- 1Institute of Microbiology and Infection, College of Life and Environmental Sciences, University of Birmingham, Birmingham, United Kingdom
- 2School of Veterinary Medicine and Science, University of Nottingham, Leicestershire, United Kingdom
- 3School of Life Sciences, University of Warwick, Coventry, United Kingdom
- 4Zeeman Institute, SBIDER: Systems Biology & Infectious Disease Epidemiology Research, Warwick Mathematics Institute, University of Warwick, Coventry, United Kingdom
AprV2 and aprB2 are variants of the apr gene of Dichelobacter nodosus, the cause of footrot in sheep. They are putative markers for severe and mild disease expression. The aim of our study was to investigate the distribution of aprV2 and aprB2 in flocks with and without footrot. Our hypotheses were that both strains are present in endemically affected flocks, with aprB2 and aprV2 associated with mild and virulent phenotypes respectively but that D. nodosus is not present in flocks without footrot. Alternatively, aprB2 persists in flocks without footrot. Despite extensive searching over 3 years only three flocks of sheep without footrot were identified. D. nodosus was not detected in these three flocks. In one further flock, only mild interdigital dermatitis was observed, and only aprB2 was detected. Twenty-four flocks with endemic footrot of all severities were sampled on three occasions and all were positive for D. nodosus and the aprV2 variant; aprB2 was detected in only 11 of these flocks. AprB2 was detected as a co-infection with aprV2 in the 22% of samples positive for aprB2 and was more likely in mild footrot phenotypes than severe. Dichelobacter nodosus serogroups were not associated with footrot phenotype. We conclude that D. nodosus, even aprB2 strains, do not persist in flocks in the absence of footrot. Our results support the hypothesis that aprB2 is associated with mild footrot phenotypes. Finally, we conclude that given the small number of flocks without footrot that were identified, footrot is highly endemic in English sheep flocks.
Introduction
Footrot is an economically important disease of sheep that reduces health, welfare and productivity (1–3). Footrot is caused by Dichelobacter nodosus (4–6), a facultative anaerobe that invades damaged interdigital epidermis (7) and has two disease presentations: interdigital dermatitis (ID) where there is inflammation of the interdigital skin and severe footrot (SFR) where the hoof horn separates from the underlying tissue (4, 6). ID and SFR vary in severity of pathology (8) ranging from minimal visible lesion (score 1) to most of the foot affected (score 4). Throughout the manuscript footrot refers to all severities of ID and SFR. Dichelobacter nodosus persists on feet with footrot for several months, but only persists on healthy feet for up to 2 weeks (9). Dichelobacter nodosus spreads between sheep via pasture, surviving off host for a few days in damp conditions (4, 9) but in dry conditions it does not persist off sheep sufficiently long for transmission to occur (9, 10). Due to the epidemiology of D. nodosus, Beveridge (4) and recently Clifton et al. (9) proposed that it should be possible to eliminate D. nodosus from flocks by eliminating all signs of footrot i.e., by eliminating ID and SFR.
Beveridge's (4) evidence led to a campaign to eliminate footrot from certain states in Australia using a flock by flock elimination program (11, 12). Elimination was successful in some flocks but there were reports that less virulent strains of D. nodosus were difficult to eradicate because they persisted in cattle which do not develop disease (13–16).
To manage these less virulent, persistent, strains of D. nodosus in New South Wales, Australia, flocks with low prevalence of foot lesions were considered to have eliminated “virulent” footrot and considered footrot free (11). Considerable research effort has since been spent trying to differentiate “benign” and “virulent” strains of D. nodosus using biochemical, and more recently genetic techniques. This is summarized in McPherson et al. (17) who concluded that no laboratory technique differentiates D. nodosus strains using the legislated flock phenotype definitions of “benign” and “virulent” footrot (17). This indicates that either the clinical definition of benign and virulent is incorrect, or that the methods do not differentiate benign and virulent strains correctly, or both.
Recently, variants of the extracellular protease apr (aprB2 and aprV2) have been proposed to differentiate benign and virulent strains of D. nodosus (18). In the field, aprV2 has been associated with severe footrot (19–21). However, in some cases, aprB2 has been detected without aprV2 in flocks with severe footrot (22), and, as stated above, aprV2/B2 did not differentiate benign and virulent footrot at flock level in a cross sectional study in Australia (17).
Footrot has been present in sheep flocks in the UK for centuries (23). It is highly endemic, in 2013 and 2014 farmers reported that >90% of flocks had footrot (ID and SFR) (24). There is no discernable spatial clustering of serogroups of D. nodosus in England, indicating homogenous mixing of strains between flocks and emphasizing the highly endemic nature of footrot (25). In the UK, the aprV2 variant dominates in flocks and feet with previous estimates that <7% of samples positive for aprV2 contain aprB2, (26–28). Since most flocks in England with footrot have both ID and SFR these findings could support the hypothesis that aprV2 is associated with “virulent” SFR and aprB2 with “benign” ID, but it does not refute the possibility that, as in Sweden (22), aprB2 is also associated with SFR.
The aim of the current study was to investigate the distribution of D. nodosus in flocks with and without footrot and the association between aprV2 and aprB2, the presence of disease and disease severity (ID and SFR). We hypothesized that D. nodosus, the cause of footrot in sheep, is not present in flocks where there is no footrot (no foot lesions on inspection of all feet) with the alternative hypothesis that aprB2 strains of D. nodosus persist in flocks with no signs of footrot (Figure 1). We also hypothesized that when footrot is endemic in a flock aprV2 strains of D. nodosus dominate over aprB2 strains. To test these hypotheses, we investigated sheep from 11 flocks where farmers reported no clinical signs of footrot and sheep from 24 flocks with a full range of footrot severities (8).
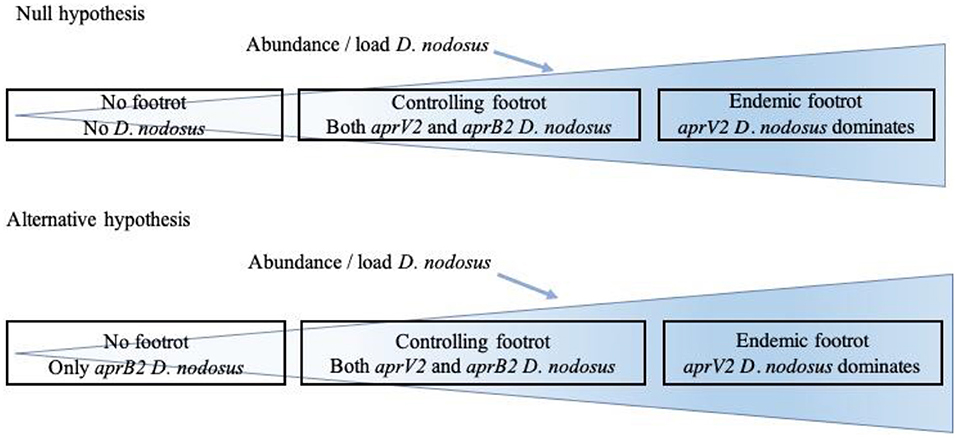
Figure 1. Relationship hypothesized between presence of clinical signs of footrot and presence of Dichelobacter nodosus, aprV2 and aprB2.
Materials and Methods
Ethical Approval
This study was carried out with approval from the Biomedical and Scientific Research Ethics Committee (BSREC REGO-2014-620) and the Animal Welfare and Ethical Review Committees (AWERB 16/13-14) at the Universities of Warwick and Birmingham, respectively.
Selection and Data Collection From Flocks Where Farmers Reported No Footrot
In 2013 (24) and 2014 (29) postal questionnaires were sent to 4,000 randomly selected lowland English sheep farms. There were 884 farmers who responded to both questionnaires and 12 of these had reported that they had not seen footrot in either year. The 12 farmers were contacted in 2016, and five reported that they had not seen footrot in their flock for at least 4 years. In a separate snowballing exercise where the goal was to identify all flocks without footrot in England (30), sheep industry experts were asked to identify flocks without footrot and to suggest other experts who might know of flocks with no clinical signs of footrot. This occurred over a period of 3 years and a further six flocks were identified. The owners of these 11 flocks agreed to participate in the study. Two of three trained researchers (EM, EN, NP) visited each flock. In four flocks <15% of sheep were present for examination; these flocks were discarded because it was not possible to examine all sheep and feet in the flock. In the remaining seven flocks (Table 1), initially the locomotion of all sheep in the flock was scored using a validated system (31). When a sheep had a locomotion score > 0 (31) all four feet were examined and scored for footrot severity (8) into ID and SFR with severity scores 1 which are a visible lesion through to 4 more than 50% of the skin/claw affected. Feet were then swabbed with a sterile cotton swab with a downward vertical wipe repeated five times. When fewer than 15 sheep were lame, all four feet of a number of non-lame ewes and rams were also scored and swabbed up to a total of 15 sheep where possible, as for the 24 flocks with footrot (see section Selection and Data Collection From 24 Flocks in England With Clinical Footrot). In four flocks where no lame sheep were observed, all feet of all sheep were examined. In three flocks all feet were swabbed, in the fourth flock lesions were observed on some feet and these were swabbed and in addition, all four feet of every fifth ewe were swabbed. The swabs were placed in amies charcoal and stored at −20°C. A total of 496 foot swabs were collected (Table 1).
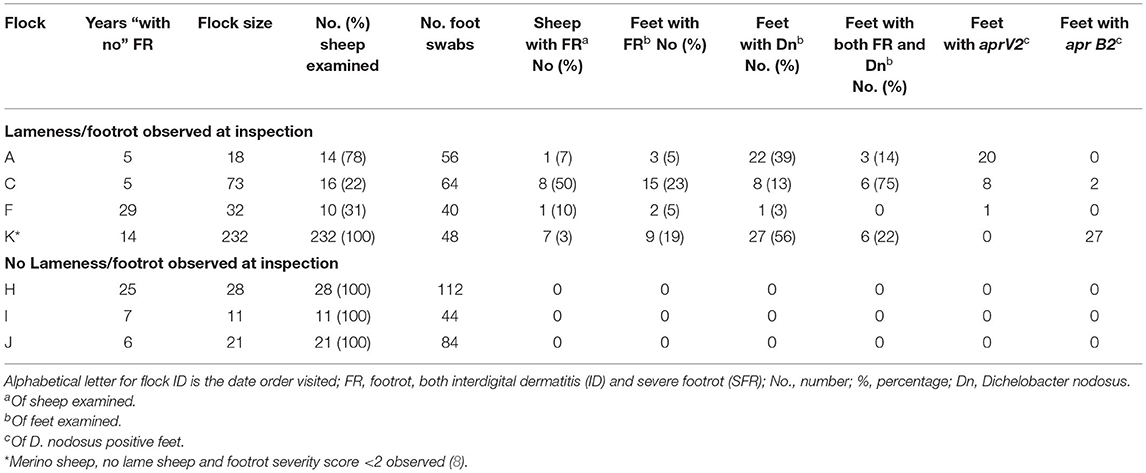
Table 1. Observation of footrot and detection of D. nodosus, aprV2 and aprB2 in seven flocks in England where farmers reported “no footrot” for at least 4 years.
Selection of and Data Collection From 24 Flocks in England With Clinical Footrot
An 18-month clinical trial was conducted on 44 flocks with footrot throughout England (32). A total of 24 farmers consented to being in this study, flocks were 100–500 breeding ewes and a prevalence of >5% lameness (Table 2). The flocks were visited on three occasions; autumn 2015 (visit 1), spring 2016 (visit 2) and autumn 2016 (visit 3). At each visit, sheep locomotion was scored as previously described. Up to 15 lame ewes, locomotion score >1 or, when there were fewer than 15 lame ewes, a combination of lame and non-lame ewes, were selected and all four feet were scored for footrot severity and swabbed. Swabs were stored in sterile cryotubes containing 300 μl sterile phosphate buffered saline (PBS) and stored at −20°C. A total of 3,506 foot swabs were collected and those from healthy feet and footrot affected feet (n = 2,338) were processed (Supplementary Table 1).
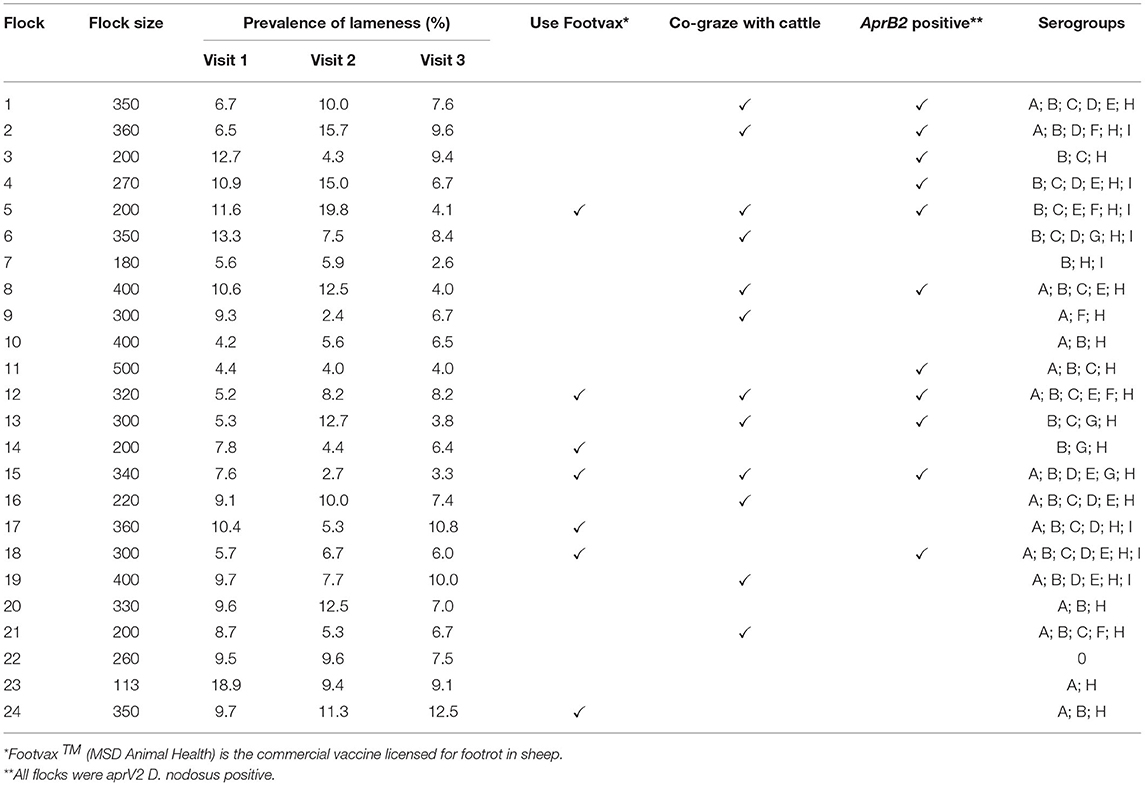
Table 2. Summary information on 24 flocks with clinical footrot: flock size, prevalence of lameness by visit, use of Footvax, co-grazing with cattle, detection of D. nodosus variant aprB2 and detection of serogroups of D. nodosus.
Laboratory Processing of Foot Swabs
Swabs were centrifuged at 1,600 g for 8 min. Swabs from the putative footrot-free farms were processed as individual samples and were not pooled prior to DNA extraction. Supernatant from swabs taken from the 24 endemically affected flocks were pooled by visit to a flock into 10 phenotypes: healthy feet from sheep where all four feet were healthy (AH), healthy feet from sheep where one or more feet had any severity of footrot (HD), and separate pools for feet with ID by severity score 1, 2, 3, or 4 and SFR by severity score 1, 2, 3, or 4 (8). There were a total of 395 pooled samples (Supplementary Table 1); 309 (78.2%) with 2–32 swabs per sample and 86 (21.8%) single swab samples. There were 12–23 pooled samples per flock (Table 3). The supernatant was removed after pooling and centrifuging the swabs and the pellets re-suspended in 500 μl of PBS. DNA was extracted as described previously (33), eluted into 45 μl aliquots and stored at −20°C.
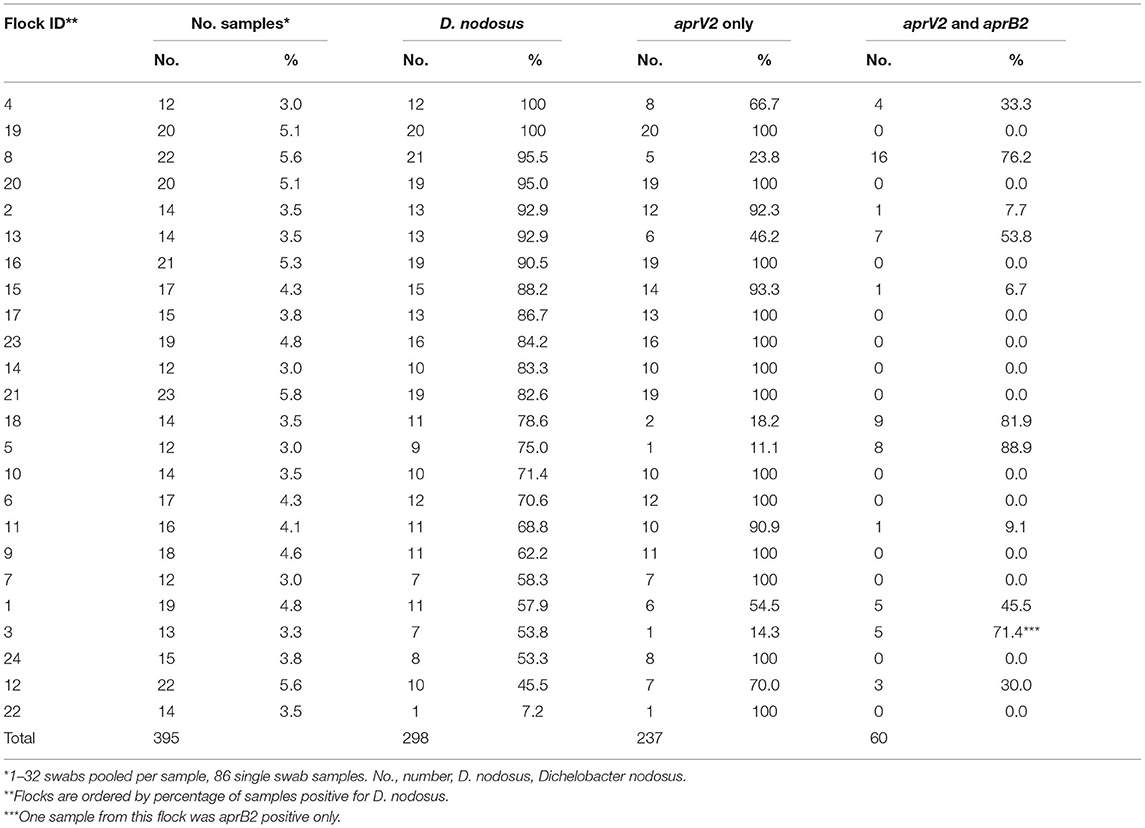
Table 3. Number and percentage of samples collected and positive for Dichelobacter nodosus, aprV2 only and both aprV2 and aprB2 for 24 flocks in England with clinical footrot.
Detection and Quantification of Dichelobacter nodosus and Protease Genes AprV2 and AprB2
Dichelobacter nodosus was detected by targeting a 61 bp sequence in the rpoD gene, and quantified by qPCR as described previously (6, 34). Samples that were positive for D. nodosus were tested for aprV2 and aprB2 using the qPCR method of Frosth et al. (22) with a modification made to the probe labeling (aprV2 probe 6FAM-BHQ1, aprB2 probe TxRd-BHQ2). The initial denaturation step was 15 min following the Klearkall master mix (LGC Group) methods. Each reaction consisted of 0.4 μM of each primer, 0.1 μM probe, 7.5 μl Klearkall master mix (LGC Group), 0.1 mg/ml−1 bovine serum albumin (BSA) solution, 4.85 μl nuclease-free water (Applied Biosystems) and 1 μl template DNA. The DNA did not amplify on 28 occasions and so the process was repeated with 2 μl DNA, a further eight samples amplified.
Detection of Dichelobacter nodosus Serogroups
Presence of D. nodosus serogroups A-I was investigated using the single serogroup target PCR protocol described in Dhungyel et al. (35). Serogroup M was not investigated because there is no serogroup M specific protocol (36). Each reaction consisted of 0.5 μM primer, 2x MyTaq Red mix (Bioline), 0.5 mg/ml−1 BSA, 16.5 μl nuclease-free water (Applied Biosystems) and 1μl template DNA. PCR products were visualized using 3% agarose stained with ethidium bromide under ultraviolet light.
Data Storage and Analysis
All data were stored in Microsoft Excel. The prevalence of lameness (defined as sheep with a locomotion score > 0) and the prevalence of footrot (sheep with any score > 0 on any foot) and the frequency of footrot lesion severity scores (28) were calculated for each flock-visit. The percentage of sheep sampled, swabs/pooled samples positive for D. nodosus, aprV2, and aprB2, and for serogroups A-I were calculated.
A binomial multivariable mixed effect model was used to investigate associations between detection of aprV2 and aprB2 in pooled samples and the explanatory variables footrot disease severity score and serogroup. The outcome was pooled sample with aprV2 vs. aprV2 and aprB2.
The equation took the form:
Where logit (aprV2 and aprB2 : aprV2) is the outcome variable, β0 is the intercept and βijk are a series of explanatory variables varying at i (sample), j (visit), and k (flock) with e the residual error at levels k and j, the residual error at “i” was assumed to follow a binomial error distribution. The model was built in a forward-stepwise process (37), using lme4 (38) in RStudio version 3.6.0 (39). The reference category for footrot severity was ID score 1 because there were few D. nodosus positive samples for AH and HD pools and so ID1 provided a stable baseline category. All severity scores were included and serogroups were added when the p-value from a likelihood ratio test comparing the model with and without the variable was ≤0.05, when p > 0.05 from the likelihood ratio test, the explanatory variable did not improve the model fit and were omitted.
Similarly, three multivariable mixed effect regression models were run using lme4 (38) with three outcomes: the log [mean (qPCR replicate) +1] for all D. nodosus, for aprV2 only and for aprV2 + aprB2. An explanatory variable of aprV2 load was forced into the aprV2 + aprB2 model to adjust for aprV2 load in the sample and remove confounding aprV2 load with explanatory variables. The explanatory variables were disease phenotype, baseline ID1, as for the binomial models and serogroup and random variables for visit (j) and flock (k), with sample (i) fitted as the unit of observation in a three-level model (37).
The equation took the form:
Where log(qPCR) is the log10 [mean (qPCR replicate load)] of the outcome variables (all D. nodosus, aprV2, aprV2, and aprB2), β0 is the intercept and βijkX are a series of exploratory variables varying at i (sample), j (visit), and k (flock) with e, the residual error at sample level. Confidence intervals and Wald's p-values were obtained from broom.mixed (40).
To investigate whether pooling samples reduced the detection of serogroups, the data were simulated assuming that the probability of detecting a particular serogroup for a given sample was dependent on both lesion severity score and flock of origin:
Where the disease state (D) and flock-level prevalence of lesion (P) were determined such that the sample results are recovered. This was done using reversible jump Markov Chain Monte Carlo (RJMCMC) (41) using the likelihood of obtaining the pooled results from the independent samples of feet within each pool. The mean and 95% credible intervals for each parameter were calculated. The model assumed independence between serogroups. The model's performance was assessed by comparing the observed results from 2,338 samples across the 24 flocks with footrot with the predicted footrot severity scores.
Results
Flocks in England Where Farmers Reported No Footrot
There were 7 flocks where farmers thought footrot was absent and where all sheep were inspected. In three flocks (H, I, and J) footrot and D. nodosus were not detected (Table 1). In three further flocks (A, C, and F) sheep were lame, footrot of all severities was observed, and D. nodosus and aprV2 were detected (Table 1); aprB2 was also detected in flock C. In flock K, a flock of 232 Merino sheep, no sheep were lame but mild interdigital lesions were observed in about 3% of sheep, although there was no necrotic smell typical of footrot (Figure 2). Only the aprB2 variant and not the aprV2 variant of D. nodosus was detected.
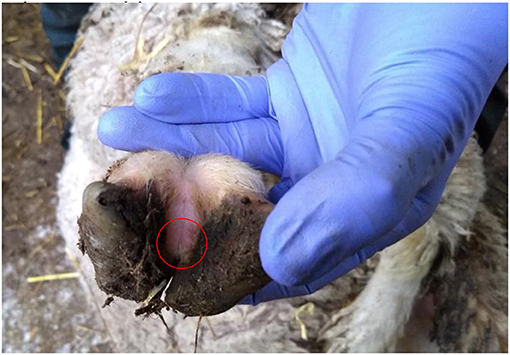
Figure 2. Photograph of a mild interdigital lesion (whitish skin in the circle) from flock K where no sheep were lame and only aprB2 variant of D. nodosus was detected.
Twenty-Four Flocks With Clinical Footrot
There were lame sheep in all 24 flocks with footrot. Footrot lesions, D. nodosus, and aprV2 were detected in all 24 flocks (Figures 3A,B). There were 298 (75.4%) D. nodosus positive samples (Table 3). AprV2 was detected in all 24 flocks and 297 of the 298 D. nodosus positive samples (Table 3) and aprB2 was detected in 11 flocks and 61 (22%) samples; aprB2 was detected without aprV2 in only one sample pooled from 2 swabs with severity score SFR 3, aprV2 was detected in this flock-visit (Figure 3A). The geometric mean load of aprV2 was typically greater than the load of aprB2 in all foot phenotypes (Figure 4) but there was a positive correlation (r = 0.097) between the log load of aprV2 and aprB2 (Figure 5).
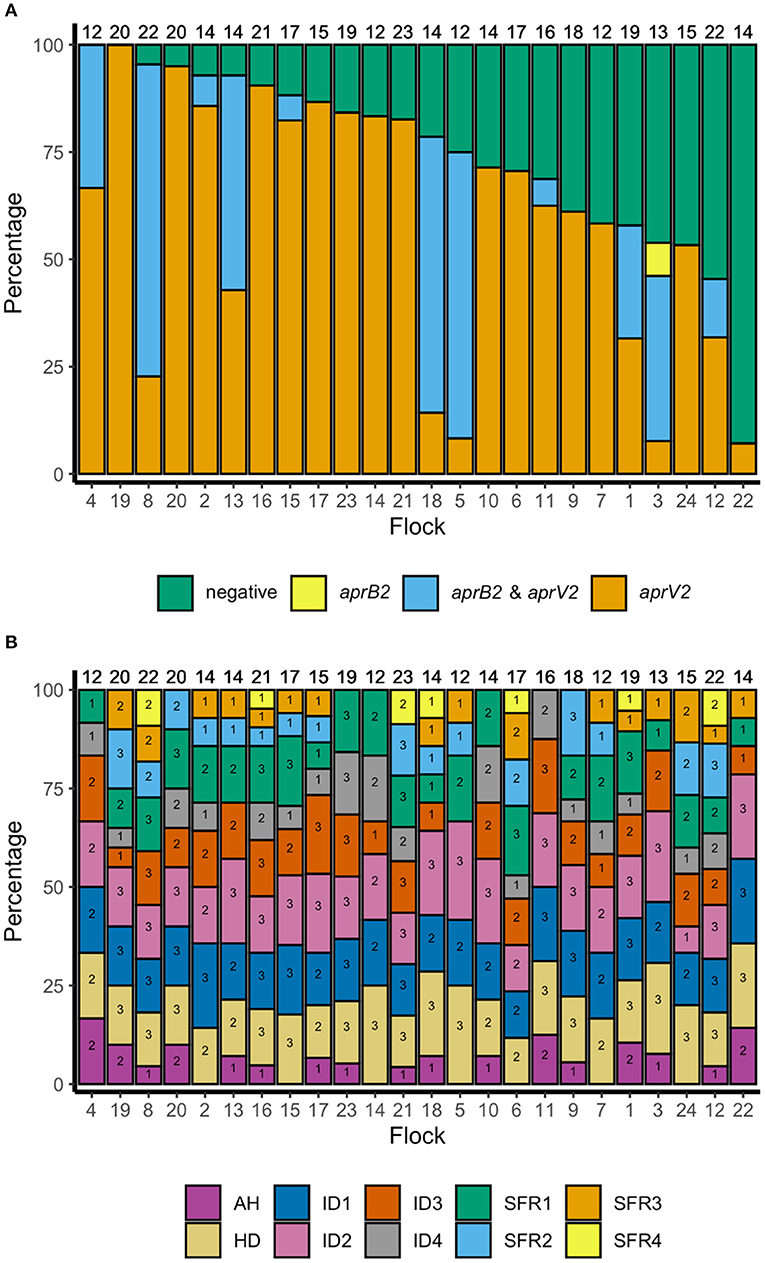
Figure 3. (A) Percentage of samples negative for D. nodosus and positive for D. nodosus by aprV2 only, aprB2 and aprV2 and aprB2 only in descending order of aprV2 only: 395 pooled samples from 24 flocks in England. Number at top of each bar = number of pooled samples over three visits, number at the bottom = flock identification. (B) Percentage of samples positive by foot phenotype: 395 pooled samples from 24 flocks in England. Top of column number = total number of pooled samples per flock for all three visits. Number within each bar = number of pooled samples per flock for all three visits by foot phenotype. AH, all healthy feet from sheep where all four feet were healthy; HD, Healthy feet from sheep where one or more feet had signs of footrot; ID, Interdigital dermatitis scores 1–4; SFR, Severe footrot scores 1–4. Number at base of bar = flock identification number, ordered as for (A).
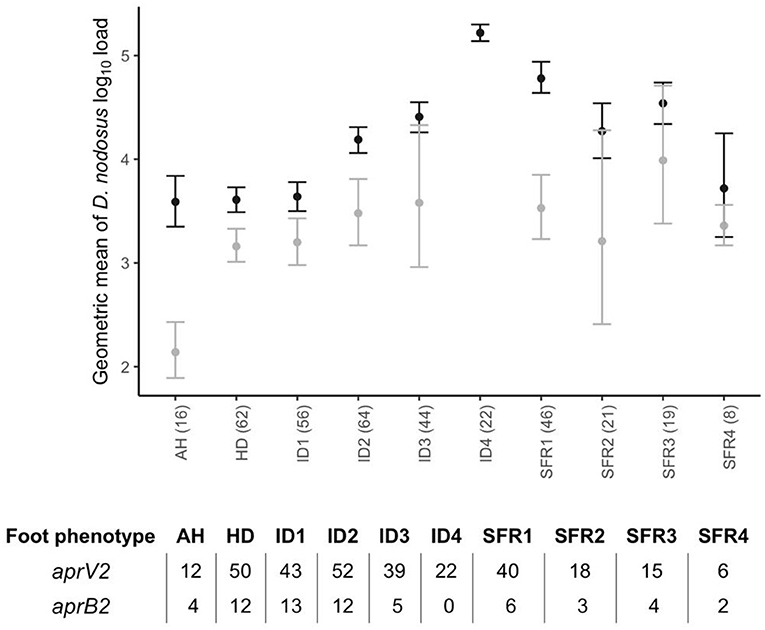
Figure 4. Geometric mean and 2 standard errors for log10 aprV2 D. nodosus load (black) and log10 aprB2 D. nodosus load (grey) by foot phenotype from 395 samples from 24 flocks in England with clinical footrot. AH, all healthy feet from sheep where all four feet were healthy; HD, Healthy feet from sheep where one or more feet had signs of footrot; ID, Interdigital dermatitis scores 1–4; SFR, Severe footrot scores 1–4. Number in brackets after each foot phenotype on the x-axis is the total number of pooled swabs positive for aprV2 or aprB2 in each foot phenotype category. The associated table shows the split between samples aprV2 or aprB2 positive out of total number positive.
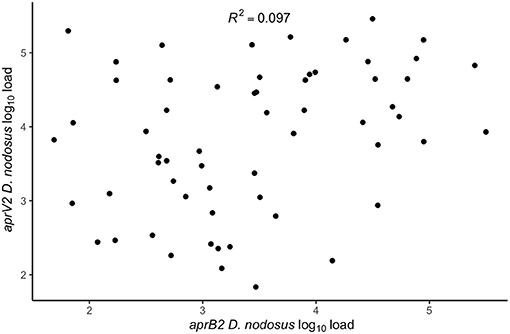
Figure 5. Within swab association between aprV2 D. nodosus log10 load and aprB2 D. nodosus log10 load from 60 samples positive for both aprV2 and aprB2 D. nodosus from 3 visits (1–3) to 24 flocks in England with clinical footrot. The correlation coefficient (R2) was calculated using the built-in lm function in R Studio Version 3.6.0 (39) to calculate a simple linear regression. P-value = 0.02.
There were 231/298 (77.5%) D. nodosus positive samples where at least one serogroup was detected. All nine serogroups (A-I) were detected in 63 combinations with a median of 5 (IQR 3–6) serogroups per flock. Serogroups H, B, and A were most frequently detected, in 38.6, 30.2, and 20.1% of samples, respectively (Supplementary Tables 2–4). In the 67 D. nodosus positive samples where serogroups were not detected, the qPCR load was below the limit of detection for serogroup PCR in 60 samples and the remaining 7 had sufficient D. nodosus load but no serogroup was detected; it is possible these were serogroup M.
In the multivariable mixed effect binomial regression model, feet with ID 3–4, or SFR of any severity score were more likely to have aprV2 only than both aprV2 and aprB2, compared with ID 1, indicating that presence of aprB2 was associated with lower footrot severity scores. Serogroups C and E were less likely to be present in samples with aprV2 alone than aprV2 and aprB2 (Table 4), other serogroups were not significantly associated with apr variant.
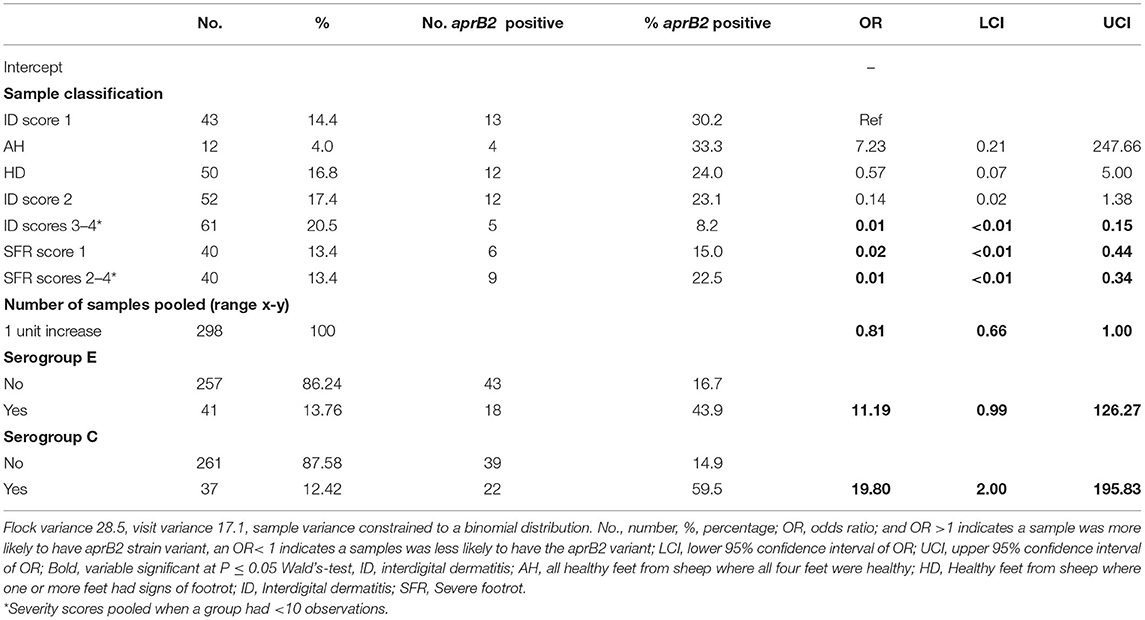
Table 4. Binomial multivariable mixed effect regression model of pooled samples that were aprV2 only vs. aprV2 and aprB2 gene variants by footrot phenotype and serogroup in 298 samples from three visits to 24 sheep flocks in England.
In the mixed effect multivariable regression models, the load of D. nodosus and aprV2 increased with severity of ID score compared with ID 1 and then plateaued once feet had SFR (Table 5; Figure 4). Load of aprB2, after adjusting for load of aprV2, was higher in feet with ID score >1 than ID 1, but did not increase with disease severity as observed for aprV2. This is also illustrated in Figure 4. A higher load of all D. nodosus was significantly associated with the six most prevalent serogroups (A, B, C, E, F, H), a higher load of aprV2 was associated with serogroups A, B, E, and H. An increasing load of aprB2 was associated with serogroup F. When serogroup positive samples from single swabs only (n = 53) were analyzed, the patterns of association were similar to those for pooled samples but confidence intervals were wider because of the reduced power of the dataset (data not shown). The simulated data indicated that pooling samples did not reduce the detection of the number of serogroups per sample (Supplementary Table 5).
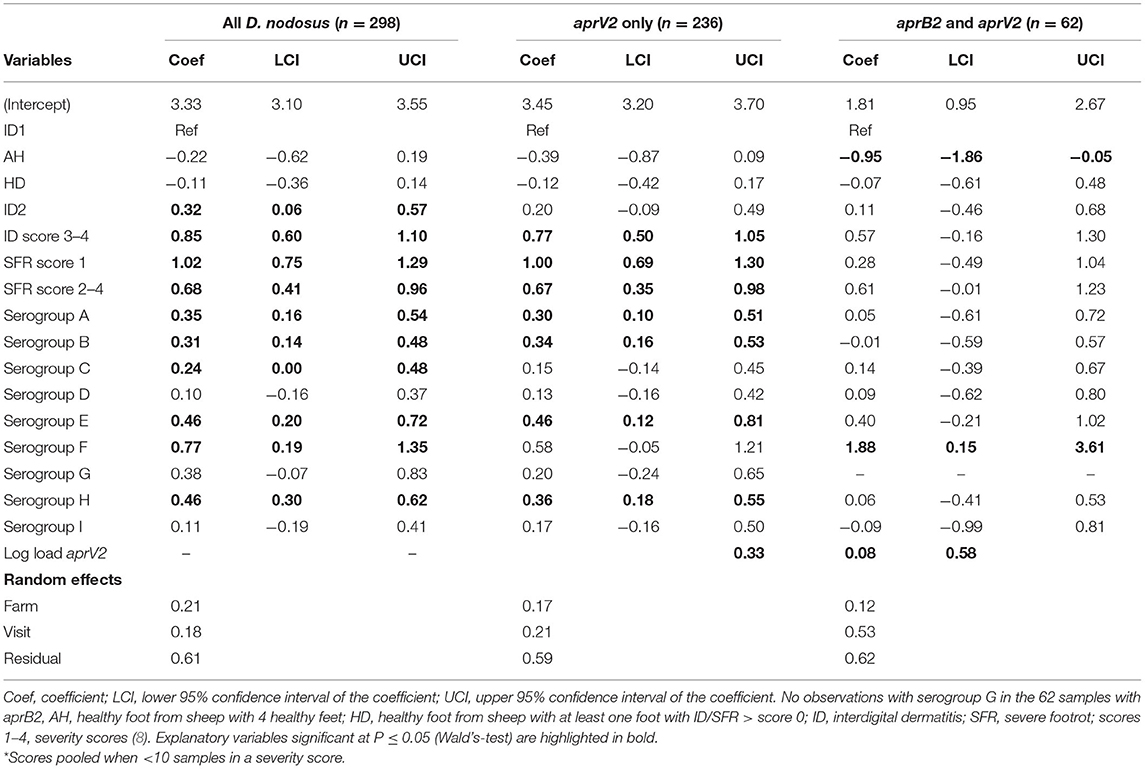
Table 5. Three multivariable mixed effects regression models of pooled samples by load of all D. nodosus, only aprV2, and aprB2 adjusted for aprV2 from 3 visits to 24 sheep flocks in England with clinical footrot.
Discussion
Our study supports previous findings that D. nodosus and footrot are highly endemic in flocks in England, with only three flocks without footrot detected over 4 years. The results from the current study, together with published work from other countries confirm our hypothesis that D. nodosus, the causative agent of footrot in sheep, is present in flocks where there is footrot and is not present where there is no footrot. As proposed by Beveridge (4) and Clifton et al. (9), D. nodosus is absent when there are no sheep with footrot. In the seven flocks that were examined that were putatively free from footrot, three flocks that had footrot 6–20 years previously had no footrot lesions and no D. nodosus, four flocks had sheep with lesions, and D. nodosus was detected in all four flocks, even flock F where only one sheep had footrot lesions. Dichelobacter nodosus was detected in all 24 endemically affected flocks. These findings do not contradict other published work as discussed below but they do add clarity to a previously confusing narrative.
One important protocol in the examination of putatively footrot-free flocks was that every foot of every sheep in the flock was examined until footrot was observed or all feet had been examined. Because mild lesions do not always cause lameness (42) it is not possible to rely on detection of lameness alone and is essential to inspect all feet to be certain that footrot was absent from a flock. Similarly, using “freedom from disease” sampling and not examining all feet would have compromised our interpretation, flock F exemplifies how this would have produced doubt into our results. A second important methodological design was the use of a strict definition of no footrot that was just that, no foot with a footrot severity score > 0, a definition that is consistent for all scoring systems across the globe (8, 43–46) but not used in many studies.
Our definition of footrot-free was stricter than that used in other studies that have suggested aprB2 can persist in flocks without footrot. In those studies not all feet were inspected and a severity score of 1 was considered negative for footrot. Studies include those from Germany (47), Australia (48–52), Switzerland (53, 54), and Norway (55). Our results from flock K and the 24 endemically affected flocks highlight the challenge of studying flocks with footrot to elucidate the relationship between strain variation and disease severity because once infected with D. nodosus, disease severity varies over time and because feet will be contaminated between sheep in the same group, elucidating the role of strain variation on disease severity is difficult.
It was extremely challenging to identify even three flocks free from all clinical signs of footrot in England. The three flocks free from footrot were small (<50 ewes) and none were from the previous study of 1,400 flocks (24, 29) where ~80% of flocks had >200 ewes. It seems intuitive that footrot is more likely to be absent in small flocks than larger flocks because they may be below the critical community size for persistence of footrot and so natural fade out of disease can occur (56). Indeed, three farmers reported no signs of footrot for 6–20 years but none had actively eliminated footrot, although active elimination of footrot, through inspection, rapid treatment and culling would require less effort in small flocks than large. Two of the three flocks were also biosecure (strict control of sheep entering the farm) and therefore more likely to prevent footrot from being reintroduced through purchase of sheep or sheep straying onto the units. There were 35,500 sheep holdings in England in 2018 (57). Surprisingly, 36% had <50 ewes (57) so it might be possible that there are more flocks without footrot in England, although our snowballing exercise did not identify them, and many of the sheep specialists involved could not recall any flocks free from footrot (personal communication). Our results further support the highly endemic nature of D. nodosus and footrot in sheep in England as highlighted in Prosser et al. (25) who reported that serogroups of D. nodosus are randomly distributed across England.
We hypothesized that when clinical footrot was endemic in a flock, aprV2 strains of D. nodosus dominate over aprB2. This was the case at a flock level in our endemically affected flocks, aprV2 was present in all 24 flocks with footrot whilst aprB2 was present in only 11 of these flocks. It was also the case that aprV2 dominated the samples from the feet with footrot, with aprB2 only present in 22% of samples. Finally, aprB2 was less likely to be present in more severe disease phenotypes than aprV2 (Table 4), with both phenotypes present in less severe phenotypes.
We hypothesized that Dichelobacter nodosus, in particular aprB2, would persist in flocks without footrot. We did not identify any such flocks and conclude that this hypothesis is false in reference to the flocks we investigated. There was one flock (K) of ~240 Merino sheep, where aprB2 was detected in the absence of aprV2 in the current study and we believe this is the first such flock detected in England to date. Flock K was unusual because Merino flocks are rare in the UK. The flock has been in the UK since 1970 and was crossbred with native UK breeds in an attempt to improve wool quality, and so had mixed with native sheep. The current owner treated the whole flock with antibiotics when purchased in 2006 and had not observed footrot since then, and they believed there was no footrot in the flock, they also returned the flock to near pure Merino. The presence of the aprB2 variant only and mild footrot lesions would fit with the proposal that aprB2 strains of D. nodosus are only capable of causing mild disease (18). However, the management of this flock would also influence the expression of footrot. The owner houses the sheep in straw bedded barns during inclement weather, including heavy rainfall, to protect the valuable fleece. This has the incidental effect of keeping feet dry, which protects from disease expression (9). The owner maintains high levels of biosecurity and the sheep are isolated from all other livestock preventing the introduction of other strains of D. nodosus.
Whilst we identified flocks without footrot and where aprV2 and aprB2 were absent, apparently after footrot had been in the flocks in the past, we do not know whether aprV2 and aprB2 strains can be eliminated from flocks with footrot. Efforts to eliminate footrot in flocks in other countries have focused on elimination of “virulent footrot” which suggests that elimination of all lesions is very difficult in many countries (47–55). From our study we conclude that aprV2 dominates in flocks where footrot is endemic, and that there is evidence that aprB2 is associated with less severe footrot. Considering these new findings hypotheses for future study would be:
1. It is easier to eliminate footrot and D. nodosus from flocks with only aprV2.
2. Where aprV2 and aprB2 are present in a flock, aprB2 will persist longer in elimination programmes.
3. It is not possible to eliminate aprB2 strains of D. nodosus.
There was no variation in disease expression by serogroup, this is consistent with reports that disease severity is not linked to serogroup (5). The current study would support the lack of association between serogroup and disease severity through epidemiological rather than microbiological study. Unlike Prosser et al. (25), pooling samples did not reduce detection of serogroups per flock, probably because more sheep were swabbed per flock-visit in the current study. We did find an association between presence of serogroups C and E and presence of aprB2 in a foot (Table 4). This might indicate that these serogroups were more likely to be aprB2 strains of D. nodosus, but since D. nodosus can seroconvert (58, 59), serogroup is unlikely to be a reliable marker of virulence.
Conclusion
We conclude that D. nodosus did not persist in the flocks without signs of footrot in this study but that these flocks were rare. In all flocks where we saw clinical signs of footrot, D. nodosus was present, including three flocks that farmers thought were free from footrot. There is no evidence from our study that D. nodosus persists when footrot is not observed in a flock for several years. The relationship between aprV2 and aprB2 strains in footrot in endemically affected flocks suggest that aprB2 is associated with milder footrot lesions. There remain questions on the feasibility of eliminating all D. nodosus from endemically diseased flocks, and whether aprB2 strains would persist for longer than aprV2 strains of D. nodosus in an elimination programme.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The animal study was reviewed and approved by the Biomedical and Scientific Research Ethics Committee University of Warwick (BSREC REGO-2014-620) and the Animal Welfare and Ethical Review Committee (AWERB 16/13-14) University of Birmingham. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
LG and KP designed the study. JW collected samples from the 24 flocks with clinical footrot with assistance from NP and EM. EM, NP, and EN collected samples from putative footrot free flocks. EN assisted in identifying footrot free flocks. EM conducted all laboratory work and statistical analysis with input from LG, KP, MK, and KL. EM and LG wrote the manuscript. All authors contributed to manuscript editing and approved the final manuscript.
Funding
This work was funded by a Biotechnology and Biological Sciences Research Council (BBSRC) Animal Health Research Club (ARC) Grant (BB/M012980). NP was funded by a BBSRC studentship with the Agricultural and Horticultural Development Board (AHDB) as the Collaborative Awards in Science and Engineering (CASE) partner. EN and KL are funded by a BBSRC Midlands Integrative Biosciences Training Partnership (MIBTP) with AHDB as the industry partner.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the farmers who participated, Jessica Taylor for technical support and Dr. Ed Smith, Dr. Rachel Clifton, Dr. Katharina Giebel, and Alan Dickins for assistance during farm visits.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.713927/full#supplementary-material
References
1. Wassink GJ, King EM, Grogono-Thomas R, Brown JC, Moore LJ, Green LE, et al. within farm clinical trial to compare two treatments (parenteral antibacterials and hoof trimming) for sheep lame with footrot. Prev Vet Med. (2010) 96:93–103. doi: 10.1016/j.prevetmed.2010.05.006
2. Winter JR, Green LE. Cost-benefit analysis of management practices for ewes lame with footrot. Vet J. (2017) 220:1–6. doi: 10.1016/j.tvjl.2016.11.010
3. Marshall DJ, Walker RI, Cullis BR, Luff MF. The effect of footrot on body-weight and wool growth of sheep. Aust Vet J. (1991) 68:45–9. doi: 10.1111/j.1751-0813.1991.tb03126.x
4. Beveridge W. Foot-rot in sheep: a transmissible disease due to infection with Fusuformis nodosus (n. sp): studies on its cause, epidemiology and control CSIRO. Austr Bull. (1941) 140:1–56.
5. Kennan RM, Dhungyel OP, Whittington RJ, Egerton JR, Rood JI. The type IV fimbrial subunit gene (fimA) of Dichelobacter nodosus is essential for virulence, protease secretion, and natural competence. J Bacteriol. (2001) 183:4451–8. doi: 10.1128/JB.183.15.4451-4458.2001
6. Witcomb LA, Green LE, Kaler J, Ul-Hassan A, Calvo-Bado LA, Medley GF, et al. A longitudinal study of the role of Dichelobacter nodosus and Fusobacterium necrophorum load in initiation and severity of footrot in sheep. Prev Vet Med. (2014) 115:48–55. doi: 10.1016/j.prevetmed.2014.03.004
7. Egerton J, Parsonson I. Benign foot-rot—a specific interdigital dermatitis of sheep associated with infection by less proteolytic strains of Fusiformis nodosus. Aust Vet J. (1969) 45:345–9. doi: 10.1111/j.1751-0813.1969.tb06606.x
8. Moore LJ, Wassink GJ, Green LE, Grogono-Thomas R. The detection and characterisation of Dichelobacter nodosus from cases of ovine footrot in England and Wales. Vet Microbiol. (2005) 108:57–67. doi: 10.1016/j.vetmic.2005.01.029
9. Clifton R, Giebel K, Liu NLBH, Purdy KJ, Green LE. Sites of persistence of Fusobacterium necrophorum and Dichelobacter nodosus: a paradigm shift in understanding the epidemiology of footrot in sheep. Sci Rep. (2019) 9:14429. doi: 10.1038/s41598-019-50822-9
10. Graham NPH. Egerton JR. Pathogenesis of ovine foot-rot: The role of some environmental factors. Austr Vet J. (1968) 44:235–40. doi: 10.1111/j.1751-0813.1968.tb09092.x
12. Buller NB, Ashley P, Palmer M, Pitman D, Richards RB, Hampson DJ. Understanding the molecular epidemiology of the footrot pathogen Dichelobacter nodosus to support control and eradication programs. J Clin Microbiol. (2010) 48:877–82. doi: 10.1128/JCM.01355-09
13. Egerton JR, Parsonson IM. Isolation of Fusiformis nodosus from cattle. Aust Vet J. (1966) 42:425–9. doi: 10.1111/j.1751-0813.1966.tb04646.x
14. Laing EA, Egerton JR. Occurrence, prevalence and transmission of Bacteroides nodosus infection in cattle. Res Vet Sci. (1978) 24:300–4. doi: 10.1016/S0034-5288(18)33037-6
15. Richards RB, Depiazzi LJ, Edwards JR, Wilkinson FC. Isolation and characterization of Bacteroides nodosus from foot lesions of cattle in Western Australia. Aust Vet J. (1980) 56:517–21. doi: 10.1111/j.1751-0813.1980.tb02577.x
16. Knappe-Poindecker M, Gilhuus M, Jensen TK, Vatn S, Jorgensen HJ, Fjeldaas T. Cross-infection of virulent Dichelobacter nodosus between sheep and co-grazing cattle. Vet Microbiol. (2014) 170:375–82. doi: 10.1016/j.vetmic.2014.02.044
17. McPherson AS, Dhungyel OP, Whittington RJ. Evaluation of genotypic and phenotypic protease virulence tests for Dichelobacter nodosus infection in sheep. J Clin Microbiol. (2017) 55:1313–26. doi: 10.1128/JCM.02403-16
18. Kennan RM, Wong W, Dhungyel OP, Han XY, Wong D, Parker D, et al. The subtilisin-like protease AprV2 is required for virulence and uses a novel disulphide-tethered exosite to bind substrates. PLos Pathog. (2010) 6:e1001210. doi: 10.1371/journal.ppat.1001210
19. Gilhuus M, Kvitle B, L'Abee-Lund TM, Vatn S, Jorgensen HJ. A recently introduced Dichelobacter nodosus strain caused an outbreak of footrot in Norway. Acta Vet Scand. (2014) 56:29. doi: 10.1186/1751-0147-56-29
20. Kennan RM, Gilhuus M, Frosth S, Seemann T, Dhungyel OP, Whittington RJ, et al. Genomic evidence for a globally distributed, bimodal population in the ovine footrot Pathogen Dichelobacter nodosus. mBio. (2014) 5:11. doi: 10.1128/mBio.01821-14
21. Stäuble A, Steiner A, Normand L, Kuhnert P, Frey J. Molecular genetic analysis of Dichelobacter nodosus proteases AprV2/B2, AprV5/B5 and BprV/B in clinical material from European sheep flocks. Vet Microbiol. (2014) 168:177–84. doi: 10.1016/j.vetmic.2013.11.013
22. Frosth S, Konig U, Nyman AK, Pringle M, Aspan A. Characterisation of Dichelobacter nodosus and detection of Fusobacterium necrophorum and Treponema spp. in sheep with different clinical manifestations of footrot. Vet Microbiol. (2015) 179:82–90. doi: 10.1016/j.vetmic.2015.02.034
23. Mohler JR, Washburn HJ. Foot-Rot of Sheep: Its Nature, Cause and Treatment. Charleston, SC: BiblioLife (1904).
24. Winter JR, Kaler J, Ferguson E, KilBride AL, Green LE. Changes in prevalence of, and risk factors for, lameness in random samples of English sheep flocks: 2004-2013. Prev Vet Med. (2015) 122:121–8. doi: 10.1016/j.prevetmed.2015.09.014
25. Prosser NS, Monaghan EM, Green LE, Purdy KJ. Serogroups of Dichelobacter nodosus, the cause of footrot in sheep, are randomly distributed across England. Sci Rep. (2020) 10:16823. doi: 10.1038/s41598-020-73750-5
26. Blanchard AM, Jolley KA, Maiden MCJ, Coffey TJ, Maboni G, Staley CE, et al. The applied development of a tiered multilocus sequence typing (MLST) scheme for Dichelobacter nodosus. Front Microbiol. (2018) 9:551. doi: 10.3389/fmicb.2018.00551
27. Maboni G, Frosth S, Aspan A, Totemeyer S. Ovine footrot: new insights into bacterial colonisation. Vet Rec. (2016) 179:228. doi: 10.1136/vr.103610
28. Smith EM, Gilbert A, Russell CL, Purdy KJ, Medley GF, Muzafar M, et al. Within-flock population dynamics of Dichelobacter nodosus. Front Vet Sci. (2017) 4:6. doi: 10.3389/fvets.2017.00058
29. Grant C, Kaler J, Ferguson E, O'Kane H, Green LE. A comparison of the efficacy of three intervention trial types: postal, group and one-to-one facilitation, prior management and the impact of message framing and repeat messages on the flock prevalence of lameness in sheep. Prev Vet Med. (2018) 149:82–91. doi: 10.1016/j.prevetmed.2017.11.013
31. Kaler J, Wassink GJ, Green LE. The inter- and intra-observer reliability of a locomotion scoring scale for sheep. Vet J. (2009) 180:189–94. doi: 10.1016/j.tvjl.2007.12.028
32. Witt J, Green L. Development and assessment of management practices in a flock-specific lameness control plan: a stepped-wedge trial on 44 English sheep flocks. Prev Vet Med. (2018) 157:125–33. doi: 10.1016/j.prevetmed.2018.06.013
33. Muzafar M, Calvo-Bado LA, Green LE, Smith EM, Russell CL, Grogono-Thomas R, et al. The role of the environment in transmission of Dichelobacter nodosus between ewes and their lambs. Vet Microbiol. (2015) 179:53–9. doi: 10.1016/j.vetmic.2015.04.010
34. Calvo-Bado LA, Oakley BB, Dowd SE, Green LE, Medley GF, Ul-Hassan A, et al. Ovine pedomics: the first study of the ovine foot 16S rRNA-based microbiome. ISME J. (2011) 5:1426–37. doi: 10.1038/ismej.2011.25
35. Dhungyel OP, Whittington RJ, Egerton JR. Serogroup specific single and multiplex PCR with pre-enrichment culture and immuno-magnetic bead capture for identifying strains of D-nodosus in sheep with footrot prior to vaccination. Mol Cell Probes. (2002) 16:285–96. doi: 10.1006/mcpr.2002.0427
36. Best N, Gwozdz J, Suter R, Rawlin G, Beddoe T. Direct serogrouping of Dichelobacter nodosus from Victorian farms using conventional multiplex polymerase chain reaction. BMC Res Notes. (2018) 11:108. doi: 10.1186/s13104-018-3229-5
38. Bates D, Machler M, Bolker BM, Walker SC. Fitting linear mixed-effects models using lme4. J Stat Softw. (2015) 67:1–48. doi: 10.18637/jss.v067.i01
39. RStudio Team. RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. (2020). Available online at: http://www.rstudio.com/ (accesse June 29, 2021).
40. Bolker B, Robinson D. broom.mixed; Tidying Methods for Mixed Models. (2020). Available online at: https://CRAN.R-project.org/package=broom.mixed (accessed April 01, 2021).
41. Green PJ. Reversible jump Markov chain Monte Carlo computation and Bayesian model determination. Biometrika. (1995) 82:711–32. doi: 10.1093/biomet/82.4.711
42. Kaler J, George TRN, Green LE. Why are sheep lame? Temporal associations between severity of foot lesions and severity of lameness in 60 sheep. Anim Welfare. (2011) 20:433–8.
43. Egerton JR, Roberts DS. Vaccination against ovine foot-rot. J Comp Pathol. (1971) 81:179–85. doi: 10.1016/0021-9975(71)90091-0
44. Stewart DJ, Claxton PD. Ovine foot rot: clinical diagnosis and bacteriology. In: Corner LA, Bagust TJ, editors. Australian Standard Diagnostic Techniques for Animal Diseases. East Melbourne, VIC: CSIRO (1993).
45. Egerton JR, Roberts DS, Parsonson IM. The aetiology and pathogenesis of ovine foot-rot: I. A histological study of the bacterial invasion. J Comp Pathol. (1969) 79:207–16. doi: 10.1016/0021-9975(69)90007-3
46. Buller N, Eamens G. Ovine Footrot—Australian and New Zealand Standard Diagnostic Procedure. Australian Government Department of Agriculture (2014). Available online at: https://www.agriculture.gov.au/animal/health/laboratories/procedures/anzsdp/ovine-footrot (accessed January 21, 2020).
47. Kraft AF, Strobel H, Hilke J, Steiner A, Kuhnert P. The prevalence of Dichelobacter nodosus in clinically footrot-free sheep flocks: a comparative field study on elimination strategies. BMC Vet Res. (2020) 16–21. doi: 10.1186/s12917-020-2243-8
48. Schwartzkoff CL, Egerton JR, Stewart DJ, Lehrbach PR, Elleman TC, Hoyne PA. The effects of antigenic-competition on the efficacy of multivalent footrot vaccines. Aust Vet J. (1993) 70:123–6. doi: 10.1111/j.1751-0813.1993.tb06101.x
49. Egerton JR, Ghimire SC, Dhungyel OP, Shrestha HK, Joshi HD, Joshi BR, et al. Eradication of virulent footrot from sheep and goats in an endemic area of Nepal and an evaluation of specific vaccination. Vet Rec. (2002) 151:290–5. doi: 10.1136/vr.151.10.290
50. Gurung RB, Dhungyel OP, Tshering P, Egerton JR. The use of an autogenous Dichelobacter nodosus vaccine to eliminate clinical signs of virulent footrot in a sheep flock in Bhutan. Vet J. (2006) 172:356–63. doi: 10.1016/j.tvjl.2005.04.032
51. Dhungyel OP, Lehmann DR, Whittington RJ. Pilot trials in Australia on eradication of footrot by flock specific vaccination. Vet Microbiol. (2008) 132:364–71. doi: 10.1016/j.vetmic.2008.05.027
52. Dhungyel O, Schiller N, Eppleston J, Lehmann D, Nilon P, Ewers A, et al. Outbreak-specific monovalent/bivalent vaccination to control and eradicate virulent ovine footrot. Vaccine. (2013) 31:1701–6. doi: 10.1016/j.vaccine.2013.01.043
53. Locher I, Greber D, Holdener K, Luchinger R, Haerdi-Landerer C, Schupbach-Regula G, et al. Longitudinal Dichelobacter nodosus status in 9 sheep flocks free from clinical footrot. Small Ruminant Res. (2015) 132:128–32. doi: 10.1016/j.smallrumres.2015.10.021
54. Greber D, Bearth G, Luchinger R, Schuepbach-Regula G, Steiner A. Elimination of virulent strains (aprV2) of Dichelobacter nodosus from feet of 28 Swiss sheep flocks: a proof of concept study. Vet J. (2016) 216:25–32. doi: 10.1016/j.tvjl.2016.06.015
55. Vatn S, Hektoen L, Hoyland B, Reiersen A, Kampen AH, Jorgensen HJ. Elimination of severe footrot from the Norwegian sheep population—a progress report. Small Ruminant Res. (2012) 106:11–3. doi: 10.1016/j.smallrumres.2012.04.012
56. Gay NJ. The theory of measles elimination: implications for the design of elimination strategies. J Infect Dis. (2004) 189(Suppl. 1):S27–35. doi: 10.1086/381592
57. AHDB. UK Sheep Yearbook 2018. Agriculture and Horticulture Development Board. (2018). Available online at: http://beefandlamb.ahdb.org.uk/wp-content/uploads/2018/10/UK-Sheep-Yearbook-.pdf (accessed November 18, 2018).
Keywords: sheep, footrot, Dichelobacter nodosus, elimination, strains, persistence
Citation: Monaghan EM, Prosser NS, Witt J, Lewis KE, Nabb E, Keeling MJ, Purdy KJ and Green LE (2021) Impact of Strain Variation of Dichelobacter nodosus on Disease Severity and Presence in Sheep Flocks in England. Front. Vet. Sci. 8:713927. doi: 10.3389/fvets.2021.713927
Received: 24 May 2021; Accepted: 22 July 2021;
Published: 16 August 2021.
Edited by:
Subhash Verma, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaReviewed by:
Salome Dürr, University of Bern, SwitzerlandAndrew McPherson, The University of Sydney, Australia
Copyright © 2021 Monaghan, Prosser, Witt, Lewis, Nabb, Keeling, Purdy and Green. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Emma M. Monaghan, ZS5tb25hZ2hhbkBiaGFtLmFjLnVr
 Emma M. Monaghan
Emma M. Monaghan Naomi S. Prosser2
Naomi S. Prosser2 Katharine E. Lewis
Katharine E. Lewis Matt J. Keeling
Matt J. Keeling Kevin J. Purdy
Kevin J. Purdy Laura E. Green
Laura E. Green