, cAMP and Ca2+ signaling, cell stress, and metabolism (28). Notably, miR-324-3p inhibition was highly and significantly correlated with cell proliferation and apoptosis. miR-324-3p, which is highly upregulated in maternal blood in the second trimester, is associated with fetal growth (29). miR-324-3p has also been reported to increase the incidence of ectopic pregnancy according to the analysis of tissues obtained from voluntarily terminated pregnancy, which has a repressive interaction with 3'-UTR of KISS1 (30). In addition, miR-324-3p overexpression in ectopic pregnancy is responsible for decreased KISS1/kisspeptin expression (31). Our previous studies have demonstrated that chi-miR-324-3p can regulate the development of ovaries in prolific goats by targeting DENND1A (data unpublished). Moreover, there is evidence that miR-324-3p plays crucial roles in ovary development and cell proliferation. Furthermore, miR-324-3p is related to SKOV3 cell viability, invasion, and migration via overexpression in ovarian cancer (32). Furthermore, a study demonstrated that miR-324-3p expression in PCOS rats is remarkably downregulated and confirmed that the overexpression of miR-324-3p represses proliferation and induces the apoptosis of GCs by targeting WNT2B (33). Using the CCK-8 assay, our results indicated that chi-miR-324-3p could inhibit goat GC proliferation, as demonstrated by the low expression of GC proliferation markers. We used goat ovaries in the follicular phase and the primary GCs as the animal material. Thus, we propose a new mechanism for the roles of chi-miR-324-3p in goat GCs.
In summary, our study reveals that chi-miR-324-3p regulates goat GC proliferation by directly targeting DENND1A. These data expand our mechanistic understanding of GC proliferation and ovary development in which miRNAs play an important role.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
The animal study was reviewed and approved by the Science Research Department (in charge of animal welfare issue) of the Institute of Animal Sciences and Chinese Academy of Agricultural Sciences (IAS-CAAS) (Beijing, China) for all the experimental procedures mentioned. Ethical approval was provided by the animal ethics committee of IASCAAS (No. IAS2019-63).
Conceptualization: YFL and MXC; Methodology: YFL, ZYZ, LT and MXC; Validation: YFL, YLC and ZYZ; Formal Analysis: YFL and MXC; Investigation: YFL, ZYZ, LT, YLC, XYH, YTJ, RL, QHH and MXC; Resources: YFL, ZYZ, LT and MXC; Data Curation: YFL, ZYZ, LT, and MXC; Writing-Original Draft Preparation: YFL and MXC; Supervision: MXC; Project Administration: YFL, QHH and MXC; Funding Acquisition: QHH and MXC. All authors have read and agreed to the published version of the manuscript.
This research was funded by the Agricultural Science and Technology Innovation Program of China (CAAS-ZDRW202106 and ASTIP-IAS13), the China Agriculture Research System of MOF and MARA (CARS-38), the Major Science and Technology Special Plan of Yunnan Province (202102AE090039), and the Basic Research Foundation Key Project of Yunnan Province (202001AS070002).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Hafez ESE, Hafez B. Folliculogenesis, egg maturation, and ovulation. In: Hafez B, Hafez E, editors. Reproduction in Farm Animals. Philadelphia, PA: Lippincott Williams and Wilkins (2000). p. 68–81. doi: 10.1002/9781119265306.ch5
2. Sharma GT, Majumdar AC. Control of follicular steroidogenesis in early-and late-luteal phase goat ovaries. Small Ruminant Res. (1999) 34:111–7. doi: 10.1016/S0921-4488(99)00006-1
3. An XP, Ma HD, Liu YH, Li F, Song YX, Li G, et al. Effects of miR-101-3p on goat granulosa cells in vitro and ovarian development in vivo via STC1. J Anim Sci Biotechnol. (2020) 11:102. doi: 10.1186/s40104-020-00506-6
4. Kumar S, Punetha M, Jose B, Bharati J, Khanna S, Sonwane A, et al. Modulation of granulosa cell function via CRISPR-Cas fuelled editing of BMPR-IB gene in goats (Capra hircus). Sci Rep. (2020) 10. doi: 10.1038/s41598-020-77596-9
5. Md Hasanur A, Jibak L, Takashi M. GDF9 and BMP15 induce development of antrum-like structures by bovine granulosa cells without oocytes. J Reprod Dev. (2018) 64:423–31. doi: 10.1262/jrd.2018-078
6. Zhu GQ, Fang C, Li J, Mo CH, Wang YJ, Li J. Transcriptomic diversification of granulosa cells during follicular development in chicken. Sci Rep. (2019) 9:5462. doi: 10.1038/s41598-019-41132-1
7. Lv XM, He CB, Huang C, Wang HB, Hua GH, Wang ZF, et al. Timely expression, and activation of YAP1 in granulosa cells is essential for ovarian follicle development. FASEB J. (2019) 33:10049–64. doi: 10.1096/fj.201900179RR
8. Li X, Wang H, Sheng Y, Wang Z. MicroRNA-224 delays oocyte maturation through targeting Ptx3 in cumulus cells. Mech Dev. (2017) 143:20–5. doi: 10.1016/j.mod.2016.12.004
9. Yin MM, Wang XR, Yao GD, Lü MR, Liang M, Sun YP, et al. Transactivation of microRNA-320 by microRNA-383 regulates granulosa cell functions by targeting E2F1 and SF-1 proteins. J Biol Chem. (2014) 289:18239–57. doi: 10.1074/jbc.M113.546044
10. Samuel G, Dessie SW, Ijaz A, Sudeep S, Md Munir H, Michael H, et al. MicroRNA expression profile in bovine granulosa cells of preovulatory dominant and subordinate follicles during the late follicular phase of the estrous cycle. PLoS ONE. (2015) 10:e0125912. doi: 10.1371/journal.pone.0125912
11. Hari Om P, Dawit T, Michael H, Samuel G, Eva H, Christiane N, et al. MicroRNA-424/503 cluster members regulate bovine granulosa cell proliferation and cell cycle progression by targeting SMAD7 gene through activin signaling pathway. J Ovarian Res. (2018) 11:34. doi: 10.1186/s13048-018-0410-3
12. Tuo H, Wang YF, Wang L, Yao BW, Li Q, Wang C, et al. MiR-324-3p promotes tumor growth through targeting DACT1 and activation of Wnt/β-catenin pathway in hepatocellular carcinoma. Oncotarget. (2017) 8:65687–98. doi: 10.18632/oncotarget.20058
13. Shi XH, Huo JZ, Gao XP, Cai H, Zhu WP. A newly identified lncRNA H1FX-AS1 targets DACT1 to inhibit cervical cancer via sponging miR-324-3p. Cancer Cell Int. (2020) 20:358. doi: 10.1186/s12935-020-01385-7
14. Fang X, Zhang J, Li CY, Liu JJ, Shi ZD, Zhou P. Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int. (2020) 20:252. doi: 10.1186/s12935-020-01321-9
15. Kulkarni R, Teves ME, Han AX, McAllister JM, Strauss JFIII. Colocalization of polycystic ovary syndrome candidate gene products in theca cells suggests novel signaling pathways. J Endocr Soc. (2019) 3:2204–23. doi: 10.1210/js.2019-00169
16. McAllister JM, Modi B, Miller BA, Biegler J, Bruggeman R, Legro RS, et al. Overexpression of a DENND1A isoform produces a polycystic ovary syndrome theca phenotype. Proc Natl Acad Sci USA. (2014) 111:E1519–27. doi: 10.1073/pnas.1400574111
17. Gammoh E, Arekat MR, Saldhana FL, Madan S, Ebrahim BH, Almawi WY. DENND1A gene variants in Bahraini Arab women with polycystic ovary syndrome. Gene. (2015) 560:30–3. doi: 10.1016/j.gene.2015.01.034
18. Bao S, Cai JH, Yang SY, Ren Y, Feng T, Jin T, et al. Association of DENND1A gene polymorphisms with polycystic ovary syndrome: a meta-analysis. J Clin Res Pediatr Endocrinol. (2016) 8:135–43. doi: 10.4274/jcrpe.2259
19. Sharma AK, Sharma RK. Effect of prostaglandins E2 and F2α on granulosa cell apoptosis in goat ovarian follicles. Iran J Vet Res. (2020) 21:97–102.
20. Yang DQ, Wang L, Lin PF, Jiang TT, Wang N, Zhao F, et al. An immortalized steroidogenic goat granulosa cell line as a model system to study the effect of the endoplasmic reticulum (ER)-stress response on steroidogenesis. J Reprod Dev. (2017) 63:27–36. doi: 10.1262/jrd.2016-111
21. Sullivan MW, Stewart-Akers A, Krasnow JS, Berga SL, Zeleznik AJ. Ovarian responses in women to recombinant follicle-stimulating hormone and luteinizing hormone (LH): a role for LH in the final stage of follicular maturation. J Clin Endocrinol Metab. (1999) 84:228–32. doi: 10.1210/jc.84.1.228
22. Girard M, Allaire PD, McPherson PS, Blondeau F. Non-stoichiometric relationship between clathrin heavy and light chains revealed by quantitative comparative proteomics of clathrin-coated vesicles from brain and liver. Mol Cell Proteomics. (2005) 4:1145–54. doi: 10.1074/mcp.M500043-MCP200
23. Welt CK, Styrkarsdottir U, Ehrmann DA, Gudmar T, Gudmundur A, Gudmundsson JA, et al. Variants in DENND1A are associated with polycystic ovary syndrome in women of European ancestry. J Clin Endocrinol Metab. (2012) 7:1342–7. doi: 10.1210/jc.2011-3478
24. Strauss JF, McAllister JM, Urbanek M. Persistence pays off for PCOS gene prospectors. J Clin Endocrinol Metab. (2012) 97:2286–8. doi: 10.1210/jc.2012-2109
25. Tasaka K, Masumoto N, Mizuki J, Ikebuchi Y, Ohmichi M, Kurachi H, et al. Rab3B is essential for GnRH-induced gonadotrophin release from anterior pituitary cells. J Endocrinol. (1998) 157:267–74. doi: 10.1677/joe.0.1570267
26. Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. (2011) 286:13791–800. doi: 10.1074/jbc.R110.217067
27. Pawel K, Amin H, Joseph AZ, Caesar ZL, Ken R, Matthew LS, et al. Epigenetic clock and methylation study of oocytes from a bovine model of reproductive aging. Aging Cell. (2021) 20:e13349. doi: 10.1111/acel.13349
28. Ignacio D, Eva CS, Raquel DT, Javier ÁM, de Rojas-de Petro ES, Domínguez-Rodríguez A, et al. miR-125a, miR-139 and miR-324 contribute to Urocortin protection against myocardial ischemia-reperfusion injury. Sci Rep. (2017) 7:8898. doi: 10.1038/s41598-017-09198-x
29. Rodosthenous RS, Burris HH, Sanders AP, Just AC, Dereix AE, Svensson K, et al. Second trimester extracellular microRNAs in maternal blood and fetal growth: an exploratory study. Epigenetics. (2017) 12:804–10. doi: 10.1080/15592294.2017.1358345
30. Romero-Ruiz A, Avendano MS, Dominguez F, Lozoya T, Molina-Abril H, Sangiao-Alvarellos S. Deregulation of miR-324/KISS1/kisspeptin in early ectopic pregnancy: mechanistic findings with clinical and diagnostic implications. Am J Obstet Gynecol. (2019) 220:e1–e17. doi: 10.1016/j.ajog.2019.01.228
31. Ghafouri-Fard S, Shoorei H, Taheri M. The role of microRNAs in ectopic pregnancy: a concise review. Noncoding RNA Res. (2020) 5:67–70. doi: 10.1016/j.ncrna.2020.04.002
32. Liu QY, Jiang XX, Tian HN, Guo HL, Guo H, Guo Y. Long non-coding RNA OIP5-AS1 plays an oncogenic role in ovarian cancer through targeting miR-324-3p/NFIB axis. Eur Rev Med Pharmacol Sci. (2020) 24:7266–75. doi: 10.26355/eurrev_202007_21881
Keywords: chi-miR-324-3p, DENND1A, granulosa cells, proliferation, goat
Citation: Liu Y, Chen Y, Zhou Z, He X, Tao L, Jiang Y, Lan R, Hong Q and Chu M (2021) chi-miR-324-3p Regulates Goat Granulosa Cell Proliferation by Targeting DENND1A. Front. Vet. Sci. 8:732440. doi: 10.3389/fvets.2021.732440
Received: 29 June 2021; Accepted: 14 October 2021;
Published: 18 November 2021.
Edited by:
Rui Su, Inner Mongolia Agricultural University, ChinaReviewed by:
Lifan Zhang, Nanjing Agricultural University, ChinaCopyright © 2021 Liu, Chen, Zhou, He, Tao, Jiang, Lan, Hong and Chu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qionghua Hong, eXhoNzE2OEAxMjYuY29t; Mingxing Chu, bXhjaHVAMjYzLm5ldA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.