- 1Doctor of Philosophy Program in Veterinary Science, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 2Department of Food Animal Clinic, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 3Research Group for Veterinary Public Health, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
- 4Department of Veterinary Biosciences and Veterinary Public Health, Faculty of Veterinary Medicine, Chiang Mai University, Chiang Mai, Thailand
Streptococcus uberis is recognized as an environmental mastitis pathogen in dairy cattle. The varied success rate of antibiotic treatment for S. uberis intramammary infection may be associated with the antimicrobial resistance (AMR) of these bacteria. This observational study aimed to analyze 228 S. uberis strains associated with bovine mastitis in northern Thailand from 2010 to 2017. AMR and AMR genes were determined by the minimum inhibitory concentration (MIC) using a microdilution method and polymerase chain reaction, respectively. The majority of S. uberis strains were resistant to tetracycline (187/228, 82.02%), followed by ceftiofur (44/228, 19.30%), and erythromycin (19/228, 8.33%). The MIC50 and MIC90 of ceftiofur in 2017 were 2–4-fold higher than those in 2010 (P < 0.01). Resistance to tetracycline and ceftiofur significantly increased between 2010 and 2017 (P < 0.05). The most common gene detected in S. uberis was tetM (199/228, 87.28%), followed by ermB (151/228, 66.23 %) and blaZ (15/228, 6.58 %). The association between tetracycline resistance and tetM detection was statistically significant (P < 0.01). The detection rates of tetM significantly increased, while the detection rates of tetO and ermB significantly decreased during 2010–2017. AMR monitoring for bovine mastitis pathogens, especially S. uberis, is necessary to understand the trend of AMR among mastitis pathogens, which can help create an AMR stewardship program for dairy farms in Thailand.
Introduction
Bovine mastitis, usually caused by an intramammary infection (IMI) of microorganisms, contributes to a major loss in dairy production by decreasing the production of milk and early culling of dairy cows (1). Streptococcus uberis commonly causes mastitis in dairy cattle worldwide (2). Although the major sources of S. uberis in dairy farms include water, soil, plant matter, bedding materials, flies, and hay (2), several studies have suggested that transmission of S. uberis can also occur between cows (3, 4).
Clinical mastitis caused by S. uberis accounts for approximately 45% of S. uberis IMIs during lactation (5). The treatment of bovine mastitis associated with S. uberis relies on the use of antimicrobial agents. Various groups of antimicrobial agents, such as macrolides, lincosamides, beta-lactams, and cephalosporins, were reported for the treatment of S. uberis IMI (6). However, the bacteriological cure rates following antimicrobial treatment of clinical mastitis caused by S. uberis are reported to vary, ranging from 64 to 91% (7–9). The variation in the success of treatment can be attributed to the virulence and antimicrobial resistance (AMR) of the pathogen.
AMR has become a worldwide problem for both human and animal health (10). In the dairy industry, antimicrobial agents have been mainly used for the treatment of bovine mastitis and in dry cow therapy (11, 12). The excessive use of antimicrobial agents in dairy herds may lead to increased AMR among mastitis pathogens (12). Therefore, monitoring AMR trends over a period of time is necessary to create an effective antimicrobial stewardship in dairy herds.
Thailand is a tropical country in Southeast Asia. In 2019, the Department of Livestock Development of Ministry of Agriculture and Cooperatives in Thailand reported the population of dairy cattle in the country to be approximately 670,000 with approximately 300,000 milking cows in 19,000 dairy farms (13). Northern Thailand is one of the dairy-intensive regions. Approximately 83,000 dairy cattle or 12.4% of the total dairy cattle population in the country are in northern Thailand. Bovine mastitis has been a major health problem in dairy cattle in this region (14).
In northern Thailand, S. uberis was reported to be a common pathogen associated with clinical and subclinical mastitis (15). Although AMR among S. uberis associated with bovine mastitis was reported in many countries (16, 17), reports in Southeast Asia, including Thailand, are limited. We aimed to investigate the AMR phenotypes and genotypes of S. uberis isolated from bovine mastitis cases in northern Thailand. The observed AMR patterns may result in an effective treatment protocol for S. uberis IMI in this region.
Materials and Methods
Sample Selection
This is an observational study that investigated phenotypic and genotypic AMR among archived isolates of S. uberis. S. uberis isolated from milk samples of cows with clinical or subclinical mastitis, submitted to the Faculty of Veterinary Medicine in Chiang Mai University (Thailand) between January 2010 and December 2017, were included in the study. All milk samples were collected and cultured as part of some previous programs during 2010–2017 from dairy cattle herds in Chiang Mai and Lamphun provinces in northern Thailand (Figure 1). These two provinces represent the most dairy-intensive region in northern Thailand, with an approximate population of 34,000 milking cows and 1,500 dairy farms (13). In this region, most dairy cattle are crossbred Holstein and raised in tied stalls. A bucket-type milking system is mostly adopted in this region. Most farms are small-holder dairy farms with 15–60 milking cows producing approximately 12 kg of milk/cow/day.
All milk samples in 2010–2011 were passively submitted for the diagnosis of IMI by local veterinarians. During 2012–2017, not only milk samples passively submitted for the diagnosis of IMI, but also milk samples of cows actively screened for clinical and subclinical mastitis as requested by owners of herds with history of high bulk milk somatic cell count, were the sources of S. uberis isolates. The mastitis screening by veterinarians was the service program provided by the university during that particular period. Clinical mastitis cases were considered for cows presenting abnormal milk and/or changes of the udder such as swelling, pain, and heat. Subclinical mastitis cases were diagnosed using the California Mastitis Test. As a part of the veterinary services, quarter milk samples were aseptically collected and submitted for the diagnosis of mastitis pathogens. The S. uberis isolates were kept frozen at −80°C in a brain heart infusion broth containing 20% glycerol until use. The sampling criteria for selecting S. uberis isolates were different based on the total number of isolates originated each year. For years with a low number of S. uberis isolates (n ≤ 10), all isolates were included in the study. For years with more than 10 isolates, 1–6 isolates per farm per isolated month were selected in order to include the highest diversity of S. uberis isolates in those years. Given these criteria, a sample of 228 isolates from a total of 442 frozen isolates was randomly selected for each month of each year as a representative collection of S. uberis isolates in each studied year. These S. uberis isolates were from 55 clinical and 173 subclinical cases of 165 cows in 105 farms.
Identification of S. uberis Isolates
All selected frozen S. uberis isolates were re-grown on blood agar (Merck®, Darmstadt, Germany) with 5% bovine blood and incubated at 37°C for 24 h. Tests such as Gram staining, catalase test, and the ability to metabolize esculin, inulin, mannitol, and salicin were used to identify S. uberis. The genomic DNAs of all isolates were extracted using a DNA extraction kit (NucleoSpin®, Düren, Germany) and confirmed to be S. uberis using polymerase chain reaction (PCR) to detect the 16S rRNA gene (Table 1), as described previously (3).
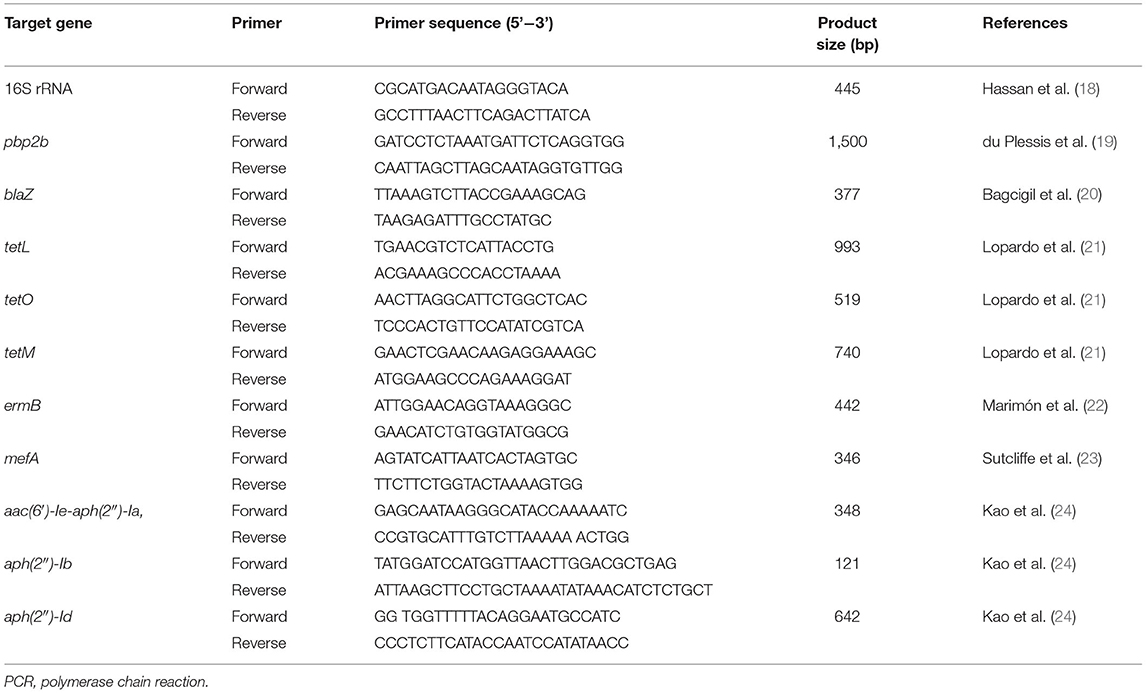
Table 1. DNA Sequences, target genes, and expected product sizes of PCR primers used for the identification of Streptococcus uberis and detection of antimicrobial resistance genes.
Determination of Antimicrobial Susceptibility of S. uberis
All S. uberis isolates were investigated for their antimicrobial susceptibility using the microdilution method recommended by the Clinical and Laboratory Standards Institute (CLSI) performance standards (25). Five antimicrobial agents were selected for testing: penicillin G, ceftiofur, erythromycin, tetracycline, and gentamicin. These antimicrobial agents were selected as representative drugs from five antimicrobial classes, namely penicillins (penicillin G), cephalosporins (ceftiofur), macrolides (erythromycin), tetracyclines (tetracycline), and aminoglycosides (gentamicin), which are commonly found in dairy herds. All antimicrobial agents were diluted in Mueller Hinton broth (MHB) according to the selected concentration ranges considered from literature reviews. The selected diluted concentration ranges were 0.0039–4 μg/mL for penicillin G, 0.0625–256 μg/mL for ceftiofur, 0.0039–8 μg/mL for erythromycin, 0.625–64 μg/mL for tetracycline, and 0.0039–16 μg/mL for gentamicin. A single colony of each S. uberis isolate was cultured in MHB. The turbidity of the inoculated MHB was adjusted to 0.5 McFarland using a McFarland spectrophotometer. Streptococcus pneumoniae ATCC® 49619 and Escherichia coli ATCC® 25922 were used as quality control strains. The minimum inhibitory concentration (MIC) breakpoint standards are listed in Table 2. The MICs of each antimicrobial agent that inhibited the visible growth of ≥50 and ≥90% of microorganisms (MIC50 and MIC90) were recorded for each antibiotic.
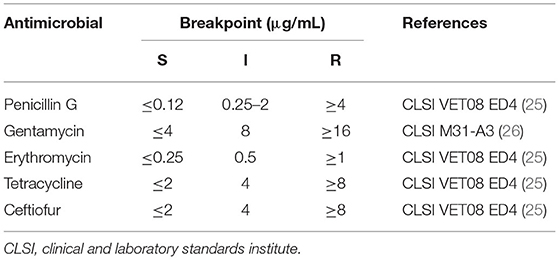
Table 2. Antimicrobial susceptibility breakpoints used to determine the antimicrobial susceptibility (S), intermediate (I), and resistance (R) of Streptococcus uberis.
Detection of AMR Genes
All S. uberis isolates were screened for AMR genes using PCR. The screened AMR genes included blaZ and pbp2b for beta-lactam resistance; tetL, tetO, and tetM for tetracycline resistance; ermB and mefA for macrolide resistance; and aac(6′)-Ie-aph(2″)-Ia, aph(2″)-Ib, and aph(2″)-Id for aminoglycoside resistance. The primers used for each gene are listed in Table 1. The PCR mixture (25 μL) contained 0.5 μL of forward and reverse primers (10 mol/L); 12.5 μL of 2X Taq Master Mix containing 1.25 U of Taq DNA polymerase, 1X ViBuffer A, 0.2 mM dNTPs, and 1.5 mM MgCl2 (MyTaq™ Red Mix; Bioline, NSW, Australia); 11 μL of DNase-free water; and 0.5 μL of DNA template (50–100 ng/μL). The tubes were placed in a thermal cycler with the following program: initial denaturation at 94°C for 5 min; 35 cycles of denaturation at 94°C for 30 s; annealing temperature as shown in Table 1, for 30 s; and an extension at 72°C for 60 s. The PCR products were determined by 2% agarose gel electrophoresis, stained with ethidium bromide, and visualized under ultraviolet illumination.
Statistical Analysis
The MIC50 and MIC90 values were descriptively reported for each antimicrobial agent. AMR and distribution of detected AMR genes were expressed as percentages. MICs, AMR, and AMR gene detection were calculated separately for S. uberis isolates each year. Temporal trends in the MIC distributions of each antimicrobial agent were analyzed using the proportional-odds cumulative logit model analysis. The trends of AMR and AMR gene detection rates over the 8-year period (2010–2017) were analyzed using the Cochran–Armitage test for trend and logistic regression model analyses, which depicted “year of isolation” as the exposure variable and “AMR” or “AMR gene detection” as the outcome variable. These analyses were based on the principles previously described by Michael et al. (27) and Aerts et al. (28). The associations between AMR and the presence of AMR genes were determined using the χ2-test and Fisher's exact test at a significance level of P < 0.05. All statistical analyses were performed using the R statistical software version 4.0.0 (29).
Results
AMR Determined by MICs
The MIC of each antimicrobial agent among all the S. uberis isolates is shown in Table 3. Most examined S. uberis isolates were resistant to tetracycline (187/228, 82.02%), followed by ceftiofur (44/228, 19.30%) and erythromycin (19/228, 8.33%). All examined S. uberis isolates were susceptible to penicillin G and gentamicin. A total of 53 isolates (23.25%) were considered to show multidrug resistance, which were defined as strains resistant to two or more antimicrobial agents (Table 4). The MIC50 and MIC90 of each antimicrobial agent among the S. uberis isolates in each year are shown in Table 5. The MIC50 and MIC90 of ceftiofur in 2017 were 2–8-fold higher than those in 2010, while the MICs of other antimicrobial agents were stable throughout the 8-year period. The proportional odds cumulative logit model analysis indicated a significant increase in MICs of ceftiofur (P < 0.001), tetracycline (P < 0.05) and penicillin G (P < 0.001) from 2010 to 2017 as shown in Table 6.
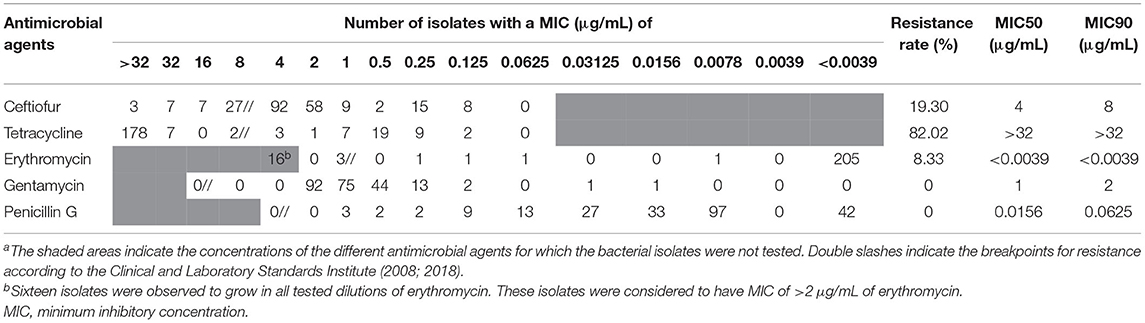
Table 3. Distribution of MIC for Streptococcus uberis (n = 228) isolates from dairy cattle with subclinical or clinical bovine mastitis in northern Thailand during 2010–2017a.
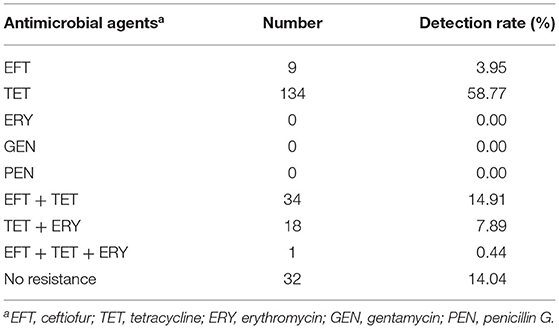
Table 4. Distribution of antimicrobial resistance patterns among Streptococcus uberis (n = 228) isolates from dairy cattle with bovine mastitis in northern Thailand during 2010–2017.
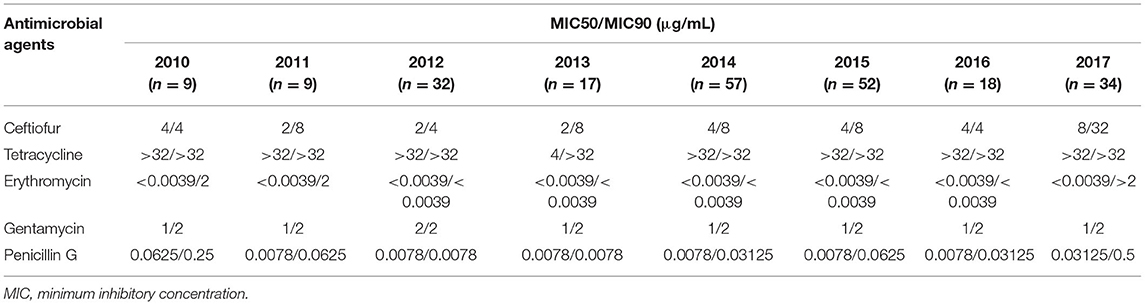
Table 5. Changes in MICs of antimicrobial agents among Streptococcus uberis (n = 228) isolates from dairy cattle with bovine mastitis in northern Thailand during 2010–2017.
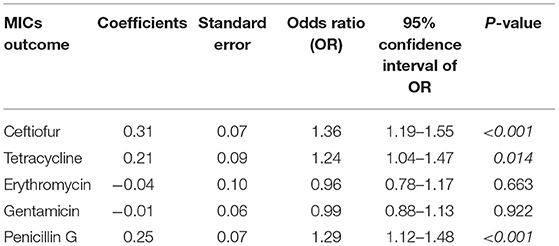
Table 6. The five proportional odds cumulative logit models depicting the association between year of isolation and the changes in the minimum inhibitory concentrations (MICs) of five antimicrobial agents among Streptococcus uberis (n = 228) associated with bovine mastitis in northern Thailand during 2010–2017.
Trends of AMR
The rates of tetracycline resistance ranged from 45 to 100%, which was the highest resistance rate observed in any year. The resistance rate to ceftiofur remained low (<25%) from 2010 to 2016, but it increased to 52.9% in 2017. The resistance rates to erythromycin, gentamicin, and penicillin G were low throughout the study period. Regarding the Cochran–Armitage trend test, the trends of AMR to tetracycline (P < 0.05) and ceftiofur (P < 0.01) were considered to be “increasing” significantly, showing an increasing resistance during 2010–2017 (Figure 2). The logistic regression model analyses showed similar results, with statistically significant resistance rates to tetracycline (P = 0.027) and ceftiofur (P < 0.001) as shown in Table 7.
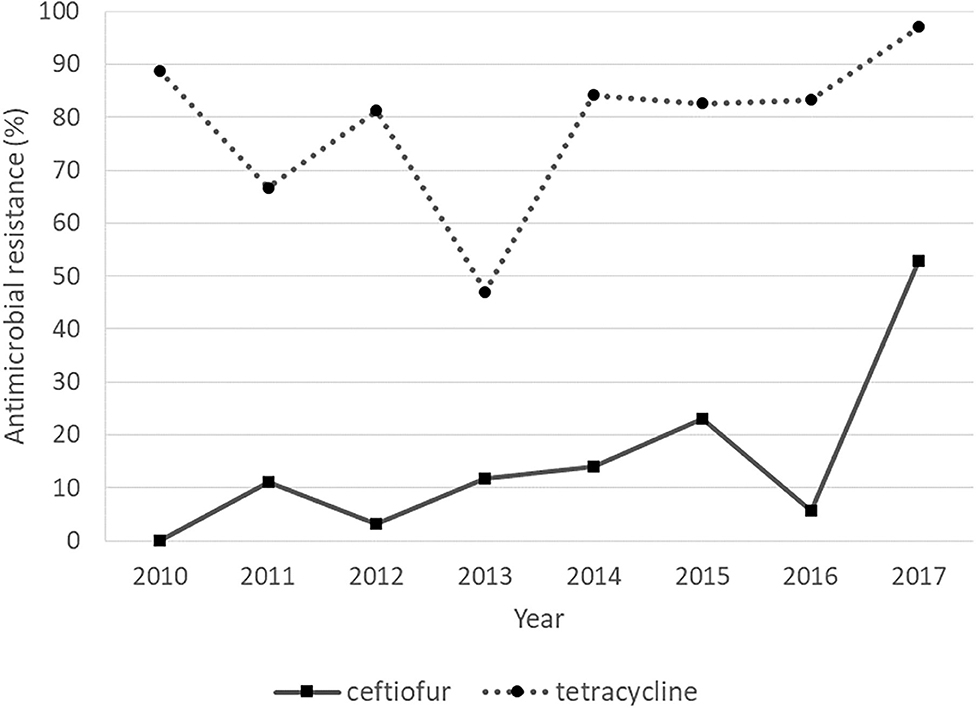
Figure 2. Antimicrobial resistance of Streptococcus uberis with “increasing” trends over a period of 8 years, from 2010 to 2017.
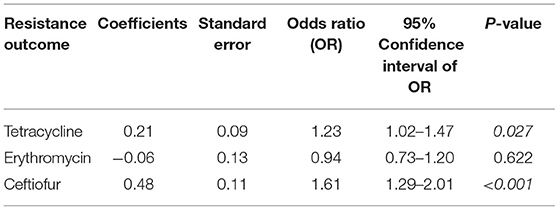
Table 7. The three logistic regression models depicting the association between year of isolation and tetracycline-, erythromycin-, and ceftiofur resistance among Streptococcus uberis (n = 228) associated with bovine mastitis in northern Thailand during 2010–2017.
Detection of AMR Genes
The distribution of AMR genes detected among S. uberis isolates from 2010 to 2017 are shown in Table 8. Among the examined AMR genes, the most commonly detected gene was tetM (199/228, 87.28%), followed by ermB (151/228, 66.23 %) and blaZ (15/228, 6.58 %). The association between tetracycline resistance and tetM detection was statistically significant (P < 0.01).
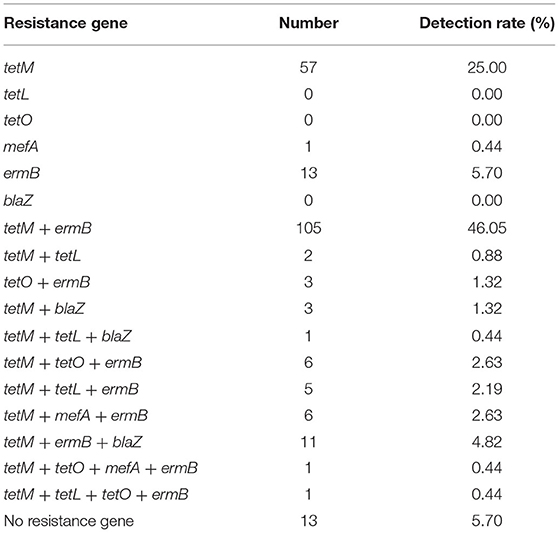
Table 8. Distribution of antimicrobial resistance gene patterns among Streptococcus uberis (n = 228) isolates from dairy cattle with bovine mastitis in northern Thailand during 2010–2017.
Trends of AMR Gene Detection Rates
Regarding the Cochran–Armitage trend test, two genes, tetM and mefA, showed a significant increase in detection rates from 2010 to 2017 (P < 0.01). The detection rates of tetM increased from 33.33% in 2010 to 97.06% in 2017, while the detection rates of mefA increased from 0% in 2010 to 29.41% in 2017. In contrast, the other two genes, tetO and ermB, showed a significant decrease in detection rates from 2010 to 2017 (P < 0.01). The detection rates of tetO were very low throughout the study period, except in 2011, when it peaked at 88.89%. The detection rates of ermB were high in 2011–2013 and gradually decreased from 100% in 2013 to 2.94% in 2017. The significant “positive trend” and “negative trend” of AMR gene detection rates are shown in Figure 3. However, when the logistic regression model analyses were performed, only the models of tetM (P = 0.002), tetO (P < 0.001), and ermB (P < 0.001) detection showed a significant association between the isolated year and detection rates as shown in Table 9.
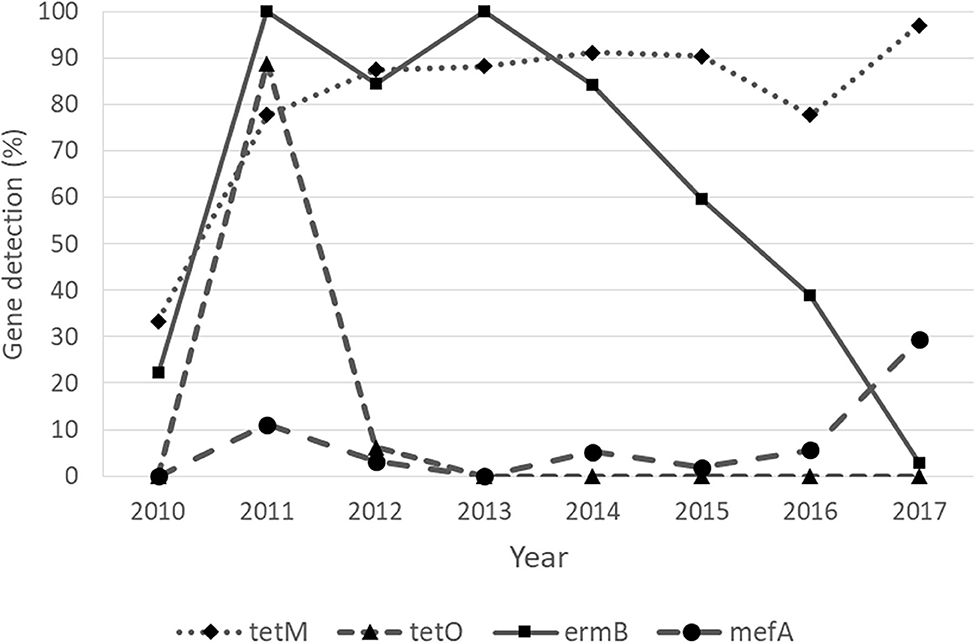
Figure 3. Detection of antimicrobial resistance genes of Streptococcus uberis with “increasing” (tetM and mefA) and “decreasing” (tetO and ermB) trends over a period of 8 years, from 2010 to 2017.
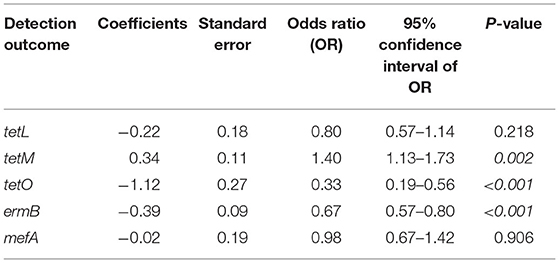
Table 9. The five logistic regression models depicting the association between year of isolation and the detection of tetL, tetM, tetO, ermB, and mefA among Streptococcus uberis (n = 228) associated with bovine mastitis in northern Thailand during 2010–2017.
Discussion
Streptococcus uberis is one of the most prevalent environmental pathogens causing bovine mastitis in many regions, including Thailand. S. uberis IMI is common during the dry period and early lactation (28). Different classes of antimicrobials were used to treat S. uberis IMI (6). The AMR of S. uberis can potentially contribute to a successful mastitis treatment.
Our MIC results showed that the S. uberis isolates were highly resistant to tetracycline (82.02%) and significantly increased from 2010 to 2017. The tetracycline resistance reported in the current study is higher than that reported in other countries, such as China (59%), Germany (42.3%), Canada (38.6%), and Sweden (12%) (30–33). Tetracycline is widely used in livestock (34); consequently, tetracycline resistance was found to be common in streptococci (35, 36). A study in 2016 reported that approximately half of dairy farmers in Chiang Mai province used oxytetracycline for treatment of sick animals in their farms (14). In addition, the antibiotic was slowly eliminated from the body. The slow degradation of tetracycline can lead to an increased selective pressure for tetracycline resistance in bacteria (37). Therefore, although tetracycline is not commonly used to treat mastitis in dairy cattle (38), tetracycline resistance was not unexpected. In the current study, the most frequent tetracycline resistance gene detected was tetM, followed by tetL and tetO. In agreement with the tetracycline resistance, a significant increasing trend of tetM detection was observed from 2010 to 2017. In contrast, the trend of tetO detection was appeared to be significantly decreased throughout the study period. These findings, together with the significant association between the presence of tetM and tetracycline resistance, indicated that tetracycline resistance in S. uberis occurs mainly through the functions of the ribosome-protected protein TetM (39). However, a peak of tetO detection up to 88.89% in 2011 was observed in the study. This finding could be a result of the limited sample size included in 2010 and 2011. This limited sample size could lead to a less variation of selected isolates, which might consequently affect the detection rates reported in the study.
The current study demonstrated that S. uberis isolates were highly susceptible to erythromycin. Similar findings were reported in other countries (32, 40). In northern Thailand, erythromycin and other macrolides, are not commonly used in dairy cattle (14). Therefore, a low erythromycin resistance and low detection rates of erythromycin resistance gene (ermB) and macrolide resistance gene (mefA) was expected to be observed in this study. However, high detection rates of ermB were observed among the S. uberis isolates from 2011 to 2013, and gradually decreased from 2013 to 2017. Although previous research suggested that the ermB gene dominates erythromycin resistance in S. uberis (41), the presence of ermB was not associated with the erythromycin resistance among S. uberis reported in the current study. Moreover, mefA was also detected at a low rate from 2010 to 2016, and increased in 2017. In agreement with the trend of mefA detection, MIC90 of erythromycin in 2017 (>2 μg/mL) was observed to be higher than that in 2016 (<0.0039 μg/mL). These findings demonstrated a possible evidence to hypothesize that the mefA gene could be responsible for erythromycin resistance among S. uberis associated with bovine mastitis in northern Thailand. However, the current study did not reveal a significant association between the presence of mefA gene and the erythromycin resistance, which might be caused by the limited sample size with low erythromycin resistance. Therefore, the mechanism of macrolide resistance in S. uberis requires further investigation.
Beta-lactam class antimicrobials are frequently used for the prevention and treatment of dairy cattle diseases. In the present study, S. uberis showed a high susceptibility to penicillin G (100%) and ceftiofur (81%). This finding is consistent with the results of various studies (31, 36, 40). Likewise, we could not detect the pbp2b gene in our sampled S. uberis. This gene was reported to play an important role in beta-lactam resistance in streptococci (42, 43). Another gene, blaZ, which is responsible for beta-lactam resistance, was also detected at a low level in our S. uberis collection, similar to the findings in other previous reports (36, 40).
From 2010 to 2017, we were able to demonstrate an increasing trend of ceftiofur resistance among S. uberis, as evidenced by both the resistance rate and MICs from 2016 to 2017. Similarly, a study in China reported an increasing trend of ceftriaxone-resistant strains of S. dysgalactiae associated with bovine mastitis (44). Although statistical significance was not observed, an increasing trend was also demonstrated with the MICs of penicillin G. These findings raised the concern of beta-lactam resistance in S. uberis in the near future. A study in 2016 reported that more than half of dairy farmers in Chiang Mai province usually used penicillin/streptomycin (52.31%) for systemic treatment of cattle diseases (14). Moreover, most dairy farmers in this region used ampicillin/cloxacillin (87.69%), followed by cephapirin (82.31%) for intramammary treatment of bovine mastitis (14). Because beta-lactam antimicrobial agents are the most commonly used antimicrobial agents for mastitis treatment, the increasing trend of the MICs of these two beta-lactams may provide critically important evidence associated with the cure rate of bovine mastitis caused by S. uberis in the future. Antibiotic usage and AMR of S. uberis in dairy farms should be continually monitored to adjust the treatment protocols for bovine mastitis in Thailand.
Conclusions
In conclusion, from 2010 to 2017, S. uberis isolates associated with bovine mastitis in Thailand were highly and increasingly resistant to tetracycline, potentially controlled by tetM. In addition, throughout the study period, we observed increasing trends in the MICs of ceftiofur, together with an increasing trend in the ceftiofur resistance rate. These findings emphasize the importance of AMR monitoring for bovine mastitis pathogens, especially S. uberis, and can serve as guidelines for effective treatment decisions. The prudent use of antimicrobial agents in dairy farms, especially tetracycline and ceftiofur, should be intensively considered and applied in this region. Understanding the trend of AMR among mastitis pathogens can help create an AMR stewardship program for dairy farms in Thailand.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
TZ, SB, and DP considered the study design. TZ and GN performed the laboratory experiments. SB performed the data analyses. SB and TZ prepared the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was funded by the Research Group for Veterinary Public Health, and the Faculty of Veterinary Medicine, Chiang Mai University, Thailand.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Miss. Thareeya Rin-ut and the Central Laboratory, Faculty of Veterinary Medicine, Chiang Mai Univerisity, for the technical guidance and supports.
References
1. Halasa T, Huijps K, Østerås O, Hogeveen H. Economic effects of bovine mastitis and mastitis management: a review. Vet Q. (2007) 29:18–31. doi: 10.1080/01652176.2007.9695224
2. Krömker V, Reinecke F, Paduch JH, Grabowski N. Bovine Streptococcus uberis intramammary infections and mastitis. Clin Microbial. (2014) 3:157. doi: 10.4172/2327-5073.1000157
3. Boonyayatra S, Tharavichitkul P, Oliver SP. Virulence-associated genes and molecular typing of Streptococcus uberis associated with bovine mastitis in northern Thailand. Turk J Vet Anim Sci. (2018) 42:73–81. doi: 10.3906/vet-1704-75
4. Abureema S, Smooker P, Malmo J, Deighton M. Molecular epidemiology of recurrent clinical mastitis due to Streptococcus uberis: evidence of both an environmental source and recurring infection with the same strain. J Dairy Sci. (2014) 97:285–90. doi: 10.3168/jds.2013-7074
5. Todhunter DA, Smith KL, Hogan JS. Environmental streptococcal intramammary infections of the bovine mammary gland. J Dairy Sci. (1995) 78:2366–74. doi: 10.3168/jds.S0022-0302(95)76864-3
6. Boonyayatra S. Treatment of bovine mastitis during lactating period. Chiang Mai Vet J. (2012) 10:87–107.
7. McDougall S. Efficacy of two antibiotic treatments in curing clinical and subclinical mastitis in lactating dairy cows. N Z Vet J. (1998) 46:226–32. doi: 10.1080/00480169.1998.36094
8. McDougall S, Agnew KE, Cursons R, Hou XX, Compton CR. Parenteral treatment of clinical mastitis with Tylosin base or penethamate hydriodide in dairy cattle. J Dairy Sci. (2007) 90:779–89. doi: 10.3168/jds.S0022-0302(07)71562-X
9. Owens WE, Ray CH, Watts JL, Yancey RJ. Comparison of success of antibiotic therapy during lactation and results of antimicrobial susceptibility tests for bovine mastitis. J Dairy Sci. (1997) 80:313–7. doi: 10.3168/jds.S0022-0302(97)75940-X
10. Roca I, Akova M, Baquero F, Carlet J, Cavaleri M, Coenen S, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. (2015) 6:22–9. doi: 10.1016/j.nmni.2015.02.007
11. Pol M, Ruegg PL. Treatment practices and quantification of antimicrobial drug usage in conventional and organic dairy farms in Wisconsin. J Dairy Sci. (2007) 90:249–61. doi: 10.3168/jds.S0022-0302(07)72626-7
12. Saini V, McClure JT, Léger D, Dufour S, Sheldon AG, Scholl DT, et al. Antimicrobial use on Canadian dairy farms. J Dairy Sci. (2012) 95:1209–21. doi: 10.3168/jds.2011-4527
13. Information and Communication Technology Center Department of Livestock Development Ministry of Agriculture and Cooperatives. Provincial Data of Dairy Farmers. (2019). Available online at: http://ict.dld.go.th/webnew/images/stories/stat_web/yearly/2562/T3-1-milk.pdf (accessed July 7, 2021).
14. Udin C. Relationship Between Antimicrobial Usage and the Presence of Escherichia coli Production Extended Spectrum Beta-Lactamase in Bulk Tank Milk in Mae-On District, Chiang Mai Province. (Dissertation/master's thesis). Chiang Mai: Chiang Mai University (2016).
15. Boonyayatra S, Thaboonpeng J, Kreausukon K, Suriyasathaporn W. Antimicrobial resistance of mastitis associated bacteria in lactating dairy cows in Chiang Mai Province. Chiang Mai Vet J. (2007) 5:135–45.
16. Rajala-Schultz PJ, Smith KL, Hogan JS, Love BC. Antimicrobial susceptibility of mastitis pathogens from first lactation and older cows. Vet Microbiol. (2004) 102:33–42. doi: 10.1016/j.vetmic.2004.04.010
17. Schmitt-Van de Leemput E, Zadoks RN. Genotypic and phenotypic detection of macrolide and lincosamide resistance in Streptococcus uberis. J Dairy Sci. (2007) 90:5089–96. doi: 10.3168/jds.2007-0101
18. Hassan AA, Khan IU, Abdulmawjood A, Lämmler C. Evaluation of PCR methods for rapid identification and differentiation of Streptococcus uberis and Streptococcus parauberis. J Clin Microbiol. (2001) 39:1618–21. doi: 10.1128/JCM.39.4.1618-1621.2001
19. du Plessis M, Bingen E, Klugman KP. Analysis of penicillin-binding protein genes of clinical isolates of Streptococcus pneumoniae with reduced susceptibility to amoxicillin. Antimicrob Agents Chemother. (2002) 46:2349–57. doi: 10.1128/AAC.46.8.2349-2357.2002
20. Bagcigil AF, Taponen S, Koort J, Bengtsson B, Myllyniemi AL, Pyörälä S. Genetic basis of penicillin resistance of S. aureus isolated in bovine mastitis. Acta Vet Scand. (2012) 54:69. doi: 10.1186/1751-0147-54-69
21. Lopardo HA, Vidal P, Jeric P, Centron D, Paganini H, Facklam RR, et al. Six-month multicenter study on invasive infections due to group B streptococci in Argentina. J Clin Microbiol. (2003) 41:4688–94. doi: 10.1128/JCM.41.10.4688-4694.2003
22. Marimón JM, Valiente A, Ercibengoa M, García-Arenzana JM, Pérez-Trallero E. Erythromycin resistance and genetic elements carrying macrolide efflux genes in Streptococcus agalactiae. Antimicrob Agents Chemother. (2005) 49:5069–74. doi: 10.1128/AAC.49.12.5069-5074.2005
23. Sutcliffe J, Grebe T, Tait-Kamradt A, Wondrack L. Detection of erythromycin-resistant determinants by PCR. Antimicrob Agents Chemother. (1996) 40:2562–6. doi: 10.1128/AAC.40.11.2562
24. Kao SJ, You I, Clewell DB, Donabedian SM, Zervos MJ, Petrin J, et al. Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in Enterococcus faecium. Antimicrob Agents Chemother. (2000) 44:2876–9. doi: 10.1128/AAC.44.10.2876-2879.2000
25. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. 4th Edn. CLSI Supplement VET08. Wayne, PA: Clinical and Laboratory Standards Institute (2018).
26. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals. CLSI Approved Standard M31—A3. Wayne, PA: Clinical and Laboratory Standards Institute (2008).
27. Michael A, Kelman T, Pitesky M. Overview of quantitative methodologies to understand antimicrobial resistance via minimum inhibitory concentration. Animals. (2020) 10:1405. doi: 10.3390/ani10081405
28. Aerts M, Faes C, Nysen R. Development of statistical methods for the evaluation of data on antimicrobial resistance in bacterial isolates from animals and food. EFSA Support Publ. (2011) 8:186. doi: 10.2903/sp.efsa.2011.EN-186
29. R Core Team. version 3.5.0. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing (2018).
30. Cameron M, Saab M, Heider L, McClure JT, Rodriguez-Lecompte JC, Sanchez J. Antimicrobial susceptibility patterns of environmental streptococci recovered from bovine milk samples in the Maritime provinces of Canada. Front Vet Sci. (2016) 3:79. doi: 10.3389/fvets.2016.00079
31. Cheng J, Qu W, Barkema HW, Nobrega DB, Gao J, Liu G, et al. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J Dairy Sci. (2019) 102:2416–26. doi: 10.3168/jds.2018-15135
32. Persson Y, Nyman AKJ, Grönlund-Andersson U. Etiology and antimicrobial susceptibility of udder pathogens from cases of subclinical mastitis in dairy cows in Sweden. Acta Vet Scand. (2011) 53:36. doi: 10.1186/1751-0147-53-36
33. Minst K, Märtlbauer E, Miller T, Meyer C. Short communication: Streptococcus species isolated from mastitis milk samples in Germany and their resistance to antimicrobial agents. J Dairy Sci. (2012) 95:6957–62. doi: 10.3168/jds.2012-5852
34. Aminov RI, Garrigues-Jeanjean N, Mackie RI. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol. (2001) 67:22–32. doi: 10.1128/AEM.67.1.22-32.2001
35. Petrovski KR, Grinberg A, Williamson NB, Abdalla ME, Lopez-Villalobos N, Parkinson TJ, et al. Susceptibility to antimicrobials of mastitis-causing Staphylococcus aureus, Streptococcus uberis and Str. dysgalactiae from New Zealand and the USA as assessed by the disk diffusion test. Aust Vet J. (2015) 93:227–33. doi: 10.1111/avj.12340
36. Ruegg PL, Oliveira L, Jin W, Okwumabua O. Phenotypic antimicrobial susceptibility and occurrence of selected resistance genes in gram-positive mastitis pathogens isolated from Wisconsin dairy cows. J Dairy Sci. (2015) 98:4521–34. doi: 10.3168/jds.2014-9137
37. Ahasan MS, Picard J, Elliott L, Kinobe R, Owens L, Ariel E. Evidence of antibiotic resistance in Enterobacteriales isolated from green sea turtles, Chelonia mydas on the Great Barrier Reef. Mar Pollut Bull. (2017) 120:18–27. doi: 10.1016/j.marpolbul.2017.04.046
38. Gruet P, Maincent P, Berthelot X, Kaltsatos V. Bovine mastitis and intramammary drug delivery: review and perspectives. Adv Drug Deliv Rev. (2001) 50:245–59. doi: 10.1016/S0169-409X(01)00160-0
39. Vélez JR, Cameron M, Rodríguez-Lecompte JC, Xia F, Heider LC, Saab M, et al. Whole-genome sequence analysis of antimicrobial resistance genes in Streptococcus uberis and Streptococcus dysgalactiae isolates from Canadian dairy herds. Front Vet Sci. (2017) 4:63. doi: 10.3389/fvets.2017.00063
40. Kaczorek E, Małaczewska J, Wójcik R, Rekawek W, Siwicki AK. Phenotypic and genotypic antimicrobial susceptibility pattern of Streptococcus spp. isolated from cases of clinical mastitis in dairy cattle in Poland. J Dairy Sci. (2017) 100:6442–453. doi: 10.3168/jds.2017-12660
41. Haenni M, Saras E, Chaussière S, Treilles M, Madec JY. ermB-mediated erythromycin resistance in Streptococcus uberis from bovine mastitis. Vet J. (2011) 189:356–8. doi: 10.1016/j.tvjl.2010.06.021
42. Piccinelli G, Carlentini G, Gargiulo F, Caruso A, De Francesco MA. Analysis of point mutations in the pbp2x, pbp2b, and pbp1a genes of Streptococcus agalactiae and their relation with a reduced susceptibility to cephalosporins. Microb Drug Resist. (2017) 23:1019–24. doi: 10.1089/mdr.2017.0013
43. Hu Y, Kan Y, Zhang Z, Lu Z, Li Y, Leng C, et al. New mutations of penicillin-binding proteins in Streptococcus agalactiae isolates from cattle with decreased susceptibility to penicillin. Microb Drug Resist. (2018) 24:1236–41. doi: 10.1089/mdr.2017.0223
44. Zhang S, Piepers S, Shan R, Cai L, Mao S, Zou J, et al. Phenotypic and genotypic characterization of antimicrobial resistance profiles in Streptococcus dysgalactiae isolated from bovine clinical mastitis in 5 provinces of China. J Dairy Sci. (2018) 101:3344–55. doi: 10.3168/jds.2017-14031
Keywords: Streptococcus uberis, bovine mastitis, antimicrobial resistance, antimicrobial resistance gene, intramammary infection
Citation: Zhang T, Niu G, Boonyayatra S and Pichpol D (2021) Antimicrobial Resistance Profiles and Genes in Streptococcus uberis Associated With Bovine Mastitis in Thailand. Front. Vet. Sci. 8:705338. doi: 10.3389/fvets.2021.705338
Received: 05 May 2021; Accepted: 14 July 2021;
Published: 17 August 2021.
Edited by:
Alejandra Victoria Capozzo, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
László Ózsvári, University of Veterinary Medicine Budapest, HungaryLucy Brunton, Royal Veterinary College (RVC), United Kingdom
Copyright © 2021 Zhang, Niu, Boonyayatra and Pichpol. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sukolrat Boonyayatra, c3Vrb2xyYXQuYkBjbXUuYWMudGg=
 Tingrui Zhang
Tingrui Zhang Guoyi Niu
Guoyi Niu Sukolrat Boonyayatra
Sukolrat Boonyayatra Duangporn Pichpol
Duangporn Pichpol