- Department of Population Health and Pathobiology, College of Veterinary Medicine, North Carolina State University, Raleigh, NC, United States
Virulent strains of Salmonella enterica subsp. enterica serovar Enteritidis (SE) harbored by poultry can cause disease in poultry flocks and potentially result in human foodborne illness. Two broiler flocks grown a year apart on the same premises experienced mortality throughout the growing period due to septicemic disease caused by SE. Gross lesions predominantly consisted of polyserositis followed by yolk sacculitis, arthritis, osteomyelitis, and spondylitis. Tissues with lesions were cultured yielding 59 SE isolates. These were genotyped by Rep-PCR followed by whole-genome sequencing (WGS) of 15 isolates which were clonal. The strain, SE_TAU19, was further characterized for antimicrobial susceptibility and virulence in a broiler embryo lethality assay. SE_TAU19 was resistant to nalidixic acid and sulfadimethoxine and was virulent to embryos with 100% mortality of all challenged broiler embryos within 3.5 days. Screening the SE_TAU19 whole-genome sequence revealed seven antimicrobial resistance (AMR) genes, 120 virulence genes, and two IncF plasmid replicons corresponding to a single, serovar-specific pSEV virulence plasmid. The pef, spv, and rck virulence genes localized to the plasmid sequence assembly. We report phenotypic and genomic features of a virulent SE strain from persistently infected broiler flocks and present a workflow for SE characterization from isolate collection to genome assembly and sequence analysis. Further SE surveillance and investigation of SE virulence in broiler chickens is warranted.
Introduction
Salmonella enterica subsp. enterica is comprised of over 2,500 serovars, including the pathogenic Salmonella enterica subsp. enterica serovar Enteritidis (SE) (1). As it relates to poultry production, SE is best known for its ability to survive on and within eggs despite causing no discernable disease in infected layers (2, 3). Because of these adaptations, SE is likely the most frequent cause of human salmonellosis globally (4), and investigations of SE today are largely focused on reducing foodborne illness. As such, the virulence features of present-day SE strains that initially colonize broilers have received relatively little attention. A fundamental understanding of SE genotypes and virulence mechanisms is essential to developing surveillance, treatment, and control approaches for broiler flocks.
Salmonella enterica subspecies enterica associated with poultry are assigned to two categories based on motility and host adaptability. First, Salmonella enterica subspecies enterica serovar Gallinarum contains two closely-related biovars, Gallinarum and Pullorum, which cause fowl typhoid and pullorum disease, respectively (1). These biovars are adapted to avian hosts, are non-motile due to mutations in flagellar genes, and have been eradicated in many developed countries but are still a significant cause of disease in countries with developing poultry production systems (5, 6). The second category holds the motile, non-host adapted serovars, including SE, which cause paratyphoid disease in poultry and encompass all foodborne Salmonella serovars. While this is the most common group associated with poultry in the present day, these serovars rarely cause symptomatic disease in older birds (1). Another subspecies, Salmonella enterica subspecies arizonae, causes an economically important disease in turkey production (7) but rarely affects broiler-type chickens (1).
In young broilers, SE is the common etiological agent of paratyphoid disease resulting in production losses (8). Salmonella enterica subsp. enterica serovar Enteritidis infection of young chicks is more likely to result in observable disease during natural infection and in challenge models whereas older birds are typically asymptomatic (8–10). Salmonella enterica subsp. enterica serovar Enteritidis can persistently colonize asymptomatic broiler and layer chickens from hatch to maturity (11, 12). Thus, a significant challenge encountered when attempting to control SE in broiler operations is egg-associated vertical transmission from breeders to broilers (13, 14). It is likely that SE can initially colonize chicks and poults during hatching and persist until opportunistically causing clinical disease. For example, an SE outbreak of broiler breeder origin caused fibrinous arthritic lesions of broilers aged 12 days (15). Similar lesions attributed to SE have also been observed in mature turkeys (16). Salmonella enterica subsp. enterica serovar Enteritidis is a significant threat to poultry production due to its potential to be vertically transmitted, persist, and cause opportunistic infections (15).
Salmonella enterica subsp. enterica serovar Enteritidis genetic characterization and surveillance can be achieved with a myriad of diagnostic and molecular methods, including plasmid analysis, pulsed-field gel electrophoresis, and detection of AMR and virulence genes (17, 18). With recent advances in whole-genome sequencing (WGS) efficiency and accessibility with concurrently decreasing costs, sequencing has replaced such methods as the standard for Salmonella surveillance, especially SE, in many countries (19–21). Additionally, WGS analysis of SE genomes can identify virulence and resistance features of field isolates, consequently adding more depth to SE genotypes. These include using mass screening software packages to identify known virulence factors and antimicrobial resistance (AMR) genes (22).
Additional identification and characterization of SE virulence features in avian hosts is warranted, especially in the context of observed clinical disease. This report describes a SE strain that not only caused disease throughout a typical 57 days growing period but was detected in two flocks grown a year apart in the same facility. Due to disease severity in broilers and subsequent isolation of SE from lesions, the aim of this study was to identify virulence and resistance features of the SE strain. After establishing lethality to broiler embryos, this strain's whole-genome sequence was screened for virulence, plasmid, and resistance genes. As virulent SE strains continue to circulate in animal and human populations, surveillance and genomic characterization are required for further virulence investigation and development of treatment and control strategies.
Materials and Methods
Flock Rearing
Broiler flocks A and B were reared 1 year apart (February to April; 2019 and 2020) for teaching and research at the North Carolina State University College of Veterinary Medicine. Approximately 5,000 mixed-sex Ross 708 broiler chicks were placed on the day of hatch on pine shavings that top-dressed a dirt surface and reared to 57 days. The 508 m2 fan ventilated, curtain-sided rearing facility included radiant brooders, forced air heaters, and paired feeder and water lines. Integrator-supplied feed containing a coccidiostat was provided with municipal water ad libitum.
Sample Collection and Preliminary Characterization
Dead broilers consisting of flock mortality and culled birds were collected twice daily for necropsy. The total number of birds submitted for postmortem examination was 357 for flock A and 274 for flock B. Septicemic bacterial infections were diagnosed based on clinical signs, postmortem findings (23), and diagnostic culture results of representative birds. Cultures were collected from samples of gross lesions, including yolk sac contents, pericardial fluid, whole spleen, liver surface, joint exudate, and caseous material from inflammatory lesions in the free thoracic vertebra. Additionally, a random sample of 24 apparently healthy broilers was selected from broiler flock A at 6 days of age for yolk sac culture. Samples were collected aseptically with Stuart swabs (BD 220144), which were transported directly to an on-site laboratory and cultured on trypticase soy agar with 5% sheep blood (BD 221261), MacConkey agar (BD 212123), Columbia CNA agar with colistin and nalidixic acid (BD 221353), and XLT-4 agar (BD 223420) followed by incubation at 37°C for 18–24 h. Colonies that did not ferment lactose on MacConkey agar and appeared with a dark black center on XLT-4 were presumptively identified as Salmonella spp. and further purified on non-selective media. Salmonella spp. was confirmed by either the Biolog GenIII Microplate™ microbial identification system as described by Borst et al. (24) or PCR for the Salmonella-specific invA gene sequence (25). All isolates were then genotyped using repetitive element-based PCR methods using the (GTG)5 primer as described by Walker et al. (26). All isolates had identical banding patterns, suggesting clonality. This was confirmed for 15 selected isolates from broiler flock A by single nucleotide polymorphism (SNP) phylogenetic analysis using WGS of sequenced isolates (below). A representative clone, SE_TAU19, that was isolated from the yolk sac of a chick from broiler flock A, was used for all downstream experiments. The minimum inhibitory concentration of 27 antimicrobial drugs was determined by microbroth dilution methods according to guidelines defined by the National Antimicrobial Susceptibility Monitoring System (NARMS) (26). These drugs included 14 antimicrobials tested by NARMS for Gram-negative pathogens (ThermoFisher CMV3AGNF) and 13 drugs specific for treatment of poultry (ThermoFisher AVIAN1F). The breakpoints for drugs in the poultry-specific formulary (clindamycin, erythromycin, novobiocin, penicillin, and tylosin tartrate) to which Salmonella spp. are known to be intrinsically resistant were not interpreted (27).The strain was designated as resistant or susceptible based on available breakpoint data from the Clinical and Laboratory Standards Institute (28). The only existing breakpoint specific for poultry is enrofloxacin (29). Otherwise, human breakpoints were used for interpretation of the data.
Embryo Lethality Assay
Fertile broiler eggs were obtained from a commercial hatchery and incubated under standard conditions (37.5°C, >50% relative humidity) to 12 days. After confirmation of viability, eight embryos were challenged via the allantoic cavity with approximately 200 CFU of SE_TAU19 as previously described (30). A positive control group of 21 embryos was challenged with the same dose of an avian pathogenic Escherichia coli (APEC) isolate, EC_06YS which is known to be virulent to broiler embryos from an unrelated study (31). Survivability was determined by candling eggs every 12 h. Dead embryos were individually cultured to verify presence of the challenge strain. A negative control group of six eggs injected with sterile PBS was also included in the experiment. All negative control eggs survived and were cultured upon termination of the experiment to verify sterility.
Whole Genome Sequencing, Assembly, and Typing
Genomic DNA of 15 SE isolates collected from broiler flock A was isolated with the Qiagen DNeasy Blood and Tissue kit (Qiagen 69504). After assessing the quality of isolated DNA with a Nanodrop 2000 spectrophotometer, double stranded DNA was quantified using a Qubit 4.0 Fluorometer (ThermoFisher Scientific, Waltham, MA) with a dsDNA high-sensitivity assay kit (ThermoFisher Q32851). DNA were sequenced on an Illumina MiSeq according to the manufacturer's instructions using manufacturer supplied reagents. The Nextera DNA Flex Library Prep kit (Illumina 20015828) was used to prepare pooled libraries with an insert size of ~350 bp prior to sequencing with a MiSeq Reagent Kit v2 (500 cycles, Illumina MS-102-2003). Genome assembly and quality check were completed with CLC Genomics Workbench (Cambridge, MA) using the de novo assembly procedure. A minimum contig length of 200 bp was specified in the parameters and scaffolding of paired-end reads was performed. The assembled genome sequence of SE_TAU19 was uploaded to SeqSero version 1.0, a web-based Salmonella serotyping tool (32). The assembled genome of SE_TAU19 was blasted in CLC Genomics Workbench for the SE-specific sdf (Salmonella difference fragment) sequence for serotype confirmation (33). Multilocus sequence typing (MLST) was conducted by uploading the assembled genome to an online database (https://cge.cbs.dtu.dk/services/MLST/) (34). Clonality was confirmed through an online SNP-calling pipeline for WGS, CSIPhylogeny (https://cge.cbs.dtu.dk/services/CSIPhylogeny/) (35), with a minimum Z-score of 1.96 and default parameters described by Wuyts et al. (36) (data not shown). The SE_TAU19 sequence was originally deposited in GenBank under the alias LB_5320 (accession number AADSUW000000000.1) as part of the FDA GenomeTrakr project (Bioproject PRJNA293224).
Identification of Virulence, Plasmid, and Resistance Genes
For additional analysis, sequencing reads were assembled and analyzed in a separate pipeline than described above. A draft genome of SE_TAU19 was generated by inputting its SRR run number (SRR8943529) in the SPAdes version 3.0-based software Shovill 1.0.4 assembler (37, 38). Default parameters were used with minimum contig length set at 200 bp and minimum contig coverage set at 5.00. The resulting contigs were screened for plasmids, virulence, and AMR genes using the following databases in ABRicate (39): Comprehensive Antimicrobial Resistance Database version 3.0.2 (CARD) (40), Virulence Factor Database (VFDB) (41), and PlasmidFinder version 2.1 (42). The assembled genome was uploaded to the ResFinder version 4.1 online database to detect chromosomal point mutations (https://cge.cbs.dtu.dk/services/ResFinder/) (43). Only sequence hits with 100% coverage and ≥98% gene identity were included. Remaining sequence hits are provided as Supplementary Material (Supplementary File 1). The plasmid sequence was assembled using plasmidSPAdes (44) with the same approach and parameters used for the genome assembly and screened independently using the above databases to identify plasmid-specific virulence genes. The presence of the detected plasmid in SE_TAU19 was confirmed by PCR amplification of a 312 bp fragment of the spvA plasmid virulence gene (45, 46) using primers: spvA-F (CAG CCT GAT GGT GGT TAA TGA) and spvA-R (CAG TGC TTC AAA TGG CGT ATA G). Pathogenicity in human hosts was predicted using the online database, PathogenFinder version 1.1 (https://cge.cbs.dtu.dk/services/PathogenFinder/) (47). The stepwise procedure used to characterize SE_TAU19 in this study is summarized in Figure 3.
Statistical Analysis
Kaplan-Meier survival curves were used to determine embryo survivability, which was analyzed by the log-rank (Mantel-Cox) test of significance (GraphPad Prism version 9, GraphPad Software, San Diego, California USA).
Results
Clinical Observations and Antimicrobial Susceptibility
Lesions from affected birds included evidence of septicemia (yolk sacculitis, splenomegaly, fibrinous pericarditis, and multifocal necrotizing and heterophilic hepatitis), and musculoskeletal infection (Figure 1). Based on culture results from daily necropsy, it was estimated that 37/357 (10%) and 42/274 (15%) of total mortality for flocks A and B was attributable to SE infection and a total of 59 isolates were collected from the flocks. Yolk sac samples from apparently healthy birds at 6 days in broiler flock A revealed that 5/20 harbored SE. Additional causes of mortality were attributed to heat stress during late growing. Turkey flocks reared 5 months prior to broiler flocks in the same facilities under similar conditions did not experience elevated mortality and SE was not isolated during routine organ cultures of deceased birds (data not shown). SE_TAU19 was resistant to 2 of 27 drugs tested: nalidixic acid, a quinolone, and sulfadimethoxine, a sulfonamide. The strain was classified as susceptible to the fluoroquinolone ciprofloxacin (MIC = 0.25 ng/μl).
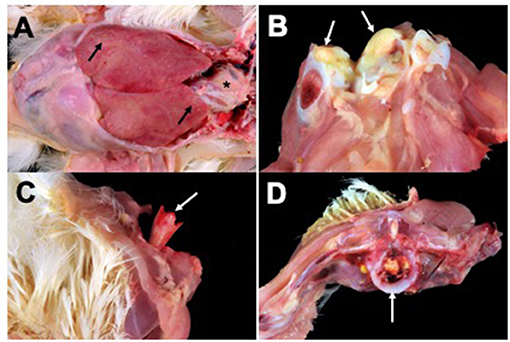
Figure 1. Broiler gross lesions from day 33 mortality attributed to septicemic disease caused by Salmonella Enteritidis (SE) strain SE_TAU19. (A) Fibrinous pericarditis (asterisk) and multifocal necrotizing and heterophilic hepatitis (black arrows). (B) Hock joint, fibrinous arthritis (white arrows). (C) Bacterial chondronecrosis with osteomyelitis (femoral head necrosis) (white arrow). (D) Vertebral osteomyelitis (white arrow).
Embryo Lethality Assay
When broiler embryos were challenged with SE_TAU19 at 12 days, over half died within 2 days, and all embryos were dead by 3.5 days post-injection (Figure 2). Embryo lethality due to infection with SE_TAU19 was similar to the virulent EC_06YS APEC positive control strain (P ≤ 0.10), which was classified as virulent based on a scheme developed by Wooley et al. (48).
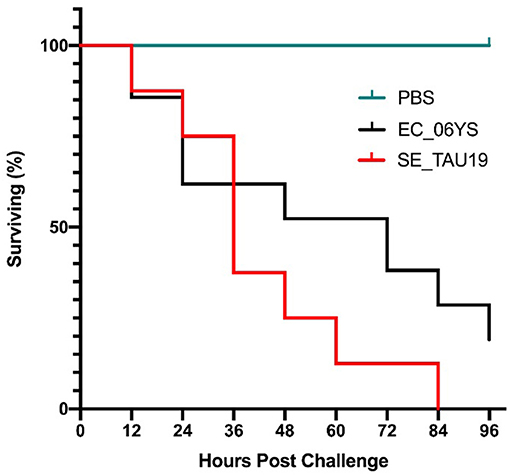
Figure 2. Survival of broiler embryos after challenge with Salmonella Enteritidis strain SE_TAU19. An avian pathogenic E. coli strain known to be virulent to broiler embryos, EC_06YS, was used to challenge 21 eggs and served as a positive control. Eight broiler embryos were challenged with SE_TAU19. Eggs were inoculated with approximately 200 CFU/egg via the intra-allantoic route at 12 days of incubation and candled every 12 h to monitor survivability. All embryos challenged with SE_TAU19 were dead by 84 h, while the six PBS-injected control eggs survived and EC_06YS achieved expected lethality.
Genome Assembly and Genotypic Characterization
Sequencing of the SE_TAU19 genome included 1,106,258 reads with 56X coverage. The de novo assembly procedure in CLC Genomics Workbench produced 57 contigs (excluding scaffolded regions) with an average length of 82,512 bp and an N50 of 304,422 bp. SE_TAU19 belongs to ST-11 based on MLST typing. SeqSero and in silico sdf gene detection confirmed the Enteriditis serotype. Screening of the SE_TAU19 genome assembly generated by Shovill using ABRicate resulted in a total of 189 gene hits: 40 genes, 2 plasmid replicons, and 147 virulence genes (Supplementary File 1). Using a threshold of 100% coverage and 98% identity, 7 AMR genes, 2 plasmid replicons, and 103 virulence genes met criteria for inclusion (Table 1). Antimicrobial resistance genes consisted of the aac(6′)-ly chromosomally encoded aminoglycoside resistance gene, the mds multidrug and metal efflux pump with its regulator, and an additional multidrug and toxic compound transporter, mdtK. Point mutation analysis detected a guanine to thymine substitution in the gyrA gene corresponding to an aspartic acid to tyrosine amino acid change for nalidixic acid and ciprofloxacin resistance. The detected plasmid replicons indicated the presence of a SE serovar-specific pSEV plasmid, a ~60 kb, non-conjugative virulence plasmid that contains both the IncFII(S)_1 and IncFIB(S)_1 incompatibility regions (49), which was confirmed by PCR (data not shown). Nine virulence genes were plasmid-borne based on screening of the plasmid assembly. These included pefA-D, spvB-C, spvR, and rck. No AMR genes were detected on the pSEV plasmid. SE_TAU19 matched 1,127 pathogenic families compared to only two matched non-pathogenic families and was therefore predicted by PathogenFinder to have a 94.2% probability of being a human pathogen.
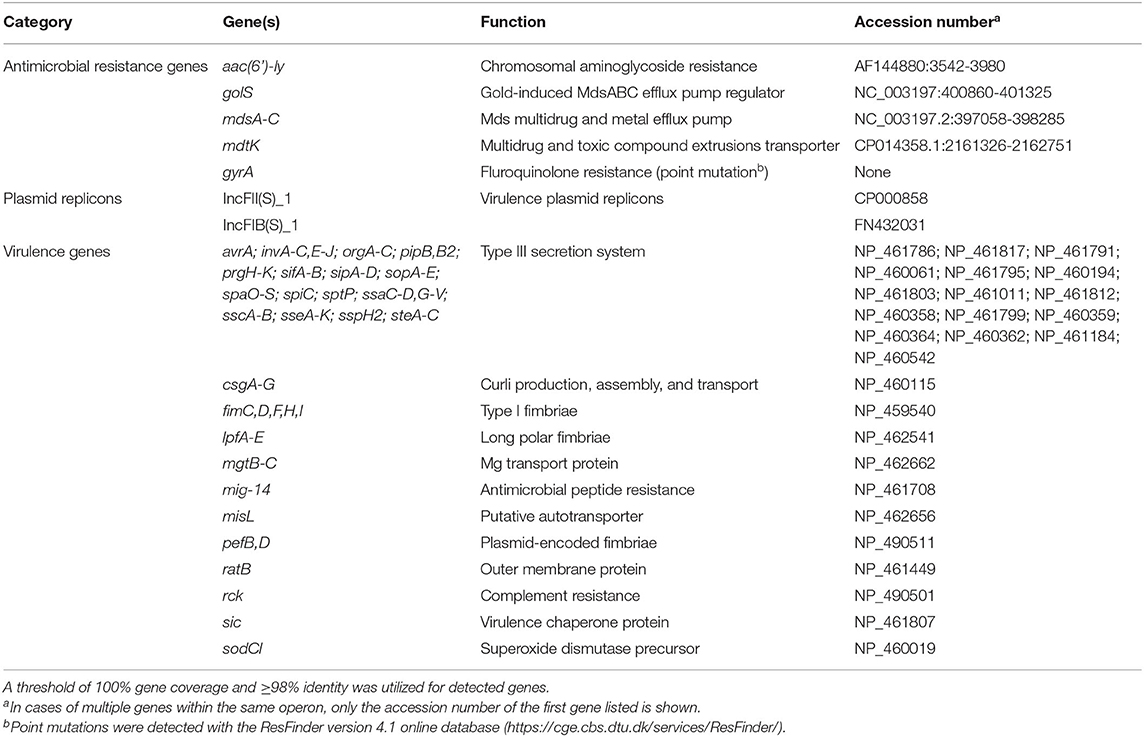
Table 1. Summary of antimicrobial resistance genes, plasmid replicons, and virulence genes detected in the draft genome sequence of Salmonella Enteritidis strain SE_TAU19 using ABRicate.
Discussion
A SE strain, SE_TAU19, caused persistent and reoccurring septicemic disease in two independent flocks of broiler chickens and was characterized using genomic analysis. This study revealed that the quinolone and sulfonamide resistant SE_TAU19 strain was lethal to broiler embryos and harbored a pSEV plasmid and typical SE virulence genes. SE_TAU19 was presumed to be vertically transmitted in the present study as is common for this pathogen (15). However, vertical transmission was not verified and existence of an on-farm reservoir host (e.g., rodents) was not eliminated. Genomic analysis of the pathogen was warranted due to septicemic disease severity, reoccurrence of disease in a subsequent broiler flock, and potential public health implications.
The SE_TAU19 serotype was determined using the assembled genome and the SeqSero online serotyping by WGS tool (32). However, this tool cannot accurately differentiate S. Gallinarum from SE due to a non-expressed fliC allele in S. Gallinarum (32). Detection of the sdf fragment, which is specific to circulating strains of SE but not S. Gallinarum (33), in combination with MLST, is needed to distinguish between S. Gallinarum and SE when relying solely on in silico serotyping methods (Figure 3). Confirmation of SE by detection of the sdf fragment is especially important when serotyping Salmonella isolated from poultry by WGS as S. Gallinarum, the agent of fowl typhoid, is a reportable disease in the United States (50).
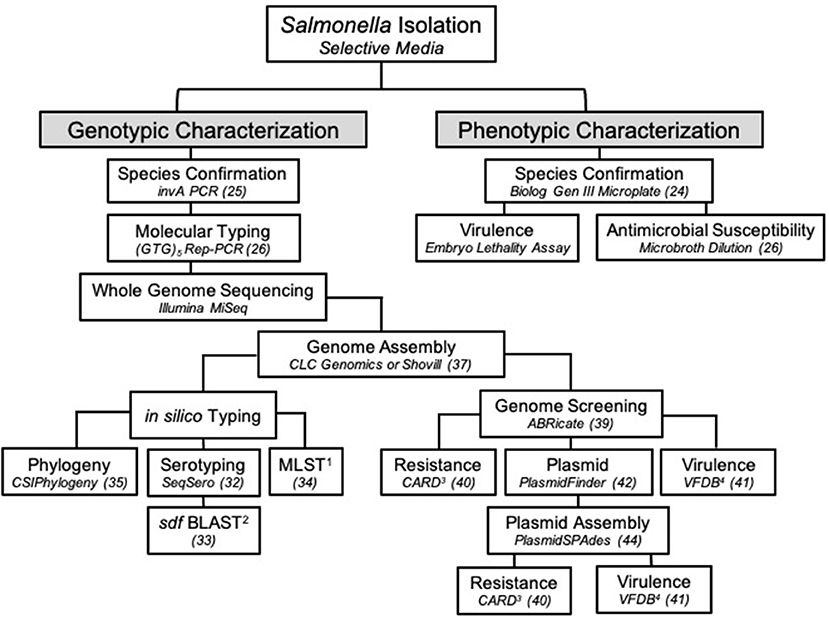
Figure 3. The workflow used to characterize Salmonella Enteritidis strain SE_TAU19 isolated from broiler chickens in this study. Specific methods or software and/or the corresponding reference are indicated in italics for each step. 1MLST, multilocus sequence typing; 2BLAST, basic local alignment search tool; 3CARD, comprehensive antibiotic resistance database; 4VFDB, virulence factor database.
SE_TAU19 was resistant to nalidixic acid and sulfadimethoxine. Nalidixic acid resistance was corroborated by detection of the gyrA mutation in our genomic analysis (Table 1). Ciprofloxacin resistance was also predicted by this genotype, but the strain was designated as susceptible based on a MIC of 0.25 ng/μl. Quinolones such as nalidixic acid and ciprofloxacin are widely used first-line therapeutics for humans with salmonellosis when antimicrobial therapy is warranted (51), and increasing incidences of therapeutic failure in treating salmonellosis with ciprofloxacin have resulted in requests to re-evaluate human breakpoints for this drug (52, 53). Nevertheless, nalidixic acid resistant SE strains like SE_TAU19 have emerged in the southeastern United States (54) and elsewhere (55) contributing to the public health threat of drug-resistant, foodborne SE. Genetic determinants of sulfadimethoxine resistance in this strain were inconclusive, as AMR gene screening did not indicate a gene corresponding to this phenotype.
In vivo virulence of SE_TAU19 was investigated in an embryo lethality assay. SE_TAU19 achieved a 100% mortality rate compared to a virulent APEC strain which achieved an 80% mortality rate (Figure 2). SE_TAU19 virulence to broiler embryos was found to be comparable to published embryo lethality in Salmonella enterica serovar Gallinarum infections (56). This approach may be useful for future SE virulence or vaccine development studies. Additionally, the embryo lethality assay was a relatively simple means to investigate in vivo virulence potential and corroborate virulence gene discoveries. The contributions of specific SE_TAU19 genes to virulence in chickens would be best determined by deletion and complementation of the desired genes in SE_TAU19 followed by challenging SPF embryos and/or chicks.
This study highlights the utility of WGS in Salmonella identification and surveillance. The value of this approach has been realized through recent epidemiological advances, such as the ability to predict foodborne outbreaks of SE (20). The threat of zoonotic transmission of SE_TAU19 was supported when an in silico virulence prediction model PathogenFinder indicated that SE_TAU19 is likely a human pathogen due to sequence hits for several well-known SE virulence genes. Additional genomic analyses of pathogens can be used to improve the identification, prevention, and research of infectious agents that diminish food animal production and threaten public health. In this case, the characterization of SE_TAU19 could aid in the detection of clones in the broiler supply chain (26) or the development of SE autogenous vaccines (57). At the time of publication, there were over 29,000 SE genome assemblies available in the NCBI Sequencing Read Archive (SRA) database (58). However, most are yet to be characterized, and published pathogen genomes rarely include clinical details regarding the nature of disease caused in livestock. In Figure 3, we present a workflow for characterizing SE isolates that encompasses genotypic and phenotypic traits. This approach can be modified as appropriate for other Salmonella serovars and similar bacterial pathogens (e.g., E. coli).
In conclusion, the present study presents two separate instances of septicemic disease in broiler chickens which were attributed to SE strain SE_TAU19 based on genetic relatedness. Genomic analysis corroborated phenotypic resistance to antimicrobials and identified multiple virulence genes that may enhance its fitness as a poultry pathogen. However, further investigation is required to determine the biological relevance of these findings. Additional characterization of virulent SE strains is warranted, and this approach can be used for future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, AADSUW000000000.1.
Ethics Statement
The animal study was reviewed and approved by NC State University Institutional Animal Care and Use Committee.
Author Contributions
GW, MS, RC, and LB conceived and designed the experiments. DH and ST assisted with genomic data collection and analysis. GW, MS, SG, FJ, and LC were responsible for animal husbandry, post-mortem examinations, and sample collection. All authors reviewed and approved the final version of the manuscript.
Funding
This work is funded through the United States Department of Agriculture Animal Plant Health Inspection Service's National Bio and Agro-defense Facility Scientist Training Program. The whole genome sequencing was completed by the FDA GenomeTrakr program funded grant 1U18FD00678801.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge Lyndy Harden and Erin Harrell for whole genome sequencing of the Salmonella isolates. We are very grateful to Ayanna Glaize and C. Annie Wang for helpful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.725737/full#supplementary-material
References
1. Gast RK, Porter RE Jr. Salmonella infections. In: Swayne DE, Boulianne M, Logue CM, McDougald LR, Nair V, Suarez DL, editors. Diseases of Poultry. 14th ed. Hoboken, NJ: John Wiley and Sons, Inc. (2020). p. 719–53.
2. Baron F, Nau F, Guérin-Dubiard C, Bonnassie S, Gautier M, Andrews SC, et al. Egg white versus Salmonella Enteritidis! A harsh medium meets a resilient pathogen. Food Microbiol. (2016) 53:82–93. doi: 10.1016/j.fm.2015.09.009
3. Guard-Petter J. The chicken, the egg and Salmonella Enteritidis. Environ Microbiol. (2001) 3:421–30. doi: 10.1046/j.1462-2920.2001.00213.x
4. Bäumler AJ, Hargis BM, Tsolis RM. Tracing the origins of Salmonella outbreaks. Science (200). 287:50–2. doi: 10.1126/science.287.5450.50
5. Shivaprasad HL. Fowl typhoid and pullorum disease. Rev Sci Tech. (2000) 19:405–24. doi: 10.20506/rst.19.2.1222
6. Xiong D, Song L, Pan Z, Jiao X. Identification and discrimination of Salmonella enterica serovar Gallinarum biovars pullorum and gallinarum based on a one-step multiplex PCR assay. Front Microbiol. (2018) 9:1719. doi: 10.3389/fmicb.2018.01718
7. Crespo R, Jeffrey JS, Chin RP, Sentíes-Cué G, Shivaprasad HL. Phenotypic and genotypic characterization of Salmonella arizonae from an integrated turkey operation. Avian Dis. (2004) 48:344–50. doi: 10.1637/7116
8. O'Brien J. Salmonella Enteritidis infection in broiler chickens. Vet Rec. (1988) 122:214. doi: 10.1136/vr.122.9.214-b
9. Gast RK. Understanding Salmonella Enteritidis in laying chickens: the contributions of experimental infections. Int J Food Microbiol. (1994) 21:107–16. doi: 10.1016/0168-1605(94)90204-6
10. Barrow PA. Experimental infection of chickens with Salmonella Enteritidis. Avian Pathol. (1991) 20:145–53. doi: 10.1080/03079459108418749
11. Gast RK, Holt PS. Persistence of Salmonella Enteritidis from one day of age until maturity in experimentally infected layer chickens. Poult Sci. (1998) 77:1759–62. 10.1093/ps/77.12.1759
12. Gorham SL, Kadavil K, Lambert H, Vaughan E, Pert B, Abel J. Persistence of Salmonella Enteritidis in young chickens. Avian Pathol. (1991) 20:433–7. doi: 10.1080/03079459108418781
13. Lei CW, Zhang Y, Kang ZZ, Kong LH, Tang YZ, Zhang AY, et al. Vertical transmission of Salmonella Enteritidis with heterogeneous antimicrobial resistance from breeding chickens to commercial chickens in China. Vet Microbiol. (2020) 240:108538. doi: 10.1016/j.vetmic.2019.108538
14. Berchieri AJ, Wigley P, Page K, Murphy CK, Barrow PA. Further studies on vertical transmission and persistence of Salmonella enterica serovar Enteritidis phage type 4 in chickens. Avian Pathol. (2001) 30:297–310. doi: 10.1080/03079450120066304
15. Oh JY, Kang MS, An BK, Song EA, Kwon JH, Kwon YK. Occurrence of purulent arthritis broilers vertically infected with Salmonella enterica serovar Enteritidis in Korea. Poult Sci. (2010) 89:2116–22. doi: 10.3382/ps.2010-00918
16. Higgins WA, Christiansen JB, Schroeder CHA. Salmonella Enteritidis infection associated with leg deformity in turkeys. Poult Sci. (1944) 23:340–1. doi: 10.3382/ps.0230340
17. Zou M, Keelara S, Thakur S. Molecular characterization of Salmonella enterica serotype Enteritidis isolates from humans by antimicrobial resistance, virulence genes, and pulsed-field gel electrophoresis. Foodborne Pathog Dis. (2012) 9:232–8. doi: 10.1089/fpd.2011.1012
18. Aktas Z, Day M, Kayacan CB, Diren S, Threlfall EJ. Molecular characterization of Salmonella Typhimurium and Salmonella Enteritidis by plasmid analysis and pulsed-field gel electrophoresis. Int J Antimicrob Agents. (2007) 30:541–5. doi: 10.1016/j.ijantimicag.2007.06.034
19. Ashton PM, Nair S, Peters TM, Bale JA, Powell DG, Painset A, et al. Identification of Salmonella for public health surveillance using whole genome sequencing. PeerJ. (2016) 4:e1752. doi: 10.7717/peerj.1752
20. Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, Jombart T, et al. Prospective use of Whole Genome Sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect. (2017) 145:289–98. doi: 10.1017/S0950268816001941
21. den Bakker HC, Allard MW, Bopp D, Brown EW, Fontana J, Iqbal Z, et al. Rapid whole-genome sequencing for surveillance of Salmonella enterica serovar Enteritidis. Emerg Infect Dis. (2014) 20:1306–14. doi: 10.3201/eid2008.131399
22. Lima NCB, Tanmoy AM, Westeel E, De Almeida LGP, Rajoharison A, Islam M, et al. Analysis of isolates from Bangladesh highlights multiple ways to carry resistance genes in Salmonella Typhi. BMC Genomics. (2019) 20:530. doi: 10.1186/s12864-019-5916-6
23. Fisher ME, Trampel DW, Griffithc RW. Postmortem detection of acute septicemia in broilers. Avian Dis. (1998) 42:452–61. doi: 10.2307/1592671
24. Borst LB, Suyemoto MM, Robbins KM, Lyman RL, Martin MP, Barnes HJ. Molecular epidemiology of Enterococcus cecorum isolates recovered from Enterococcal Spondylitis outbreaks in tte southeastern united states. Avian Pathol. (2012) 41:479–85. doi: 10.1080/03079457.2012.718070
25. Rahn K, De Grandis SA, Clarke RC, McEwen SA, Galán JE, Ginocchio C, et al. Amplification of an invA Ggene sequence of Salmonella typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Mol Cell Probes. (1992) 6:271–9. doi: 10.1016/0890-8508(92)90002-f
26. Walker GK, Suyemoto MM, Borst LB, Brake J. Repetitive element–based polymerase chain reaction genotyping improves efficiency of Salmonella surveillance in a model broiler production system. Poult Sci. (2020) 99:2684–9. doi: 10.1016/j.psj.2019.12.048
27. St Amand JA, Otto SJG, Cassis R, Annett Christianson CB. Antimicrobial resistance of Salmonella enterica serovar Heidelberg isolated from poultry in Alberta. Avian Pathol. (2013) 42:379–86. doi: 10.1080/03079457.2013.811465
28. CLSI Supplement. M100: Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. (2020).
29. Li J, Hao H, Cheng G, Wang X, Ahmed S, Shabbir MAB, et al. The effects of different enrofloxacin dosages on clinical efficacy and resistance development in chickens experimentally infected with Salmonella Typhimurium. Sci Rep. (2017) 7:11676. doi: 10.1038/s41598-017-12294-7
30. Borst LB, Suyemoto MM, Keelara S, Dunningan SE, Guy JS, Barnes HJ, et al. Chicken embryo lethality assay for pathogenic Enterococcus cecorum. Avian Dis. (2014) 58:244–8. doi: 10.1637/10687-101113-Reg.1
31. Walker GK, Suyemoto MM, Gall S, Chen L, Thakur S, Borst LB. The role of Enterococcus faecalis during co-infection with avian pathogenic Escherichia coli in avian colibacillosis. Avian Pathol. (2020) 49:589–99. doi: 10.1080/03079457.2020.1796926
32. Zhang S, Yin Y, Jones MB, Zhang Z, Kaiser BLD, Dinsmore BA, et al. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol. (2015) 53:1685–92. doi: 10.1128/JCM.00323-15
33. Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, et al. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar Enteritidis. Appl Environ Microbiol. (2001) 67:4984–91. doi: 10.1128/AEM.67.11.4984-4991.2001
34. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. (2012) 50:1355–61. doi: 10.1128/JCM.06094-11
35. Kaas RS, Leekitcharoenphon P, Aarestrup FM, Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. (2014) 9:e104984. doi: 10.1371/journal.pone.0104984
36. Wuyts V, Denayer S, Roosens NHC, Mattheus W, Bertrand S, Marchal K, et al. Whole genome sequence analysis of Salmonella Enteritidis PT4 outbreaks from a national reference laboratory's viewpoint. PLoS Curr. (2015) 7:ecurrents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc. doi: 10.1371/currents.outbreaks.aa5372d90826e6cb0136ff66bb7a62fc
37. SeeMann T. Shovill. (2019). Available online at: https://github.com/tseemann/shovill (accessed July 25, 2021).
38. Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. (2012) 19:455–77. doi: 10.1089/cmb.2012.0021
39. SeeMann T. ABRicate: Mass Screening of Contigs for Antimicrobial Resistance or Virulence Genes. (2018). https://github.com/tseemann/abricate (accessed July 25, 2021).
40. McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, et al. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. (2013) 57:3348–57. doi: 10.1128/AAC.00419-13
41. Chen L, Zheng D, Liu B, Yang J, Jin QVFDB. 2016: Hierarchical and refined dataset for big data analysis - 10 years on. Nucleic Acids Res. (2016) 44:D694–7. doi: 10.1093/nar/gkv1239
42. Carattoli A, Zankari E, Garcìa-Fernandez A, Larsen M, Lund O, Villa L, et al. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–38903. doi: 10.1128/AAC.02412-14
43. Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. (2012) 67:2640–4. doi: 10.1093/jac/dks261
44. Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. PlasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics. (2016) 32:3380–7. doi: 10.1093/bioinformatics/btw493
45. Guiney DG, Fang FC, Krause M, Libby S. Plasmid-mediated virulence genes in non-typhoid Salmonella serovars. FEMS Microbiol Lett. (1994) 124:1–9. doi: 10.1111/j.1574-6968.1994.tb07253.x
46. Gebreyes WA, Thakur S, Dorr P, Tadesse DA, Post K, Wolf L. Occurrence of spvA virulence gene and clinical significance for multidrug-resistant Salmonella strains. J Clin Microbiol. (2009) 47:777–80. doi: 10.1128/JCM.01660-08
47. Cosentino S, Voldby Larsen M, Møller Aarestrup F, Lund O. PathogenFinder - distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE. (2013) 8:e77302. doi: 10.1371/journal.pone.0077302
48. Wooley ARE, Gibbs PS, Brown TP, Maurer JJ. Chicken embryo lethality assay for determining the virulence of avian Escherichia coli isolates. Avian Dis. (2000) 44:318–24. doi: 10.2307/1592546
49. Rodríguez I, Guerra B, Mendoza MC, Rodicio MR. puo-SeVR1 is an emergent virulence-resistance complex plasmid of Salmonella enterica serovar Enteritidis. J Antimicrob Chemother. (2011) 66:218–20. doi: 10.1093/jac/dkq386
50. United States Department of Agriculture. Reportable Diseases in the Untied States - 2018 Annual Report. (2021). Available online at: https://www.aphis.usda.gov/aphis/ourfocus/animalhealth/monitoring-and-surveillance/sa_nahss/status-reportable-disease-us (accessed July 25, 2021).
51. Mølbak K, Gerner-Smidt P, Wegener HC. Increasing quinolone resistance in Salmonella enterica serotype Enteritidis. Emerg Infect Dis. (2002) 8:514–5. doi: 10.3201/eid0805.010288
52. Kumar VA. Revised ciprofloxacin breakpoints for Salmonella: is it time to write an obituary? J Clin Diagnostic Res. (2013) 7:2467–9. doi: 10.7860/JCDR/2013/7312.3581
53. Ryan MP, Dillon C, Adley CC. Nalidixic acid-resistant strains of Salmonella showing decreased susceptibility to fluoroquinolones in the midwestern region of the Republic of Ireland due to mutations in the gyrA gene. J Clin Microbiol. (2011) 49:2077–9. doi: 10.1128/JCM.02574-10
54. Velasquez CG, MacKlin KS, Kumar S, Bailey M, Ebner PE, Oliver HF, et al. Prevalence and antimicrobial resistance patterns of Salmonella isolated from poultry farms in Southeastern United States. Poult Sci. (2018) 97:2144–52. doi: 10.3382/ps/pex449
55. Voss-Rech D, Potter L, Vaz CSL, Pereira DIB, Sangioni LA, Vargas ÁC, et al. Antimicrobial resistance in nontyphoidal Salmonella isolated from human and poultry-related samples in Brazil: 20-year meta-analysis. Foodborne Pathog Dis. (2017) 14:116–24. doi: 10.1089/fpd.2016.2228
56. Zhang JF, Wei B, Cha SY, Shang K, Jang HK, Kang M. The use of embryonic chicken eggs as an alternative model to evaluate the virulence of Salmonella enterica serovar Gallinarum. PLoS ONE. (2020) 15:e238630. doi: 10.1371/journal.pone.0238630
57. Abd El-Ghany WS.A., El-Shafii S, Hatem MEE., Dawood R. A trial to prevent Salmonella Enteritidis infection in broiler chickens using autogenous bacterin compared with probiotic preparation. J Agric Sci. (2012) 4:e25. doi: 10.5539/jas.v4n5p91
Keywords: Salmonella Enteritidis, poultry, genomics, antimicrobial resistance, broilers
Citation: Walker GK, Suyemoto MM, Hull DM, Gall S, Jimenez F, Chen LR, Thakur S, Crespo R and Borst LB (2021) Genomic Characterization of a Nalidixic Acid-Resistant Salmonella Enteritidis Strain Causing Persistent Infections in Broiler Chickens. Front. Vet. Sci. 8:725737. doi: 10.3389/fvets.2021.725737
Received: 15 June 2021; Accepted: 10 August 2021;
Published: 01 September 2021.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Alexey V. Rakov, Somov Institute of Epidemiology and Microbiology, RussiaNarayan Paudyal, Nepal Agricultural Research Council, Nepal
Mahdi Askari Badouei, Ferdowsi University of Mashhad, Iran
Copyright © 2021 Walker, Suyemoto, Hull, Gall, Jimenez, Chen, Thakur, Crespo and Borst. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Luke B. Borst, bGJib3JzdEBuY3N1LmVkdQ==
 Grayson K. Walker
Grayson K. Walker M. Mitsu Suyemoto
M. Mitsu Suyemoto Dawn M. Hull
Dawn M. Hull Siddhartha Thakur
Siddhartha Thakur Rocio Crespo
Rocio Crespo