- State Key Laboratory of Veterinary Etiological Biology, National Foot-and-Mouth Disease Reference Laboratory, Lanzhou Veterinary Research Institute, Chinese Academy of Agriculture Science, Lanzhou, China
The porcine reproductive and respiratory syndrome virus (PRRSV) is the pathogen causing epidemics of porcine reproductive and respiratory syndrome (PRRS), and is present in every major swine-farming country in the world. Previous studies have demonstrated that PRRSV infection leads to a range of consequences, such as persistent infection, secondary infection, and co-infection, and is common among pigs in the field. In recent years, coinfection of PRRSV and other porcine pathogens has occurred often, making it more difficult to define and diagnose PRRSV-related diseases. The study of coinfections may be extremely suitable for the current prevention and control in the field. However, there is a limited understanding of coinfection. Therefore, in this review, we have focused on the epidemiology of PRRSV coinfection with other pathogens in swine, both in vivo and in vitro.
Introduction
The porcine reproductive and respiratory syndrome virus (PRRSV) is a single-stranded positive-sense RNA virus with a capsid, and it belongs to the family Arteriviridae (1). The PRRSV genome is ~15 kb in length and contains at least 10 open reading frames (ORFs). ORF1a and ORF1b encode RNA replicase and at least 16 non-structural proteins. ORF2–ORF7 encode glycoproteins (GP2–GP5), membrane protein M, and nucleocapsid protein N (2, 3). Porcine reproductive and respiratory syndrome (PRRS) caused by PRRSV, also known as porcine blue ear disease, is a highly immunosuppressive disease (4, 5). It causes subclinical, lethal, and persistent infections (6). Clinical symptoms of PRRS depend on the infecting strain, growth stage, immune status, secondary or co-infecting pathogens, environmental conditions, and disease management level (7).
In 1987, PRRSV first became prevalent in the United States, and then gradually appeared in swine-farming countries worldwide (8). It was described with the following two genotypes: PRRSV-1 (alternatively Betaarterivirus suid 1), of European origin, and PRRSV-2 (alternatively Betaarterivirus suid 2) of North American origin (9). The prevalent PRRSV strains in China are mainly the PRRSV-2. Although the outbreak of the African swine fever virus (ASFV) in 2018 masked PRRS, it is still difficult to get rid of completely (10, 11). For this reason, it is necessary to understand the evolutionary trends of PRRSV in China.
The disease was initially reported in China in 1996, and since then the highly pathogenic PRRSV (HP-PRRSV) strain has caused outbreaks in several provinces and led to serious economic losses (12, 13). After 2012, the new recombinant strains GM2 and NADC30-like appeared in China (14, 15). After 2015, HP-PRRSV and NADC30-like became prevalent (13). The emergence of NADC30-like strains may be due to the recombination of North American NADC30 strains and Chinese HP-PRRSV strains (16). Although not as highly pathogenic as HP-PRRSV, NADC30-like strains are known for their high incidence of recombination with other viral strains, resulting in altered virulence. Moreover, there might be outbreaks of NADC30-like strain in vaccinated swine (17). Furthermore, coinfection of NADC30-like strains and HP-PRRSV is common in piglets with porcine respiratory disease complex (PRDC) (18). It is demonstrated that recombination caused by coinfection with different strains or genotypes makes the epidemiology of PRRSV strains more complex and diverse. Moreover, this may be one of the reasons for the different results of PRRSV coinfection with other pathogens. Therefore, it is necessary to review the prevalence of PRRSV.
Coinfection has become a common phenomenon in current swine farms, and PRRSV is one of the key pathogens (19). The PRRSV coinfection viruses include porcine circovirus 2 (PCV2), pseudorabies virus (PRV), swine influenza virus (SIV), classical swine fever virus (CSFV), hepatitis E virus (HEV), porcine parvovirus (PPV), porcine group A rotavirus (PARV) and porcine epidemic diarrhea virus (PEDV) (20, 21). PRRSV can cause co-infection/secondary infection with bacteria such as Haemophilus parasuis (HPS), Streptococcus suis (SS), and Actinobacillus pleuropneumoniae (APP) (22–24). Moreover, coinfection of PRRSV and Toxoplasma gondii (T. gondii) or Mycoplasma hyopneumoniae (Mhp) often occurs (25, 26). However, there is a limited understanding of it. The current research is mainly on the interaction between host and pathogens, which limits the study of coinfection (19). Therefore, in this review, we discuss the interaction of PRRSV and the above-mentioned pathogens as systematically as possible. The aim is to provide a reference for the prevention and control of related diseases, the production of vaccines, and research methods for coinfection.
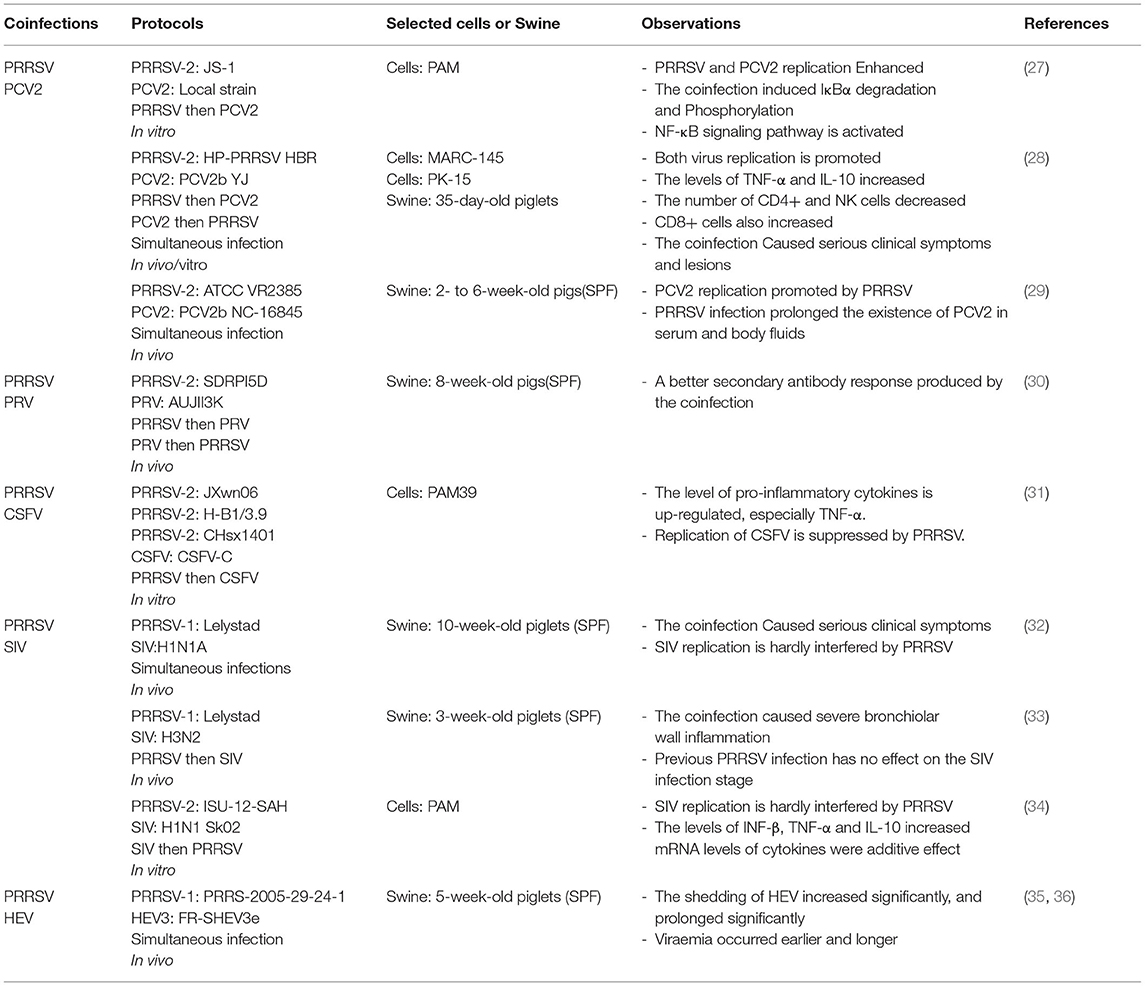
Co-infection of PRRSV and Viruses
Although coinfection is important in this field, there are limited studies on coinfection of PRRSV and other viruses. The current single host-pathogen research method and the extremely high mutation rate of PRRSV itself may be the factors that limit the study of coinfection. Moreover, in limited coinfection experimental studies, different PRRSV strains, infection sequence, animal models, and susceptible cells affect the observation results (Table 1).
Co-infection of PRRSV and PCV2
PCV is a naked, circular, single-stranded DNA virus belonging to the family Circoviridae. It can be divided into four types: PCV1, PCV2, PCV3, and PCV4 (37, 38). It is the main pathogen causing porcine circovirus diseases (PCVDs) and porcine circovirus 2-systemic disease (PCV2-SD) (39, 40). A series of disease syndromes caused by PCV2 are collectively known as porcine circovirus-associated disease (PCVAD) (41). Its symptoms mainly include PCV2-associated respiratory disease, low growth performance, and postweaning multisystemic wasting syndrome (PMWS) (42, 43). Samples of 159 sick and dead swine collected from eight provinces and cities in China were evaluated, and it was demonstrated that the coinfection rates of PRRSV, CSFV, PCV2, and PCV3 reached 15.72%. Meanwhile, the coinfection rate of PRRSV and PCV2 reached 10.69% (44). Another study on lesions of proliferative and necrotizing pneumonia (PNP) showed that coinfection with PRRSV and PCV2 occurred more often in cases, including in weaned piglets, with a coinfection rate of 42% (45). Furthermore, PRRSV was detected in 51.9% of the PMWS cases (46). A high rate of coinfection involving PRRSV and PCV2 was observed in the swineherd where the lesions occurred. Therefore, it is assumed that coinfection of PRRSV and PCV2 may be one of the main causes of PCVAD.
Both PRRSV and PCV2 target the host's immune cells, disrupting their immune function and leading to increased susceptibility to primary and secondary pathogens, which may affect host growth performance and the incidence and lethality of associated diseases (47). It has been reported that the inoculation of PCV2 1 week after HP-PRRSV resulted in the highest viral load, the most severe clinical signs, and the highest mortality (60%) in piglets (28). In further studies, it was found that a PCV2 infection in swine infected with HP-PRRSV enhanced the replication of both viruses and led to more severe clinical signs and lesions (28). Furthermore, this result seems to be verified in an in vitro coinfection model, based on changes in the levels of PRRSV N gene and PCV2 Cap gene in porcine alveolar macrophage (PAM) cells. It was concluded that the coinfection model enhanced the replication of PRRSV and PCV2 (27). Moreover, the levels of p-IκBα and p65 proteins in the nucleus were significantly increased after coinfection. The co-infection could induce NF-κB translocation from the cytoplasm to nucleus, which leads to activation of the NF-κB pathway (27). Cumulatively, these results demonstrate synergistic effects during PRRSV and PCV2 coinfection (28, 48).
Based on the Cap gene sequence of PCV2, five subtypes can be classified: PCV2a, PCV2b, PCV2c, PCV2d, and PCV2e (49). Some studies have reported that PRRSV can also affect infection with PCV2 subtypes (mainly PCV2a and PCV2b) by prolonging viremia and in vivo shedding of PCV2 (29). Coinfection of different subtypes of PCV2 with PRRSV also increased the mutation rate of PRRSV in successive generations. The mutation rates of ORF5 and ORF6 in swine co-infected with PRRSV and PCV2b were significantly higher than in those infected with PRRSV alone. The ORF7 mutation rate in swine coinfected with PRRSV and PCV2a was also significantly higher (50). Therefore, it is shown that coinfection with different subtypes of PRRSV and PCV2 can cause a higher mutation rate of PRRSV, which may be one of the reasons for difficulty in complete clearance of PRRSV.
Co-infection of PRRSV and PRV
PRV is a linear, double-stranded DNA virus with an envelope, also known as porcine herpesvirus type I, belonging to the family Herpesviridae (51). PRV causes neurological and febrile symptoms and is one of the major causes of infectious diseases affecting the global pig industry (52). It has an excessively broad host range, infecting both domestic and wild animals, and causes different symptoms (53). Infected animals typically develop fever, sneezing, coughing, and vomiting. This is occasionally accompanied by typical neurological signs such as twitching, aggressiveness, and lack of coordination. PRV can infect swine of all ages, with a 100% lethality rate in piglets up to 15 days of age (54). However, the adult tolerant swine can establish and maintain latent infection in the peripheral nervous system. In the event of external stimulation or immune deficiency, the latently infected viral genome can be reactivated. This causes massive viral replication in epithelial tissues and outgrowth to other tissues, as well as outgrowing the virus, making the host a dangerous source of infection again (55).
Previous surveys have also shown a high proportion of latent PRV infections in swine (53). It has been found that the current cause of infection in swine farms is the continuous mutation of PRV strains. Although the traditional Bartha-K61 vaccine provides complete protection against the classical strain (SC), it does not protect against the currently prevalent mutated strain (HeN1) (56). Co-infection of PRRSV and PRV is also common, with a co-infection rate of 36% (21). Moreover, it is common in intensive fattening swine in Japan (57). However, there are few studies on the co-infection of the two viruses in vivo and vitro, and the interaction between them needs to be further explored. Moreover, the latent infection of PRV in adult-tolerant pigs may be one of the reasons for the high coinfection rate of the two viruses.
Co-infection of PRRSV and CSFV
CSFV, a single-stranded positive-sense RNA virus with a capsid belongs to the Flaviviridae family, is the pathogen that causes classical swine fever (58). It has been suggested that CSFV includes three different subtypes; however there is no international consensus on their classification system (59). CSFV contains a large ORF, 3′-UTR and 5′-UTR, encoding four structural proteins: C, Erns, E1, and E2. It also encodes eight nonstructural proteins: Npro, P7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B (60). Established multiplex real time PCR (MRT-PCR) was used to analyze 69 clinical samples from three provinces in China, showing that the coinfection rate of PRRSV and CSFV reached 4.4% (20). Overall, the coinfection rate of PRRSV and CSFV in China is about 0–7.7% (20). Furthermore, PRRSV infection affects CSFV vaccination. The Chinese (C) strain vaccine is considered to be the most effective in inducing protective immunity against CSFV (61), but in the field, immunization often fails owing to infection by immunosuppressive pathogens such as PRRSV.
It is clear that the coinfection rate of PRRSV and CSFV in intensive swine farms is low, but PRRSV infection in the field can cause CSFV vaccination failure. Moreover, current vaccination strategies against both viruses still use two single modified live vaccines, leading to mutual interference (62). Therefore, it is speculated that the PRRSV strain interacts with the CSFV vaccine strains. In a study, it was found that PRRSV infection caused the upregulation of pro-inflammatory cytokines in vitro, particularly TNF-α. The results showed that the replication of CSFV-C was inhibited, which also provided an explanation for the failure of CSFV vaccination caused by PRRSV (31). However, this study does not have the data of in vivo experiments. Moreover, the cell used by authors is PAM39, a cell line based on 3D4/21. This is not the primary culture cell. It is still difficult to choose a model suitable for the study of coinfection. Therefore, the absence of suitable host cells limits its study.
Co-infection of PRRSV and SIV
SIV is any strains of the influenza virus family prevalent in swine, including influenza C and subtypes of influenza A, namely H1N1, H1N2, H2N1, H2N3, H3N1, and H3N2 (63, 64). Among these, the H1N1 SIV type A is an influenza virus with pandemic potential (65). Both SIV and PRRSV can cause subclinical infection; the symptoms of infection in swine mainly include high temperature, fever, cough, and dyspnea, which are difficult to distinguish clinically (66–68). Therefore, coinfection caused by SIV and PRRSV is even more difficult to prevent and control.
In a study involving 636 SIV-positive cases, 109 PRRSV-positive samples were detected, and the coinfection rate reached 17% (69). The study on coinfection of PRRSV and SIV was reported as early as 1996. The results showed that the coinfection group had more severe clinical symptoms and growth retardation than the single infection group. However, there was little effect on SIV replication (32). This is consistent with the results of another study which showed that the coinfection group suffered more severe inflammation of the bronchial wall and that preinfection with PRRSV did not affect the stage of SIV infection (33). In contrast, a different clinical outcome was found in another study (70). The phenomenon may be related to different strains and the immune status of swine. Moreover, the growth kinetics of the two viruses were studied in PAM cells. Results showed that no matter which virus first infected, PRRSV or SIV could slightly inhibit the replication of the later infected virus (34). This is consistent with the results of in vivo experimental studies (32). Further, the mRNA levels of IFN-β, IL-10, and TNF-α were found to increase significantly after coinfection and had an additive effect (34). However, this study only involved assessment of the expression of host cytokines. Similarly, such defects are common in all coinfection studies. Moreover, this is limited by the current level of research. Therefore, a clear understanding of the pathogenic mechanism of single infection is necessary for the study of co-infection. The latest research showed that PRRSV-1 changes the relationship between SIV and its main target cells, and the interaction between them could also affect vaccination (71). This undoubtedly provides a new research method for studying the interaction between the two viruses, and also indicates the complexity of the interaction between the two viruses in coinfection. To sum up, it can be inferred that the results of coinfection are also related to other factors, such as different virus strains, sequence of infection and susceptible cells.
Co-infection of PRRSV and Other Viruses
In addition to the above-mentioned viruses, coinfection could also occur between PRRSV and other viruses, such as HEV, PPV, PARV, and PEDV (36, 72). HEV is a non-enveloped single-stranded positive-sense RNA virus, which belongs to the Hepeviridae family (73). It can be classified in four different genotypes and 24 subtypes based on its nucleotide sequence (74). Its transmission is common between humans and pigs, in which genotypes three and four are found in both pigs and humans (75, 76). Human infections with HEV can cause acute liver failure or chronic infections (77). Although pigs infected with HEV have no pathogenicity, they may enhance the pathogenicity of other porcine viruses (77). Coinfection of HEV and PRRSV can lead to long-term shedding of HEV and even chronic infection. After coinfection with PRRSV and HEV, the shedding of HEV was delayed by 1.9 times, and the specific immune response was delayed by 1.6 times compared with that of HEV infection alone (35). It is suggested that PRRSV has an important effect on the infection dynamics of HEV. Moreover, in another study, it was found that viremia occurred earlier and longer after co-infection of HEV and PRRSV (36). Thus, this may indicate that coinfection regulates the duration of viremia in HEV. However, another study found that there was no significant correlation between PRRSV-positive and HEV-positive status in plasma, tonsil and cecal content samples (78).
PPV is an important virus causing reproductive disorders in pigs. With the development of molecular technology, the detection of several new types of PPV have increased in the past few years. Such as the porcine parvovirus type 2 (PPV2), also known as Cnvirus (CnP-PARV4), porcine parvovirus type 3 (PPV3), and porcine parvovirus type 4 (PPV4) (79). Moreover, many studies have proved the possibility of coinfection of emerging PPV and PCV2, especially in PRDC (80). Although coinfection between PRRSV and emerging PPV remains to be studied, PRRSV, as one of the key pathogens in PRDC, has a great possibility of co-infection. In a study on the co-infection of PRRSV and other pathogens in the intestinal tract, it was found for the first time that PRRSV and porcine group a rotavirus (PARV) were co-infected, up to 52.4%. Furthermore, the coinfection rate of PPV and PEDV reached 33.3%, and the coinfection rate of PRRSV with more than two viruses also reached 33.3% (72). However, there are few studies on the co-infection of PRRSV, PARV, and PEDV in vivo and in vitro. The current research can only prove the co-existence of these viruses in the host, but cannot show that there is a complex interaction between them.
Multiple infections are common in intensive pig farms in addition to single and co-infections of related pathogens (20, 44). However, studies on the interaction of PRRSV-associated multiple infections and their pathogenic mechanisms are rare. Most relevant studies were based on MRT-PCR methods constructed for multiple pathogens and remain only in the stage of diagnostic testing. Therefore, coinfection studies between two pathogens associated with PRRSV are becoming increasingly important, contributing to the study of PRRSV-associated multiple infections.
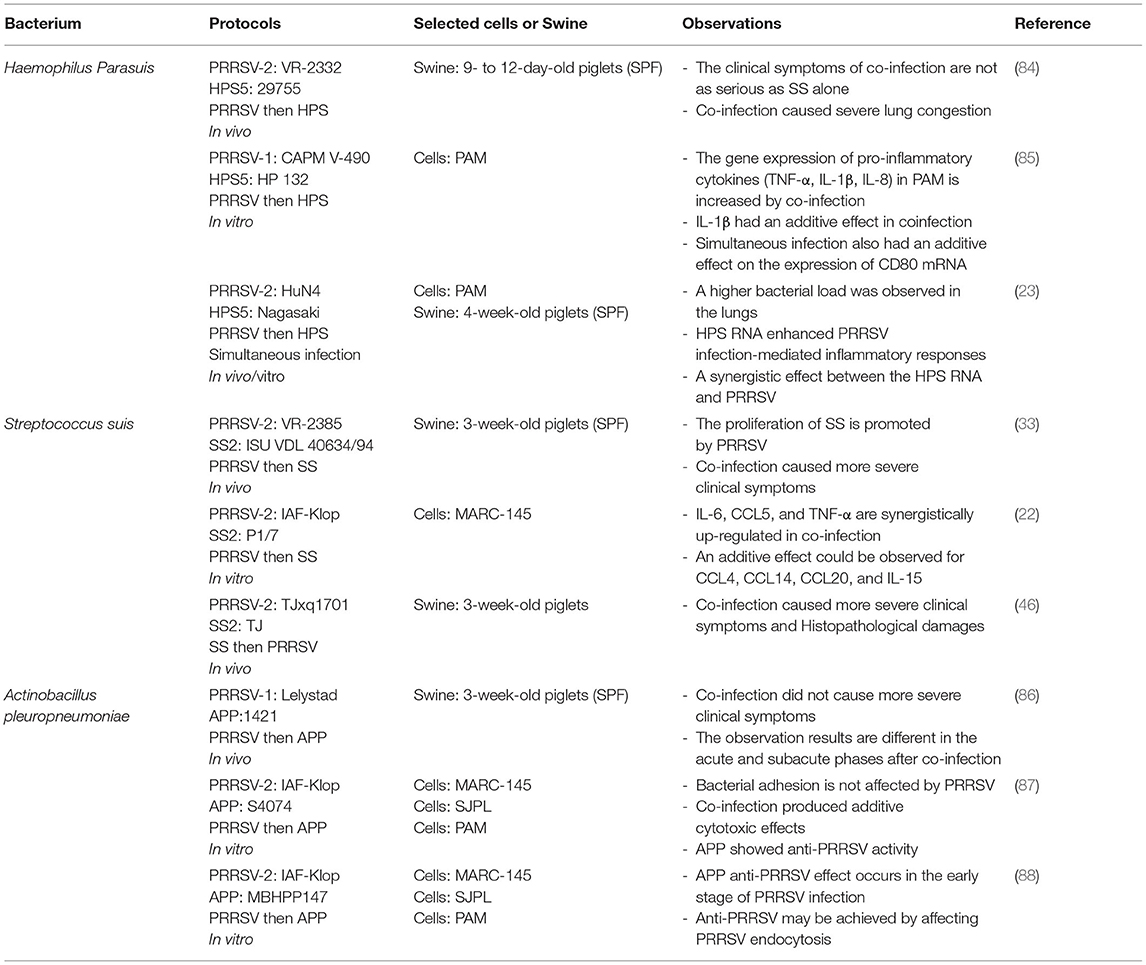
Co-infection of PRRSV and Bacteria
It is well-known that viral infections can induce an ideal environment for bacterial secondary infection through different mechanisms, such as disruption of the epithelial barrier, modulation of the expression of receptors involved in bacterial adhesion, and alteration of the host immune response (81–83). In addition, the results of secondary infections caused by different PRRSV strains seem to be inconsistent, which creates a great deal of confusion in the study of PRRSV-bacterial coinfection (Table 2).
Co-infection of PRRSV and HPS
HPS is a gram-negative bacterium and a common pathogen causing respiratory disease in swine. It can invade, and cause severe systemic disease under suitable conditions, such as fibrinous polyserositis, arthritis, and meningitis (89).
PRRSV infection can predispose swine to secondary infection by destroying PAM cells and inducing inflammation of the nasal mucosa (90). The high detection rate of HPS in PRRSV-infected pig farms implies that PRRSV infection increases the susceptibility for HPS (91). Indeed, studies have demonstrated that PRRSV can increase secondary HPS invasion. However, in vivo and in vitro studies appear to have produced different results. In 1997, a study of coinfection in vivo has reported that there were no more serious clinical symptoms in the co-infection group (84). However, an in vitro study showed that a strong pro-inflammatory immune response is triggered by co-infection in PAM cells (85). Besides, HP-PRRSV was found to promote HPS proliferation in blood and tissues in co-infection studies, which could explain the susceptibility of HPS in PRRSV-positive pig populations (92). Further, changes in cytokines have been reported in other studies. It is found that co-infection of PRRSV with HPS can increase the expression of pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-8 in PAM cells. At the protein level, it is also confirmed that co-infection has an additive effect on IL-1β (85). In addition, recent related studies have shown that transfection of HPS RNA enhances HP-PRRSV-mediated inflammatory responses in co-infection (23), which expands on previous studies examining only the effect of PRRSV on HPS. In summary, the co-infection of PRRSV and HPS cannot be explicitly defined as a cooperative promotion, since different results were found in the co-infection group. Moreover, the reasons for these results are closely related to a better immune system in vivo. In the current general trend of the epidemic of recombinant PRRSV strains, their high recombination rate and widespread transmission increase the morbidity and mortality of HPS. Therefore, research dedicated to the mechanisms of co-infection between the currently prevalent strains and HPS is more appropriate for the prevention and control of HPS outbreaks.
Co-infection of PRRSV and SS
SS is a gram-positive bacterium that is believed to be responsible for various clinical disease syndromes in swine (93). SS can be divided into type I and type II. Its symptoms mainly include arthritis, meningitis, pneumonia, septicemia, endocarditis, polyserositis, abortion, and abscess (94). Moreover, SS II can infect humans, causing meningitis, septicemia, and endocarditis (95).
PRRSV and SS coinfection usually causes disease progression, leading to increased morbidity and mortality (87). In the early days, most studies were based on the invasion of individual SS to investigate the pathogenic mechanisms, and the studies on coinfection were scarce. Since 2,000, there has been an increasing number of studies on PRRSV and SS coinfection. Many studies have shown that PRRSV infection can increase susceptibility to SS and cause more severe clinical symptoms (86, 96, 97). One of the studies showed that PRRSV infection suppressed cell immune function of PAM, thereby affecting their ability to clear SS, leading to a wider spread of SS in tissues. Furthermore, it exacerbates the development of related diseases, among which swine inoculated with the HP-PRRSV strain are most severely infected by SS (86). Another study reported the expression levels of inflammatory factors in bone marrow dendritic cells (BMDCs) infected by the two pathogens. The results showed that secondary infection of SS after PRRSV infection resulted in an additive effect of CCL4, CCL14, CCL20, and IL-15, with significant synergistic upregulation of IL-6, CCL5, and TNF-α (87). There are many studies with similar results, suggesting that secondary infection with SS enhances the inflammatory response mediated by PRRSV infection (97, 98). Furthermore, recent studies have reported that early infection with SS type II increased the virulence of the HP-PRRSV MLV-like strain (TJxq1701), causing an excessive inflammatory response and tissue damage, resulting in higher morbidity and mortality in piglets (88). It is suggested that recombination between PRRSV modified live vaccine strains and other PRRSV strains leads to the emergence of PRRSV MLV-like strains and secondary infection with SS causes more severe clinical signs. Also, there appears to be a cooperative promotion between PRRSV and SS. This provides a reference for the prevention and control of PRRSV as well as coinfection studies.
Co-infection of PRRSV and APP
APP is a gram-negative bacterium which can cause highly contagious respiratory disease (99). APP can be broadly classified into two types, namely, biotype 1 and biotype 2. According to the polysaccharide antigen on its surface, biotype 1 has 12 serotypes, and biotype 2 has 6 serotypes (100). Swine of all ages can be infected with APP; however, the clinical symptoms are different according to their APP strains and infection cycle (101).
As early as 1997, an in vivo study was carried out on the coinfection of PRRSV and APP. It was found that the experimental results were completely inconsistent between the acute and subacute stages after co-infection. It was observed that secondary infection with APP in the acute phase could lead to more severe disease, but it did not have any effect during the subacute phase (33). This is consistent with results from another in vitro study, in which pre-infection with PRRSV did not affect the adhesion capacity of APP (22). In addition, other studies have found that pre-infection of in vitro cell lines with APP, completely blocked PRRSV infection. Even the supernatant of APP cultures was sufficient to significantly block PRRSV infection, presumably in association with heat-resistant APP metabolites (22). Further, its anti-PRRSV mechanism has been investigated in recent studies. It is found that this blocking effect may be achieved by activating cofilin and causing actin depolymerization, which in turn affects PRRSV endocytosis (46). In summary, PRRSV had little effect on APP while APP possessed anti-PRRSV infection activity, suggesting an inhibitory effect between them. However, studies on the underlying molecular mechanisms do not seem to have explored in depth the molecular associations between the two pathogens, such as how APP metabolites are associated with cofilin in PRRSV. Such a problem seems to be a limitation in every coinfection study. Thus, whether future studies can find prevention and control strategies based on the molecular link between the two pathogens remains to be further investigated.
Coinfection With Other Pathogens
In addition to coinfection with the above-mentioned viruses and bacteria, PRRSV can co-infect with other pathogens, such as T. gondii, Mhp. T. gondii is a specific intracellular protozoan parasite that can invade any nucleated host cell (102). Among these, the pig is one of the intermediate hosts. T. gondii can also cause opportunistic pathogen infection (103). PRRSV is a common pathogen that causes coinfection with T. gondii. Through the evaluation of 372 samples from 9 provinces in China, it was found that the coinfection rate was 1.61% (26). This is the first time that coinfection of PRRSV and T. gondii has been reported in China, but there is no further explanation of the pathological manifestations after co-infection. A case of co-infection was also found in South Korea. The pathological manifestations in the lungs were alveolar macrophage necrosis, hemorrhage, multiple necrosis, and diffuse interstitial pneumonia. It was found that T. gondii infection can lead to more serious pathological changes, and it is speculated that T. gondii promotes PRRSV infection. However, there are few studies on parasites and PRRSV, especially on the related mechanisms.
Mhp infection can destroy the ciliated epithelium of the respiratory tract, thereby adversely affecting the mucosal system. The main clinical symptoms are chronic cough, and coinfection is often accompanied by other clinical symptoms such as fever and growth impairment. Mhp is also one of the main causes of PRDC, which is often isolated with PRRSV. It is suggested that Mhp plays an important role in PRRSV infections. Studies on related coinfection have been conducted for a long time. Some studies have pointed out that Mhp infection can increase the production of pro-inflammatory factors, which in turn leads to an increase in the incidence of PRRSV (25). Through bioinformatics analysis of differential genes in PAM cells after coinfection, some researchers found that inflammatory factors such as CCL-4, IL-1β, IL-1α, and CCL-2 were significantly up-regulated after coinfection (104). It is suggested that excessive inflammatory reaction is one of the causes of severe lung lesions after coinfection, which confirms the results of previous studies (105). This provides a reference for the study of the mechanism of coinfection and acts as a guide for the prevention and control of PRDC.
Perspectives
As shown in this review, the emergence of new strains such as NADC30-like, represents a high mutation stage of the PRRSV epidemic in China, and PRRSV-2 appears to become more common (106, 107). Not only the NADC30-like strain itself is highly recombinant, but its coinfection with other pathogens increased the mutation rate of PRRSV. This affects the molecular outcome of the virus itself and the host, such as the evolution of the virus, clinical symptoms, susceptibility to antiviral therapy, and lethality of the virus infecting the host. As one of the main strains in the current epidemic, HP-PRRSV is also susceptible to coinfection and recombination with other strains (10). The continuous evolution of PRRSV has made the study of pathogenic mechanisms strenuous as well as the clinical signs of PRRS and co-infection with other porcine pathogens difficult to diagnose. Therefore, in addition to the study of the pathogenic mechanism, it is necessary to control the virus mutation caused by co-infection.
However, the high mutation rate of PRRSV, the variable epidemiological trends, the complexity of the interactions among different PRRSV strains and other porcine pathogens, the different molecular consequences of co-infection, and numerous abiotic factors limit the study of co-infection. Moreover, after reviewing the above in vivo and in vitro experiments and field co-infection rate, we briefly summarized the interactions, influencing factors and molecular consequences between PRRSV and other porcine pathogens (Figure 1). Different strains, infection sequence, cell selection, animal selection, immune status and individual differences, and even pig sex and different ages may cause the different co-infection results. Further, the unclear pathogenic mechanism of single virus also greatly limits the relevant co-infection research. Therefore, in addition to studying the interactions between PRRSV and other porcine pathogens during co-infection, more research is needed on how these alter the host immune response and influence the effectiveness of vaccination. Hence, further research on the recombination of PRRSV is warranted. These studies may provide a new strategy for the prevention and control of PRRSV co-infection associated diseases.
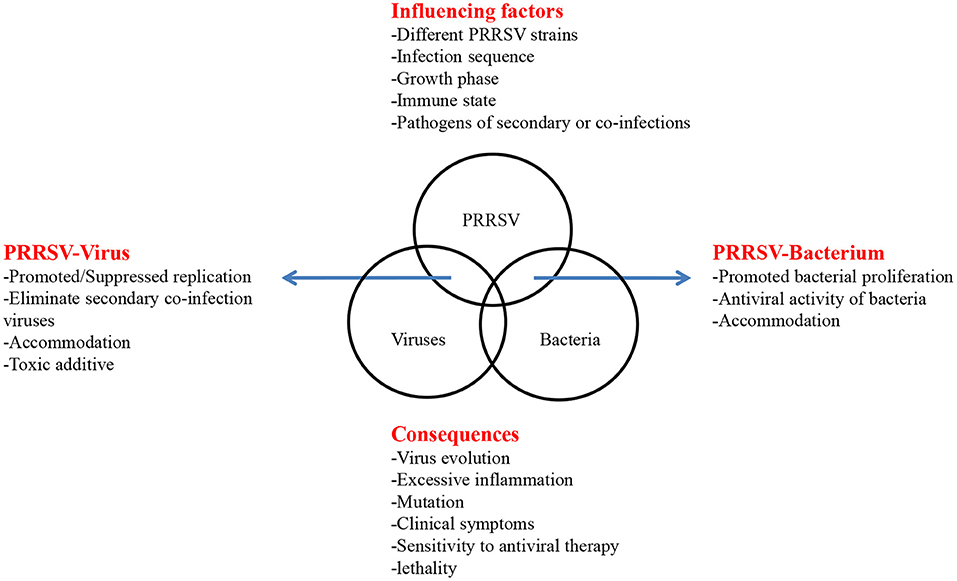
Figure 1. Influencing factors and consequences of PRRSV co-infection. The interaction of coinfection is listed by arrows on both sides. In the top box, factors that may affect coinfections are listed. In the below box, some consequences of coinfections are listed.
Author Contributions
DZhao, BY, and KZ conceived and designed the study. DZhao and XL wrote the original draft of the manuscript. KZ, HZ, XY, CS, DZhan, XS, TZ, HC, XC, and YH wrote sections of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from Gansu major science and technology projects (20ZD7NA006) and Joint Project of National Natural Science Foundation of China (31941002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to thank the editors and reviewers for their valuable comments and suggestions that helped improve the quality of this manuscript.
References
1. Ma Z, Yu Y, Xiao Y, Opriessnig T, Wang R, Yang L, et al. Sustaining interferon induction by a high-passage atypical porcine reproductive and respiratory syndrome virus strain. Sci Rep. (2016) 6:36312. doi: 10.1038/srep36312
3. Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, et al. Porcine reproductive and respiratory syndrome virus (PRRSV): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. (2016) 4:129–54. doi: 10.1146/annurev-animal-022114-111025
4. Meng XJ, Paul PS, Halbur PG, Lum MA. Characterization of a high-virulence US isolate of porcine reproductive and respiratory syndrome virus in a continuous cell line, ATCC CRL11171. J Vet Diagn Invest. (1996) 8:374–81. doi: 10.1177/104063879600800317
5. Drew TW. A review of evidence for immunosuppression due to porcine reproductive and respiratory syndrome virus. Vet Res. (2000) 31:27–39. doi: 10.1051/vetres:2000106
6. Snijder EJ, Kikkert M, Fang Y. Arterivirus molecular biology and pathogenesis. J Gen Virol. (2013) 94:2141–63. doi: 10.1099/vir.0.056341-0
7. Albina E. Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol. (1997) 55:309–16. doi: 10.1016/S0378-1135(96)01322-3
8. Rosendal T, Dewey C, Friendship R, Wootton S, Young B, Poljak Z. Spatial and temporal patterns of porcine reproductive and respiratory syndrome virus (PRRSV). Genotypes in Ontario, Canada, 2004-2007. BMC Vet Res. (2014) 10:83. doi: 10.1186/1746-6148-10-83
9. Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bao Y, et al. Reorganization and expansion of the nidoviral family arteriviridae. Arch Virol. (2016) 161:755–68. doi: 10.1007/s00705-015-2672-z
10. Zhao K, Ye C, Chang XB, Jiang CG, Wang SJ, Cai XH, et al. Importation and recombination are responsible for the latest emergence of highly pathogenic porcine reproductive and respiratory syndrome virus in China. J Virol. (2015) 89:10712–6. doi: 10.1128/JVI.01446-15
11. Kvisgaard LK, Larsen LE, Hjulsager CK, Botner A, Rathkjen PH, Heegaard PMH, et al. Genetic and biological characterization of a porcine reproductive and respiratory syndrome virus 2 (PRRSV-2). Causing significant clinical disease in the field. Vet Microbiol. (2017) 211:74–83. doi: 10.1016/j.vetmic.2017.10.001
12. Bai W, Wang Z, Sun P, Zhang J, Bao H, Cao Y, et al. The molecular characteristic analysis of PRRSV GSWW/2015 strain and its pathogenicity to pigs. BMC Vet Res. (2018) 14:240. doi: 10.1186/s12917-018-1548-3
13. Jiang Y, Li G, Yu L, Li L, Zhang Y, Zhou Y, et al. Genetic diversity of porcine reproductive and respiratory syndrome virus (PRRSV). From 1996 to 2017 in China. Front Microbiol. (2020) 11:618. doi: 10.3389/fmicb.2020.00618
14. Wenhui L, Zhongyan W, Guanqun Z, Zhili L, JingYun M, Qingmei X, et al. Complete genome sequence of a novel variant porcine reproductive and respiratory syndrome virus (PRRSV). Strain: evidence for recombination between vaccine and wild-type PRRSV strains. J Virol. (2012) 86:9543. doi: 10.1128/JVI.01341-12
15. Lu WH, Tun HM, Sun BL, Mo J, Zhou QF, Deng YX, et al. Re-emerging of porcine respiratory and reproductive syndrome virus (lineage 3). And increased pathogenicity after genomic recombination with vaccine variant. Vet Microbiol. (2015) 175:332–40. doi: 10.1016/j.vetmic.2014.11.016
16. Zhang Q, Jiang P, Song Z, Lv L, Li L, Bai J. Pathogenicity and antigenicity of a novel NADC30-like strain of porcine reproductive and respiratory syndrome virus emerged in China. Vet Microbiol. (2016) 197:93–101. doi: 10.1016/j.vetmic.2016.11.010
17. Zhou L, Kang R, Yu J, Xie B, Chen C, Li X, et al. Genetic characterization and pathogenicity of a novel recombined porcine reproductive and respiratory syndrome virus 2 among Nadc30-Like, Jxa1-Like, Mlv-Like strains. Viruses. (2018) 10:551. doi: 10.3390/v10100551
18. Qin S, Ruan W, Yue H, Tang C, Zhou K, Zhang B. Viral communities associated with porcine respiratory disease complex in intensive commercial farms in Sichuan province, China. Sci Rep. (2018) 8:13341. doi: 10.1038/s41598-018-31554-8
19. Saade G, Deblanc C, Bougon J, Marois-Crehan C, Fablet C, Auray G, et al. Coinfections and their molecular consequences in the porcine respiratory tract. Vet Res. (2020) 51:80. doi: 10.1186/s13567-020-00807-8
20. Liu S, Zhao Y, Hu Q, Lv C, Zhang C, Zhao R, et al. A multiplex RT-PCR for rapid and simultaneous detection of porcine teschovirus, classical swine fever virus, and porcine reproductive and respiratory syndrome virus in clinical specimens. J Virol Methods. (2011) 172:88–92. doi: 10.1016/j.jviromet.2010.12.023
21. Zhou H, Pan Y, Liu M, Han Z. Prevalence of porcine pseudorabies virus and its coinfection rate in heilongjiang province in China from 2013 to 2018. Viral Immunol. (2020) 33:550–4. doi: 10.1089/vim.2020.0025
22. Levesque C, Provost C, Labrie J, Hernandez Reyes Y, Burciaga Nava JA, Gagnon CA, et al. Actinobacillus pleuropneumoniae possesses an antiviral activity against porcine reproductive and respiratory syndrome virus. PLoS ONE. (2014) 9:e98434. doi: 10.1371/journal.pone.0098434
23. Li J, Wang S, Li C, Wang C, Liu Y, Wang G, et al. Secondary haemophilus parasuis infection enhances highly pathogenic porcine reproductive and respiratory syndrome virus (HP-PRRSV). Infection-mediated inflammatory responses. Vet Microbiol. (2017) 204:35–42. doi: 10.1016/j.vetmic.2017.03.035
24. Li J, Wang J, Liu Y, Yang J, Guo L, Ren S, et al. Porcine reproductive and respiratory syndrome virus NADC30-like strain accelerates streptococcus suis serotype 2 infection in vivo and in vitro. Transbound Emerg Dis. (2019) 66:729–42. doi: 10.1111/tbed.13072
25. Thacker EL, Halbur PG, Ross RF, Thanawongnuwech R, Thacker BJ. Mycoplasma hyopneumoniae potentiation of porcine reproductive and respiratory syndrome virus-induced pneumonia. J Clin Microbiol. (1999) 37:620–7. doi: 10.1128/JCM.37.3.620-627.1999
26. Wang S, Zhang M, Liu XC, Lin T, Yang HC, Yuan SS, et al. Investigation on the co-infections of toxoplasma gondii with PRRSV, CSFV or PCV-2 in swine in part of China. J Integr Agric. (2015) 14:1838–44. doi: 10.1016/S2095-3119(15)61044-9
27. Sun N, Sun P, Lv H, Sun Y, Guo J, Wang Z, et al. Matrine displayed antiviral activity in porcine alveolar macrophages co-infected by porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Sci Rep. (2016) 6:24401. doi: 10.1038/srep24401
28. Fan P, Wei Y, Guo L, Wu H, Huang L, Liu J, et al. Synergistic effects of sequential infection with highly pathogenic porcine reproductive and respiratory syndrome virus and porcine circovirus type 2. Virol J. (2013) 10:265. doi: 10.1186/1743-422X-10-265
29. Sinha A, Shen HG, Schalk S, Beach NM, Huang YW, Meng XJ, et al. Porcine reproductive and respiratory syndrome virus (PRRSV). Influences infection dynamics of porcine circovirus type 2 (PCV2). subtypes PCV2a and PCV2b by prolonging PCV2 viremia and shedding. Vet Microbiol. (2011) 152:235–46. doi: 10.1016/j.vetmic.2011.05.005
30. Albina E, Piriou L, Hutet E, Cariolet R, L'Hospitalier R. Immune responses in pigs infected with porcine reproductive and respiratory syndrome virus (PRRSV). Vet Immunol Immunopathol. (1998) 61:49–66. doi: 10.1016/S0165-2427(97)00134-7
31. Chen D, Liu X, Xu S, Chen D, Zhou L, Ge X, et al. TNF-alpha induced by porcine reproductive and respiratory syndrome virus inhibits the replication of classical swine fever virus C-strain. Vet Microbiol. (2019) 234:25–33. doi: 10.1016/j.vetmic.2019.05.007
32. Van Reeth K, Nauwynck H, Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol. (1996) 48:325–35. doi: 10.1016/0378-1135(95)00145-X
33. Pol JM, van Leengoed LA, Stockhofe N, Kok G, Wensvoort G. Dual infections of PRRSV/influenza or PRRSV/Actinobacillus pleuropneumoniae in the respiratory tract. Vet Microbiol. (1997) 55:259–64. doi: 10.1016/S0378-1135(96)01323-5
34. Dobrescu I, Levast B, Lai K, Delgado-Ortega M, Walker S, Banman S, et al. In vitro and ex vivo analyses of co-infections with swine influenza and porcine reproductive and respiratory syndrome viruses. Vet Microbiol. (2014) 169:18–32. doi: 10.1016/j.vetmic.2013.11.037
35. Salines M, Barnaud E, Andraud M, Eono F, Renson P, Bourry O, et al. Hepatitis E virus chronic infection of swine co-infected with porcine reproductive and respiratory syndrome virus. Vet Res. (2015) 46:55. doi: 10.1186/s13567-015-0207-y
36. Salines M, Demange A, Stephant G, Renson P, Bourry O, Andraud M, et al. Persistent viremia and presence of hepatitis E virus RNA in pig muscle meat after experimental co-infection with porcine reproductive and respiratory syndrome virus. Int J Food Microbiol. (2019) 292:144–9. doi: 10.1016/j.ijfoodmicro.2018.12.023
37. Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. (2016) 13:184. doi: 10.1186/s12985-016-0642-z
38. Zhang HH, Hu WQ, Li JY, Liu TN, Zhou JY, Opriessnig T, et al. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, Hunan province, China. Transbound Emerg Dis. (2020) 67:1057–61. doi: 10.1111/tbed.13446
39. Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, et al. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J Gen Virol. (1998) 79 (Pt 9):2171–9. doi: 10.1099/0022-1317-79-9-2171
40. Liu Y, Li J, Yang J, Zeng H, Guo L, Ren S, et al. Emergence of different recombinant porcine reproductive and respiratory syndrome viruses, China. Sci Rep. (2018) 8:4118. doi: 10.1038/s41598-018-22494-4
41. Opriessnig T, Halbur PG. Concurrent infections are important for expression of porcine circovirus associated disease. Virus Res. (2012) 164:20–32. doi: 10.1016/j.virusres.2011.09.014
42. Segales J. Porcine circovirus type 2 (PCV2). Infections: clinical signs, pathology and laboratory diagnosis. Virus Res. (2012) 164:10–9. doi: 10.1016/j.virusres.2011.10.007
43. Opriessnig T, Langohr I. Current state of knowledge on porcine circovirus type 2-associated lesions. Vet Pathol. (2013) 50:23–38. doi: 10.1177/0300985812450726
44. Chen N, Huang Y, Ye M, Li S, Xiao Y, Cui B, et al. Co-infection status of classical swine fever virus (CSFV), porcine reproductive and respiratory syndrome virus (PRRSV). And porcine circoviruses (PCV2 and PCV3). in eight regions of China from 2016 to 2018. Infect Genet Evol. (2019) 68:127–35. doi: 10.1016/j.meegid.2018.12.011
45. Drolet R, Larochelle R, Morin M, Delisle B, Magar R. Detection rates of porcine reproductive and respiratory syndrome virus, porcine circovirus type 2, and swine influenza virus in porcine proliferative and necrotizing pneumonia. Vet Pathol. (2003) 40:143–8. doi: 10.1354/vp.40-2-143
46. Hernandez Reyes Y, Provost C, Traesel CK, Jacques M, Gagnon CA. Actinobacillus pleuropneumoniae culture supernatant antiviral effect against porcine reproductive and respiratory syndrome virus occurs prior to the viral genome replication and transcription through actin depolymerization. J Med Microbiol. (2018) 67:249–64. doi: 10.1099/jmm.0.000659
47. Niederwerder MC, Jaing CJ, Thissen JB, Cino-Ozuna AG, McLoughlin KS, Rowland RR. Microbiome associations in pigs with the best and worst clinical outcomes following co-infection with porcine reproductive and respiratory syndrome virus (PRRSV). And porcine circovirus type 2 (PCV2). Vet Microbiol. (2016) 188:1–11. doi: 10.1016/j.vetmic.2016.03.008
48. Harms PA, Sorden SD, Halbur PG, Bolin SR, Lager KM, Morozov I, et al. Experimental reproduction of severe disease in CD/CD pigs concurrently infected with type 2 porcine circovirus and porcine reproductive and respiratory syndrome virus. Vet Pathol. (2001) 38:528–39. doi: 10.1354/vp.38-5-528
49. Jiang CG, Wang G, Tu YB, Liu YG, Wang SJ, Cai XH, et al. Genetic analysis of porcine circovirus type 2 in China. Arch Virol. (2017) 162:2715–26. doi: 10.1007/s00705-017-3414-1
50. Yin SH, Xiao CT, Gerber PF, Beach NM, Meng XJ, Halbur PG, et al. Concurrent porcine circovirus type 2a (PCV2a). Or PCV2b infection increases the rate of amino acid mutations of porcine reproductive and respiratory syndrome virus (PRRSV). during serial passages in pigs. Virus Res. (2013) 178:445–51. doi: 10.1016/j.virusres.2013.09.007
51. Peng Z, Ouyang T, Pang D, Ma T, Chen X, Guo N, et al. Pseudorabies virus can escape from CRISPR-Cas9-mediated inhibition. Virus Res. (2016) 223:197–205. doi: 10.1016/j.virusres.2016.08.001
52. Mettenleiter TC. Immunobiology of pseudorabies (Aujeszky's disease). Vet Immunol Immunopathol. (1996) 54:221–9. doi: 10.1016/S0165-2427(96)05695-4
53. Gu J, Hu D, Peng T, Wang Y, Ma Z, Liu Z, et al. Epidemiological investigation of pseudorabies in Shandong Province from 2013 to 2016. Transbound Emerg Dis. (2018) 65:890–8. doi: 10.1111/tbed.12827
54. Pedersen K, Bevins SN, Baroch JA, Cumbee JC Jr, Chandler SC, et al. Pseudorabies in feral swine in the United States, 2009-2012. J Wildl Dis. (2013) 49:709–13. doi: 10.7589/2012-12-314
55. Klupp BG, Hengartner CJ, Mettenleiter TC, Enquist LW. Complete, annotated sequence of the pseudorabies virus genome. J Virol. (2004) 78:424–40. doi: 10.1128/JVI.78.1.424-440.2004
56. An TQ, Peng JM, Tian ZJ, Zhao HY, Li N, Liu YM, et al. Pseudorabies virus variant in Bartha-K61-vaccinated pigs, China, 2012. Emerg Infect Dis. (2013) 19:1749–55. doi: 10.3201/eid1911.130177
57. Shibata I, Yazawa S, Ono M, Okuda Y. Experimental dual infection of specific pathogen-free pigs with porcine reproductive and respiratory syndrome virus and pseudorabies virus. J Vet Med B Infect Dis Vet Public Health. (2003) 50:14–9. doi: 10.1046/j.1439-0450.2003.00605.x
58. Rios L, Coronado L, Naranjo-Feliciano D, Martinez-Perez O, Perera CL, Hernandez-Alvarez L, et al. Deciphering the emergence, genetic diversity and evolution of classical swine fever virus. Sci Rep. (2017) 7:17887. doi: 10.1038/s41598-017-18196-y
59. Beer M, Goller KV, Staubach C, Blome S. Genetic variability and distribution of classical swine fever virus. Anim Health Res Rev. (2015) 16:33–39. doi: 10.1017/S1466252315000109
60. Meyers G, Thiel HJ. Molecular characterization of pestiviruses. Adv Virus Res. (1996) 47:53–118. doi: 10.1016/S0065-3527(08)60734-4
61. van Oirschot JT. Vaccinology of classical swine fever: from lab to field. Vet Microbiol. (2003) 96:367–84. doi: 10.1016/j.vetmic.2003.09.008
62. Gao F, Jiang Y, Li G, Zhou Y, Yu L, Li L, et al. Porcine reproductive and respiratory syndrome virus expressing E2 of classical swine fever virus protects pigs from a lethal challenge of highly-pathogenic PRRSV and CSFV. Vaccine. (2018) 36:3269–77. doi: 10.1016/j.vaccine.2018.04.079
63. Meiners C, Loesken S, Doehring S, Starick E, Pesch S, Maas A, et al. Field study on swine influenza virus (SIV). Infection in weaner pigs and sows. Tierarztl Prax Ausg G Grosstiere Nutztiere. (2014) 42:351–9. doi: 10.15653/TPG-131130
64. Baudon E, Peyre M, Peiris M, Cowling BJ. Epidemiological features of influenza circulation in swine populations: a systematic review and meta-analysis. PLoS ONE. (2017) 12:e0179044. doi: 10.1371/journal.pone.0179044
65. Jiang P, Zhou N, Chen X, Zhao X, Li D, Wang F, et al. Integrative analysis of differentially expressed microRNAs of pulmonary alveolar macrophages from piglets during H1N1 swine influenza a virus infection. Sci Rep. (2015) 5:8167. doi: 10.1038/srep08167
66. Wernike K, Hoffmann B, Dauber M, Lange E, Schirrmeier H, Beer M. Detection and typing of highly pathogenic porcine reproductive and respiratory syndrome virus by multiplex real-time rt-PCR. PLoS ONE. (2012) 7:e38251. doi: 10.1371/journal.pone.0038251
67. Yang K, Tian Y, Zhou D, Duan Z, Guo R, Liu Z, et al. A multiplex RT-PCR assay to detect and discriminate porcine reproductive and respiratory syndrome viruses in clinical specimens. Viruses. (2017) 9:205. doi: 10.3390/v9080205
68. Chen N, Ye M, Xiao Y, Li S, Huang Y, Li X, et al. Development of universal and quadruplex real-time RT-PCR assays for simultaneous detection and differentiation of porcine reproductive and respiratory syndrome viruses. Transbound Emerg Dis. (2019) 66:2271–8. doi: 10.1111/tbed.13276
69. Choi YK, Goyal SM, Joo HS. Retrospective analysis of etiologic agents associated with respiratory diseases in pigs. Can Vet J. (2003) 44:735–7.
70. Van Reeth K, Nauwynck H, Pensaert M. Clinical effects of experimental dual infections with porcine reproductive and respiratory syndrome virus followed by swine influenza virus in conventional and colostrum-deprived pigs. J Vet Med B Infect Dis Vet Public Health. (2001) 48:283–92. doi: 10.1046/j.1439-0450.2001.00438.x
71. Saade G, Menard D, Hervet C, Renson P, Hue E, Zhu J, et al. Porcine reproductive and respiratory syndrome virus interferes with swine influenza a virus infection of epithelial cells. Vaccines. (2020) 8:508. doi: 10.3390/vaccines8030508
72. Zhao G, Zhang L, Li C, Zhao J, Liu N, Li Y, et al. Identification of enterobacteria in viscera of pigs afflicted with porcine reproductive and respiratory syndrome and other viral co-infections. Microb Pathog. (2020) 147:104385. doi: 10.1016/j.micpath.2020.104385
73. Di Bartolo I, Ponterio E, Castellini L, Ostanello F, Ruggeri FM. Viral and antibody HEV prevalence in swine at slaughterhouse in Italy. Vet Microbiol. (2011) 149:330–8. doi: 10.1016/j.vetmic.2010.12.007
74. Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. (2006) 16:5–36. doi: 10.1002/rmv.482
75. Okamoto H. Genetic variability and evolution of hepatitis E virus. Virus Res. (2007) 127:216–28. doi: 10.1016/j.virusres.2007.02.002
76. Wang H, Karlsson M, Lindberg M, Nystrom K, Norder H. Hepatitis E virus strains infecting swedish domestic pigs are unique for each pig farm and remain in the farm for at least 2 years. Transbound Emerg Dis. (2019) 66:1314–23. doi: 10.1111/tbed.13153
78. Frossard JP, Grierson S, Cheney T, Steinbach F, Choudhury B, Williamson S. UK pigs at the time of slaughter: investigation into the correlation of infection with PRRSV HEV. Viruses. (2017) 9:110. doi: 10.3390/v9060110
79. Afolabi KO, Iweriebor BC, Obi LC, Okoh AI. Prevalence of porcine parvoviruses in some South African swine herds with background of porcine circovirus type 2 infection. Acta Trop. (2019) 190:37–44. doi: 10.1016/j.actatropica.2018.10.010
80. Lagan Tregaskis P, Staines A, Gordon A, Sheridan P, McMenamy M, Duffy C, et al. Co-infection status of novel parvovirus's (PPV2 to 4). with porcine circovirus 2 in porcine respiratory disease complex and porcine circovirus-associated disease from 1997 to 2012. Transbound Emerg Dis. (2020) 68:1979–94. doi: 10.1111/tbed.13846
81. McCullers JA. The co-pathogenesis of influenza viruses with bacteria in the lung. Nat Rev Microbiol. (2014) 12:252–62. doi: 10.1038/nrmicro3231
82. Almand EA, Moore MD, Jaykus LA. Virus-bacteria interactions: an emerging topic in human infection. Viruses. (2017) 9:58–68. doi: 10.3390/v9030058
83. Kumar N, Sharma S, Barua S, Tripathi BN, Rouse BT. Virological and immunological outcomes of coinfections. Clin Microbiol Rev. (2018) 31:e00111-7. doi: 10.1128/CMR.00111-17
84. Solano GI, Segales J, Collins JE, Molitor TW, Pijoan C. Porcine reproductive and respiratory syndrome virus (PRRSv). Interaction with haemophilus parasuis. Vet Microbiol. (1997) 55:247–57. doi: 10.1016/S0378-1135(96)01325-9
85. Kavanova L, Prodelalova J, Nedbalcova K, Matiasovic J, Volf J, Faldyna M, et al. Immune response of porcine alveolar macrophages to a concurrent infection with porcine reproductive and respiratory syndrome virus and haemophilus parasuis in vitro. Vet Microbiol. (2015) 180:28–35. doi: 10.1016/j.vetmic.2015.08.026
86. Thanawongnuwech R, Brown GB, Halbur PG, Roth JA, Royer RL, Thacker BJ. Pathogenesis of porcine reproductive and respiratory syndrome virus-induced increase in susceptibility to streptococcus suis infection. Vet Pathol. (2000) 37:143–52. doi: 10.1354/vp.37-2-143
87. Auray G, Lachance C, Wang Y, Gagnon CA, Segura M, Gottschalk M. Transcriptional analysis of PRRSV-infected porcine dendritic cell response to streptococcus suis infection reveals up-regulation of inflammatory-related genes expression. PLoS ONE. (2016) 11:e0156019. doi: 10.1371/journal.pone.0156019
88. Sun YF, Jiang X, Zhang A, Ma JF, Yu XX, Li LA, et al. Early infection of streptococcus suis serotype 2 increases the virulence of highly pathogenic porcine reproductive and respiratory syndrome MLV-like virus in pigs. Res Vet Sci. (2020) 130:68–72. doi: 10.1016/j.rvsc.2020.02.010
89. Oliveira S, Pijoan C. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol. (2004) 99:1–12. doi: 10.1016/j.vetmic.2003.12.001
90. Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, et al. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332). In North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest. (1992) 4:117–26. doi: 10.1177/104063879200400201
91. Zhao Z, Qin Y, Lai Z, Peng L, Cai X, Wang L, et al. Microbial ecology of swine farms and PRRS vaccine vaccination strategies. Vet Microbiol. (2012) 155:247–56. doi: 10.1016/j.vetmic.2011.09.028
92. Yu J, Wu J, Zhang Y, Guo L, Cong X, Du Y, et al. Concurrent highly pathogenic porcine reproductive and respiratory syndrome virus infection accelerates haemophilus parasuis infection in conventional pigs. Vet Microbiol. (2012) 158:316–21. doi: 10.1016/j.vetmic.2012.03.001
93. Staats JJ, Feder I, Okwumabua O, Chengappa MM. Streptococcus suis: past and present. Vet Res Commun. (1997) 21:381–407. doi: 10.1023/A:1005870317757
94. Elliott SD. Streptococcal infection in young pigs. I. An immunochemical study of the causative agent (PM streptococcus). J Hyg. (1966) 64:205–12. doi: 10.1017/S0022172400040468
95. Shneerson JM, Chattopadhyay B, Murphy MF, Fawcett IW. Permanent perceptive deafness due to streptococcus suis type II infection. J Laryngol Otol. (1980) 94:425–7. doi: 10.1017/S0022215100089040
96. Galina L, Pijoan C, Sitjar M, Christianson WT, Rossow K, Collins JE. Interaction between streptococcus suis serotype 2 and porcine reproductive and respiratory syndrome virus in specific pathogen-free piglets. Vet Rec. (1994) 134:60–64. doi: 10.1136/vr.134.3.60
97. Xu M, Wang S, Li L, Lei L, Liu Y, Shi W, et al. Secondary infection with Streptococcus suis serotype 7 increases the virulence of highly pathogenic porcine reproductive and respiratory syndrome virus in pigs. Virol J. (2010) 7:184. doi: 10.1186/1743-422X-7-184
98. Brockmeier SL, Loving CL, Palmer MV, Spear A, Nicholson TL, Faaberg KS, et al. Comparison of asian porcine high fever disease isolates of porcine reproductive and respiratory syndrome virus to United States isolates for their ability to cause disease and secondary bacterial infection in swine. Vet Microbiol. (2017) 203:6–17. doi: 10.1016/j.vetmic.2017.02.003
99. Chiers K, De Waele T, Pasmans F, Ducatelle R, Haesebrouck F. Virulence factors of actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet Res. (2010) 41:65. doi: 10.1051/vetres/2010037
100. Schaller A, Djordjevic SP, Eamens GJ, Forbes WA, Kuhn R, Kuhnert P, et al. Identification and detection of actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet Microbiol. (2001) 79:47–62. doi: 10.1016/S0378-1135(00)00345-X
101. Bosse JT, Janson H, Sheehan BJ, Beddek AJ, Rycroft AN, Kroll JS, et al. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Micro Infect. (2002) 4:225–35. doi: 10.1016/S1286-4579(01)01534-9
102. Kochanowsky JA, Koshy AA. Toxoplasma gondii. Curr Biol. (2018) 28:R770–1. doi: 10.1016/j.cub.2018.05.035
103. Sullivan WJ Jr, Jeffers V. Mechanisms of toxoplasma gondii persistence and latency. FEMS Microbiol Rev. (2012) 36:717–33. doi: 10.1111/j.1574-6976.2011.00305.x
104. Li B, Du L, Xu X, Sun B, Yu Z, Feng Z, et al. Transcription analysis on response of porcine alveolar macrophages to co-infection of the highly pathogenic porcine reproductive and respiratory syndrome virus and mycoplasma hyopneumoniae. Virus Res. (2015) 196:60–9. doi: 10.1016/j.virusres.2014.11.006
105. Thanawongnuwech R, Thacker B, Halbur P, Thacker EL. Increased production of proinflammatory cytokines following infection with porcine reproductive and respiratory syndrome virus and Mycoplasma hyopneumoniae. Clin Diagn Lab Immunol. (2004) 11:901–8. doi: 10.1128/CDLI.11.5.901-908.2004
106. Peng Z, Zhao T, Liang W, Song W, Gao Z, Tang X, et al. RT-PCR detection of porcine reproductive and respiratory syndrome virus based on the ORF5 gene in mainland China, 2012-2015. Acta Virol. (2017) 61:336–40. doi: 10.4149/av_2017_312
Keywords: porcine reproductive and respiratory syndrome virus, porcine respiratory disease complex, co-infection, secondary infection, pathogens
Citation: Zhao D, Yang B, Yuan X, Shen C, Zhang D, Shi X, Zhang T, Cui H, Yang J, Chen X, Hao Y, Zheng H, Zhang K and Liu X (2021) Advanced Research in Porcine Reproductive and Respiratory Syndrome Virus Co-infection With Other Pathogens in Swine. Front. Vet. Sci. 8:699561. doi: 10.3389/fvets.2021.699561
Received: 11 May 2021; Accepted: 02 August 2021;
Published: 26 August 2021.
Edited by:
Massimo Amadori, Istituto Zooprofilattico Sperimentale Lombardia ed Emilia Romagna (IZSLER), ItalyReviewed by:
Tongling Shan, Chinese Academy of Agricultural Sciences (CAAS), ChinaJianzhong Zhu, Yangzhou University, China
Copyright © 2021 Zhao, Yang, Yuan, Shen, Zhang, Shi, Zhang, Cui, Yang, Chen, Hao, Zheng, Zhang and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haixue Zheng, aGFpeHVlemhlbmdAMTYzLmNvbQ==; Keshan Zhang, emhhbmdrZXNoYW5AY2Fhcy5jbg==
 Dengshuai Zhao
Dengshuai Zhao Bo Yang
Bo Yang Xingguo Yuan
Xingguo Yuan Haixue Zheng
Haixue Zheng