- Hebei Key Laboratory of Animal Physiology, Biochemistry and Molecular Biology, College of Life Sciences, Hebei Normal University, Shijiazhuang, China
DNA methylation, mediated by DNA methyltransferases (Dnmts), is a typical epigenetic process that plays an important role in affecting organism acclimatization and adaptation to environmental changes. However, information about Dnmts and their associations with the cold tolerance of ticks remains meager. Hence, in the present study, the Dnmts in important vector ticks Dermacentor silvarum and Haemaphysalis longicornis were cloned and identified, and their functions in cold response were further explored. Results showed that the length of DsDnmt and DsDnmt1 in D. silvarum, and HlDnmt1 and HlDnmt in H. longicornis were 1,284, 549, 1,500, and 1,613 bp, respectively. Bioinformatics in protein analysis revealed that they were all unstable hydrophilic proteins and were mainly characterized with Dcm (DNA cytosine methyltransferase domain), Dnmt1-RFD (DNA methyltransferase replication foci domain), zf-CXXC (zinc finger-CXXC domain), and BAH (Bromo adjacent homology domain). The relative expression of these Dnmts was reduced after cold treatment for 3 days (P < 0.05), and increased with the extension of treatment. Western blot revealed that Dnmt1 decreased first and then increased significantly (P < 0.05) in both tick species, whereas other Dnmts fluctuated at varying degrees. RNA interference significantly silenced the genes Dnmts (P < 0.01), and mortality increased significantly (P < 0.05), when exposed to sub-lethal temperature, underscoring the important roles of Dnmts during the cold response of D. silvarum and H. longicornis. The above results lay the foundation for further understanding of the epigenetic regulation of DNA methylation in cold acclimatization and adaptation of ticks.
Introduction
Ticks are obligate blood-sucking ectoparasitic arthropods, which can spread a variety of pathogens and cause zoonotic diseases (1). They are considered as the most important vector of pathogens of domestic and wild animals, and only second to mosquitoes as vectors of human disease (2). With social and economic development and changes in the ecological environment, humans have more contact with ticks which has led to an increase in the incidence of tick-borne diseases (1). Ticks and tick-borne diseases have raised public health issues and posed a serious threat to animal husbandry development. Recently, ticks have been implicated as the vector of Songling virus (SGLV) (3) and Alongshan virus (ALSV), both associated with human febrile illness (4, 5), and severe fever with thrombocytopenia syndrome virus (SFTSV), the causative agent of severe fever with thrombocytopenia syndrome (SFTS) (6).
The tick Dermacentor silvarum Olenev is widely distributed in Russia, Mongolia, and the north of China (7, 8), and has been implicated as the vector and reservoir for the maintenance of tick-borne encephalitis virus through transstadial and transovarial transmission in China (9). Rickettsia raoultii, R. slovaca, and R. heilongjiangensis (spotted fever group rickettsiae), along with Ehrlichia chaffeensis (the causative agent of human monocyclic Ehrlichia), have been identified in D. silvarum (10, 11). D. silvarum is also the primary vector of two babesiosis-causing parasites, Babesia equi and B. caballi; and the North Asian tick typhus pathogen known as R. sibirica (1). The tick Haemaphysalis longicornis Neuman, is one of the most predominant tick species in Asia countries and a serious pest of livestock in the Australasian and Western Pacific Regions (12, 13). H. longicornis has expanded rapidly to many states in the USA since its first identification in New Jersey (14–16). It has been implicated as a vector of several pathogenic agents including the causative agent (Theileria sergenti and T. buffeli) of hemolytic anemia in wild and domestic cattle (17), causative agent (T. orientalis) of bovine theileriosis in Australia, and New Zealand (18), and causative agent (Babesia spp.) of livestock babesiosis (19). Moreover, it can transmit many spotted fever group rickettsiae (SFGR) such as R. japonica, R. heilongjiangensis, R. raoultii, and Candidatus Rickettsia tarasevichiae (12, 20–22). More recently, the severe fever with thrombocytopenia syndrome virus (SFTSV) was also identified in H. longicornis from South Korea, Japan, and China, which has caused great damages to livestock production and human health (6, 23, 24).
Many terrestrial arthropods are frequently faced with temperatures low and sustained enough to freeze their body fluids and therefore, have implemented some strategies to mitigate this risk (25, 26). Some of these strategies include the modification of body fluid constituents to prevent ice crystallization (27), cryoprotective dehydration (28), freeze avoidance, behavioral avoidance, and freeze tolerance which involves the conversion of about 82% of body water into internal ice (26, 29). Although freeze-tolerant arthropods utilize many potentially protective molecules, there is no obvious molecule that appears to be sufficient to support this cold-tolerance strategy (26). Climate variables are important factors affecting tick distribution (30), which also influence tick survival, development, and activity during off-host periods (31, 32). Hence, the ambient temperature, especially the low temperatures in winter, could play a significant role in determining the host-seeking behavior of ticks. A complex physiological and biochemical process is involved in the response of ticks to low temperatures, including changes in the intracellular environment and substance synthesis, as well as the expression and regulation of some temperature-stress-related genes (33, 34).
DNA methylation is an epigenetic modification that is heritable and reversible, which could play some roles in the mediation of rapid plastic responses of organisms to environmental stress (35, 36). The methylation of DNA at the fifth position of cytosine (5 mC) and the sixth position of adenine (6 mA) influences gene expression by the regulation of the chromatin architecture and gene transcription (37, 38). The capacity of organisms to adapt to environmental perturbations is increased by DNA methylation (39). This mechanism is mediated by a vital group of evolutionarily conserved enzymes known as DNA methyltransferases (Dnmts), which catalyze the methylation of DNA at cytosine residue to form 5-methylcytosine (40). Though mounting evidence has highlighted that variation in DNA methylation patterns could contribute to the acclimation and adaptation of organisms to different environmental stresses (41–43), the relationship between Dnmts and the cold tolerance of ticks remains poorly understood.
In the present study, the DNA methyltransferases in D. silvarum and H. longicornis were cloned and identified, and their functions in the cold tolerance of ticks were further explored via RNA interference. Western-blot methods were used to investigate the expression dynamics of DNA methyltransferases of two ticks under cold stress. The findings of this study provide insight into the regulatory role of DNA methylation in the cold response of these two tick species, and lay the foundation for further investigation on the epigenetic and molecular regulatory mechanisms underlying the cold tolerance of ticks.
Materials and Methods
Ticks
The unfed adult ticks of Dermacentor silvarum and Haemaphysalis longicornis were collected by flag-dragging from the vegetation of Xiaowutai National Nature Reserve, Zhangjiakou City, Hebei Province (114°47'−115°28'E, 39°50'– 40°6'N). They were brought back to the laboratory and put on the ears of the New Zealand white rabbits for feeding, and cultured in an environmental chamber (26 ± 1°C, relative humidity 75 ± 5%, 16 h of light: 8 h in darkness) during the non-parasitic period (44). The second-generation adult ticks that molted for two weeks were used in subsequent tests. All experiments were approved by the Animal Ethics Committee of Hebei Normal University (Protocol Number: IACUC-176810).
RNA Extraction and cDNA Synthesis
Total RNA was extracted from ticks (each group contains 10 D. silvarum or 20 H. longicornis) using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's protocol. RNA quantification was carried out using Nano Drop® ND-1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA), with a ratio of A260/A280 typically above 2.0, and the RNA quality was evaluated by 1% agarose gel electrophoresis. Then, 2.0 μg of each extracted RNA was used in a 20.0 μL reaction volume with an oligo (dT)18 primer, and reverse transcription was performed to synthesize cDNA following the instructions provided with the TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd, Beijing, China). Primers (Table 1) were designed using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) and Primer Premier version 5.0 for Windows (Premier Biosoft International, Palo Alto, CA, USA), based on an alignment of several sequences obtained from the National Center of Bioinformatics Information (NCBI) website (http://www.ncbi.nlm.nih.Gov) and transcriptomic assembled sequences (Bio-Project PRJNA565692). The PCR conditions include an initial 2 min denaturation at 94°C, 35 cycles of 30 s at 94°C, 30 s at the melting temperature Tm (58–60°C) of the gene-specific primers, and 30 s at 72°C, and then a final extension at 72°C for 10 min carried out on an Applied Biosystems® Veriti® 96-Well Thermal Cycler (Life Technologies Ltd, Marsiling, Singapore). The amplified fragments were verified and separated on a 1% agarose gel. Bands with expected sizes were excised and purified by EG101-01 EasyPure Quick Gel Extraction Kit (TransGen, Beijing, China) according to the manufacturer's protocol. The purified products were sequenced and used for subsequent analysis.
Bioinformatics Analysis of DNA Methyltransferase
Sequence alignment and identity analyses were carried out using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) and BLASTn (http://www.ncbi.nlm.nih.gov/BLAST). The identification of Open reading frames (ORFs) was carried out using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/orfg.cgi), and a conserved domain search using the Conserved Domain Database (CDD) v.3.16 of NCBI was performed (45). The complete genome sequence of the DNA methyltransferases of D. silvarum and H. longicornis were deposited in the NCBI nucleotide database under the accession numbers: MW528395 (DsDnmt), MW528396 (DsDnmt1), MW528397 (HlDnmt1), and MW528398 (HlDnmt). The physicochemical properties of DNA methyltransferase proteins including the molecular weights and theoretical isoelectric points (pI) were calculated using the Compute pI/Mw tool (http://web.expasy.org /protparam/) and DiANNA website (http://bioinformatics.bc.edu/clotelab/DiANNA/). The hydrophobic properties (hydrophobicity) of the putative DNA methyltransferases were determined using Hphob/Kyte&Doolittle model (46). The prediction of the protein tertiary structures was carried out with the Swiss-Model web server (http://swissmodel.expasy.org/).
To reconstruct the evolutionary relationships of the DNA methyltransferase genes, phylogenetic analysis was performed using Molecular Evolutionary Genetics Analysis (MEGA) software version 6.06 (47) based on the amino acid sequences retrieved from the NCBI protein database. Alignment of sequences was carried out with the ClustalW method, and the phylogenetic tree was obtained by the Neighbor-Joining method (48) with complete gap deletion and a bootstrap value of 100% in 1,000 replicates. Distances were computed using the Poisson correction method (49) and were in units of the number of amino acid substitutions per site.
Synthesis of dsRNA of Target Genes
The primers (Table 1) for RNA interference of DNA methyltransferase genes were designed using DNAMAN (Lynnon Biosoft, San Ramon, CA, USA) and Primer Premier Version 5.0 for Windows (Premier Biosoft International, Palo Alto, CA, USA). Oligonucleotide primers comprising T7 promoter sequences at the 5′ end for in vitro transcription and synthesis of double-strand RNA (dsRNA) were used to amplify cDNA encoding the DNA methyltransferases. All the oligonucleotide primers used for this study were synthesized by InvitrogenTM (Beijing, China). In vitro dsRNA synthesis was carried out using the T7 RiboMAXTM Express RNAi System (Promega, Madison, USA). The transcription reaction contained 1 μL of cDNA as the transcription template, 7.0 μL of RNase-free water, and 2 μL of T7 enzyme mix in 10 μL Express 2x Buffer culminating to a final volume of 20.0 μL.
The reactions were carried out at 37°C for 30 min, 70°C for 10 min, and allowed to stand for 20 min at room temperature. After adding 2 μl of RQ1 RNase-Free DNase and 2 μL of diluted RNase A solution, the reaction was incubated at 37°C for 30 min. Then, 4.4 μL Sodium Acetate and 110 μL 95% ethanol was added and incubated on ice for 5 min, followed by centrifugation at 16,000 g for 5 min at 4°C. The precipitate was added 500 μL of 70% ethanol (pre-cooled at −20°C), resuspended and centrifuged at 10,000 g for 5 min at 4°C, and the precipitate was dried at room temperature for 15 min. Finally, the dsRNA pellet was resuspended in diethylpyrocarbonate (DEPC)-treated water. The concentration and quality of dsRNA were determined by Nano Drop® ND-1000 Spectrophotometer and 1% agarose gel electrophoresis. The dsRNA was then stored at −80°C until use.
RNA Interference
The procedure for the injection of dsRNA into 40 female ticks (20 D. silvarum and 20 H. longicornis) was performed according to (50). After surface-sterilization of the ticks, 2 μL of dsRNA solutions were injected into the hemocoel of unfed female ticks through the third and fourth coxa at a concentration of 1 μg/tick using a 10 μL MicroliterTM Syringes (Hamilton, Nevada, USA). The control groups were injected with dsRNA-GFP. After injection, the ticks were placed in the environmental incubator (temperature 26 ± 1°C, relative humidity 75 ± 5%, 16 h of light: 8 h in darkness) for 24 h. Each treatment was carried out in three replicates.
The total RNA was extracted from the ticks using TRIzol reagent, and their cDNA was synthesized as described previously. The mRNA expression of the target genes after knockdown with dsRNA was evaluated by quantitative real-time PCR (qRT-PCR), performed using an Mx3005P qPCR system (Agilent Technologies, Santa Clara, USA) using TransStart® Top Green qPCR SuperMix (TransGen Biotech) according to the manufacturer's instructions. The real-time PCR assays were prepared using a 96 well plate (Axygen, California, USA) with 10 μL of 2 × TransStart® Top Green qPCR SuperMix, 1.0 μL of the cDNA template, 0.4 μL of each forward and reverse gene-specific primers (Table 1), and 0.4 μL of Passive Reference DyeII in a final volume of 20.0 μL. The conditions of the thermocycler used were set at 94°C for 30 s, followed by 40 cycles of 94°C for 5 s and 60°C for 30 s. Then, 95°C for 1 min, 55°C for 30 s, and a final 95°C for 30 s. Each sample was assessed in triplicate (technical replicates). Control without the cDNA was included in all batches. Actin was used as the reference gene given that it is a housekeeping gene that is constitutively expressed under various temperature stress conditions (40). The relative expressions of the DNA methyltransferase genes were calculated by a 2−ΔΔCT method (51).
Cold Tolerance of Ticks After RNAi
After confirmation of the knockdown of DNA methyltransferase genes, D. silvarum and H. longicornis ticks were exposed to a sub-lethal temperature of −22 and −14°C, respectively, for 2 h in a low-temperature thermostatic bath (Meixiang Instrument Co. Ltd., Shanghai, China). The temperature of −22°C was selected given that the discriminating temperatures (resulting in 20% survival) for the females and males were −21.7 and −22.6°C, respectively (52). Then the ticks were removed from the thermostatic bath, and the mortality rate was calculated. Ticks were considered dead if they cannot coordinate their appendage after stimulated by CO2.
Relative Expression of DNA Methyltransferase Proteins Under Different Cold Treatments
The unfed adult D. silvarum or H. longicornis were divided into 6 groups with each group comprising 20 ticks. Three groups kept at the normal environment chamber (26°C) served as control; whereas the other three groups (experimental group) were exposed to low temperature (4°C) for 3, 6, and 9 days, respectively. After treatment, the ticks were grounded to powder in liquid nitrogen and transferred to a pre-cooled 1.5 mL Eppendorf (EP) tube containing 1 mL of sterile phosphate-buffered saline (PBS, pH 7.4) and 10 μL of phenylmethylsulfonyl fluoride (PMSF) protein inhibitor. Then, it was vortexed and centrifuged at 13,000, rpm for 10 min at 4°C. The supernatant was transferred to a pre-cooled 1.5 mL EP tube. Protein concentration was measured using BCA (Bicinchoninic Acid) Protein Assay Kit (CWBIO, Haimen, Jiangsu, China), following the manufacturer's recommendations.
The extracted protein samples were dissolved in 0.1 M Tris-HCl (pH 6.8) containing 2% SDS, 5% 2-mercaptoethanol, 10% glycerol, and 0.05 % bromophenol blue, followed by 2 min boiling. The SDS-PAGE was performed on 12% separating gels with 4% stacking gels containing 0.1% SDS using PAGE Gel Preparation Kit (Epizyme, USA). A 0.05 M Tris-glycine buffer (pH 8.3) containing 0.1% SDS was used to perform electrophoresis at a constant voltage of 120 V until the tracking dye reached the end of the gel. Subsequently, the electro-transfer onto polyvinylidene difluoride (PVDF) membrane was carried out at 22V for 25 min using a Trans-Blot SD apparatus (BIO-RAD, Hercules, CA, USA). The membrane was blocked for 2.5 h with 5% skim milk in 1xTBST (Tris-buffered saline with 0.1% Tween® 20 detergent) solution, washed three times with 1xTBST in 15 min, and then incubated overnight with a monoclonal anti-DNMT antibody (1: 2000) (GeneTex, TX, USA). After three washes with 1xTBST in 15 min, the membrane was incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG for 2 h at room temperature, the signal was detected by ultra-sensitive enhanced chemiluminescent (ECL) substrate using SuperSignalTM West Femto Trial Kit (Thermo Scientific, Waltham, MA, USA), visualized and analyzed with an Image Lab Software-controlled Gel DocTM XR+ System (BIO-RAD).
Statistical Analysis
The differences among different treatment groups were compared with one way ANOVA test, and Tukey's test was used for post hoc analysis. A significant difference is defined as P < 0.05. Data analysis was carried out using Graph Prism version 8.0.2 for Windows (GraphPad Software, Inc., San Diego, USA).
Results
Cloning of DNA Methyltransferase Genes
The cDNA obtained from the total RNA of D. silvarum and H. longicornis were used as a template with specific primers for PCR amplification of the target DNA methyltransferase gene. The amplified products subjected to agarose gel electrophoresis showed single, clear, and bright bands (Supplementary Figure 1). The target fragments of D. silvarum DNA methyltransferase gene 1 were 493, 456, 552, and 539 bp, while the target fragment for DNA methyltransferase gene 2 was 549 bp. The target fragments of H. longicornis DNA methyltransferase gene 1 were 433, 851, and 526 bp, whereas the target fragments for gene 2 were 389, 517, 470, and 467 bp. After the blast, the two DNA methyltransferases of D. silvarum showed the highest sequence identity with the DNA cytosine methyltransferase (Dnmt) of Ixodes ricinus (65%) and DNA (cytosine-5)-methyltransferase (Dnmt1) of Ixodes scapularis (64%), whereas the two DNA methyltransferases of H. longicornis showed the highest sequence identity with the DNA methyltransferase 1 (Dnmt1) (81%) and DNA (cytosine-5)-methyltransferase PliMCI (Dnmt) (77%) of Ixodes scapularis. Thus, the DNA methyltransferase of D. silvarum and H. longicornis were named DsDnmt (1,284 bp), DsDnmt1 (549 bp), HlDnmt1 (1,500 bp), and HlDnmt (1,613 bp), respectively. The multiple sequence alignments indicate that the deduced amino acid sequences of DsDnmt, DsDnmt1, HlDnmt1, and HlDnmt showed high sequence identity to their corresponding sequences (Supplementary Figure 2).
Sequence Analysis
The DsDnmt had a relative molecular mass of 391957.00 Daltons with a base ratio of A+T at 5.56% and a base ratio of G+C at 54.44%. The predicted full length of its open reading frame (ORF) was 1,128 nucleotide (nt) ranging from 67 to 1,194 base pairs. DsDnmt1 had a relative molecular mass of 166573.00 Daltons with the base ratio of A+T <44.99% and G+C more than 55.01%. Its coding sequence (CDS) had a full length of 549 nt (1 to 549 bp). The sequence lengths of HlDnmt1 and HlDnmt were 1,500 and 1,613 bp, respectively, with multiple replications start sites. The total ORF length of HlDnmt1 was 1,464 nt (17 to 1,480 bp) with a relative molecular mass of 458465.00 Daltons and a base ratio of 37.87 and 62.13% for A+T and G+C, respectively. The relative molecular mass of HlDnmt mRNA was 491394.00 Daltons with a base ratio of 38.19% for A+T and 61.81% for G+C. The total length of the CDS was 1,611 nt and it ranges from 1 to 1,611 bp.
Protein Composition Analysis
Analysis of Amino Acid Composition
The predicted protein sequence of DsDnmt constitutes 375 amino acids, of which there were 19 types of amino acids. Leu accounted for 12.27%, followed by Ser, Pro, and Arg which accounted for 8.27, 7.47, 7.20, and 7.47%, respectively, whereas the least content was methionine which accounted for 1.87%. The total number of positively charged residues (Arg+Lys) was 38 and negatively charged residues (Asp+Glu) was 42. The predicted protein sequence of DsDnmt1 fragment contains 183 amino acids, of which can be categorized into 20 types. The protein encoded by the DsDnmt1 fragment contains 183 amino acids, of which there are 20 types of amino acids, most of which was Leu accounting for 13.11%. Gly accounted for 9.29%, while each of Ser and Arg, accounted for 8.20% and with the least content being Trp which accounted for 0.55%. The total number of positively and negatively charged residues are 22 and 12, respectively. The predicted protein sequence of HlDnmt1 contains 487 amino acids, of which there are 20 types of amino acids. The most abundant amino acid was Ala accounting for 10.25%, followed by Leu, Arg, Glu, and Lys, accounting for 9.84, 9.63, 8.20, and 7.99% respectively, whereas the least abundant was Cys accounting for 0.82%. The residues that were positively charged were 86 amino acids while the negatively charged residues were 66 amino acids. There were 537 amino acids in the predicted protein sequence of HlDnmt, of which can be categorized into 20 types, with Val being the most abundant accounting for 8.75%. Asp, Glu, and Ala accounted for 8.57, 8.19, and 8.01%, respectively, whereas the least abundant, is Trp, which accounts for 1.30%. A total of 62 amino acids were positively charged while a total of 90 were negatively charged.
Prediction of Amino Acid Hydrophobicity
The hydrophobic properties of the DNA methyltransferases were determined with the Hphob/Kyte&Doolittle model by a window size of 9 amino acids. The maximum hydrophobic value for DsDnmt was 2.078, located at the 50th and 121st amino acids which were Ala and Phe, respectively, whereas the minimum was −2.911 at the 111th amino acid (Gln). For DsDnmt1, the maximum hydrophobic value was 2.233, which was the 172nd amino acid (Ala), while the minimum was −2.489, the 92nd amino acid (His). The maximum hydrophobic value for HlDnmt1 was 1.722 at the 412th amino acid (Ala) and the minimum value was −3.844 at the 223rd amino acid (Arg). HlDnmt had 2.044 as its maximum hydrophobic value at the 445th amino acid (Leu) and −2.900 as its minimum hydrophobic value at the 338th amino acid (Arg). The total average hydrophobicity of DsDnmt, DsDnmt1, HlDnmt1, and HlDnmt proteins are −0.348, −0.239, −0.728, and −0.459, respectively, suggesting that the four DNA methyltransferases were hydrophilic proteins (Supplementary Figure 3).
Prediction of the Protein Physical and Chemical Properties
The physicochemical properties of DNA methyltransferase protein were predicted through the ProtParam Expasy website (http://web.expasy.org/protparam/) and DiANNA website (http://bioinformatics.bc.edu/clotelab/DiANNA/). All four putative proteins were predicted to have a nuclear localization (Table 2). The instability index for all four studied proteins was higher than 40, indicating that they were unstable, whereas the predicted disulphide bonds of the proteins might help to maintain their molecular shape, stability, and substrate specificity (Table 2).
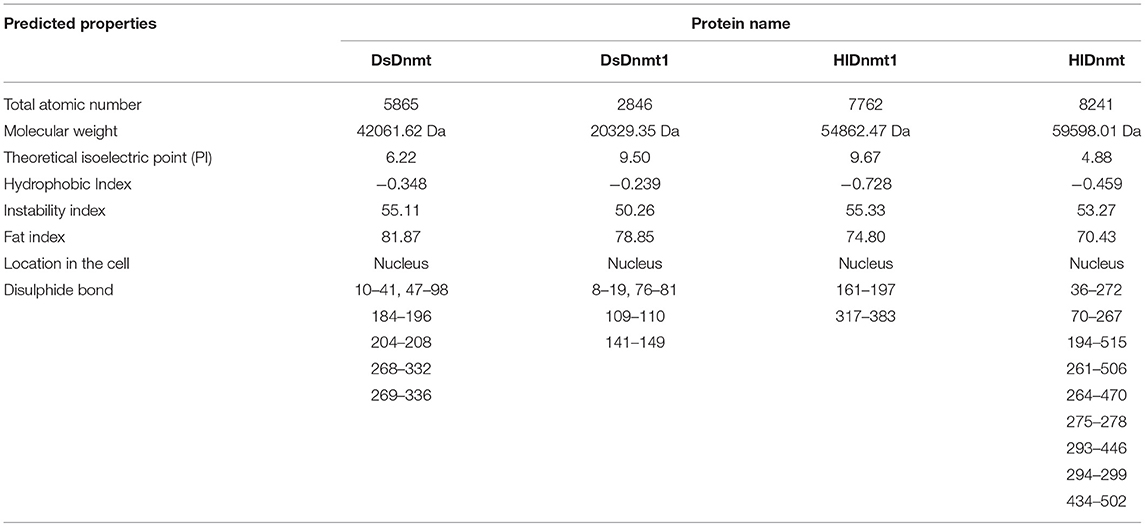
Table 2. Predicted properties of the DNA methyltransferase protein of Dermacentor silvarum and Haemaphysalis longicornis.
Phylogenetic Relationship
The evolutionary relationship between the DNA methyltransferases of D. silvarum and other species as well as those of DNA methyltransferases of H. longicornis were constructed (Figure 1). Results revealed that the DNA methyltransferases of D. silvarum and H. longicornis clustered the arthropod species into their corresponding taxonomic classes. The putative DsDnmt and DsDnmt1 share the same node with the sequences of the closely related I. ricinus (QCB92136.1), I. scapularis (EEC18555.1), and I. ricinus (QCB92132.1), I. scapularis (EEC00472.1), respectively. Similarly, the putative HlDnmt1 and HlDnmt share the same node with the sequence of the closely related I. scapularis (XP029828587.1, XP029831274.1). Ticks belong to a relatively independent branch (arachnids) and are shown to be closely related to other arachnids than the insects. Vertebrates such as mammals, birds, and fishes were placed in an independent branch indicating that their DNA methyltransferases are less closely related to those of ticks than mollusks and other marine organisms.
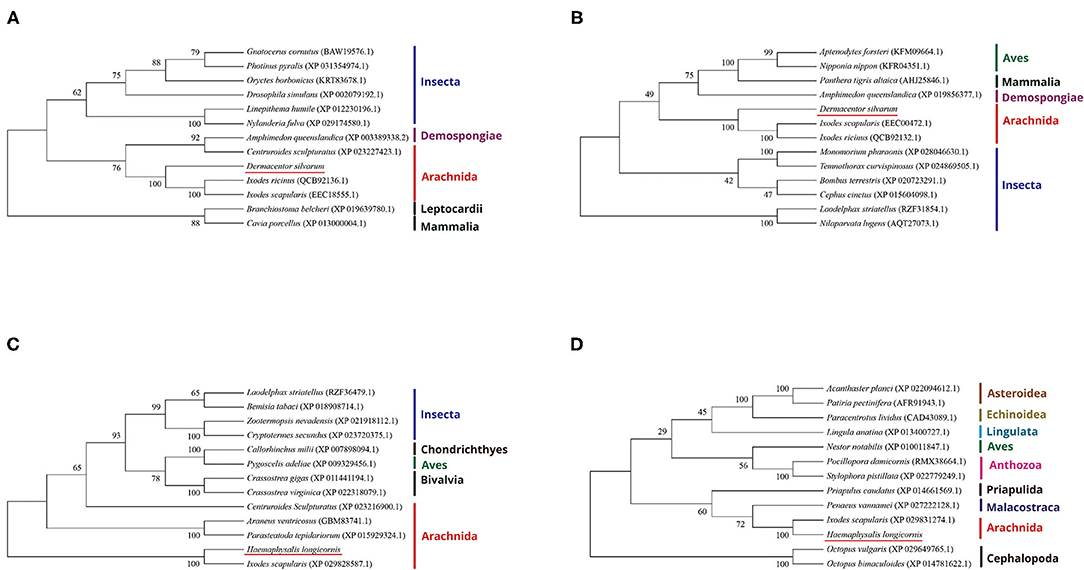
Figure 1. Phylogenetic reconstruction of DNA methyltransferases based on the neighbor-joining method. The listed species are colored according to their classification to particular taxonomic classes. (A) Illustration of the phylogenetic relationship of DsDnmt sequences to Dnmts of other species; (B) Illustration of the phylogenetic relationship of DsDnmt1 sequences to Dnmts of other species; (C) Illustration of the phylogenetic relationship of HlDnmt1 sequences to Dnmts of other species; (D) Illustration of the phylogenetic relationship of HlDnmt sequences to Dnmts of other species.
Conserved Domain Analysis
The conserved domain search was performed using the Conserved Domain Database (CDD) v.3.16 of NCBI with default parameters and a standard display mode. The result revealed that putative DsDnmt and DsDnmt1 contain DNA cytosine methylase (Dcm) domain (interval 24–369 bp and interval 5–172 bp, respectively) with multiple substrates- and DNA-binding sites (Figure 2). HlDnmt1 contains DMAP1 (DNA methyltransferase 1-associated protein) domain (interval 241–401 bp) and SANT domain (interval 121–201 bp). The putative HlDnmt has Bromo Adjacent Homology (BAH) domains (interval 370–498 bp), Dnmt1-RFD (cytosine-specific DNA methyltransferase replication foci domain) at an interval of 19–148 bp, and Zinc finger (zf-CXXC) domain (interval 253–299 bp) which is present in a typical structure of Dnmt (Figure 2). The open reading frames of DsDnmt1 and HlDnmt are probably incomplete and it is speculated that they lack other domains at the N- and C-terminal, respectively.
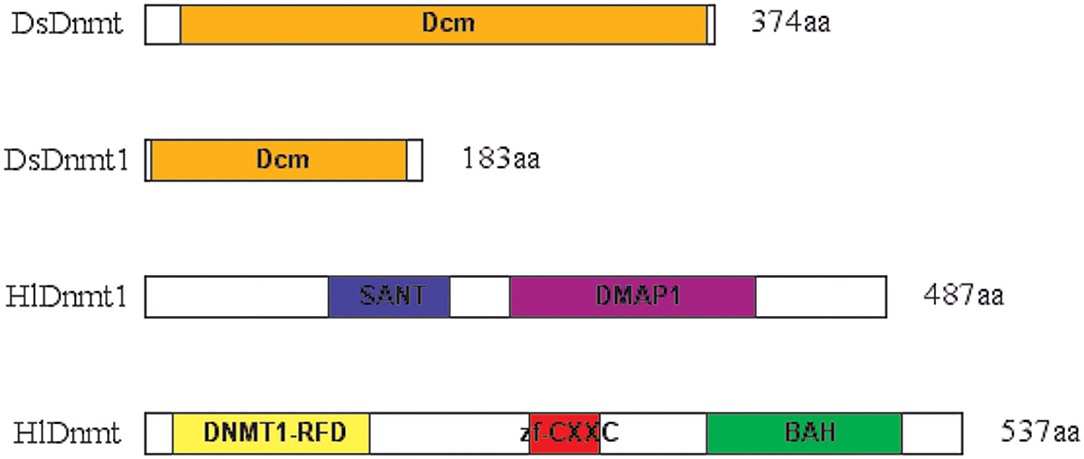
Figure 2. Schematic presentation of conserved domains identified for the putative DNA methyltransferases of D. silvarum and H. longicornis. Putative DsDnmt and DsDnmt1: orange—Dcm domain; putative HlDnmt1: blue—SANT domain, purple—DMAP1 domain; putative HlDnmt: yellow—DNA methyltransferase replication foci domain (DNMT1-RFD), red—CXXC zinc finger domain, green—Bromo adjacent homology (BAH) domain.
Protein Secondary Structure Prediction
The prediction of the secondary structure of the DNA methyltransferase proteins was carried out and the results showed that the four DNA methyltransferases have mixed secondary structures (Supplementary Table 1). α-helix and random coiled loop account for a larger proportion of the structure, whereas β-sheet strand accounts for a relatively small proportion, which provides an important structural basis for protein functional implementation and conformation. Furthermore, the DNA binding sites and protein binding sites of the DNA methyltransferase proteins were predicted (Supplementary Table 2).
Protein Tertiary Structure (3D Structure) Prediction
The tertiary structures of the four DNA methyltransferase proteins were predicted by Swiss-Model (http://swissmodel.expasy.org/). The reliability range of the global model quality estimation (GMQE) is from 0 to 1. The GMQE values of DsDnmt, DsDnmt1, HlDnmt1, and HlDnmt proteins were 0.65, 0.79, 0.10, and 0.74, respectively. The tertiary structure prediction models of DsDnmt, DsDnmt1, and HlDnmt have higher confidence, except for HlDnmt1, possibly due to the lower homology of their amino acid sequences in the software database. The predicted DsDnmt contains one Ca2+ and one SAH (S-adenosyl-homocysteine) binding site, where SAH is the binding site of the methyl donor molecule SAM (S-adenosyl-L-methionine molecule); DsDnmt1 contains two Zn2+ binding sites. HlDnmt1 contains one Ca2+ binding site, whereas HlDnmt contains five Zn2+ and one SAH binding site (Figure 3).
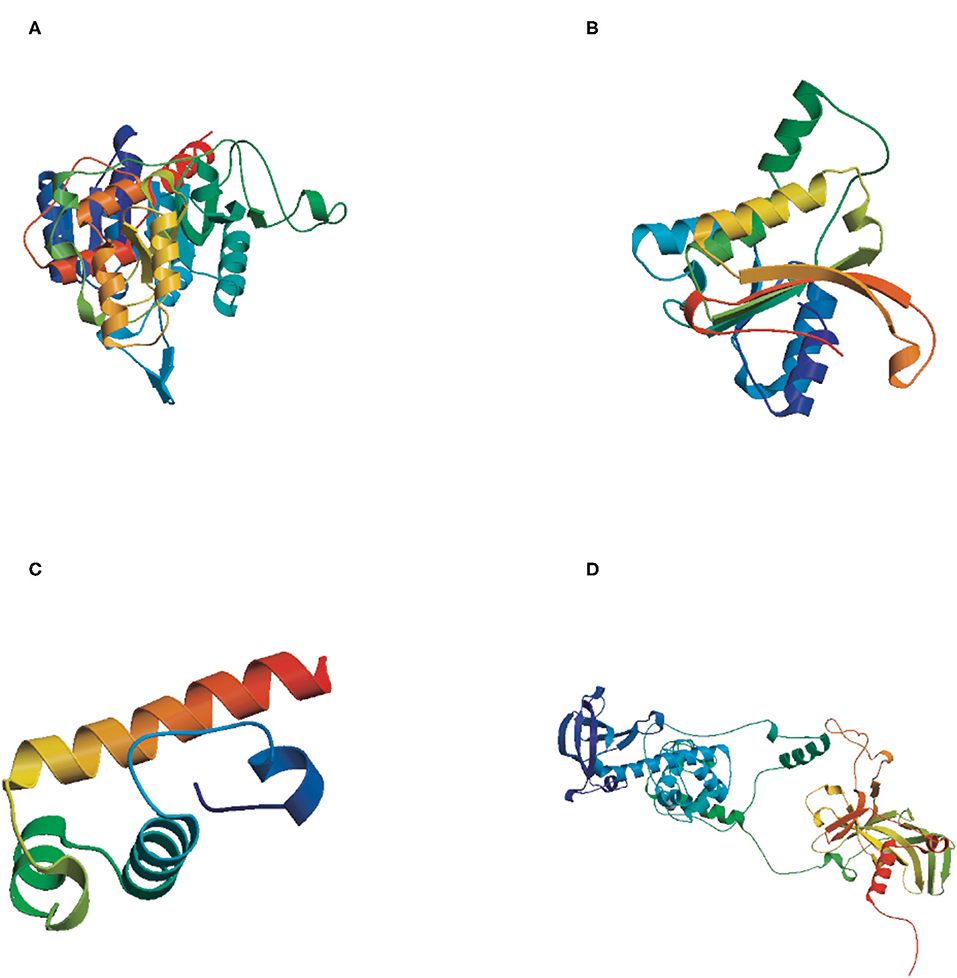
Figure 3. The tertiary structure (3D) of putative DNA methyltransferase proteins predicted by Swiss-Model. (A) DsDnmt, GMQE value = 0.65; (B) DsDnmt1, GMQE value = 0.79; (C) HlDnmt1, GMQE value = 0.10; (D) HlDnmt, GMQE value = 0.74.
Relative Expression of DNA Methyltransferase Genes Under Cold Treatments
The relative expression levels of HlDnmt1, DsDnmt1, and DsDnmt decreased significantly after 3 days of cold treatment (P < 0.01), but increased with the extension of cold treatment time/days (Figures 4A–C). HlDnmt expression decreased significantly after 3 days of cold treatment (P < 0.05), but had a sharp significant increase (P < 0.01) after 9 days of cold treatment (Figure 4D).
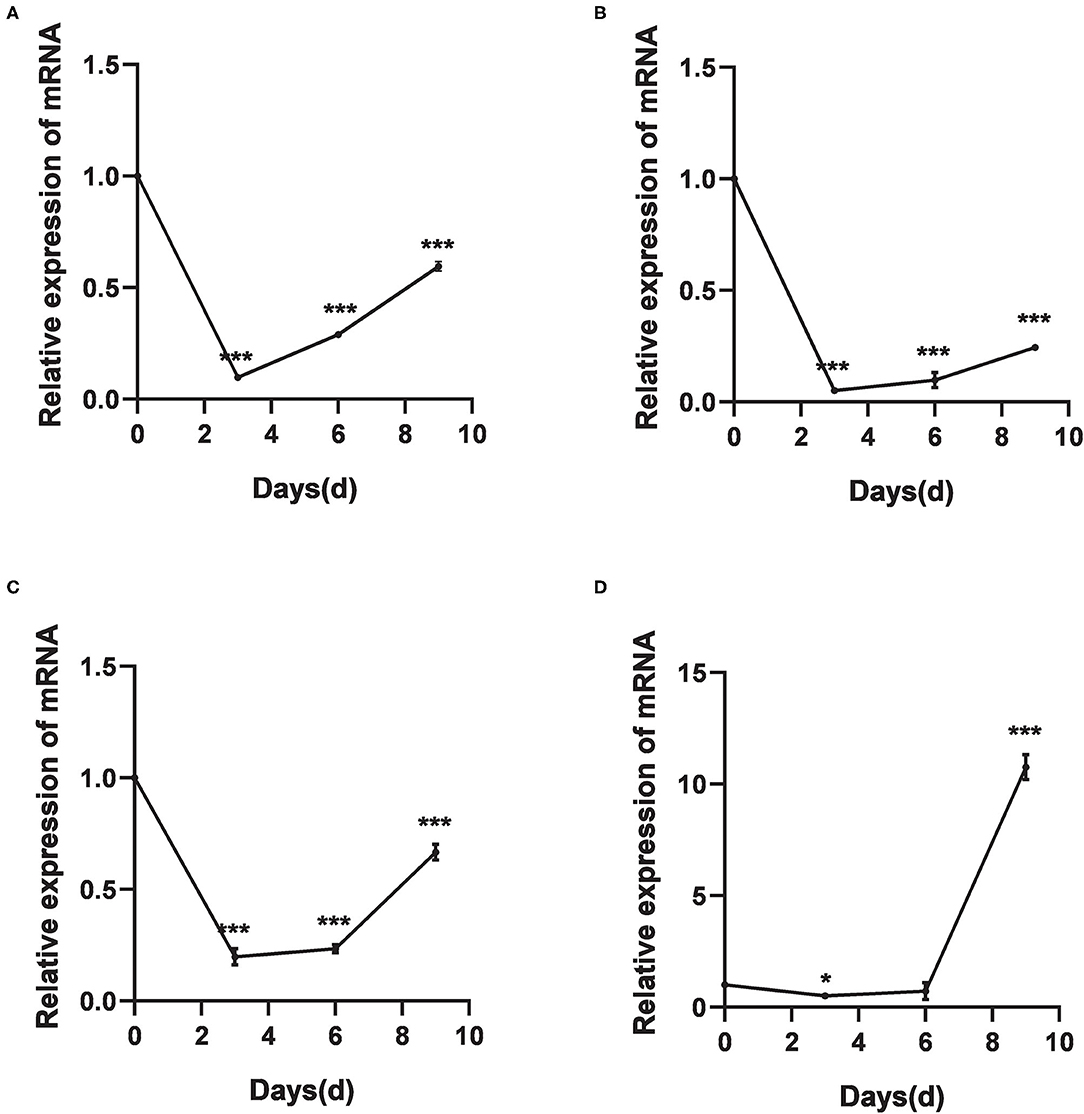
Figure 4. The relative expression of DNA methyltransferase mRNAs after cold treatments vis-à-vis the untreated group. (A–D) Relative expression of DsDnmt (A), DsDnmt1 (B), HlDnmt1 (C), HlDnmt (D) measured using qRT-PCR. The results are expressed as the means (n = 3) ± SEM. The asterisks above bars indicate significant difference based on one-way ANOVA followed by Turkey's test, *P < 0.05, ***P < 0.01.
The Efficiency of RNA Interference of DNA Methyltransferase
The relative expressions of the DNA methyltransferase gene were determined after RNAi. Results showed that the expression of the DNA methyltransferase genes was decreased significantly (P < 0.01) (Figure 5). Analysis of gene silencing efficiency showed that the average silencing efficiency of DsDnmt and DsDnmt1 genes were 74.79 and 73.78%, respectively, whereas that of HlDnmt1 and HlDnmt genes were 82.42 and 83.72%, respectively, indicating that the dsRNA injection of DNA methyltransferase gene had a significant silencing effect on the expression of the target gene.
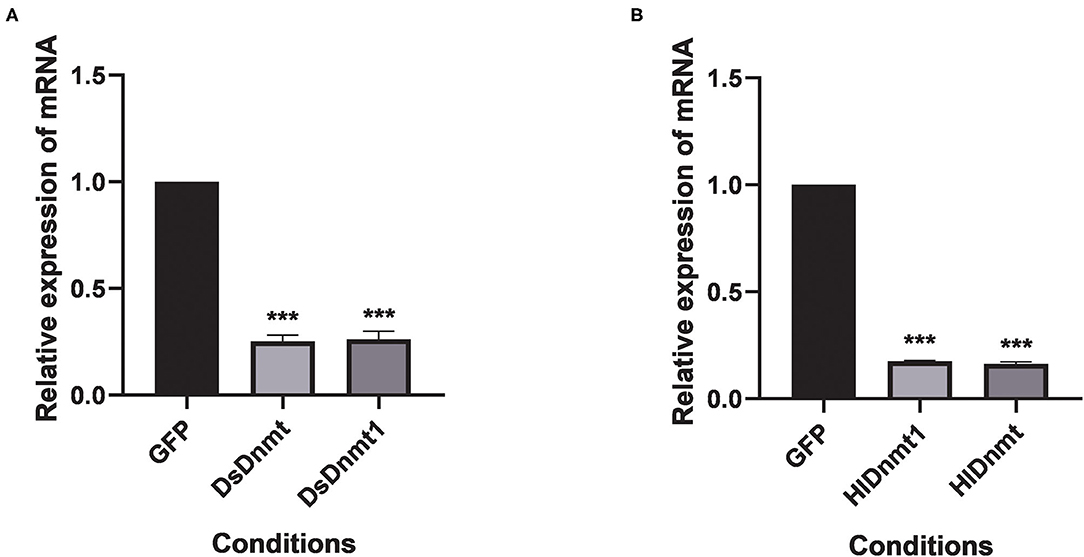
Figure 5. Effect of double-stranded RNA treatment on expression of Dnmts in D. silvarum (A) and H. longicornis (B). The expressions of Dnmts were significantly decreased post 24 h after the injection of respective dsRNAs compared with expression in the controls. The results are expressed as the means (n = 3) ± SEM. The asterisks above bars indicate significant difference based on one way ANOVA followed by Turkey's test, ***P < 0.01. GFP represents the green fluorescent protein gene.
Cold Tolerance of D. silvarum and H. longicornis After RNAi
After the interference of DNA methyltransferases, the changes in cold tolerance of D. silvarum and H. longicornis were analyzed, and the role of DNA methyltransferase in the cold response of ticks was determined. The mortality rates of both tick species increased significantly (P < 0.05) (Figure 6). The resultant mortality rate from the knockdown of DsDnmt, DsDnmt1, and HlDnmt was more than 60%, while the resultant mortality from the knockdown of HlDnmt1 was over 70%, indicating the important function of DNA methyltransferase in the cold tolerance of D. silvarum and H. longicornis.
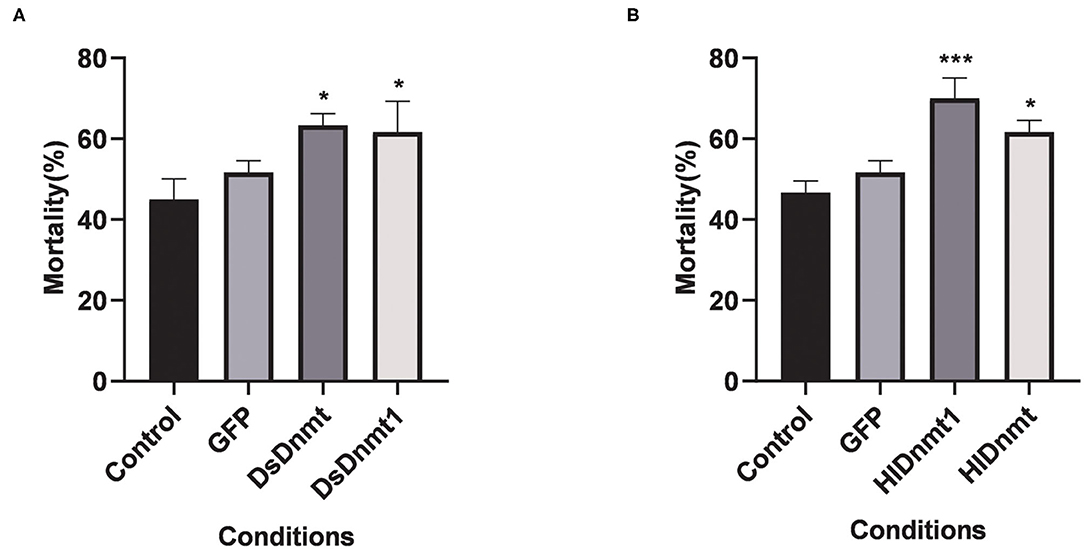
Figure 6. The effects on the cold tolerance of ticks after the knockdown of Dnmts. (A) The mortality rate of D. silvarum after cold treatment post DsDnmt and DsDnmt1 knockdown. The mortality rate increased significantly in the ticks with Dnmts' knockdown in relation to the controls. (B) The mortality rate of H. longicornis after cold treatment post HlDnmt1 and HlDnmt knockdown H. longicornis. The mortality rate increased significantly in the ticks with Dnmts' knockdown in relation to the controls. The results are expressed as the means (n = 3) ± SEM. The asterisks above bars indicate significant difference based on one-way ANOVA followed by Turkey's test, *P < 0.05, ***P < 0.01.
Relative Expression of DNA Methyltransferase Proteins Under Different Cold Treatments
A total of 5 DNA methyltransferase-related proteins were selected (Dnmt1, Dnmt2, Dnmt3A, Dnmt3B, and Dnmt3L) as the target proteins which were determined depending on the commercially available specific antibody products. The results showed that the relative expression levels of various DNA methyltransferase proteins in the ticks increased or decreased to varying degrees after cold treatment. In D. silvarum, the protein expression of Dnmt1 and Dnmt2 changed significantly (P < 0.05). After cold treatment for 3 days, the protein expression was down-regulated, but with the extension of treatment, the expression was up-regulated. After cold treatment, the expression of Dnmt3A increased significantly (P < 0.05), and then gradually decreased. Dnmt3L showed a similar trend of change as Dnmt3A, but not to a significant level (Figure 7). In H. longicornis, the expression of Dnmt1 decreased after cold treatment for 3 days, and then gradually increased significantly (P < 0.05) after cold treatment for 9 days. Dnmt3B showed a similar trend as Dnmt1. The protein expression of Dnmt3L up-regulated initially after cold treatment before down-regulating with time, just as in D. silvarum ticks (Figure 8).
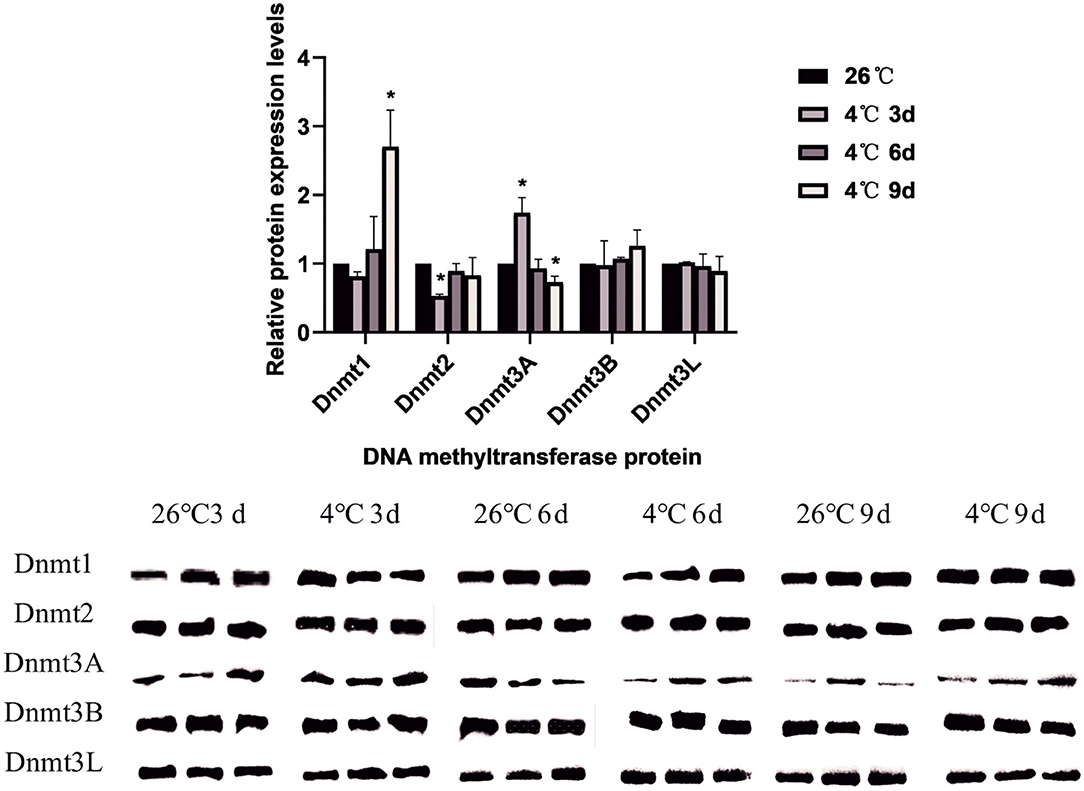
Figure 7. The relative protein expression of Dnmts in D. silvarum after cold treatment. The relative level of the expression of DNA methyltransferase proteins were calculated, and the expression in the control group served as the reference, which was set at 1. The results are expressed as the means (n = 3) ± SEM. The asterisks above bars indicate a significant difference based on one-way ANOVA followed by Turkey's test, *P < 0.05.
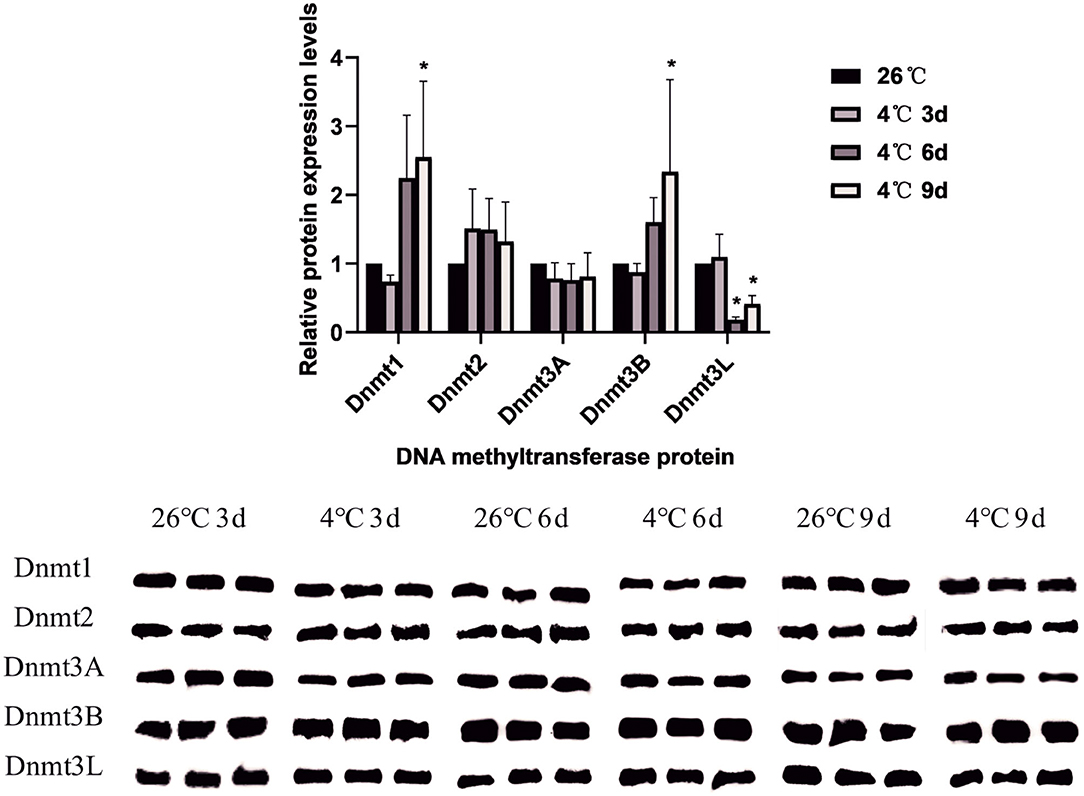
Figure 8. The relative protein expression of Dnmts in H. longicornis after cold treatment. The relative level of the expression of DNA methyltransferase proteins were calculated, and the expression in the control group served as the reference, which was set at 1. The results are expressed as the means (n = 3) ± SEM. The asterisks above bars indicate a significant difference based on one-way ANOVA followed by Turkey's test, *P < 0.05.
Discussion
In the present study, four DNA methyltransferase genes were characterized from D. silvarum and H. longicornis, and phylogenetic analysis revealed that they were clustered with the arthropod species into their corresponding taxonomic classes. The putative DsDnmt and DsDnmt1 share the same node with the sequences of the closely related species (I. ricinus and I. scapularis). Similarly, the putative HlDnmt1 and HlDnmt share the same node with the sequences of closely related I. scapularis. Ticks belong to a relatively independent branch and are closely related to other arachnids than the insects. Vertebrates such as mammals, birds, and fishes were placed in an independent branch indicating that their DNA methyltransferases are less closely related to those of ticks than mollusks and other marine organisms. Overall, these results confirm that DNA methyltransferases of ticks are good candidates for phylogenetic analysis at higher levels. However, a previous study could not find a common ancestry between putative I. ricinus DNA methyltransferase (IrDAMT) and other arachnid species, but it was rather placed in a sister branch of the cluster of Maxillopoda, Branchiopoda, and insect (38). Additionally, HlDnmt results suggest that they could be less suitable for a detailed study of the evolutionary relationship among arachnids/arthropods as none of its orthologs were found (apart from I. scapularis), presumably because of their non-existence in a publicly available database.
All the four putative proteins were predicted to have a nuclear localization, which is consistent with their hypothetical assignment to the methyltransferase group of enzymes. This was corroborated by the three putative I. ricinus methyltransferases—IrDNMT1, IrDNMT3, and IrDAMT (38). The instability index is a measure used to determine whether proteins will be stable in a test tube (53), and the index values above 40 are an indicator for unstable proteins, which is true for the four DNA methyltransferases in the present study. The conserved domain search revealed that putative DsDnmt and DsDnmt1 contain DNA cytosine methylase (Dcm) domain with multiple substrates- and DNA-binding sites. At the Dcm domain target sites (CCAGG and CCTGG), a methyl group is added to the 5th position of the cytosine base in the context of CpG dinucleotide by C-5 cytosine-specific DNA methylases (C5 Mtase); this plays a vital role in gene expression by either inhibiting the binding of transcription factors or the synthesis of methyl binding protein and related chromatin remodeling factors (54, 55). HlDnmt1 contains the DMAP1 (DNA methyltransferase 1-associated protein) domain and SANT domain. DMAP1 possesses intrinsic transcriptional inhibitory activity and could play an important role in the development and maintenance of the integrity of methylation via interaction with Dnmt1 (56), whereas the SANT domain contains a putative DNA-binding site (57). Moreover, some interesting findings showed that the SANT domain was involved in the activities of ATP-dependent chromatin remodeling enzymes, as well as in the histone acetyltransferase and deacetylase complexes (57–59). The putative HlDnmt has BAH domain, Dnmt1-RFD at the interval of 19–148 bp, and Zinc finger-CXXC (zf-CXXC) domain. BAH domain is responsible for linking DNA methylation, replication, and transcriptional regulation (37, 60). The functions of the BAH domain in chromatin biology are versatile including taking part in the protein-protein interaction that mediates gene silencing, recognition of methylated histones, and direct interaction with nucleosome (61). Dnmt1-RFD is located at the longer N-terminal region of Dnmt1 which targets the replication foci (62). Specifically, it ensures that the Dnmt1 protein is targeted toward the replication foci in a non-catalytic fashion, which allows it to methylate the correct residues, and has been associated with the conversion of critical histone lysine moieties (63). Of note, Dnmt1-RFD binds to HDACs (histone deacetylases) and DMAP1 in a manner that can mediate transcriptional repression (63). The zf-CXXC domain contains eight conserved cysteine residues (present in the typical structure of Dnmts) which bind to Zn2+ ions and non-methylated CpG dinucleotides (64). Although it was generally acknowledged that methylated CpGs (cytosine guanine dinucleotides) are specifically recognized by proteins that encode MBDs (methyl-CpG-binding domains) (65, 66), it has also been proposed that non-methylated CpG dinucleotides could present a protein-binding site (64). This was made possible by the identification of non-methylated CGBP (CpG-binding protein) whose DNA-binding capacity is dependent on a cysteine-rich zf-CXXC domain (67).
Temperature is one of the most important factors that greatly influence the development, growth, geographical distribution, abundance, and physiology of organisms (68, 69). Organisms respond to changes in environmental temperature by activating stress response mechanisms. Studies suggest that epigenetic enzymes play a vital role in arthropod cold hardiness (70). To the best of our knowledge, this is the first report of the response of DNA methyltransferases of D. silvarum and H. longicornis ticks to cold stress. Several studies with other species have demonstrated the relationship between DNA methylation, a vital environment-induced epigenetic phenomenon, and variations in temperature (40, 71–73). For example, there was a significant change in the expression level of Dnmt3 in the Senegalese sole (fish) after short-term exposure to embryonic temperatures (42). In the present study, the first 3 days of cold exposure were marked with a significant decrease in the expression levels of all the Dnmt genes but increased significantly with the prolongation of days of cold treatment. A similar trend was observed in the armyworm (Mythimna separata) where a decreased pattern of expression was observed after short exposures to various temperatures (73). A sharp significant increase in the expression levels of HlDnmt was observed after 9 days of cold exposure (4°C). This was corroborated by the significant up-regulation of the expression levels of Bemisia tabaci DNA methyltransferase genes (BtDnmt3) after exposure to low temperatures ranging from 0 to 20°C (74). A possible explanation could be that for the first few days of cold exposure, the ticks employ other physiological strategies such as cryoprotective dehydration (28), the modification of body fluid constituents (27), and the utilization of numerous potentially protective molecules in their body (26) before the full activation of the activities of the Dnmts as the cold stress prolongs. Additionally, the same pattern of expression was observed via western blot where the relative expression of various DNA methyltransferase proteins in the ticks decreased or increased to varying degrees after cold treatment. The protein expression level was down-regulated after 3 days of cold treatment but was up-regulated with the extension of treatment time for both the Dnmts of D. silvarum and H. longicornis. Similarly, a study observed that cold and subzero temperature exposure in Eurosta solidaginis (goldenrod gall fly) resulted in the upregulation of several DNA methyltransferases (70). The down-regulation and up-regulation due to temperature variations is a confirmation of the thermal plasticity of Dnmt expression which is reflected in the corresponding protein expression. Ticks are remarkably plastic and can inhabit diverse ecological niches, from the tropics to the polar regions of the planet (2, 75). Like insects, ticks are ectotherms that are sensitive to external temperature variations (76). The cold hardiness of ticks that enables them to overwinter could be a function of epigenetic modifications such as DNA methylation that might occur rapidly in response to large-scale temperature changes (40).
Due to the vital role of DNA methyltransferases in different biological processes, the knockdown of DsDnmt, DsDnmt1, HlDnmt1, and HlDnmt was carried out to identify their functions by assessing the mortality rate after the exposure of D. silvarum and H. longicornis ticks to sublethal low temperatures of −22 and −14°C, respectively, for 2 h. Interestingly, the mortality rates of the two tick species significantly increased which underscores the impact of DNA methyltransferases on the cold tolerance of ticks. This result was corroborated by the resultant significant mortality of armyworms (whose MsDnmt1 genes have been knocked down) after exposure to a low temperature (73). Previous studies have demonstrated the effect of extreme temperatures on arthropod development which include reduced fecundity, deformity, and mortality (77–79). It is noteworthy that ticks, which are widely distributed globally, can adapt to different climatic regions and they might possess unique adaptability mechanisms. One of these adaptability mechanisms could be mediated by DNA methyltransferases. DNA methylation, a vital epigenetic mechanism induced by the environment, can serve as a potential link between phenotypic variability and temperature variation, which is reprogrammed by Dnmts (40).
Conclusion
This study characterized DNA methyltransferases in D. silvarum and H. longicornis, and the results suggest that they contribute to the cold tolerance of ticks. The above findings highlighted the functional implications of DNA methyltransferases during cold stress of ticks and thus could advance our understanding of the epigenetic regulation of cold acclimation and adaptation of ticks.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
DA wrote the manuscript. MiZ, XS, SZ, MeZ, TW, and DA conducted the literature, performed the experiments, and prepared all the figures and tables. ZY, AM, and JL reviewed and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the National Natural Science Foundation of China (No. 32071510), the Youth Top Talent Support Program of Hebei Province to ZY (2018-2023), and the Natural Science Foundation of Hebei Province (C2019205064).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
DA acknowledges the support of the China Scholarship Council (CSC) for doctoral research.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.726731/full#supplementary-material
Supplementary Figure 1. Agarose gel illustrating the results of PCR amplification of DNA methyltransferase genes. (A) Gene 1 (DsDnmt) fragments of D. silvarum; (B) Gene 2 (DsDnmt1) fragment of D. silvarum; (C) Gene 1 (HlDnmt1) fragments of H. longicornis; (D) Gene 2 (HlDnmt) fragments of H. longicornis.
Supplementary Figure 2. Amino acid sequence homologous alignment results of DNA methyltransferases. (A) DsDnmt; (B) DsDnmt1; (C) HlDnmt1; (D) HlDnmt.
Supplementary Figure 3. Hydrophobicity prediction of the amino acid sequence of DNA methyltransferases. (A) DsDnmt; (B) DsDnmt1; (C) HlDnmt1; (D) HlDnmt.
Supplementary Table 1. The secondary structure prediction of the DNA methyltransferase proteins of Dermacentor silvarum and Haemaphysalis longicornis.
Supplementary Table 2. The binding sites prediction of the DNA methyltransferase protein.
Abbreviations
Leu, Leucine; Ser, serine; Pro, proline; Arg, arginine; Asp, aspartic acid; Glu, glutamic acid; Lys, lysine; Gly, glycine; Ala, alanine; Trp, tryptophan; Cys, cysteine; Val, valine; Gln, glutamine; Phe, phenylalanine; His, histidine.
References
1. Yu Z, Wang H, Wang T, Sun W, Yang X, Liu J. Tick-borne pathogens and the vector potential of ticks in China. Parasit Vectors. (2015) 8:24. doi: 10.1186/s13071-014-0628-x
2. De La Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. (2008) 13:6938–46. doi: 10.2741/3200
3. Ma J, Lv X, Zhang X, Han S, Wang Z, Li L, et al. Identification of a new orthonairovirus associated with human febrile illness in China. Nat Med. (2021) 27:434–9. doi: 10.1038/s41591-020-01228-y
4. Kuivanen S, Levanov L, Kareinen L, Sironen T, Jääskeläinen AJ, Plyusnin I, et al. Detection of novel tick-borne pathogen, Alongshan virus, in Ixodes ricinus ticks, south-eastern Finland, 2019. Euro Surveill. (2019) 24:1900394. doi: 10.2807/1560-7917.ES.2019.24.27.1900394
5. Wang Z, Wang W, Wang N, Qiu K, Zhang X, Tana G, et al. Prevalence of the emerging novel Alongshan virus infection in sheep and cattle in Inner Mongolia, northeastern China. Parasit Vectors. (2019) 12:450. doi: 10.1186/s13071-019-3707-1
6. Yun S-M, Lee Y-J, Choi W, Kim H-C, Chong S-T, Chang K-S, et al. Molecular detection of severe fever with thrombocytopenia syndrome and tick-borne encephalitis viruses in ixodid ticks collected from vegetation, Republic of Korea, 2014. Ticks Tick Borne Dis. (2016) 7:970–8. doi: 10.1016/j.ttbdis.2016.05.003
8. Yu Z, Jia Q, Wang T, Dong N, Yang X, Wang H, et al. Gene expression profiling of the unfed nymphal Dermacentor silvarum (Acari: Ixodidae) in response to low temperature. Syst Appl Acarol. (2017) 22:2178–89. doi: 10.11158/saa.22.12.10
9. Lu Z, Broker M, Liang G. Tick-borne encephalitis in mainland China. Vector Borne Zoonotic Dis. (2008) 8:713–20. doi: 10.1089/vbz.2008.0028
10. Tian ZC, Liu GY, Shen H, Xie JR, Luo J, Tian MY. First report on the occurrence of Rickettsia slovaca and Rickettsia raoultii in Dermacentor silvarum in China. Parasit Vectors. (2012) 5:19. doi: 10.1186/1756-3305-5-19
11. Wen J, Jiao D, Wang JH, Yao DH, Liu ZX, Zhao G, et al. Rickettsia raoultii, the predominant Rickettsia found in Dermacentor silvarum ticks in China-Russia border areas. Exp Appl Acarol. (2014) 63:579–85. doi: 10.1007/s10493-014-9792-0
12. Jiang J, An H, Lee JS, O'guinn ML, Kim HC, Chong ST, et al. Molecular characterization of Haemaphysalis longicornis-borne rickettsiae, Republic of Korea and China. Ticks Tick Borne Dis. (2018) 9:1606–13. doi: 10.1016/j.ttbdis.2018.07.013
13. Agwunobi DO, Hu Y, Yu Z, Liu J. Cymbopogon citratus essential oil-induced ultrastructural and morphological changes in the midgut, cuticle and Haller's organ of the tick Haemaphysalis longicornis (Acari: Ixodidae). Syst Appl Acarol. (2020) 25:2047–62. doi: 10.11158/saa.25.11.10
14. Rainey T, Occi JL, Robbins RG, Egizi A. Discovery of Haemaphysalis longicornis (Ixodida: Ixodidae) Parasitizing a sheep in New Jersey, United States. J Med Entomol. (2018) 55:757–9. doi: 10.1093/jme/tjy006
15. Duncan KT, Sundstrom KD, Saleh MN, Little SE. Haemaphysalis longicornis, the Asian longhorned tick, from a dog in Virginia, USA. Vet Parasitol Reg. (2020) 20:100395. doi: 10.1016/j.vprsr.2020.100395
16. United States Department of Agriculture. National Haemaphysalis longicornis (Asian longhorned tick) Situation Report. (2020). Available online at: https://www.aphis.usda.gov/animal_health/animal_diseases/tick/downloads/longhorned-tick-sitrep.pdf (accessed February 21, 2021).
17. Chen Z, Liu Q, Liu J-Q, Xu B-L, Lv S, Xia S, et al. Tick-borne pathogens and associated co-infections in ticks collected from domestic animals in central China. Parasit Vectors. (2014) 7:237. doi: 10.1186/1756-3305-7-237
18. Heath A. Biology, ecology and distribution of the tick, Haemaphysalis longicornis neumann (Acari: Ixodidae) in New Zealand. NZ Vet J. (2016) 64:10–20. doi: 10.1080/00480169.2015.1035769
19. Ikadai H, Sasaki M, Ishida H, Matsuu A, Igarashi I, Fujisaki K, et al. Molecular evidence of Babesia equi transmission in Haemaphysalis longicornis. Am J Trop Med Hyg. (2007) 76:694–7. doi: 10.4269/ajtmh.2007.76.694
20. Tabara K, Kawabata H, Arai S, Itagaki A, Yamauchi T, Katayama T, et al. High incidence of rickettsiosis correlated to prevalence of Rickettsia japonica among Haemaphysalis longicornis tick. J Vet Med Sci. (2011) 73:507–10. doi: 10.1292/jvms.10-0369
21. Liu H, Li Q, Zhang X, Li Z, Wang Z, Song M, et al. Characterization of rickettsiae in ticks in northeastern China. Parasit Vectors. (2016) 9:498. doi: 10.1186/s13071-016-1764-2
22. Noh Y, Lee YS, Kim H-C, Chong S-T, Klein TA, Jiang J, et al. Molecular detection of Rickettsia species in ticks collected from the southwestern provinces of the Republic of Korea. Parasit Vectors. (2017) 10:20. doi: 10.1186/s13071-016-1955-x
23. Wu XB, Na RH, Wei SS, Zhu JS, Peng HJ. Distribution of tick-borne diseases in China. Parasit Vectors. (2013) 6:119. doi: 10.1186/1756-3305-6-119
24. Yun SM, Lee WG, Ryou J, Yang SC, Park SW, Roh JY, et al. Severe fever with thrombocytopenia syndrome virus in ticks collected from humans, South Korea, 2013. Emerg Infect Dis. (2014) 20:1358–61. doi: 10.3201/eid2008.131857
25. Somme L. The physiology of cold hardiness in terrestrial arthropods. Eur J Entomol. (1999) 96:1–10.
26. Toxopeus J, Sinclair BJ. Mechanisms underlying insect freeze tolerance. Biol Rev Camb Philos Soc. (2018) 93:1891–914. doi: 10.1111/brv.12425
27. Sformo T, Kohl F, Mcintyre J, Kerr P, Duman JG, Barnes BM. Simultaneous freeze tolerance and avoidance in individual fungus gnats, Exechia nugatoria. J Comp Physiol B. (2009) 179:897–902. doi: 10.1007/s00360-009-0369-x
28. Elnitsky MA, Hayward SAL, Rinehart JP, Denlinger DL, Lee RE Jr. Cryoprotective dehydration and the resistance to inoculative freezing in the Antarctic midge, Belgica antarctica. J Exp Biol. (2008) 211:524–30. doi: 10.1242/jeb.011874
29. Ramløv H, Bedford J, Leader J. Freezing tolerance of the New Zealand alpine weta, Hemideina maori Hutton [Orthoptera; Stenopelmatidae]. J Therm Biol. (1992) 17:51–4. doi: 10.1016/0306-4565(92)90019-C
30. Estrada-Peña A, Horak IG, Petney T. Climate changes and suitability for the ticks Amblyomma hebraeum and Amblyomma variegatum (Ixodidae) in Zimbabwe (1974-1999). Vet Parasitol. (2008) 151:256–67. doi: 10.1016/j.vetpar.2007.11.014
31. Oorebeek M, Kleindorfer S. Climate or host availability: what determines the seasonal abundance of ticks? Parasitol. Res. (2008) 103:871–5. doi: 10.1007/s00436-008-1071-8
32. Eisen RJ, Eisen L, Ogden NH, Beard CB. Linkages of weather and climate with Ixodes scapularis and Ixodes pacificus (Acari: Ixodidae), enzootic transmission of Borrelia burgdorferi, and Lyme disease in North America. J Med Entomol. (2016) 53:250–61. doi: 10.1093/jme/tjv199
33. Neelakanta G, Sultana H, Fish D, Anderson JF, Fikrig E. Anaplasma phagocytophilum induces Ixodes scapularis ticks to express an antifreeze glycoprotein gene that enhances their survival in the cold. J Clin Invest. (2010) 120:3179–90. doi: 10.1172/JCI42868
34. Wang J, Zhang R-R, Gao G-Q, Ma M-Y, Chen H. Cold tolerance and silencing of three cold-tolerance genes of overwintering Chinese white pine larvae. Sci Rep. (2016) 6:34698. doi: 10.1038/srep34698
35. Johannes F, Porcher E, Teixeira FK, Saliba-Colombani V, Simon M, Agier N, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. (2009) 5:e1000530. doi: 10.1371/journal.pgen.1000530
36. Reinders J, Wulff BB, Mirouze M, Marí-Ordóñez A, Dapp M, Rozhon W, et al. Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev. (2009) 23:939–50. doi: 10.1101/gad.524609
37. Yarychkivska O, Shahabuddin Z, Comfort N, Boulard M, Bestor TH. BAH domains and a histone-like motif in DNA methyltransferase 1 (DNMT1) regulate de novo and maintenance methylation in vivo. J Biol Chem. (2018) 293:19466–75. doi: 10.1074/jbc.RA118.004612
38. Kotsarenko K, Vechtova P, Hammerova Z, Langova N, Malinovska L, Wimmerova M, et al. Newly identified DNA methyltransferases of Ixodes ricinus ticks. Ticks Tick Borne Dis. (2020) 11:101348. doi: 10.1016/j.ttbdis.2019.101348
39. Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. (2011) 14:267–74. doi: 10.1016/j.pbi.2011.03.004
40. Dai T-M, Lü Z-C, Liu W-X, Wan F-H, Hong X-Y. The homology gene BtDnmt1 is essential for temperature tolerance in invasive Bemisia tabaci mediterranean cryptic species. Sci Rep. (2017) 7:3040. doi: 10.1038/s41598-017-03373-w
41. Steward N, Ito M, Yamaguchi Y, Koizumi N, Sano H. Periodic DNA methylation in maize nucleosomes and demethylation by environmental stress. J Biol Chem. (2002) 277:37741–6. doi: 10.1074/jbc.M204050200
42. Campos C, Valente LM, Conceição LE, Engrola S, Fernandes JM. Temperature affects methylation of the myogenin putative promoter, its expression and muscle cellularity in Senegalese sole larvae. Epigenetics. (2013) 8:389–97. doi: 10.4161/epi.24178
43. Skjærven KH, Hamre K, Penglase S, Finn RN, Olsvik PA. Thermal stress alters expression of genes involved in one carbon and DNA methylation pathways in Atlantic cod embryos. Comp Biochem Physiol A Mol Integr Physiol. (2014) 173:17–27. doi: 10.1016/j.cbpa.2014.03.003
44. Liu J, Liu Z, Zhang Y, Yang X, Gao Z. Biology of Dermacentor silvarum (Acari: Ixodidae) under laboratory conditions. Exp Appl Acarol. (2005) 36:131–8. doi: 10.1007/s10493-005-1271-1
45. Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, Deweese-Scott C, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res. (2011) 39:D225–9. doi: 10.1093/nar/gkq1189
46. Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. (1982) 157:105–32. doi: 10.1016/0022-2836(82)90515-0
47. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. (2007) 24:1596–9. doi: 10.1093/molbev/msm092
48. Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. (1987) 4:406–25.
49. Zuckerkandl E, Pauling L. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving Genes and Proteins. New York, NY: Academic Press (1965). p. 97–166.
50. Hernandez EP, Kusakisako K, Talactac MR, Galay RL, Hatta T, Fujisaki K, et al. Glutathione S-transferases play a role in the detoxification of flumethrin and chlorpyrifos in Haemaphysalis longicornis. Parasit Vectors. (2018) 11:460. doi: 10.1186/s13071-018-3044-9
51. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. (2001) 25:402–8. doi: 10.1006/meth.2001.1262
52. Wang T, Yang X, Jia Q, Dong N, Wang H, Hu Y, et al. Cold tolerance and biochemical response of unfed Dermacentor silvarum ticks to low temperature. Ticks Tick Borne Dis. (2017) 8:757–63. doi: 10.1016/j.ttbdis.2017.05.010
53. Gasteiger E, Hoogland C, Gattiker A, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy server. In: Walker JM, editor. The Proteomics Protocols Handbook. Totowa, NJ: Humana Press Inc., Springer (2005). p. 571–607.
54. Marinus MG, Løbner-Olesen A. DNA methylation. EcoSal Plus. (2009) 3:1–37. doi: 10.1128/ecosal.4.4.5
55. Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. (2013) 38:23–38. doi: 10.1038/npp.2012.112
56. Negishi M, Chiba T, Saraya A, Miyagi S, Iwama A. Dmap1 plays an essential role in the maintenance of genome integrity through the DNA repair process. Genes Cells. (2009) 14:1347–57. doi: 10.1111/j.1365-2443.2009.01352.x
57. Aasland R, Stewart AF, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. (1996) 21:87–8. doi: 10.1016/0968-0004(96)30009-1
58. Humphrey GW, Wang Y, Russanova VR, Hirai T, Qin J, Nakatani Y, et al. Stable histone deacetylase complexes distinguished by the presence of SANT domain proteins CoREST/kiaa0071 and Mta-L1. J Biol Chem. (2001) 276:6817–24. doi: 10.1074/jbc.M007372200
59. Boyer LA, Langer MR, Crowley KA, Tan S, Denu JM, Peterson CL. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. (2002) 10:935–42. doi: 10.1016/S1097-2765(02)00634-2
60. Callebaut I, Courvalin JC, Mornon JP. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. (1999) 446:189–93. doi: 10.1016/S0014-5793(99)00132-5
61. Yang N, Xu RM. Structure and function of the BAH domain in chromatin biology. Crit Rev Biochem Mol Biol. (2013) 48:211–21. doi: 10.3109/10409238.2012.742035
62. Liu Y, Oakeley EJ, Sun L, Jost JP. Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res. (1998) 26:1038–45. doi: 10.1093/nar/26.4.1038
63. Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. (2000) 25:269–77. doi: 10.1038/77023
64. Long HK, Blackledge NP, Klose RJ. ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans. (2013) 41:727–40. doi: 10.1042/BST20130028
65. Meehan RR, Lewis JD, Mckay S, Kleiner EL, Bird AP. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. (1989) 58:499–507. doi: 10.1016/0092-8674(89)90430-3
66. Lewis JD, Meehan RR, Henzel WJ, Maurer-Fogy I, Jeppesen P, Klein F, et al. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. (1992) 69:905–14. doi: 10.1016/0092-8674(92)90610-O
67. Voo KS, Carlone DL, Jacobsen BM, Flodin A, Skalnik DG. Cloning of a mammalian transcriptional activator that binds unmethylated CpG motifs and shares a CXXC domain with DNA methyltransferase, human trithorax, and methyl-CpG binding domain protein 1. Mol Cell Biol. (2000) 20:2108–21. doi: 10.1128/MCB.20.6.2108-2121.2000
68. Colinet H, Sinclair BJ, Vernon P, Renault D. Insects in fluctuating thermal environments. Annu Rev Entomol. (2015) 60:123–40. doi: 10.1146/annurev-ento-010814-021017
69. Rodrigues YK, Beldade P. Thermal plasticity in insects' response to climate change and to multifactorial environments. Front Ecol Evol. (2020) 8:271. doi: 10.3389/fevo.2020.00271
70. Williamson SM. Epigenetic underpinnings of freeze tolerance in the goldenrod gall fly Eurosta solidaginis and the goldenrod gall moth Epiblema scudderiana [Dissertation/master's thesis]. Carleton University, Ottawa, ON, Canada (2017).
71. Campos C, Valente LM, Fernandes JM. Molecular evolution of zebrafish dnmt3 genes and thermal plasticity of their expression during embryonic development. Gene. (2012) 500:93–100. doi: 10.1016/j.gene.2012.03.041
72. Yan XP, Liu HH, Liu JY, Zhang RP, Wang GS, Li QQ, et al. Evidence in duck for supporting alteration of incubation temperature may have influence on methylation of genomic DNA. Poult Sci. (2015) 94:2537–45. doi: 10.3382/ps/pev201
73. Wang Y, Wang F, Hong D-K, Gao S-J, Wang R, Wang J-D. Molecular characterization of DNA methyltransferase 1 and its role in temperature change of armyworm Mythimna separata Walker. Insect Biochem Physiol. (2020) 103:e21651. doi: 10.1002/arch.21651
74. Dai TM, Lü ZC, Wang YS, Liu WX, Hong XY, Wan FH. Molecular characterizations of DNA methyltransferase 3 and its roles in temperature tolerance in the whitefly, Bemisia tabaci Mediterranean. Insect Mol Biol. (2018) 27:123–32. doi: 10.1111/imb.12354
75. Cabezas-Cruz A, Estrada-Peña A, Rego RO, De La Fuente J. Tick-pathogen ensembles: do molecular interactions lead ecological innovation? Front Cell Infect Microbiol. (2017) 7:74. doi: 10.3389/fcimb.2017.00074
76. Moran EV, Alexander JM. Evolutionary responses to global change: lessons from invasive species. Ecol Lett. (2014) 17:637–49. doi: 10.1111/ele.12262
77. Woods HA, Hill RI. Temperature-dependent oxygen limitation in insect eggs. J Exp Biol. (2004) 207:2267–76. doi: 10.1242/jeb.00991
78. Kobori Y, Hanboonsong Y. Effect of temperature on the development and reproduction of the sugarcane white leaf insect vector, Matsumuratettix hiroglyphicus (Matsumura) (Hemiptera: Cicadellidae). J Asia-Pacif Entomol. (2017) 20:281–4. doi: 10.1016/j.aspen.2017.01.011
Keywords: DNA methyltransferase, epigenetics, Dermacentor silvarum, Haemaphysalis longicornis, cold tolerance
Citation: Agwunobi DO, Zhang M, Shi X, Zhang S, Zhang M, Wang T, Masoudi A, Yu Z and Liu J (2021) DNA Methyltransferases Contribute to Cold Tolerance in Ticks Dermacentor silvarum and Haemaphysalis longicornis (Acari: Ixodidae). Front. Vet. Sci. 8:726731. doi: 10.3389/fvets.2021.726731
Received: 17 June 2021; Accepted: 05 August 2021;
Published: 26 August 2021.
Edited by:
Nicola Pugliese, University of Bari Aldo Moro, ItalyReviewed by:
G. Y. Liu, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, ChinaQuan Liu, Foshan University, China
Copyright © 2021 Agwunobi, Zhang, Shi, Zhang, Zhang, Wang, Masoudi, Yu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhijun Yu, eXV6aGlqdW4mI3gwMDA0MDtoZWJ0dS5lZHUuY24=; Jingze Liu, bGl1amluZ3plJiN4MDAwNDA7aGVidHUuZWR1LmNu
†These authors have contributed equally to this work
 Desmond Onyeka Agwunobi
Desmond Onyeka Agwunobi Miao Zhang
Miao Zhang Xinyue Shi
Xinyue Shi Shiqi Zhang
Shiqi Zhang Meng Zhang
Meng Zhang Tongxuan Wang
Tongxuan Wang Abolfazl Masoudi
Abolfazl Masoudi Zhijun Yu
Zhijun Yu Jingze Liu
Jingze Liu