- College of Animal Science and Technology, Guangxi University, Nanning, China
Enterovirus G (EV-G) infects porcine populations worldwide and the infections are generally asymptomatic, with the insertion of the papain-like cysteine protease gene (PLCP) increasing the potential public health threats. However, the genetic and pathogenic characteristics of EV-G itself are not fully understood as yet. In the present study, one EV-G strain, named CH/17GXQZ/2017, was isolated and purified from piglets with diarrheic symptoms from the Guangxi Province, China. This strain produced stable cytopathic effects on Marc-145 cells with a titer of 5 × 106 PFU/mL. The spherical enterovirus particles with diameters of 25–30 nm were observed by using transmission electron microscopy. The whole genome sequence of the CH/17GXQZ/2017 strain consists of 7,364 nucleotides, and the phylogenetic tree based on the amino acid sequences of VP1 indicated this strain was clustered to the G1 genotype. Seven-day-old piglets were inoculated orally with the CH/17GXQZ/2017 strain in order to evaluate its pathogenicity. Although none of the infected piglets died during the experiment, clinical neurological symptoms were observed manifesting as mild hyperemia and Nissl bodies vacuolization in the cerebrum. In addition, the infection with the CH/17GXQZ/2017 strain decelerated the weight gain of suckling piglets significantly. This study demonstrates that CH/17GXQZ/2017 is pathogenic to neonatal piglets and advance knowledge on the biological characteristics, evolution and pathogenicity of EV-G.
Introduction
Enterovirus species include viruses that infect humans (species A~D), bovine (species E and F), swine (species G) and non-human primates (A, B, D, H, and J) (1, 2). Porcine enteroviruses (PEV) were originally divided into 13 serotypes (PEV-1 to−13) (3, 4), and then subdivided into three genera Teschovirus, Sapelovirus and Enterovirus (5, 6). PEV-1 to 7 and PEV-11 to 13 were reclassified to the genus Teschovirus, PEV-8 to the genus Sapelovirus (5, 7), and PEV-9 and PEV-10 were reclassified as Enterovirus G (EV-G) (8, 9). EV-G is the member of the family Picornaviridae of the order Picornavirales, which currently comprises 20 genotypes (EV-G-1 to−20) (10– 14) (https://www.picornaviridae.com/sg3/enterovirus/ev-g/ev-g.htm).
EV-G is a non-enveloped virus with a small positive-sense single stranded RNA genome. The genome of EV-G is ~7,400–7,500 nucleotides (nt) and possesses a single open reading frame (ORF) flanked by untranslated regions. The ORF is processed proteolytically into four structural proteins (VP1, VP2, VP3, and VP4) and seven non-structural proteins (2Apro, 2B, 2C, 3A, 3B, 3Cpro, and 3Dpol) (14–17). The UTR at both ends of the genome are required for enteroviral RNA initiation. The 5′-UTR is ~700–825 nt in length, containing secondary structural elements which are important for RNA replication as well as an internal ribosomal entry site (IRES) for the initiation of translation (18). The 3′-UTR consists of 75 to 100 nt and contains complex cis-acting elements essential for RNA replication (19).
EV-G infects porcine populations worldwide and the infection is generally asymptomatic (20). Although piglets inoculated orally with EV-G positive fecal samples can cause flaccid paralysis of the hind limbs (21), there is little evidence to confirm the correlation between EV-G infection and clinical diarrhea (11). Novel EV-G1 variants carrying the porcine torovirus (ToV) papain-like cysteine protease gene (PLCP) in the 2C/3A junction region of their genomes were first found in fecal samples from pigs with diarrheic diseases in the USA (22). There is increasing evidence to suggest that the lateral acquisition of the PLCP gene might represent a novel host immune control strategy for EV-Gs to establish a pathogenic potential under certain circumstances, and the recombination between EV-G and ToV implies there is a risk of cross-species transmission (23).
The prototype of EV-G was isolated in 1973 and 1975 from the UK (24). It is relatively small and spherical with a diameter of ~25–30 nm. It occurs without cysts and the nucleocapsid has an icosahedral symmetry (25). Most of the complete genome sequences of EV-G were obtained by next generation sequencing (NGS) (22, 26, 27), whereas studies regarding virus isolation is limited. Recently, the insertion of PLCP were shown to cause changes in the innate immune signal of EV-G, but the evolutionary genetic and pathogenic characteristics of EV-G itself are not yet fully understood. In the present study, one EV-G strain, named CH/17GXQZ/2017, was isolated and purified. The genetic characteristics as well as the pathogenesis of the CH/17GXQZ/2017 strain in 7-day-old piglets was investigated.
Materials and Methods
Specimen Collection and Cell Cultures
In November 2017, stool specimens were obtained from piglets with diarrhea were collected from a farm in Qinzhou City, Guangxi Province, China. Marc-145 cells were grown in Minimum Essential Medium (MEM) supplemented with 10% fetal bovine serum (FBS) (Biological Industries, Kibbutz Beit Haemek, Israel) and antibiotics (100 units/mL of penicillin and 100 units /mL of streptomycin) at 37°C in a humidified incubator in an atmosphere of 5% CO2.
Detection of Samples
The stool samples were diluted 5 times in phosphate buffered saline (PBS) and the supernatants were collected after being vortexed and centrifuged at 12,000 × g at 4°C for 10 min. Total RNA was extracted from the supernatants by using a Viral DNA/RNA Miniprep Kit (Axygen Scientific, Union City, USA). RT-PCR was used to detect porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine rotavirus (PoRV), porcine delta coronavirus (PDCoV), porcine kobuvirus (PKV) and EV-G, in all the samples collected and only EV-G was found. The samples were then passed through 0.22 μm filters (Millipore, Billerica, MA, USA) and the filtrates were used for virus isolation.
Virus Isolation
EV-G was propagated in Marc-145 cells and the virus particles were isolated using trypsin as previously described (13), with minor modifications. Briefly, the monolayers of Marc-145 cells were grown in six-well plates. Growth media were then removed and washed three times with PBS before inoculation. Five hundred microliter of sample inoculum was adsorbed for 60 min at 37°C, EV-G growth medium consisting of MEM supplemented with antibiotics (100 units/mL of penicillin and 100 μg/mL of streptomycin) and 0.5 μg/mL of trypsin was added without removing the inoculum. Cells were incubated at 37°C in a 5% CO2 atmosphere, and examined daily for cytopathic effects (CPE). When CPE appeared in more than 80% of cells (~5 days after inoculation), the cells were harvested and subjected to three freeze-thaw cycles and then stored at −80°C.
Plaque Assay
The EV-G isolates were purified by using a plaque method in Marc-145 cells. The cell monolayers were infected with 200 μL of a 10-fold dilution of virus particles for 1 h at 37°C, and shaken gently every 15 min. Then the virus inoculum was removed and washed three times with PBS, following which it was overlaid with 2 mL of MEM containing 1% agarose and 0.5 μg/mL trypsin. The overlaid medium was solidified by keep the cell plates at room temperature (~20°C) for 20 min. The plates were then inverted and incubated at 37°C in a 5% CO2 incubator. After 4 days of incubation, the cells were fixed in 10% formaldehyde for 12 h, and the cells were stained with crystal violet for the plaques to be visualized. The strain of EV-G, subsequently named CH/17GXQZ/2017, was successfully obtained after three series of purification by the plaque assay.
Immune Serum Preparation and Indirect Immunofluorescence Assay
Since most picornaviruses are transmitted via the oral route (28), and it was demonstrated that antibody levels in serum were higher in mice immunized with live virus orally compared to peritoneally (29), the EV-G immune serum was prepared by oral administration of the live virus. Three-month-old Kunming mice were orally inoculated with 5 × 105 PFU of EV-G strain CH/17GXQZ/2017 in a volume of 100 μL as previously described (29, 30) with a few modifications. Serum was collected at 21 days post-infection (dpi) and stored at −80°C. Non-immune sera were obtained from naïve mice. This was subsequently used for the IFA and immunohistochemistry (IHC) test.
Marc-145 cell monolayers were inoculated with EV-G CH/17GXQZ/2017 strain at a multiplicity of infection (MOI) of 0.01. At 48 h post-inoculation (hpi), cells were washed three times with PBS, followed by fixation in cold acetone at −4°C for 30 min. The cells were washed three times with PBS and then blocked with 1% BSA (Roche, Mannheim, Germany) for 60 min at room temperature (~20°C). After being washed with PBS, the cells were incubated with the mouse anti-EV-G serum as prepared above (1:100) for 1 h at 37°C. Then the cells were washed with PBS three times followed by incubation with Alexa Fluor® 488 conjugated goat anti-mouse IgG (H + L) (Abcam Inc., USA) (1:4,000) for 1 h at 37°C. The cells were subsequently washed five times with PBS. Finally, images were captured using an inverted fluorescence microscope (Nikon, Tokyo, Japan).
Viral Replication Kinetics in Marc-145 Cells
Marc-145 cell monolayers grown in 12-well plates were infected with EV-G CH/17GXQZ/2017 strain at 0.01 MOI. After adsorption at 37°C for 60 min, MEM containing 0.5 μg/mL trypsin was added, and the plates were incubated at 37°C with 5% CO2. Cell supernatants were harvested in triplicates at 6, 12, 24, 36, 48, 60, 72, and 84 hpi, by centrifugation at 12,000 × g for 10 min at 4°C, and used for virus titration by using the plaque assay. Virus titration at each time points was carried out in three replicates.
Transmission Electron Microscope Observation of the Virus
Marc-145 cells were infected with EV-G CH/17GXQZ/2017 strain at 0.01 MOI, and when the CPE was more than 80%, the cell culture supernatants were collected by centrifugation at 10,000 rpm at 4°C for 1 h. The supernatants were filtered through 0.22 μm filters and mixed with 10% polyethylene glycol 8,000 (Solarbio, Beijing, China), precipitated and stirred gently at 4°C overnight. The virus particles were precipitated by centrifugation at 12,000 rpm at 4°C for 2 h, and re-suspended with 1 mL of Tris-buffered saline solution (Solarbio, Beijing, China). Then negative staining with 2% phosphotungstic acid was performed and these were imaged using a transmission electron microscope (Hitachi TEM system, Japan, HT7800).
Sequencing and Data Analysis
The whole genome of EV-G stain CH/17GXQZ/2017 was obtained from three overlapping fragments which were amplified by using three pairs of primers. The primers have been described previously with some of modifications (10) (Table 1). The viral genomic RNA was extracted from the EV-G stock samples using the Axy Prep™ Viral DNA/RNA Miniprep Kit (OMEGA, USA) according to the manufacturer's instructions (31). The cDNA was synthesized in a total volume of 25 μL containing 16 μL of RNA solution, 2 μL of dNTP mixture, 0.5 μL of reverse transcriptase M-MLV, 0.5 μL of RNasin inhibitor, 1 μL of oligo (dT) or the reverse primer, and 5 μL of 5 × reverse transcriptase M-MLV buffer. The mixture was incubated at 42°C for 60 min and then chilled on ice to destroy the RNA secondary structure. PCR was performed using the PrimeSTAR Max DNA Polymerase (TaKaRa, Dalian, China). The size of PCR products was checked by electrophoresis in 1.0% agarose gel after staining with GelRedTM (Biotum Inc., USA) and visualization by using an UVItec imager (Bio-Rad Laboratories Inc., USA). The expected DNA band was purified and cloned into pEASY-Blunt Cloning vector (TransGen, Beijing, China) and sequenced by Beijing Genomics Institute, Shenzhen.
The chromatograms were checked and edited by the SeqMan program. The nucleotide sequences were aligned by the MegAlign software (Lasergene, DNASTAR Inc., USA). A phylogenetic tree was constructed by using the maximum likelihood method in MEGA5.0 software.
Quantitative Real-Time PCR
Primers were designed (forward primer: 5′-CTTGTGCATCCGGTTATGCC-3′ and reverse primer: 5′-GCAAGCAGGCACAGAGAACGC-3′) using Primer Premier 5.0 software. The recombinant plasmid was constructed by cloning the PCR fragments into a pMD18-T vector (TaKaRa, Dalian, China) and calculating the resulting copy numbers. A 10-fold series of diluted recombinant plasmids in triplicates with known copy numbers were used as templates to generated a standard curve for quantitative PCR. The reaction mixture for qRT-PCR contained 2 μL cDNA, 10 μL TB GreenTM Premix Ex TaqTM II (Takara, Dalian, China), 1.0 μL (10 pmol/μL) of each primer and ddH2O. Reactions were performed in triplicates. The amplification conditions were as follows: 95°C for 2 min, 40 cycles of 95°C for 15 s, 60°C for 1 min and a melting-curve analysis was included in the end. The qRT-PCR was performed in 96-well plates by using a LightCycler 96 (Roche, Mannheim, Germany).
Experimental Infection of 7-Day-Old Piglets
A total of eight 7-day-old piglets (Duroc × Landrace × Yorkshire) which were nucleotide negative for foot-and-mouth disease virus (FMDV), classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), PEDV, TGEV, PoRV, and EV-G were purchased from a commercial farm. Piglets of either sex were randomly assigned to two groups of four piglets. The piglets in the infected group (n = 4) were orally inoculated with 2 mL of the EV-G CH/17GXQZ/2017 strain with titers of 5 × 106 PFU/mL. The same amount of cell culture media was inoculated to the piglet in mock group and these were used as controls. Milk powder (Shenzhen Premixinve Nutrition Co. Ltd., China) were used to feed the animals four times a day and they were allowed water, ad libitum. Clinical observations, weight and rectal temperatures were recorded daily from 0 to 7 dpi.
Fecal swabs were collected at hpi of 0, 12, 24, 36, 48, 60, 72, 84, 96, 108, 120, 132, 144, 156, and 168. Pigs were euthanized at 7 dpi and the tissues of the brain, spinal cord, stomach, duodenum, jejunum, ileum, cecum and colon were collected at necropsy. Tissues were homogenized at a concentration of 0.1 g/mL which were used for viral load detection by qPCR. Some tissue samples were also immediately fixed in 10% neutral buffered formalin for subsequent histological examination. According to previous studies (32) with minor modification, the fixed brain tissue sections were trimmed, processed and embedded in paraffin wax, stained with Nissl and then analyzed for histopathological changes. Diluted mouse anti-EV-G serum (1:100) was used for the detection of EV-G antigen with goat anti-mouse IgG conjugated to horseradish peroxidase (HRP) (Servicebio, China) (1:200) being used as secondary antibody. The animal study was reviewed and approved by the Animal Care and Welfare Committee of Guangxi University (Permission Number: GXU2020-021).
Statistical Analysis
All the values are expressed as the means ± standard deviations (SD) or standard error of the means (SEM) of four piglets (33). Data were analyzed using the two-tailed t-test. Statistically significant was set to a P-value of 0.05.
Results
Isolation and Purification of EV-G Strain
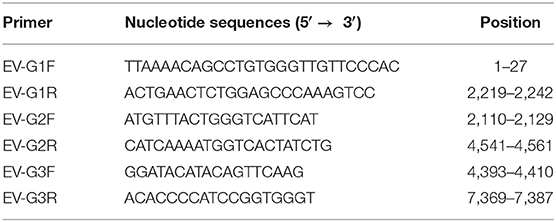
At passage 4, typical CPE of Marc-145 cells was characterized by cell shrinking into round structures. In addition, cell layer splitting and shedding were observed at 48 hpi (Figure 1B). The virus was purified by subjecting the samples to three series of plaque assays (Figure 1D). Neither CPE nor viral plaques were observed in the mock cells (Figures 1A,C) One strain of EV-G, named CH/17GXQZ/2017, was successfully isolated, and the estimated titer was 5 × 106 PFU/mL.
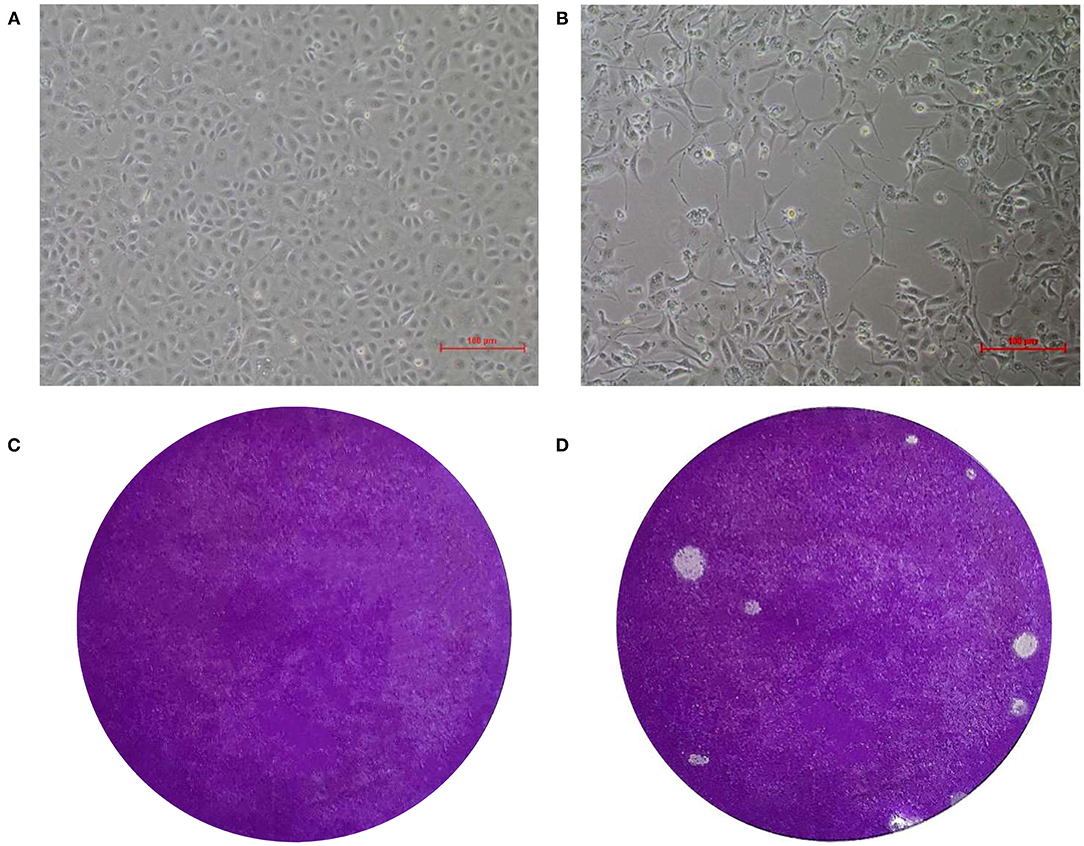
Figure 1. CPE and plaque formation of the EV-G CH/17GXQZ/2017 strain. Neither CPE nor viral plaques were observed in the mock cells (A,C). CPE caused by the EV-G CH/17GXQZ/2017 strain in Marc-145 cells was characterized by cell shrinkage, rounding and detachment at 48 hpi (200 × magnification) (B). Viral plaques of EV-G isolates of CH/17GXQZ/2017 developed at 4 dpi (D).
Immunofluorescence Assay and Biological Characteristics of the EV-G CH/17GXQZ/2017 Strain
Specific green fluorescence was seen in Marc-145 cells infected with the EV-G CH/17GXQZ/2017 strain by using an IFA (Figure 2B). This indicated that the EV-G CH/17GXQZ/2017 strain could react with the specific mouse anti-EV-G serum (Figure 2A). No green fluorescence was observed using normal mouse serum control or in the mock cells.
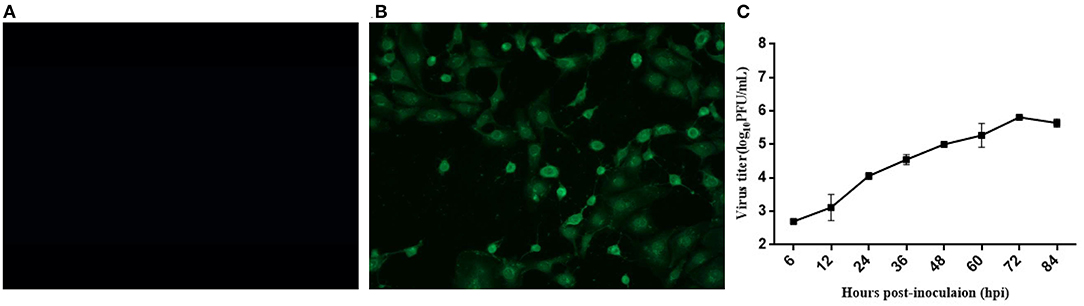
Figure 2. Immunofluorescence assay and growth kinetics of the EV-G CH/17GXQZ/2017 strain in Marc-145 cells. Negative control (A). IFA staining in Marc-145 cells infected with the EV-G CH/17GXQZ/2017 strain at 48 hpi with mouse anti-EV-G serum (1:100) and Alexa Fluor® 488 conjugated goat anti-mouse IgG (H + L) (1:4,000) which were used as primary and secondary antibodies, respectively. This shows that the specific green fluorescence was mainly localized in the cytoplasm of infected cells (200 × magnification) (B). Replication kinetics of the EV-G CH/17GXQZ/2017 strain in Marc-145 cells was determined by plaque assays. Marc-145 cells were inoculated with the EV-G CH/17GXQZ/2017 strain at 0.01 MOI, and the cell culture supernatants were harvested at 6, 12, 24, 36, 48, 60, 72, and 84 hpi, separately. The virus titers are expressed as PFU per milliliter and are shown as the mean values with standard deviations from three replicates (C).
Marc-145 cells were infected with the EV-G CH/17GXQZ/2017 strain at 0.01 MOI and the growth curve of the virus was determined (Figure 2C). The mean titers of three independent measurements at each time point at 6, 12, 24, 36, 48, 60, 72, and 84 hpi were measured. The number of viral plaques of strain CH/17GXQZ/2017 continued to rise from 6 to 72 hpi, and reached a peak at 72 hpi with a titer of 6.5 × 105 PFU/mL.
A typical Picornavirales morphology was observed for the EV-G CH/17GXQZ/2017 strain when the virus particles were observed through TEM. They appeared small and spherical and without cysts. Their average diameter was ~25–30 nm (Figure 3).
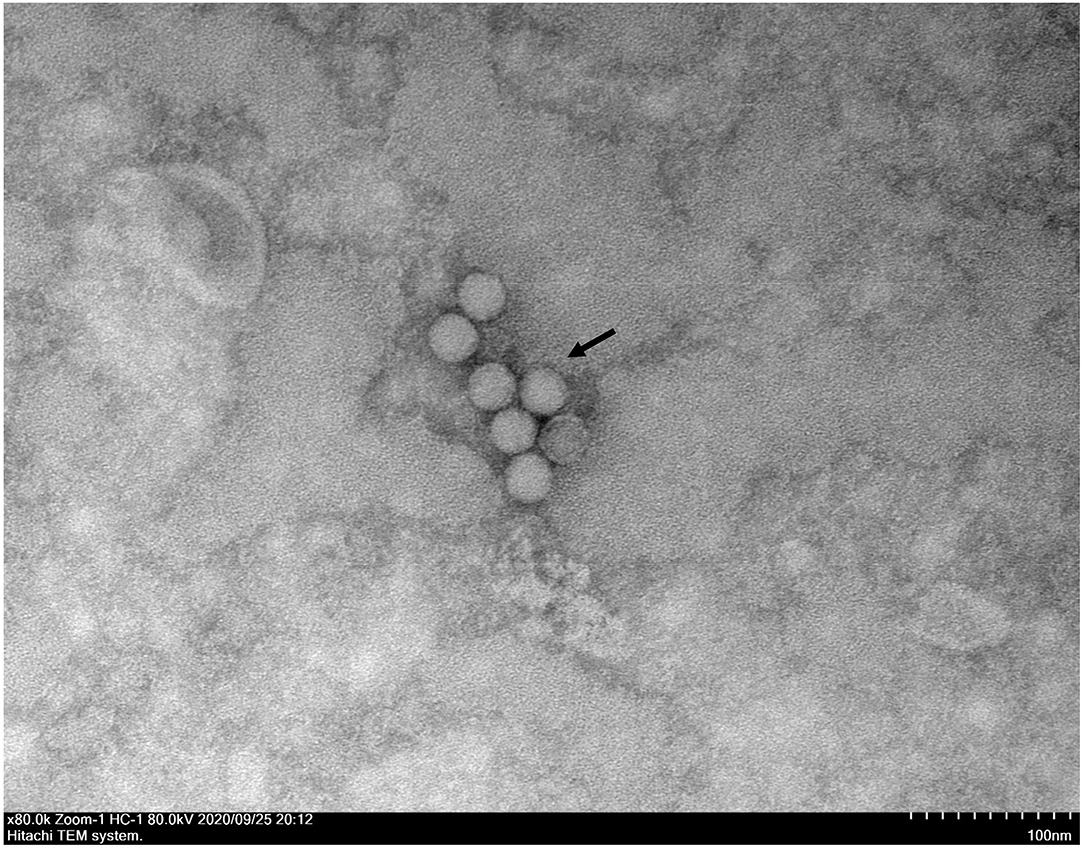
Figure 3. A transmission electron microscopic image of purified EV-G particles. Spherical virions with a diameter of 25–30 nm were observed and marked with a black arrow. The sample was negatively stained with 2% phosphotungstic acid. Scale bar indicates 100 nm divisions.
Whole Genome Amplification of the EV-G CH/17GXQZ/2017 Strain
The complete genome sequence of the EV-G CH/17GXQZ/2017 strain was successfully obtained from three overlapping fragments by RT-PCR analysis (Figure 4). The complete genome size of the CH/17GXQZ/2017 strain was 7,364-nt and consisted of 821-nt of 5′-UTR, 72-nt of 3′-UTR and 6,471-nt of ORF, the ORF maps between positions 822 and 7,292 and encoded a 2,157-amino-acid polyprotein. The CH/17GXQZ/2017 strain sequence was deposited in the GenBank database under the Accession Number, MT274669.
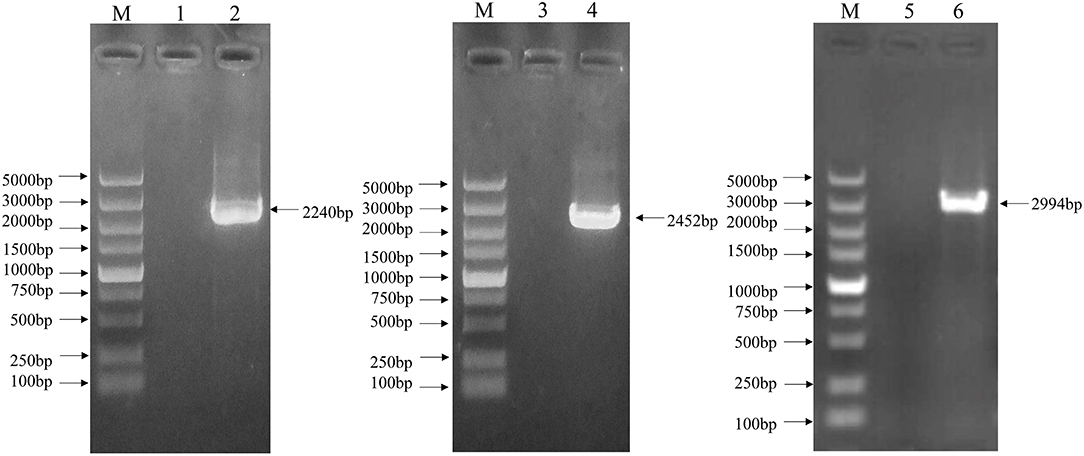
Figure 4. The complete genome identification of the EV-G CH/17GXQZ/2017 strain by RT-PCR. Amplification of the complete genome of the EV-G CH/17GXQZ/2017 strain. Lane M, DNA marker DL5000; Lanes 1, 3, and 5, negative controls; Lanes 2, 4, and 6, three overlapping PCR products of viral genome from the EV-G CH/17GXQZ/2017 strain.
Sequence Analysis of the EV-G CH/17GXQZ/2017 Strain
The nucleotide and amino acid sequence identities of the 5′-UTR, VP4, VP3, VP2, VP1, 2A, 2B, 2C, 3A, 3B, 3C, 3D, and 3′-UTR of CH/17GXQZ/2017 were compared with the strains of UKG/410/73, KOR/KUN-1811/2018/G1-PLCP and EVG/Porcine/USA/Texas2/2014/G1-PLpro (Table 2). The genome organization of CH/17GXQZ/2017 is similar to the prototype the EV-G UKG/410/73 strain, and it shows whole genome nucleotide similarities of 79.8, 77.9, and 77.2% to the strains of UKG/410/73, KOR/KUN-1811/2018/G1-PLCP and EVG/Porcine/USA/Texas2/2014/G1-PLpro, respectively.
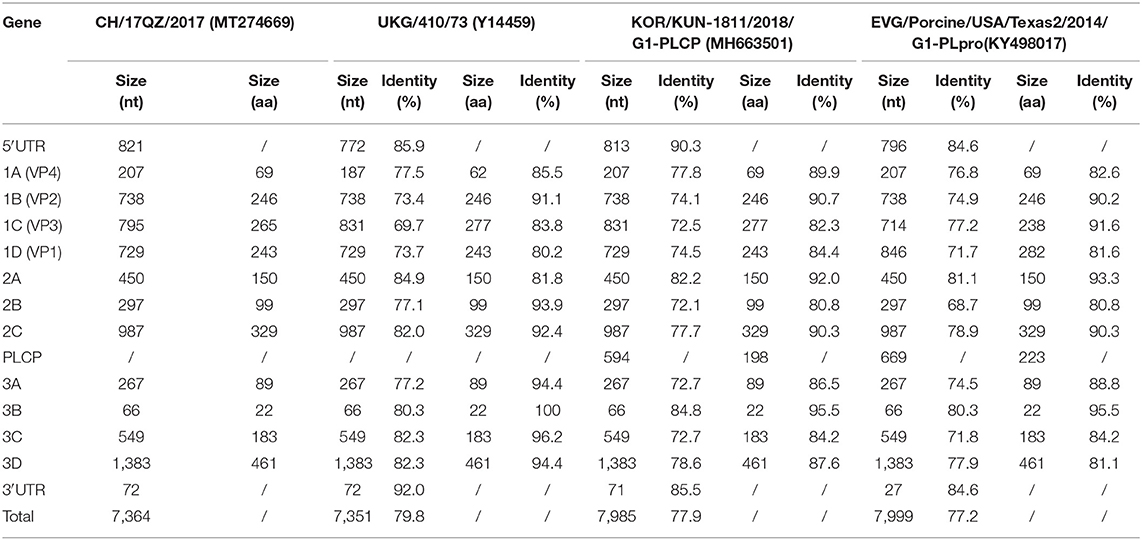
Table 2. Nucleotide and amino acid identities of CH/17GXQZ/2017 when compared with EV-G reference strains.
Compared to strain UKG/410/73, the strain of CH/17GXQZ/2017 had only 80.2% amino acid homology in the VP1 gene, and it had 36 nucleotide deletions in the area of 2488~2529bp in the VP3 gene and 21 nucleotide insertions in the area of 1009~1028bp in the VP4 gene, respectively. The 3B gene of EV-G was found to be relatively conservative, and it exhibited the highest amino acid homology of up to 100% when compared with UKG/410/73, 95.5% identity when compared to the strains of KOR/KUN-1811/2018/G1-PLCP and EVG/Porcine/USA/Texas2/2014/G1-PLpro, respectively.
The strain of KOR/KUN-1811/2018/G1-PLCP and EVG/Porcine/USA/Texas2/2014/ G1-PLpro had 594 and 669 bp PLCP gene insertions at the 2C/3A junction region, respectively. Although there was no PLCP insertion in the strain of CH/17GXQZ/2017, it is noteworthy that the length of 5′-UTR of CH/17GXQZ/2017 was slightly longer than all three reference strains, due to the continuous insertion of the thirty-two A bases at position of 687–718 bp.
Phylogenetic Analysis of the EV-G CH/17GXQZ/2017 Strain
EV-G classification is solely based on sequence identities of the VP1 gene and the phylogenetic tree based on the VP1 gene amino acid sequences showed that the CH/17GXQZ/2017 strain was clustered into the G1 subtype (Figure 5A). Phylogenetic analysis based on the complete genome nucleotide sequences revealed that the four strains (KY498017, MH663501, LC316782, and LC316775) were clustered to a separate group of G1 type due to the insertions of PLCP (Figure 5B). This indicated that the strain of CH/17GXQZ/2017 had a closer genetic relationship with the other three G1 type strains, USA/13-03212/2013 (KF985175), CHN/Ch-ah-f1/2010 (HM131607), and EVG/Porcine/JPN/Iba464-3-1/2015/G1 (LC316790), with the overall nucleotide similarities of 82.9, 82.8, and 81.8%, respectively.
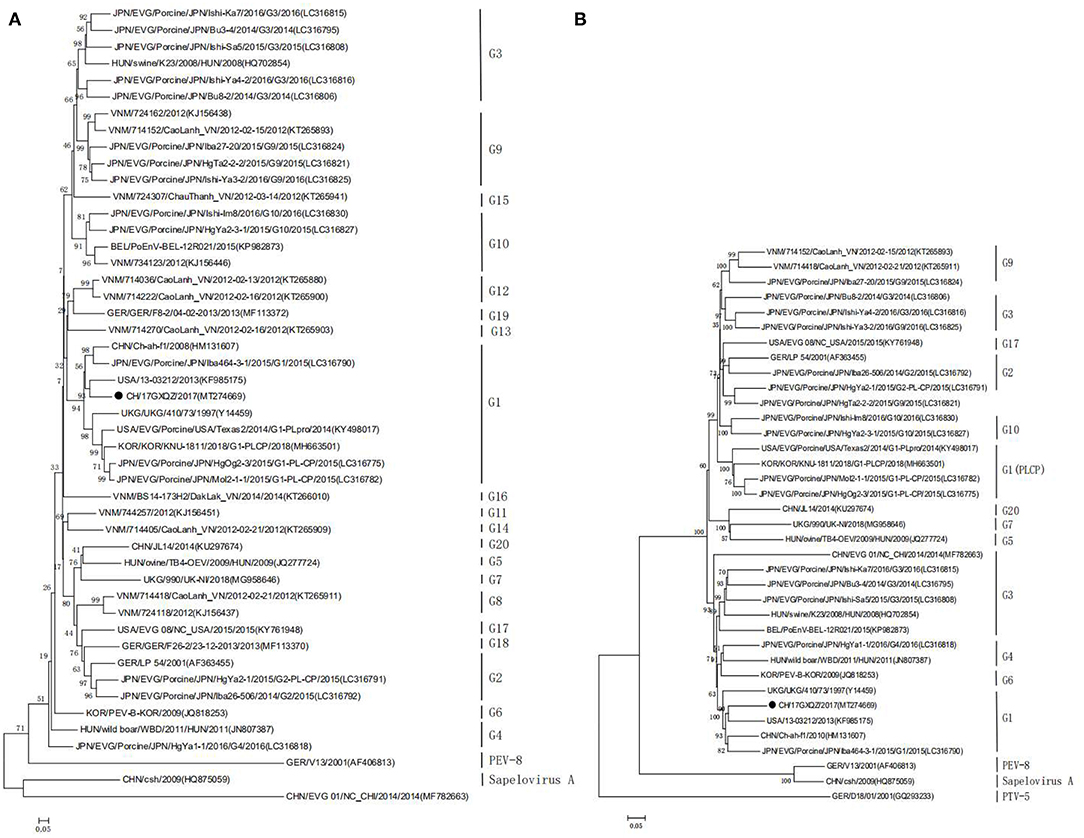
Figure 5. Phylogenetic analyses of the EV-G CH/17GXQZ/2017 strain. Phylogenetic analyses based on the amino acid sequences (A) of VP1 and the complete genome sequences (B) of the EV-G CH/17GXQZ/2017 strain with reference EV-G sequences obtained from the GenBank. The phylogenetic trees were constructed using the maximum likelihood method in MEGA 5.0 software. Bootstrap analysis was carried out on 1,000 replicate datasets and values are indicated adjacent to the branching points. The genetic distances were calculated using the Tamura-Nei model. The EV-G CH/17GXQZ/2017 strain obtained in this study is indicated by solid circles. The scale bars indicate the amino acid or nucleotide substitutions at each site.
Pathogenicity of the EV-G CH/17GXQZ/2017 Strain on 7-Day-Old Piglets
In the virus inoculated group, all four piglets exhibited trembling motions of the hind limbs at 2 dpi, which persisted for 1–2 days. Two piglets developed neurological symptoms which manifested as excitation and they spun around in circles at 5–6 dpi. One piglet developed mild diarrhea at 4–7 dpi. No piglets exhibited any clinical signs in the mock group.
All piglets had normal rectal temperatures at 0–7 dpi (Figure 6A). Piglets in the CH/17GXQZ/2017 infected group gained less weight than those in the mock group. The weight gain rate in piglets from the infected group was significant less than those in the mock group at 1, 2, 6, and 7 dpi, with 3.5, 7, 11.5, and 11% less weight gain rate, respectively (Figure 6B).
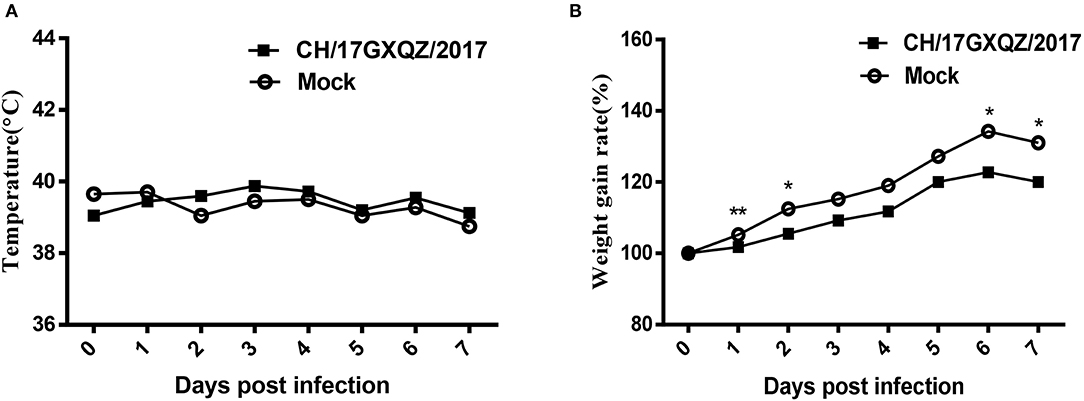
Figure 6. Trends of rectal temperatures and weight gain of 7-day-old piglets inoculated with EV-G. Normal rectal temperatures were present in both the infected and mock groups (A). The weight gain rates of piglets were analyzed, those in the infected group was significant less than in pigs from the mock group at 1, 2, 6, and 7 dpi. Each point on the graph represents the average values from the four pigs. * and ** indicate P < 0.05 and P < 0.01, respectively, between the two groups (B).
Fecal swab samples were collected from which the viral copies were detected by qRT-PCR. The results showed that genomic copies of EV-G increased rapidly between 12 and 48 h and the viral load was at 104.9-105.3 copies/mL within 48–168 h (Figure 7A). EV-G genomic copies in the tissue samples of the brain, spinal marrow, stomach, duodenum, jejunum, ileum, cecum and colon were detected, and these showed that the genomic copies of EV-G in cecum and colon were 105.6 and 105.1 copies/mL, respectively, which were magnitudes higher than those in the small intestine (Figure 7B).
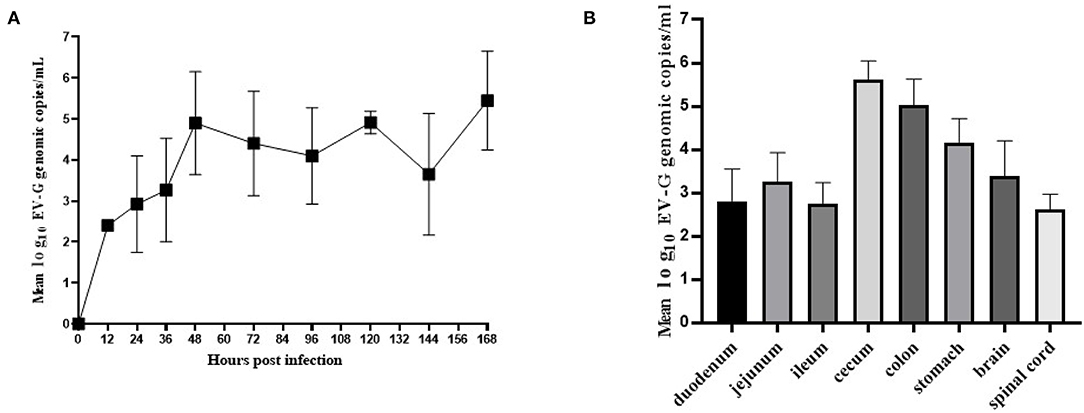
Figure 7. The viral shedding in fecal swabs and the viral distribution in different tissues in EV-G infected piglets. The fecal swabs were collected from piglets at 0–168 hpi. The EV-G shedding in fecal swab samples from infected piglets were detected (A). Intestinal segments and other tissues (stomach, brain, spinal cord) were collected at 7 dpi. The EV-G viral RNA loads were detected (B). Each point or histogram shown are the mean values ± SD (error bars) from four piglets.
Brain necropsy examinations of the piglets was performed and mild hyperemia was observed in the cerebrum of infected piglets. Histopathological examination showed that the degeneration and necrosis of the neurons led to an increase in nearby glial cells and vacuolization as well as fusion of the Nissl bodies. There was also uneven dyeing of the Nissl bodies and the cell nuclei were hyperchromatic (Figure 8). However, no cerebrum lesion or pathological change were found in the mock group.
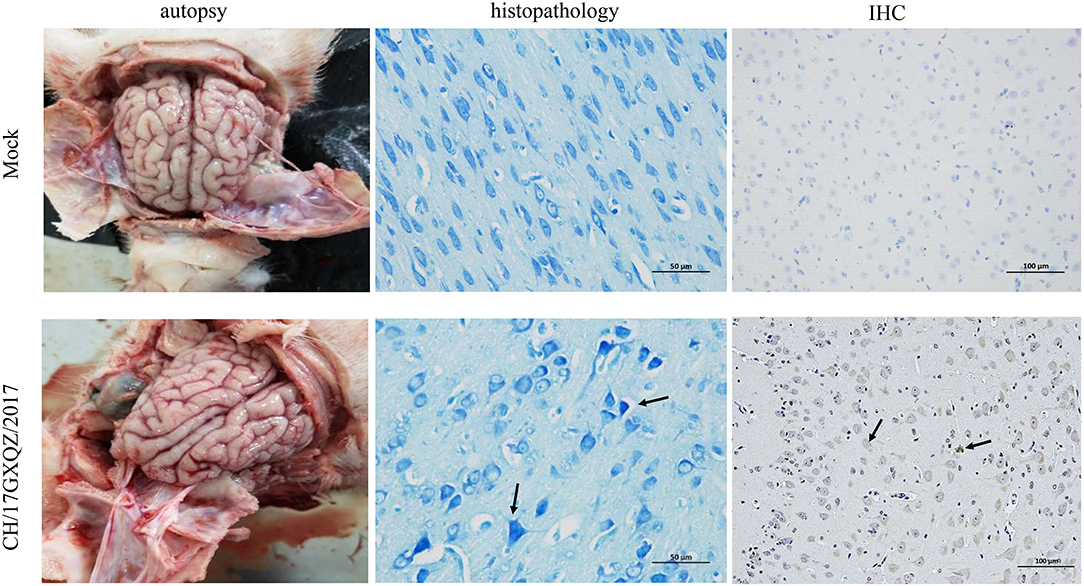
Figure 8. Macroscopic, histologic lesions and immunohistochemistry of the cerebrum as caused by EV-G. Macroscopic observations of the brain of one representative piglet from the mock and the inoculated groups. The microscopic lesions observed in the cerebrum of piglets at 400 × magnification with Nissl staining of a section in one representative piglet from the mock and infected groups, and the black arrows in micrograph indicate Nissl bodies vacuolization, fusion and uneven dyeing. Immunohistochemical staining of cerebrum sections from the mock and EV-G-infected groups. EV-G antigen was detected (marked with black arrows) in the brain of EV-G-inoculated pigs. The neuronal cells of EV-G-inoculated piglets were stained brown and observed at 200 × magnification.
According the results of IHC, the EV-G antigen appeared in the cytoplasm and cell nuclei of neuronal cells in the infected pigs. The antigen labeling was observed as brownish-yellow granules with a diffuse distribution (Figure 8), and no antigenic marker was found in the mock group.
Discussion
In recent years, a natural recombination insertion of the PLCP gene in enterovirus was found in the United States, South Korea, Japan, Belgium and China. Most of recombinant EV-G-PLCP strains were detected from fecal samples taken from these animals indicating that EV-G-PLCP was the potential causative agent for porcine diarrhea. A previous study has shown that when the PLCP gene was inserted into the EV-G genome, it could potentially suppress the host cellular innate immune response (16). It was reported that the recombinant events which occurred in EV-G1, EV-G2, and EV-G17 genotypes are rare, but they seem to occur more frequently in EV-G1 genotype, and this might contribute significantly evolution of the virus (23). Therefore, the finding of PLCP gene insertions in EV-G can increase the potential public health threats. There was no insertion of the PLCP gene in the CH/17GXQZ/2017 strain in this study, but successive inserts of 32 A bases at a downstream region of the IRES in the 5′-UTR of CH/17GXQZ/2017 strain were found. The 5′-UTR of EV-G is important for RNA replication due to the fact that it contains secondary structural elements, although it is not clear what the effects of continuous insertion of A bases at the 5′-UTR of enterovirus would be, and this phenomenon deserves to be tested and studied further.
The VP1 gene is important for EV-G genotyping, because the main part of the VP1 protein is exposed to the surface of the capsid and it is the main component that determines the antigenicity of the virus (http://www.picornaviridae.com/) (34). A phylogenetic tree was constructed based on the EV-G VP1 amino acid sequences, and it showed that the CH/17GXQZ/2017 strain clustered into the G1 genotype with EV-G1-PLCP strains. However, the G1-PLCP recombinant strains were clustered to a separate branch based on the analysis of the complete genome sequences. Based on the whole genome sequences, the strain of CH/17GXQZ/2017 in this study had the highest sequence homology, 82.9%, with the USA strain of 13-03212. The homology value was not particularly high, but this may be a reflection of the limited amount of complete sequences uploaded in the GenBank (35).
EV-G can be propagated in BHK-21, Vero, ST, and Marc-145 cells in the presence of trypsin (4). The first EV-G strain which was isolated in the United States used Marc-145 cells until 2013 (13). The Ch-ah-f1 strain is basically considered to be the representative strain in China, but the virus has yet to be isolated (10). EV-G-PLCP recombinant viruses were originally obtained using ST cells in 2015 (16). By analyzing the growth characteristics of the CH/17GXQZ/2017 strain on Marc-145 cells, we found its growth trend to be similar to that described previously (16), with the replication cycle being relatively long and the virus titer reaching its peak at 72 hpi. However, studies on the pathogenicity of EV-G in piglets are limited. In a previous study, 2-week-old piglets inoculated orally with the EV-G1 positive fecal samples showed no clinical signs, but the virus did cause pathological changes in the cerebrum and the lungs (21). In this study, the CH/17GXQZ/2017 strain was isolated and purified, which may be beneficial in order to provide more reliable data for the animal pathogenicity experiments of EV-G.
Some enteroviruses have been shown to have the ability to infect the central nervous system and cause various effects such as paralysis and ataxia (36). In our study, piglets infected with the EV-G CH/17GXQZ/2017 strain showed some clinical neurological symptoms, such as excitement, trembling of the hind-limbs and spinning in circles. In addition, samples of the brain and spinal cord from all four infected piglets were qRT-PCR-positive for EV-G and pathological lesions were observed in the brain. EV-G antigen was also detected in the brain by immunohistochemistry. The above results all indicated that EV-G had neurotropic effects, which is consistent with a previous report (21).
EV-G is prevalent and widespread in the general pig population in the middle and eastern China (21), and the infections tended to occur early, usually within the first week after birth (37). Pigs at >20 weeks of age had a lower frequency of EV-G infection compared to pigs of <15 weeks of age, which may be due to acquired immunity as the animals age (21). When 7-day-old piglets were infected with CH/17GXQZ/2017, it caused neurological symptoms and a reduced body weight. In addition, the majority of tissues from the infected piglets carried EV-G which could be detected at 7 dpi. The CH/17GXQZ/2017 strain was isolated from piglets with diarrhea, but mild diarrhea was only observed in one of the four piglets infected with this strain. Although no piglets died after being infected with the EV-G CH/17GXQZ/2017 strain during the entire experimental infection process, it is worth noting that EV-G infection can significantly affect the weight gain of suckling piglets which is particularly concerning for pig farmers.
In conclusion, an EV-G strain, named CH/17GXQZ/2017, was isolated and purified in this study. Several techniques were used to identify the biological characteristics of infection with this strain of EV-G including CPE, IFA, replication kinetics and TEM. The phylogenetic trees based on the amino acid sequences of VP1 indicated that the CH/17GXQZ/2017 strain was clustered to the G1 genotype. The pathogenicity of the CH/17GXQZ/2017 strain was determined in 7-day-old piglets, and clinical neurological symptoms manifested as mild hyperemia and Nissl bodies vacuolization in the cerebrum. In addition, infection with the CH/17GXQZ/2017 strain significantly decelerated weight gain in suckling piglets. This study provides a reference for the biological characteristics, evolution and pathogenicity of EV-G.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Welfare Committee of Guangxi University.
Author Contributions
KO, XM, and CY conceived and designed the experiment. XM, CY, YLu, HW, QQ, RC, ZC, and YLuo performed the experiments. KO, XM, CY, YC, ZW, and WH analyzed the data. KO and XM wrote the paper. All authors have seen and approved the final manuscript.
Funding
This work was supported by the Scientific Research Foundation of Guangxi University (Grant Number XGZ170239) and the One-Hundred Talent Program of Guangxi.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Dev Sooranna, Imperial college of London for English language edits of the manuscript. We thank Xueting Liu, Ruomu Wang, Tingting Yuan, and Yaxin Qu for excellent technical assistance.
References
1. Knowles NJ, Hovi T, Hyypiä T, King AMQ, Lindberg AM, Pallansch MA, et al. Virus taxonomy: classification and nomenclature of viruses. In: King AMQ, Adams MJ, Carstens EB, editor. Virus Taxonomy Classification and Nomenclature of Viruses Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Academic Press (2012). p. 855–80.
2. Nikonov OS, Chernykh ES, Garber MB, Nikonova EY. Enteroviruses: classification, diseases they cause, and approaches to development of antiviral drugs. Biochem Biokhimiia. (2017) 82:1615–31. doi: 10.1134/S0006297917130041
3. Auerbach J, Prager D, Neuhaus S, Loss U, Witte KH. Grouping of porcine enteroviruses by indirect immunofluorescence and description of two new serotypes. Zentralbl Veterinarmed B. (1994) 41:277–82. doi: 10.1111/j.1439-0450.1994.tb00228.x
4. Knowles NJ, Buckley LS, Pereira HG. Classification of porcine enteroviruses by antigenic analysis and cytopathic effects in tissue culture: description of 3 new serotypes. Arch Virol. (1979) 62:201–8. doi: 10.1007/BF01317552
5. Krumbholz A, Dauber M, Henke A, Birch-Hirschfeld E, Knowles NJ, Stelzner A, et al. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J Virol. (2002) 76:5813–21. doi: 10.1128/JVI.76.11.5813-5821.2002
6. Boros A, Nemes C, Pankovics P, Biro H, Kapusinszky B, Delwart E, et al. Characterization of a novel porcine enterovirus in wild boars in Hungary. Arch Virol. (2012) 157:981–6. doi: 10.1007/s00705-012-1255-5
7. Tseng CH, Tsai HJ. Sequence analysis of a duck picornavirus isolate indicates that it together with porcine enterovirus type 8 and simian picornavirus type 2 should be assigned to a new picornavirus genus. Virus Res. (2007) 129:104–14. doi: 10.1016/j.virusres.2007.06.023
8. Kaku Y, Sarai A, Murakami Y. Genetic reclassification of porcine enteroviruses. J Gen Virol. (2001) 82:417–24. doi: 10.1099/0022-1317-82-2-417
9. Zell R, Dauber M, Krumbholz A, Henke A, Birch-Hirschfeld E, Stelzner A, et al. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J Virol. (2001) 75:1620–31. doi: 10.1128/JVI.75.4.1620-1631.2001
10. Zhang W, Yang S, Shen Q, Ren L, Shan T, Wei J, et al. Complete genome sequence of a novel porcine enterovirus strain in China. J Virol. (2012) 86:7008–9. doi: 10.1128/JVI.00711-12
11. Van Dung N, Anh PH, Van Cuong N, Hoa NT, Carrique-Mas J, Hien VB, et al. Prevalence, genetic diversity and recombination of species G enteroviruses infecting pigs in Vietnam. J Gen Virol. (2014) 95:549–56. doi: 10.1099/vir.0.061978-0
12. Tsuchiaka S, Naoi Y, Imai R, Masuda T, Ito M, Akagami M, et al. Genetic diversity and recombination of enterovirus G strains in Japanese pigs: high prevalence of strains carrying a papain-like cysteine protease sequence in the enterovirus G population. PLoS ONE. (2018) 13:e0190819. doi: 10.1371/journal.pone.0190819
13. Anbalagan S, Hesse RA, Hause BM. First identification and characterization of porcine enterovirus G in the United States. PLoS ONE. (2014) 9:e97517. doi: 10.1371/journal.pone.0097517
14. Bunke J, Receveur K, Oeser AC, Fickenscher H, Zell R, Krumbholz A. High genetic diversity of porcine enterovirus G in Schleswig-Holstein, Germany. Arch Virol. (2018) 163:489–93. doi: 10.1007/s00705-017-3612-x
15. Ao D, Sun SQ, Guo HC. Topology and biological function of enterovirus non-structural protein 2B as a member of the viroporin family. Vet Res. (2014) 45:87. doi: 10.1186/s13567-014-0087-6
16. Shang P, Misra S, Hause B, Fang Y. A naturally occurring recombinant enterovirus expresses a torovirus deubiquitinase. J Virol. (2017) 91:e00450–17. doi: 10.1128/JVI.00450-17
17. Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Ann Rev Genet. (1993) 27:353–436. doi: 10.1146/annurev.ge.27.120193.002033
18. Beales LP, Rowlands DJ, Holzenburg A. The internal ribosome entry site (IRES) of hepatitis C virus visualized by electron microscopy. RNA (New York, NY). (2001) 7:661–70. doi: 10.1017/S1355838201001406
19. Zoll J, Heus HA, van Kuppeveld FJM, Melchers WJG. The structure-function relationship of the enterovirus 3′-UTR. Virus Res. (2009) 139:209–16. doi: 10.1016/j.virusres.2008.07.014
20. Doherty M, Todd D, McFerran N, Hoey EM. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J Gen Virol. (1999) 80:1929–41. doi: 10.1099/0022-1317-80-8-1929
21. Yang S, Wang Y, Shen Q, Zhang W, Hua X. Prevalence of porcine enterovirus 9 in pigs in middle and eastern China. Virol J. (2013) 10:99. doi: 10.1186/1743-422X-10-99
22. Knutson TP, Velayudhan BT, Marthaler DG. A porcine enterovirus G associated with enteric disease contains a novel papain-like cysteine protease. J Gen Virol. (2017) 98:1305–10. doi: 10.1099/jgv.0.000799
23. Lee S, Lee C. First detection of novel enterovirus G recombining a torovirus papain-like protease gene associated with diarrhoea in swine in South Korea. Transbound Emerg Dis. (2018) 66:1023–8. doi: 10.1111/tbed.13073
24. Knowles NJ. Isolation and identification of porcine enteroviruses in Great Britain, 1979 to 1980. Br Vet J. (1983) 139:19–22. doi: 10.1016/S0007-1935(17)30584-5
25. Lukashev AN. Role of recombination in evolution of enteroviruses. Rev Med Virol. (2005) 15:157–67. doi: 10.1002/rmv.457
26. Imai R, Nagai M, Oba M, Sakaguchi S, Ujike M, Kimura R, et al. A novel defective recombinant porcine enterovirus G virus carrying a porcine torovirus papain-like cysteine protease gene and a putative anti-apoptosis gene in place of viral structural protein genes. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. (2019) 75:103975. doi: 10.1016/j.meegid.2019.103975
27. Conceição-Neto N, Theuns S, Cui T, Zeller M, Yinda CK, Christiaens I, et al. Identification of an enterovirus recombinant with a torovirus-like gene insertion during a diarrhea outbreak in fattening pigs. Virus Evol. (2017) 3(2):vex024. doi: 10.1093/ve/vex024
28. Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, et al. ICTV virus taxonomy profile: picornaviridae. J Gen Virol. (2017) 98:2421–2. doi: 10.1099/jgv.0.000911
29. Kang BS, Yahikozawa H, Koh CS, Kim BS. Oral administration of live virus protects susceptible mice from developing Theiler's virus-induced demyelinating disease. Virology. (2007) 366:185–96. doi: 10.1016/j.virol.2007.04.017
30. Wu TC, Wang YF, Lee YP, Wang JR, Liu CC, Wang SM, et al. Immunity to avirulent enterovirus 71 and coxsackie A16 virus protects against enterovirus 71 infection in mice. J Virol. (2007) 81:10310–5. doi: 10.1128/JVI.00372-07
31. Lu Y, Su X, Du C, Mo L, Ke P, Wang R, et al. Genetic diversity of porcine epidemic diarrhea virus with a naturally occurring truncated ORF3 gene found in Guangxi, China. Front Vet Sci. (2020) 7:435. doi: 10.3389/fvets.2020.00435
32. Fang Q, Wang C, Liu H, Wu Q, Liang S, Cen M, et al. Pathogenic characteristics of a porcine astrovirus strain isolated in China. Viruses. (2019) 11:1156. doi: 10.3390/v11121156
33. Ouyang K, Shyu DL, Dhakal S, Hiremath J, Binjawadagi B, Lakshmanappa YS, et al. Evaluation of humoral immune status in porcine epidemic diarrhea virus (PEDV) infected sows under field conditions. Vet Res. (2015) 46:140. doi: 10.1186/s13567-015-0285-x
34. Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. Crystal structure of human enterovirus 71. Science. (2012) 336:1274. doi: 10.1126/science.1218713
35. Moon HJ, Song D, Seon BH, Kim HK, Park SJ, An DJ, et al. Complete genome analysis of porcine enterovirus B isolated in Korea. J Virol. (2012) 86:10250. doi: 10.1128/JVI.01548-12
36. Huang HI, Shih SR. Neurotropic enterovirus infections in the central nervous system. Viruses. (2015) 7:6051–66. doi: 10.3390/v7112920
Keywords: Enterovirus G, virus isolation, phylogenetic analysis, pathogenicity, piglets
Citation: Mi X, Yang C, Lu Y, Wang H, Qin Q, Chen R, Chen Z, Luo Y, Chen Y, Wei Z, Huang W and Ouyang K (2021) Isolation, Identification, and Evaluation of the Pathogenicity of a Porcine Enterovirus G Isolated From China. Front. Vet. Sci. 8:712679. doi: 10.3389/fvets.2021.712679
Received: 21 May 2021; Accepted: 29 June 2021;
Published: 22 July 2021.
Edited by:
Zhenhai Chen, Yangzhou University, ChinaCopyright © 2021 Mi, Yang, Lu, Wang, Qin, Chen, Chen, Luo, Chen, Wei, Huang and Ouyang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kang Ouyang, b3V5YW5na2FuZ0BneHUuZWR1LmNu
 Xue Mi
Xue Mi Chunjie Yang
Chunjie Yang Hejie Wang
Hejie Wang Ying Chen
Ying Chen Zuzhang Wei
Zuzhang Wei Weijian Huang
Weijian Huang Kang Ouyang
Kang Ouyang