- 1Laboratory of Metabolic Manipulation of Herbivorous Animal Nutrition, College of Animal Science and Technology, Yangzhou University, Yangzhou, China
- 2Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou University, Yangzhou, China
- 3College of Animal Science and Veterinary Medicine, Heilongjiang Bayi Agricultural University, Daqing, China
- 4Jiangsu Coastal Area, Institute of Agricultural Sciences, Yancheng, China
This study investigated the effects of tannic acid (TA)-treated corn on changes in ruminal fermentation characteristics and the composition of the ruminal bacterial community in vitro. Ruminal fluid was obtained from three rumen-fistulated goats fed a 60:40 (forage/concentrate) diet. The batch cultures consisted of 25 ml of strained rumen fluid in 25 ml of an anaerobic buffer containing 0.56 g of ground corn, 0.24 g of soybean meal, 0.10 g of alfalfa, and 0.10 g of oat grass. Ground corn (2 mm) was steeped in an equal quantity (i.e., in a ratio of 1:1, w/v) of water alone (Con), 15 (TA15), 25 (TA25), and 35 g/l (TA35) TA solution for 12 h. After incubation for 24 h, TA-treated corn linearly increased (P <0.05) ruminal pH and the molar proportion of acetate, but linearly reduced (P <0.05) total volatile fatty acids and the molar proportion of butyrate compared with the Con treatment. Illumina MiSeq sequencing was used to investigate the profile changes of the ruminal microbes. A principal coordinates analysis plot based on weighted UniFrac values revealed that the structure of the ruminal bacterial communities in the control group was different from that of the TA-treated corn groups. The results of changes in the rumen bacterial communities showed that TA-treated corn linearly enriched (P <0.05) Rikenellaceae_RC9_gut_group, but linearly reduced (P <0.05) Ruminococcaceae_NK4A214_group, Ruminococcus_2, and unclassified_o__Clostridiales. Functional prediction of ruminal microbiota revealed that the TA-treated corn linearly decreased ruminal microbiota function of utilizing starch through pyruvate metabolism. In conclusion, TA-treated corn can modulate the rumen fermentation characteristics, microbial composition, and metabolic pathways, which may be potentially useful for preventing the occurrence of ruminal acidosis.
Introduction
In the current intensive rearing system, ruminants are often fed a high-grain diet or easily fermentable carbohydrates to maximize energy intake and support fattening and milk production. Excessive amounts of carbohydrates cause volatile fatty acid (VFA) accumulation, ruminal pH depression, and ruminal microbiota dysbiosis, which could increase the risk of developing subacute rumen acidosis (SARA) accompanied with laminitis, mastitis, liver abscesses, and digestive tract damage (1, 2). Due to the negative effect of high-grain diets, which decreased the long-term production of ruminants, several dietary strategies such as sodium bicarbonate buffer (3), dicarboxylic acids (4), probiotics (5), and thiamine (6) have been used to prevent the occurrence of SARA. In addition, substantial research efforts have been made over the years to develop grain processing technologies to modulate the rumen degradability of grain, aiming to improve grain feed efficiency and its health benefits (7–9).
Recently, organic acids saved as a modifier have been proposed for use in chemical processing of easily fermentable cereal grains to change ruminal starch degradation site and patterns and then alleviate the negative effects of SARA [as reviewed by (10)]. More specially, studies by Iqbal et al. (11), Iqbal et al. (12), and Iqbal et al. (13) showed that feeding barley grain steeped in 0.5% lactic acid increased starch resistance to microbial fermentation, lowered ruminal concentration of VFA, lactic acid, and lipopolysaccharide (LPS) in dairy cows. Treatment of barley or corn grain with other organic acids, such as citric acid, also increases ruminal-resistant starch (14–17). As for tannic acid (TA), Deshpande et al. (18) reported that at room temperature, the binding of TA to starches (legume starches and potato starch) and starch fractions (amylose and amylopectin) lowered the in vitro digestibility by 8.8–17.0% with porcine pancreatic α-amylase. The protective effect of TA against ruminal degradation in cereal grains has been scarcely explored. A study by Martínez-Moya et al. (8) using TA to protect barley meal against ruminal degradation showed limited microbial hydrolysis of endosperm cell walls in TA-treated barley. It proved that TA might affect slowing ruminal disappearance of barley, which benefits to improve starch utilization by ruminants. Corn is a starch-rich grain that is widely used as animal feed in China. A high intake of corn-based diet increases the starch digestibility in the rumen. Thus, feeding high-corn diet can induce SARA in dairy cows and goats (19–21). Accordingly, we chose TA as a modifier of the fermentation characteristics of corn. We hypothesized that ground corn steeped in TA solution would reduce the rate of starch ruminal digestion under high-corn substrates, which will improve the ruminal pH and reduce the risk of SARA. Most studies to date have focused on the effects of organic acid-treated cereal grains on the rumen fermentation pattern; however, limited information was reported from changes in ruminal microbial point of view. Thus, the present study aimed to investigate the effects of TA-treated corn with different concentrations on the fermentation characteristics, the ruminal microbial composition, the correlations between microbial communities and ruminal variables as well as the effects of TA-treated corn on the ruminal microbial metabolic pathways.
Materials and Methods
All animal experiments were conducted in line with the Animal Protection Law based on the Guidelines for the Care and Use of Laboratory Animals approved by the Ethics Committee of Yangzhou University (SXXY2015-0054).
Corn Sample Processing
Ground corn grain was passed through a 2-mm mesh sieve and then treated with four different tannic acid concentrations (purity 98%, Macklin Biochemical Technology Co., Ltd, Shanghai, China): 0, 15, 25, and 35 g tannic acid/kg corn. In the TA-treated corn, 100 ml of an aqueous solution of tannic acid with concentrations of 1.5, 2.5, and 3.5 g tannic acid/100 ml were thoroughly mixed with 100 g of ground corn, respectively. In the control corn, 100 g of ground corn was soaked in an equal volume (100 ml) of distilled water. After 10 min of stirring, all the treatments were soaked for 12 h and then oven dried at 45°C before being mixed with other substrates.
In vitro Batch Fermentations
The effects of TA-steeped corn on rumen microbial fermentation and bacterial communities were evaluated in in vitro batch culture of rumen fluid supplied with a 20:80 forage/concentrate diet [17.49 CP, 21.01% NDF, 12.44% ADF, and 34.94% starch, dry matter (DM) basis] to simulate in vivo SARA induced by high concentrate diet. The substrate (DM basis) consisted of 560 mg ground corn, 240 mg soybean, 100 mg alfalfa, and 100 mg oat grass. Corn in the control group and TA groups were steeped in water and tannic acid solution for 12 h, respectively. Ruminal fluid was obtained from three rumen-fistulated goats (30.25 ± 1.25 kg of body weight) fed a 60:40 forage/concentrate diet (Table 1). The animals were housed in individual pens with free access to water and fed twice daily at 0800 h and 1,800 h. Rumen fluid was obtained immediately before morning feeding and strained through four layers of cheesecloth, mixed in a 1-to-1 proportion with phosphate-bicarbonate buffer (22), purged with anaerobic-grade CO2 (<2 ppm O2), and standardized at pH 7.0 with 3 N HCl (23). Immediately after inoculation, the bottles were gassed with CO2 before sealing with rubber stoppers. Incubations were conducted at 39°C in a shaking water bath. There were nine replicate bottles in both the TA treatment groups and the control group. The fermentation process was conducted in 100-ml sterile glass bottles containing 50 ml of culture fluid with 1 g of the diet ground through a 2-mm screen. After 24 h, the culture fluid of each bottle was filtered through four layers of cheesecloth and the pH was determined immediately. Filtered liquor samples for VFAs, lactate acid, and microbial DNA extraction were collected for analyses.
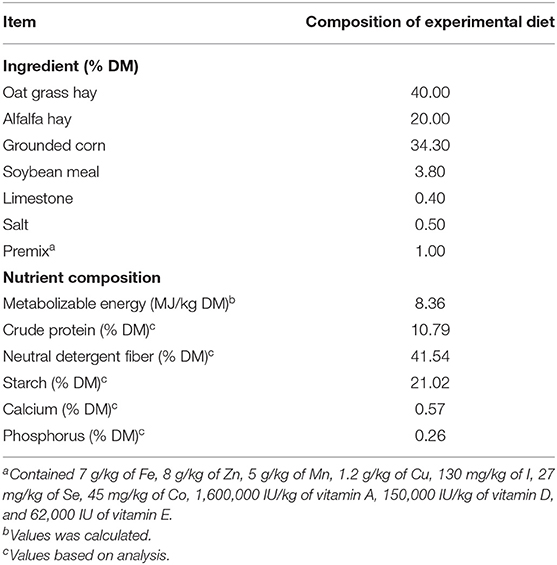
Table 1. Ingredients and nutritional composition of basal experimental diet offered to rumen fluid donor goats.
Chemical Analysis
The chemical analysis of the diet for rumen fluid donor goats was analyzed for DM (ISO 6496) and CP (ISO 15670), according to the Association of Analytical Chemists (24). Neutral detergent fiber contents were determined according to the procedure of Van Soest (25). Starch content was determined according to the procedure of Hall (26). Calcium and phosphorus were measured by inductively coupled plasma emission spectroscopy using an Atom Scan 25 Plasma Spectroscopy (Thermo Jarrell Ash Corp., Grand Junction, CO) after acid digestion.
Rumen Fluid Sampling and Analysis
The ruminal pH values in each bottle was determined at the end of the 24-h fermentation with a pH meter (FE20, METTLER TOLEDO, USA). The VFA concentrations were analyzed according to the gas chromatography (GC-14B, Shimadzu, Japan) method proposed by Qin et al. (27) with slight modifications. Briefly, 1 ml ruminal fluid supernatant was added to 0.2 ml metaphosphoric acid solution (25%) and vortexed for 30 s. Then, samples were centrifuged at 12,000 × g for 15 min at 4°C. Samples were filtered by a 0.45-μm filter and then transferred into a capillary column GC (30 m ×1.0 μm ×0.53 mm, Agilent, Netherlands; column temperature, 150°C; injector temperature, 200°C; and detector temperature, 200°C). The concentration of lactate acid was analyzed following the method described by Barker and Summerson (28).
DNA Extraction and Illumina MiSeq Sequencing, Sequence Processing, and Analysis
Microbial DNA was extracted using the FastDNA® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA) according to manufacturer's protocol. The final DNA concentration and purification were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) by a thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 min of denaturation at 95°C, 27 cycles of 30 s at 95°C, 30 s for annealing at 55°C, 45 s for elongation at 72°C, and a final extension at 72°C for 10 min. Polymerase chain reactions were performed in triplicate 20-μl mixture containing 4 μl of 5x FastPfu Buffer, 2 μl of 2.5 mM dNTPs, 0.8 μl of each primer (5 μM), 0.4 μl of FastPfu Polymerase, and 10 ng of template DNA. The resulted PCR products were extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) and quantified using QuantiFluor TM-ST (Promega, USA) according to the manufacturer's protocol. Amplicon libraries were generated using NEXTFLEX® Rapid DNA-Seq Kit (Bioo Scientific, USA) following the manufacturer's recommendations. The paired-end sequence (2 ×300 bp) was conducted on an Illumina MiSeq platform (Illumina, San Diego, USA).
Raw fastq files were quality-filtered by Trimmomatic and merged by FLASH with the following criteria: (i) the reads were truncated at any site receiving an average quality score <20 over a 50-bp sliding window. (ii) Sequences whose overlap being longer than 10 bp were merged according to their overlap with mismatch no more than 2 bp. (iii) Sequences of each sample were separated according to barcodes (exactly matching) and primers (allowing two-nucleotide mismatching), and reads containing ambiguous bases were removed. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1) (29) with a novel greedy algorithm that performs chimera filtering and OTU clustering simultaneously. The taxonomy of each 16S rRNA gene sequence was analyzed by RDP Classifier against the Silva (SSU138) 16S rRNA database using a confidence threshold of 70% (30). OTUs were used to generate rarefaction curves and Shannon–Wiener curves. The bacterial community diversity by calculating the diversity estimator indices of Shannon–Wiener, Simpson, Chao1, and abundance-based coverage estimator (ACE) (31) using Mothur version v.1.30.2 (32). The weighted UniFrac distance method was used to perform a principal coordinate analysis (PCoA), and an analysis of similarity (ANOSIM) in QIIME with 999 permutations (33) was conducted to assess significant differences between samples.
Functional Prediction of Ruminal Microbiome
Functional capacity of the ruminal microbial community was predicted using the PICRUSt (phylogenetic investigation of communities by reconstruction of unobserved states) software package (http://picrust.github.io/picrust) (34). The significant difference in KEGG (Kyoto Encyclopedia of Genes and Genomes) ortholog (KO) abundances among treatments was identified and analyzed using the KEGG Mapper pathway search function.
Statistical Analyses
Data of ruminal fermentation profile, the rumen bacterial communities and their functional changes, were analyzed using one-way analysis of variance (ANOVA) procedure of SAS (SAS Inst. Inc., Cary, NC). The differences among treatment means were determined using Duncan's multiple range tests. Correlations between bacterial communities and ruminal fermentation parameters as well as correlations between bacterial communities and altered metabolic pathways were assessed by Pearson's correlation test. P-values below 0.05 were regarded as statistically significant.
Results
In vitro Ruminal Fermentation Parameters
The ruminal pH value, VFA concentrations, and individual VFA profiles are presented in Table 2. TA-treated corn linearly increased (P <0.05) in ruminal pH and linearly decreased (P <0.05) the total VFA concentration. With the respect to individual VFA, TA-treated corn linearly increased (P <0.05) the molar proportion of acetate and the ratio of acetate to propionate, but linearly decreased (P <0.05) the molar proportion of butyrate. The molar proportion of propionate was the highest in TA15 treatment, followed by TA35, TA25 and Con treatments. The molar proportion of valerate was linearly decreased (P <0.05) in TA treatments. The molar proportion of isovalerate was the highest in TA25 treatment, and there were no significant effect (P > 0.05) among TA15, TA35, and Con treatments. In the present study, the result showed that lactate and isobutyrate were not affected (P > 0.05) by TA treatments.
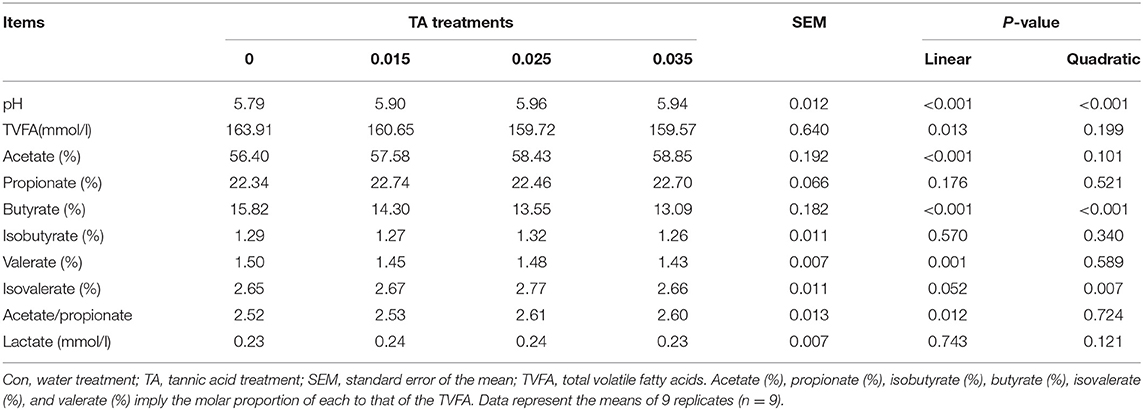
Table 2. Effects of tannic acid concentration treatments of ground corn on in vitro fermentation characteristics and lactate concentration.
Diversity, Richness, and Composition of Bacterial Communities in Rumen Fluid
A total of 2,197,647 reads were obtained for the bacterial 16S rRNA genes by sequencing, with a mean length of 413 bp. Quality filtering at 97% similarity resulted in 837,216 high-quality sequences, which clustered in 1,670 OTUs. A Venn diagram revealed that the numbers of unique OTU for the Con, TA15, TA25, and TA35 treatments were 29, 35, 49, and 41, respectively. Moreover, the Venn profile also revealed that 1,171 OTUs (around 70% of total OTUs) were shared among the four treatments, revealing the presence of a core microbiota (Figure 1A). The rarefaction curve showed that the sequencing work was relatively comprehensive in covering the bacterial diversity, as the rarefaction curves tended to approach saturation (Figure 1B). The Shannon curves indicated that dataset from the diversity analysis was large enough to reflect the bacterial diversity information of samples (Figure 1C).
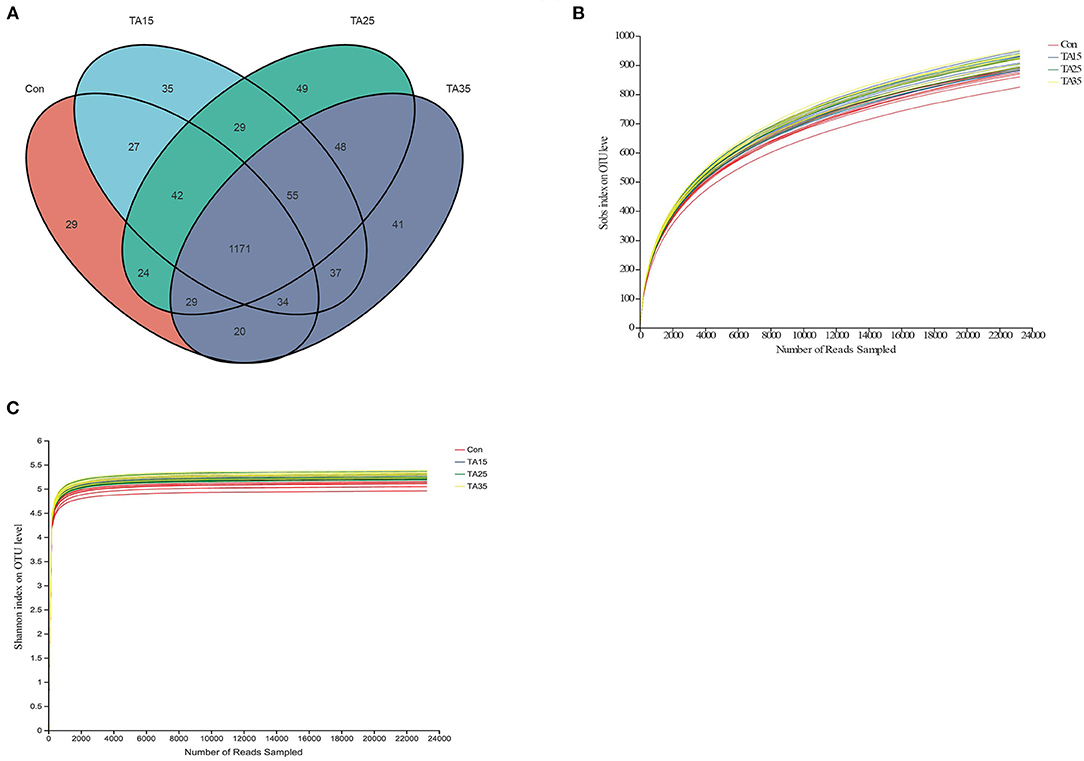
Figure 1. Ruminal microbial operational taxonomic units (OTUs) among different groups. CON, corn steeped in water; TA15, corn steeped in tannic acid solution of 15 g/l; TA25, corn steeped in tannic acid solution of 25 g/l; TA35, corn steeped in tannic acid solution of 35 g/l. (A) Venn diagram of ruminal bacterial OTUs. (B) Bacterial rarefaction curves and (C) Shannon curves based on OTUs used to assess the depth of coverage for each sample.
As shown in Table 3, there were no significant differences (P > 0.05) in Good's coverage and ACE index among the groups. TA treatments significantly increased the OTU numbers and Shannon index, but significantly decreased (P <0.05) the Simpson index compared with the Con group. The results of PCoA with weighted UniFrac distances indicated that the Con group largely separated from the TA treatments. The PCoA axis 1 accounted for 57.55% of the variation, and the PCoA axis 2 accounted for 12.97% of the variation (Figure 2).
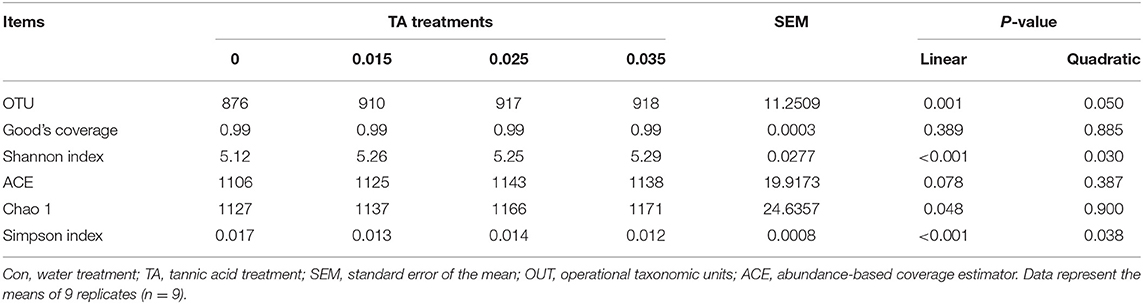
Table 3. Effects of tannic acid concentration treatments of ground corn on the number of observed species and alpha diversity of bacterial community structures.
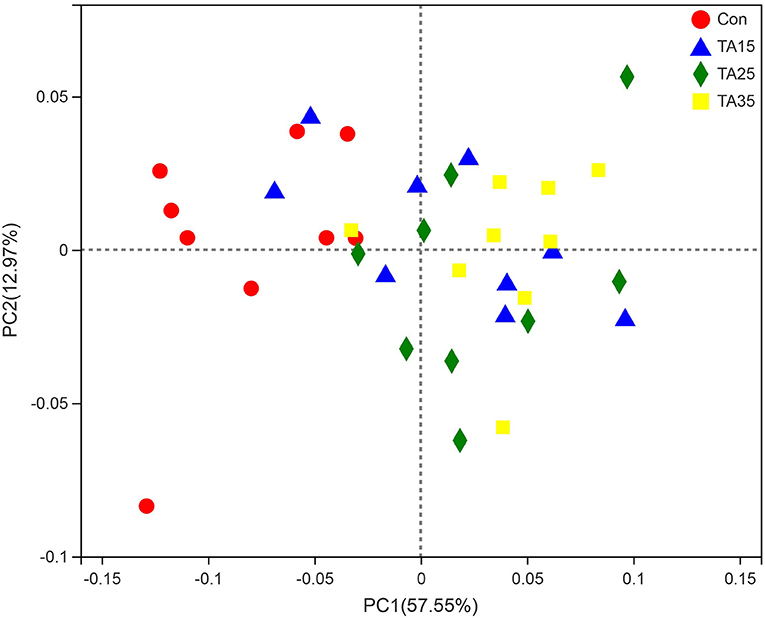
Figure 2. Principal coordinate analysis (PCoA) of bacterial community structures of the ruminal microbiota in CON (red circles), TA15 (blue triangle), TA25 (green diamond), and TA35 (yellow square) groups. PCoA plots were constructed using the weighted UniFrac method.
Overall, 22 phyla were identified across all samples. Bacteroidetes and Firmicutes were the two dominant phyla (Figure 3). The proportions of Verrucomicrobia, Tenericutes, Synergistetes, Spirochaetes, Lentisphaerae, Cyanobacteria, Chloroflexi, WPS-2, Planctomycetes, and Elusimicrobia phyla were less than 1% of total microbial community, while Nitrospirae, Gemmatimonadetes, Fibrobacteres, Armatimonadetes, and Acidobacteria phyla were not consistently present in all of the ruminal samples.
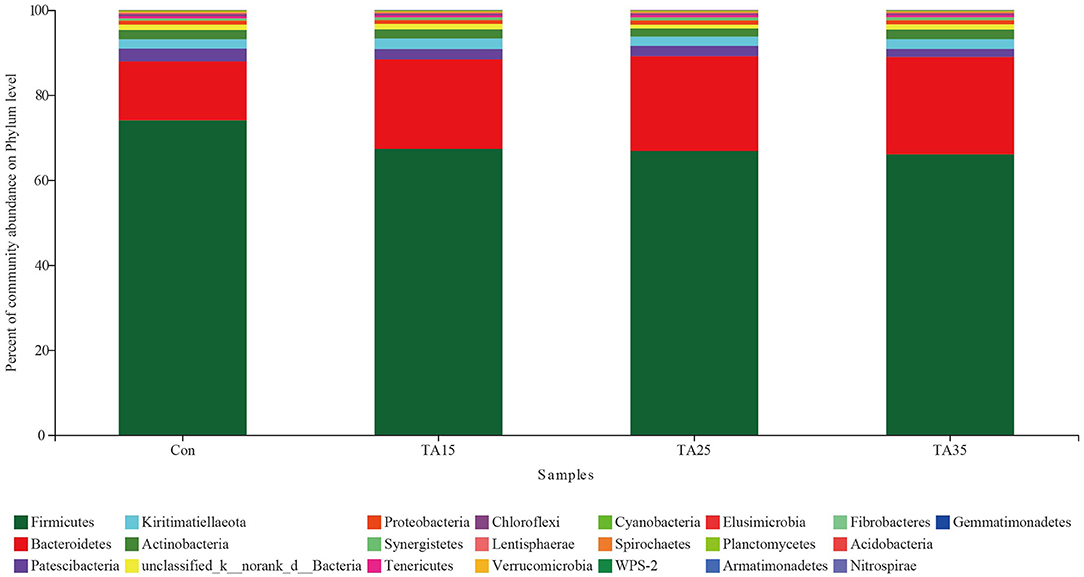
Figure 3. Percentage composition of the total phyla in rumen fluid. CON, corn steeped in water; TA15, corn steeped in tannic acid solution of 15 g/l; TA25, corn steeped in tannic acid solution of 25 g/l; TA35, corn steeped in tannic acid solution of 35 g/l.
Changes in the Rumen Bacterial Communities
The 10 most relatively abundant bacterial phyla are shown in Table 4. The relative abundance of Firmicutes was the highest in the Con treatment, and the lowest in the TA35 treatment (P <0.05), but there was no significant difference (P > 0.05) between the TA15, TA25, and TA35 treatments. The relative abundance of Bacteroidetes was lower in the Con treatment (P <0.05) than the TA15, TA25, and TA35 treatments. The relative abundance of Patescibacteria was the highest in the control treatment, followed by the TA15 and TA25 treatments, and the lowest in the TA35 treatment (P <0.05). There were no significant shifts (P > 0.05) detected in the relative abundance of Kiritimatiellaeota, Actinobacteria, unclassified_k__norank_d__Bacteria, Proteobacteria, Synergistetes, Tenericutes, and Chloroflexi.
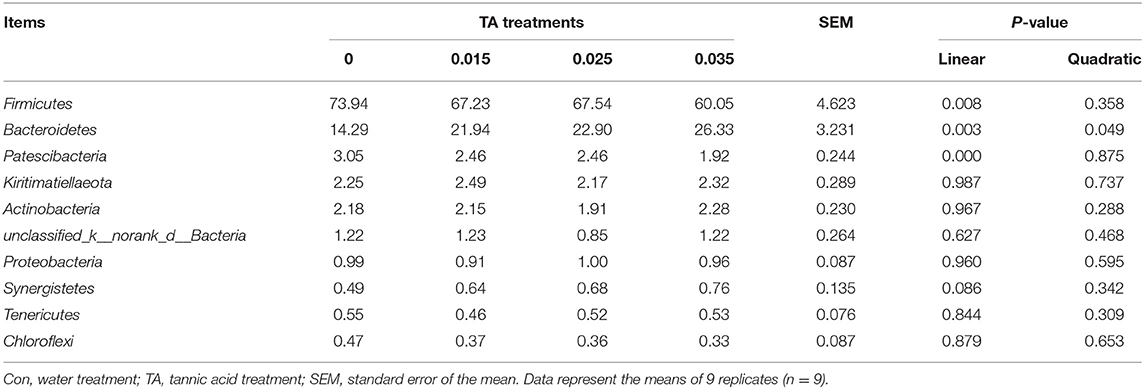
Table 4. Effects of tannic acid concentration treatments of ground corn on the 10 most relatively abundant bacterial phyla.
The 10 most relatively abundant bacterial genera are shown in Table 5. The relative abundance of Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214_group, Ruminococcus_2, and unclassified_ o__ Clostridiales was higher (P <0.05) in the Con treatment than the TA15, TA25, and TA35 treatments. Moreover, the relative abundance of Rikenellaceae_RC9_gut_group was lower (P <0.05) in the Con treatment than the TA15, TA25, and TA35 treatments. The relative abundance of Succiniclasticum and Quinella increased with TA treatment, where it reached a significant level between TA25, TA35, and the Con treatment. The relative abundance of Prevotella_1, norank_f__Muribaculaceae, and Family_XIII_AD3011_group was not affected (P > 0.05) by any treatments.
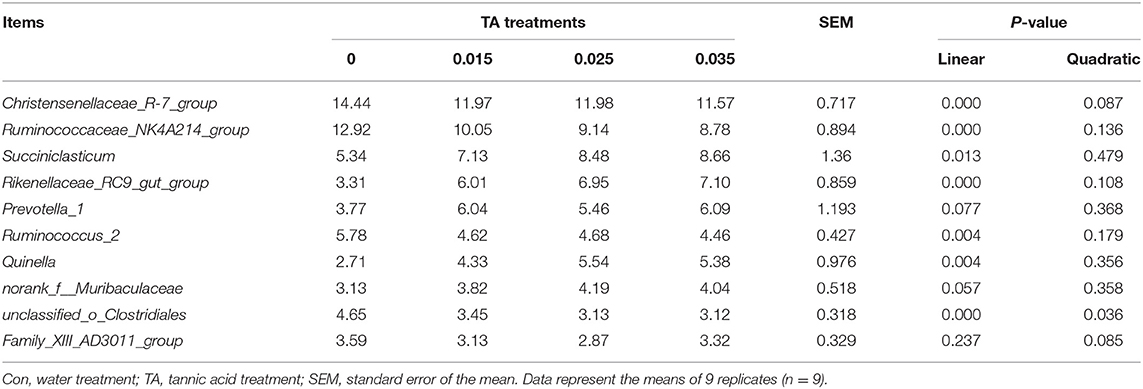
Table 5. Effects of tannic acid concentration treatments of ground corn on the 10 most relatively abundant bacterial genera.
Correlations Between Bacterial Communities and Ruminal Variables
A correlation heat map showed that the relationships between ruminal variables and the 10 most relatively abundant bacterial genera were different (Figure 4). The ruminal pH value was positively correlated (P <0.05) with Rikenellaceae_RC9__gut__group, Succiniclasticum, and Quinella and negatively correlated (P <0.05) with Ruminococcus_2, Ruminococcaceae_NK4A214_group, Christensenellaceae_R-7_group, and unclassified_o__Clostridiales. The molar proportion of total volatile fatty acids (TVFA) showed a significant negative correlation (P <0.05) with Rikenellaceae_RC9__gut__group, Succiniclasticum, and Quinella and a positive correlation (P <0.05) with Ruminococcaceae_NK4A214_group and Ruminococcus_2. The molar proportion of butyrate was negatively correlated (P <0.05) with norank_f_Muribaculaceae, Rikenellaceae_RC9__gut__group, and Quinella, but was positively correlated (P <0.05) with Ruminococcus_2, Ruminococcaceae_NK4A214_group, Christensenellaceae_R-7_group, and unclassified_o __Clostridiales. The molar proportion of propionate demonstrated a significantly negative correlation (P <0.05) with Ruminococcus_2, Ruminococcaceae_NK4A214_group, Christensenellaceae_R-7_group, and unclassified_o__Clostridiales and a positive correlation (P <0.05) with Succiniclasticum and Quinella. The molar proportion of acetate was negatively correlated (P <0.05) with Ruminococcaceae_NK4A214_group and unclassified_o__Clostridiales, but was positively correlated (P <0.05) with Rikenellaceae_RC9__gut__group and norank_f_Muribaculaceae. The molar proportion of isobutyrate revealed a significantly positive correlation (P <0.05) with Quinella and a significantly negative correlation (P <0.05) with Family_XIII__AD3011__group. The molar proportion of isovalerate was negatively related (P <0.05) with Family_XIII__AD3011__group and unclassified_o__Clostridiales, but was positively related (P <0.05) with Rikenellaceae_RC9__gut__group and Quinella. No significant correlations (P > 0.05) were found between the relative abundance of genera and valerate or lactate.
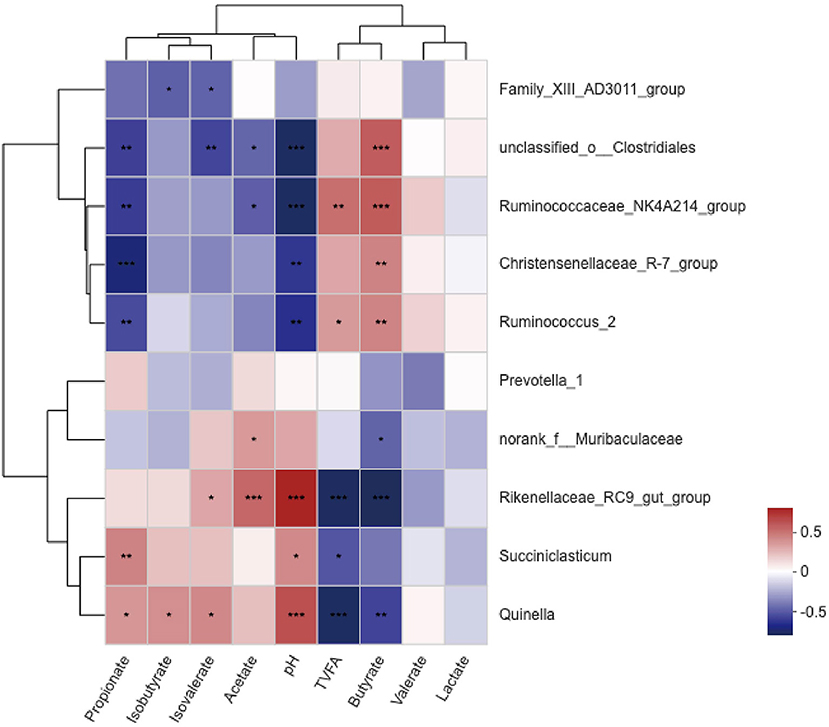
Figure 4. Correlation analyses between the 10 most relatively abundant bacterial genera and ruminal fermentation parameters. The blue represents a negative correlation between the abundance of the species and the VFA concentration, the red color represents a positive correlation, and the white shows that the correlation was not significant (*0.01 < P ≤ 0.05, **0.001 < P ≤ 0.01, ***P ≤ 0.001).
Metabolic Functions Altered in Carbohydrates of Ruminal Bacteria
The significance changed in carbohydrate metabolic pathways of the rumen bacterial microbiota in level 3 KEGG (Table 6). There were no significant differences (P > 0.05) in glycolysis/gluconeogenesis, amino sugar and nucleotide sugar metabolism, butanoate metabolism, galactose metabolism, propanoate metabolism, inositol phosphate metabolism, and ascorbate and aldarate metabolism among the treatments. The relative abundance of pyruvate metabolism and pentose phosphate pathway was higher in the Con treatment than the TA15, TA25, and TA35 treatments (P <0.05). The relative abundance of fructose and mannose metabolism, citrate cycle (TCA cycle), and glyoxylate and dicarboxylate metabolism was lower in the Con treatment than the TA15, TA25, and TA35 treatments (P <0.05). The relative abundance of starch and sucrose metabolism as well as pentose and glucuronate interconversions decreased with TA treatment, where it reached a significant level between the TA25, TA35, and the Con treatments. The relative abundance of C5-branched dibasic acid metabolism increased with TA treatment, where it reached a significant level between the TA35 and the Con treatments.
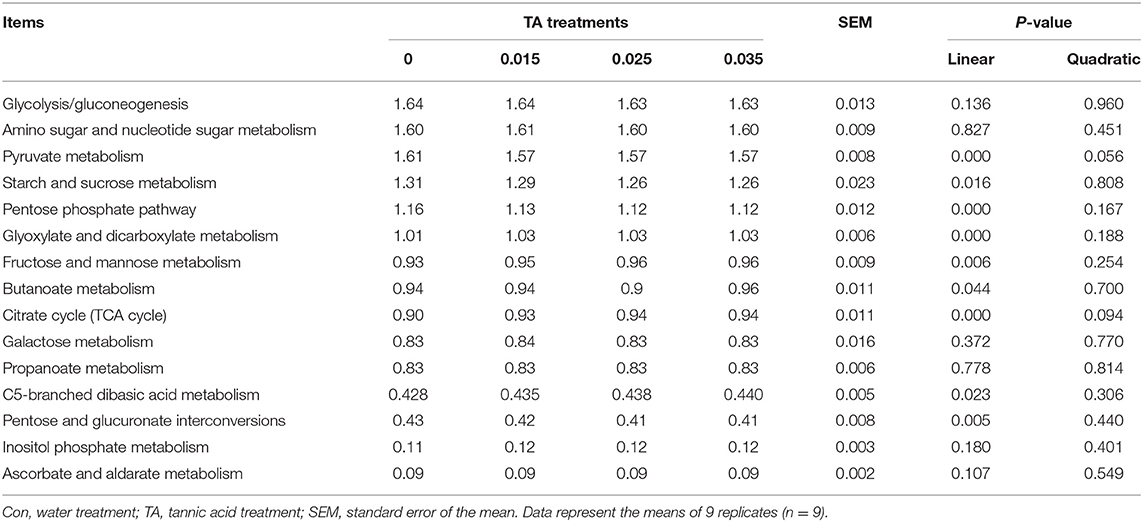
Table 6. Effects of tannic acid concentration treatments of ground corn on the altered abundances of carbohydrate metabolic pathways (%) in level 3 KEGG.
Correlations Between Bacterial Genera and Altered Carbohydrate Metabolic Pathway
In order to explore the relationships between bacterial genera and carbohydrate metabolic pathways, a Pearson correlation analysis was carried out (Figure 5). As shown in Table 7, Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214_group, Succiniclasticum, Rikenellaceae_RC9_gut_group, Prevotella_1, Ruminococcus_2, Quinella, unclassified_o_Clostridiales, and Family_XIII_AD3011_group were associated with multiple metabolic pathways. For example, starch and sucrose metabolism as well as pyruvate metabolism were negatively correlated with Succiniclasticum, Quinella, and Rikenellaceae_RC9_gut_group (P <0.05), but positively correlated with Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214_group, Ruminococcus_2, and unclassified_o_Clostridiales (P <0.05).
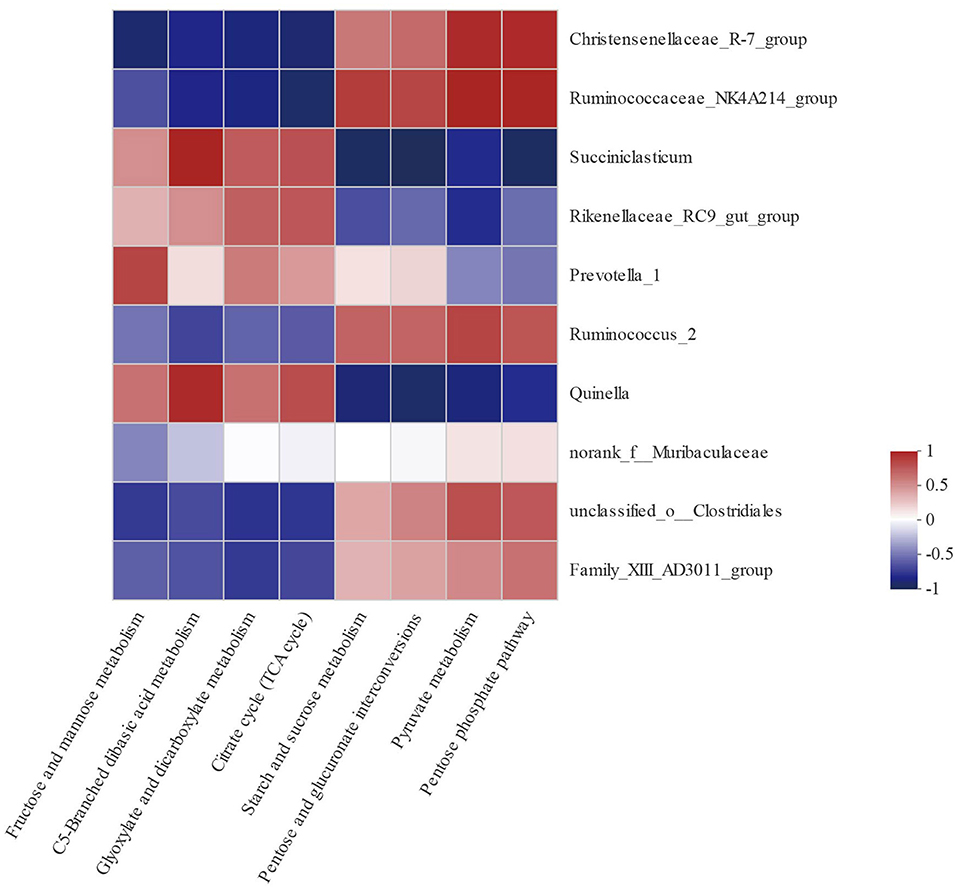
Figure 5. Correlation analyses between the 10 most relatively abundant bacterial genera and altered carbohydrate metabolic pathway. The blue represents a negative correlation between the abundance of the species and the metabolic pathways, the red color represents a positive correlation, and the white shows that the correlation was not significant.
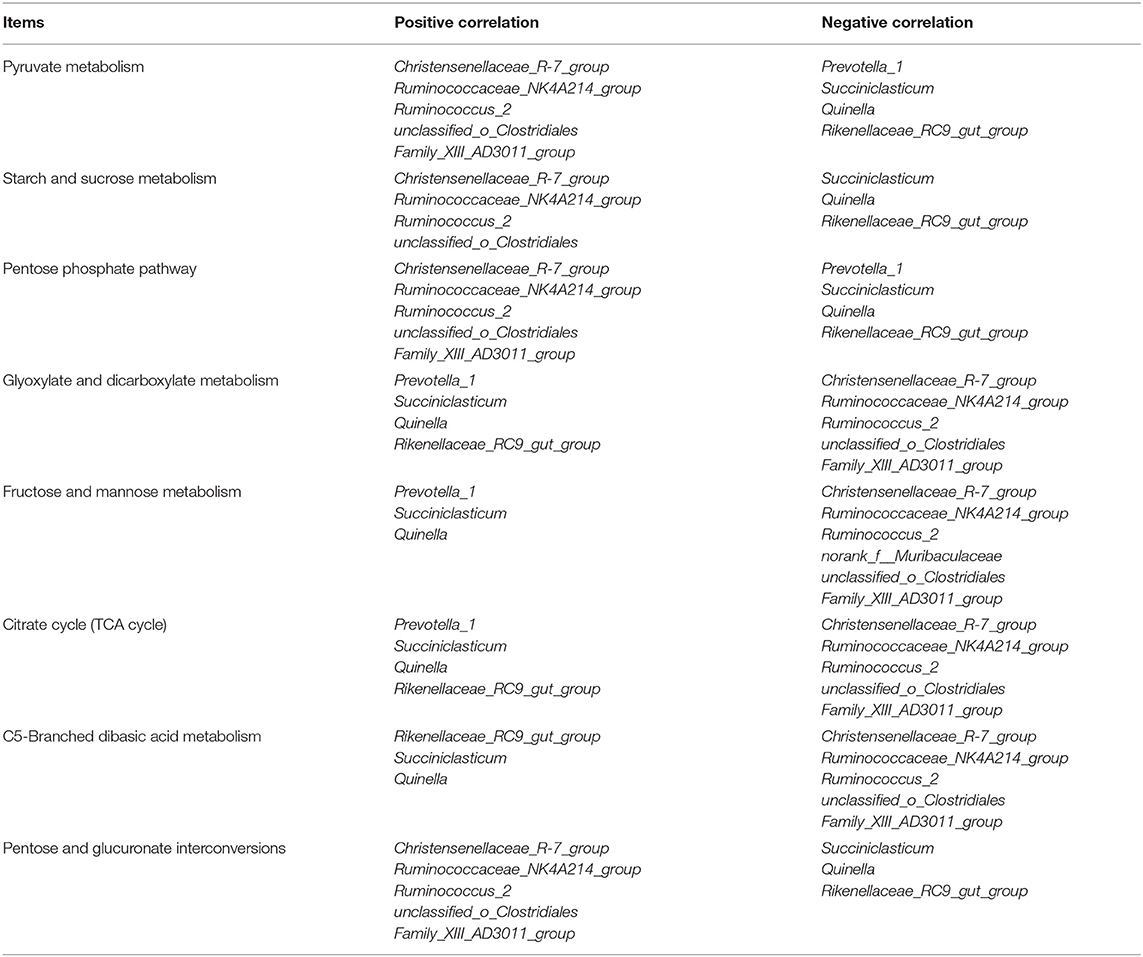
Table 7. Significantly positive and negative correlations between the 10 most relatively abundant bacterial genera and altered carbohydrate metabolic pathway.
Discussion
In the present study, the results of ruminal fermentation showed that the TA-treated corn substrates significantly altered the ruminal fermentation profile in vitro. The TVFA concentration was significantly decreased by TA-treated corn accompanied by an increase in the ruminal pH value, suggesting the potential effect of tannic acid pre-treated corn in alleviating ruminal environment disorder by slowing the rate of starch degradation. Ruminal butyric subacute ruminal acidosis was induced in sheep with an intraruminal supply of ground corn, whereas lactic acidosis and propionic subacute ruminal acidosis were respectively provoked by wheat and beet pulp (35). The significant decrease in molar proportion of butyrate in TA treatments explains to some extent the potential effect of TA-treated corn in alleviating ruminal acidosis induced by high corn grain. Furthermore, the significant decrease in molar proportion of butyrate in TA treatments may be an explanation for the increasing acetate in response to TA-treated corn, because acetyl-CoA acted as substrate for producing acetate and butyrate in rumen. In addition, some studies have shown that tannins increased the proportion of acetate (36, 37) and decreased the proportion of butyrate (36). Ruminal fermentation characteristics are directly affected by the ruminal microbiota, where varied concentrations of VFA and other metabolites are dependent on the microbial population. In order to understand the relationship of TA pre-treated corn and ruminal fermentation profile, high-throughput sequencing was used to investigate the response of bacterial community to high tannic acid pre-treated corn substrates.
A number of recent studies demonstrated that high grain diet altered the community of the ruminal bacterial microbiota, decreasing the abundance of Bacteroidetes and increasing the abundance of Firmicutes under low ruminal pH (38–40). In the present study, the results of the PCoA showed that the tannic acid pre-treated corn substrates significantly altered the composition and structure of the rumen bacteria. At the phyla level, the high-grain substrates had a higher abundance of Firmicutes and lower Bacteroidetes, while the abovementioned phyla change was reversed by TA-treated corn substrates, indicating TA-treated corn substrates could stabilize the bacterial community in the rumen. Besides, we found that TA-treated corn substrates significantly decreased the relative abundance of Patescibacteria in respect to the Con treatment. However, little information about this phylum has been reported in the literature, but the reason for alteration in the status of this phylum remains unclear.
Significantly, shifts in the genera Succiniclasticum, Ruminococcus_2, Ruminococcaceae_NK4A214_group, Quinella, and Rikenellaceae_RC9_gut_group were found in the present study. Succiniclasticum only ferments succinate to propionate but not other carbohydrates; amino acids; or mono-, di-, and other tricarboxylic acids (41). As the primary succinate-utilizing bacteria, Succiniclasticum accounted for 5.34% of total bacterial community in the Con group and decreased significantly compared with the TA25 and TA35 groups. Therefore, the increase of Succiniclasticum needs more pyruvate to metabolize succinate for producing propionate. The above observation fits with the results that there is more molar proportion of propionate in TA treatment corn substrate group. Meanwhile, a positive correlation between the relative abundance of Succiniclasticum and the molar proportion of propionate was found in the current study. As for acetate-producing bacteria, we observed that there was a decrease in the relative abundance of Ruminococcus_2 and Ruminococcaceae_NK4A214_group in TA treatments. It is somewhat surprising that the molar proportion of acetate was still increased in the TA groups. The high abundance of Rikenellaceae_RC9_gut_group may be the possible explanation for this finding. Collins et al. (42) and Su et al. (43) reported that acetate is a major product of glucose fermentation in the family Rikenellaceae. These explanations became plausible as the significantly positive correlation between the relative abundance of Rikenellaceae_RC9_gut_group and the molar proportion of acetate was observed. The decreased abundance of Christensenellaceae_R-7_group in TA-treated corn groups demonstrated that the TA-treated corn inhibits growth of this genera. Further research should be undertaken to investigate why TA treatment reduced the relative abundance of this genera. Lactic acid is produced as the major fermentation product by Quinella, when sugars do not limit growth. When sugars are limited and growth is slow, acetate and propionate are the major fermentation products (44). Lactate can also be produced by Clostridiales, Ruminococcaceae, Lachnospiraceae, and Butyrivibrio in the rumen (45). In the current study, the significant changes in the relative abundance of Quinella, Ruminococcaceae_NK4A214_group, and unclassified_o__Clostridiales should have caused a shift in lactate concentration. Surprisingly, lactate concentration was not found different among the groups. Meanwhile, no correlation was found between bacterial genera and lactate. The results above might be explained by the fact that as the intermediate product of carbohydrate metabolism, lactate can be metabolized by lots of bacterial genera for their growth. This combination of competitive process may lead to the disappearing of change in lactate concentration. Therefore, causes for this phenomenon are not well-understood and need to be further studied to get a more precise explanation. Among various relevant studies, high-grain diet supplementation increased, decreased, or had no effect on the amount of rumen Prevotella (46–49). Our result showed that the relative abundance of Prevotella_1 was not affected by TA treatments compared with the Con group. The members of Prevotella are characterized by various metabolic potentials and possess multiple degradation activities, including those of starch, fiber, hemicellulose, protein, and sugar (50, 51). These results suggested that Prevotella spp. may be more tasteful than expected due to its multiple activities leading to adaptive responses to the rumen substrate changes, and these changes did not exhibit specific patterns.
Our analysis using PICRUSt found that lots of carbohydrate metabolic pathways in level 3 KEGG were differential. The metabolites of starch and sucrose metabolism, pentose phosphate pathway, glyoxylate and dicarboxylate metabolism, as well as fructose and mannose metabolism (carbohydrate metabolism) can enter glycolysis/gluconeogenesis and pyruvate metabolism pathways (52–55). Importantly, TA-treated corn reduced pyruvate metabolism as well as starch and sucrose metabolism compared with the control group. Meanwhile, pyruvate metabolism as well as starch and sucrose metabolism were negatively correlated with Christensenellaceae_R-7_group, Ruminococcaceae_NK4A214_group, Ruminococcus_2, and unclassified_o_Clostridiales. These results indicated that TA- treated corn could weaken starch and pyruvate metabolism of microbiota. In rumen, pyruvate produced by starch can be utilized to produce acetate, butyrate (acetate and butyrate pathway), and propionate (succinate pathway and acrylate pathway). The decrease in pyruvate metabolism leads to the reduction of VFA and increase ruminal pH produced by ruminal microbiota in TA treatments, which may be attributed to the slow degradation starch. Furthermore, the results presented earlier seem to indicate that tannic acid pre-treated corn has a potential to improve ruminal environment by slowing the rate of starch degradation.
Conclusion
The present study investigated the effects of TA-treated corn on the rumen fermentation characteristics and microbial composition in vitro. In summary, TA-treated corn discriminatively altered the ruminal fermentation, microbial composition, and diversity and modulated the carbohydrate metabolic pathways of microbial metabolism. These alterations may help us for a better understanding of the mechanism of TA-treated corn to regulate rumen environment disorder induced by high-grain substrate.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA768092.
Author Contributions
FZ and HW designed the research. FZ, XZ, and YQ conducted the research. FZ, YZ, HS, and HZ analyzed the data. FZ wrote the manuscript. ME revised the language. All authors contributed to the article and approved the submitted version.
Funding
The authors would like to acknowledge the funding received to conduct this study from the project funded by the National Natural Science Foundation of China (NSFC No. 31872988 and No. 31572429), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX18_2375), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors thank the guidance of all the members of Laboratory of Metabolic Manipulation of Herbivorous Animal Nutrition.
References
1. Plaizier JC, Krause DO, Gozho GN, McBride BW. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J. (2008) 176:21–31. doi: 10.1016/j.tvjl.2007.12.016
2. Kleen JL, Hooijer GA, Rehage JM, Noordhuizen JPT. Subacute ruminal acidosis (sara): a review. J Vet Med A Physiol Pathol Clin Med. (2003) 50:406–14. doi: 10.1046/j.1439-0442.2003.00569.x
3. Mao SY, Huo WJ, Liu JH, Zhang RY, Zhu WY. In vitro effects of sodium bicarbonate buffer on rumen fermentation, levels of lipopolysaccharide and biogenic amine, and composition of rumen microbiota. J Sci Food Agric. (2017) 97:1276–85. doi: 10.1002/jsfa.7861
4. Vyas D, Beauchemin KA, Koenig KM. Using organic acids to control subacute ruminal acidosis and fermentation in feedlot cattle fed a high-grain diet. J Anim Sci. (2015) 93:3950–8. doi: 10.2527/jas.2015-9009
5. Henning PH, Horn CH, Leeuw KJ, Meissner HH, Hagg FM. Effect of ruminal administration of the lactate-utilizing strain megasphaera elsdenii (me) ncimb 41125 on abrupt or gradual transition from forage to concentrate diets. Anim Feed Sci Technol. (2010) 157:20–9. doi: 10.1016/j.anifeedsci.2010.02.002
6. Zhang H, Peng AL, Zhao FF, Yu LH, Wang MZ, Osorio JS, et al. Thiamine ameliorates inflammation of the ruminal epithelium of Saanen goats suffering from subacute ruminal acidosis. J Dairy Sci. (2020) 103:1931–43. doi: 10.3168/jds.2019-16944
7. Montaño MF, Chai W, Zinn-Ware TE, Zinn RA. Influence of malic acid supplementation on ruminal pH, lactic acid utilization, and digestive function in steers fed high-concentrate finishing diets. J Anim Sci. (1999) 77:780–4. doi: 10.2527/1999.773780x
8. Martínez-Moya TF, Moyano FJ, Díaz-López M, Barroso FG, Alarcón FJ. Use of tannic acid to protect barley meal against ruminal degradation. J Sci Food Agric. (2005) 85:1371–8. doi: 10.1002/jsfa.2100
9. Schmidt J, Tóth T, Fábián J. Rumen fermentation and starch degradation by holstein steers fed sodium-hydroxide or formaldehyde-treated wheat. Acta Vet Hung. (2006) 54:201–12. doi: 10.1556/AVet.54.2006.2.7
10. Humer E, Zebeli Q. Grains in ruminant feeding and potentials to enhance their nutritive and health value by chemical processing. Anim Feed Sci Technol. (2017) 226:133–51. doi: 10.1016/j.anifeedsci.2017.02.005
11. Iqbal S, Zebeli Q, Mazzolari A, Bertoni G, Dunn SM, Yang W. Feeding barley grain steeped in lactic acid modulates rumen fermentation patterns and increases milk fat content in dairy cows. J Dairy Sci. (2009) 92:6023–32. doi: 10.3168/jds.2009-2380
12. Iqbal S, Zebeli Q, Mazzolari A, Dunn SM, Ametaj BN. Feeding rolled barley grain steeped in lactic acid modulated energy status and innate immunity in dairy cows. J Dairy Sci. (2010) 93:5147–56. doi: 10.3168/jds.2010-3118
13. Iqbal S, Terrill SJ, Zebeli Q, Mazzolari A, Dunn SM, Yang W. Treating barley grain with lactic acid and heat prevented sub-acute ruminal acidosis and increased milk fat content in dairy cows. Anim Feed Sci Technol. (2012) 172:141–9. doi: 10.1016/j.anifeedsci.2011.12.024
14. Harder H, Khol-Parisini A, Zebeli Q. Modulation of resistant starch and nutrient composition of barley grain using organic acids and thermal cycling treatments. Starch-Starke. (2015) 67:654–62. doi: 10.1002/star.201500040
15. Harder H, Khol-Parisini A, Metzler-Zebeli BU, Klevenhusen F, Zebeli Q. Treatment of grain with organic acids at 2 different dietary phosphorus levels modulates ruminal microbial community structure and fermentation patterns in vitro. J Dairy Sci. (2016) 98:8107–20. doi: 10.3168/jds.2015-9913
16. Kazemi-Bonchenari M, Salem AZM, López S. Influence of barley grain particle size and treatment with citric acid on digestibility, ruminal fermentation and microbial protein synthesis in holstein calves. Animal. (2017) 11:1295–302. doi: 10.1017/S1751731116002810
17. Shen YZ, Ding LY, Chen LM, Xu JH, Zhao R, Yang WZ, et al. Feeding corn grain steeped in citric acid modulates rumen fermentation and inflammatory responses in dairy goats. Animal. (2019) 13:301–8. doi: 10.1017/S1751731118001064
18. Deshpande SS, Salunkhe DK. Interactions of tannic acid and catechin with legume starches. J Food Sci. (2010) 47:2080–1. doi: 10.1111/j.1365-2621.1982.tb12956.x
19. Yang WZ, Beauchemin KA, Koenig K, Rode LM. Effects of barley, hulless barley, and corn in concentrates on site and extent of digestion by lactating cows. J Dairy Sci. (1997) 80:2885–95. doi: 10.3168/jds.S0022-0302(97)76253-2
20. Huo WJ, Zhu WY, Mao SY. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J Microbiol Biotechnol. (2014) 30:669–80. doi: 10.1007/s11274-013-1489-8
21. Liu JH, Bian GR, Zhu WY, Mao SY. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of toll-like receptor genes in goats. Front Microbiol. (2015) 6:167. doi: 10.3389/fmicb.2015.00167
22. McDougall EI. Studies on ruminant saliva. 1. the composition and output of sheep's saliva. Biochem J. (1948) 43:99–109. doi: 10.1042/bj0430099
23. Busquet M, Calsamiglia S, Ferret Kamel AC. Plant extracts affect in vitro rumen microbial fermentation. J Dairy Sci. (2006) 89:761–71. doi: 10.3168/jds.S0022-0302(06)72137-3
24. AOAC (Ed.). Official Methods of Analysis. Arlington, VA: Association of Official Analytical Chemists (AOAC) (1995).
25. Van Soest PJ, Robertson JB, Lewis BA. Methods for dietary fiber, neutral-detergent fiber and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci. (1991) 74:3583–97. doi: 10.3168/jds.S0022-0302(91)78551-2
26. Hall MB. Neutral detergent-soluble carbohydrates nutritional relevance and analysis (a laboratory manual). In: Department of Animal Sciences, Institute of Food and Agricultural Sciences, University of Florida Bull. Vol. 339. Gainesville, FL: Bulletin. (2001). p. V-1–V-9.
27. Qin WL. Determination of rumen volatile fatty acids by means of gas chromatography (in Chinese, with English abstract). J Nanjing Agric College. (1982) 4:110–6.
28. Barker SB, Summersson WH. The colorimetric determination of lactic acid in biological materials. J Biol Chem. (2020) 138:535–54. doi: 10.1016/S0021-9258(18)51379-X
29. Edgar RC, Haas BJ, Clemente J, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. (2011) 27:2194–200. doi: 10.1093/bioinformatics/btr381
30. Amato KR, Yeoman CJ, Kent A, Righini N, Carbonero F, Estrada A. Habitat degradation impacts black howler monkey (alouatta pigra) gastrointestinal microbiomes. ISME J. (2013) 7:1344–53. doi: 10.1038/ismej.2013.16
31. Chao A, Bunge J. Estimating the number of species in a stochastic abundance model. Biometrics. (2002) 58:531–9. doi: 10.1111/j.0006-341X.2002.00531.x
32. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. (2009) 75:7537–41. doi: 10.1128/AEM.01541-09
33. Team C. Team rdc. R: A Language and Environment for Statistical Computing. Vienna: R foundation for statistical computing (2012).
34. Langille MGI, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16s rrna marker gene sequences. Nat Biotechnol. (2013) 31:814–21. doi: 10.1038/nbt.2676
35. Lettat A, Nozière P, Silberberg M, Morgavi DP, Berger C, Martin C. Experimental feed induction of ruminal lactic, propionic, or butyric acidosis in sheep. J Anim Sci. (2010) 88:3041–6. doi: 10.2527/jas.2010-2926
36. Hassanat F, Benchaar C. Assessment of the effect of condensed (acacia and quebracho) and hydrolysable (chestnut and valonea) tannins on rumen fermentation and methane production in vitro. J Sci Food Agric. (2013) 93:332–9. doi: 10.1002/jsfa.5763
37. Zhou K, Bao Y, Zhao G. Effects of dietary crude protein and tannic acid on rumen fermentation, rumen microbiota and nutrient digestion in beef cattle. Arch Anim Nutr. (2019) 73:30–43. doi: 10.1080/1745039X.2018.1545502
38. Khafipour E, Li SC, Plaizier JC, Krause DO. Rumen microbiome composition determined using two nutritional models of subacute ruminal acidosis. Appl Environ Microbiol. (2009) 75:7115–24. doi: 10.1128/AEM.00739-09
39. Fan WJ, Li HP, Zhu HS, Sui SP, Chen PG, Deng Y. NF-κB is involved in the LPS-mediated proliferation and apoptosis of MAC-T epithelial cells as part of the subacute ruminal acidosis response in cows. Biotechnol Lett. (2016) 38:1839–49. doi: 10.1007/s10529-016-2178-0
40. Zhang RY, Liu JH, Jiang LS, Mao SY. Effect of high-concentrate diets on microbial composition, function, and the VFAs formation process in the rumen of dairy cows. Anim Feed Sci Technol. (2020) 269:114619. doi: 10.1016/j.anifeedsci.2020.114619
41. Fred AR. Bergey's Manual of Systematics of Archaea and Bacteria. (2015). Available online at: https://doi.org/10.1002/9781118960608.gbm00707 (accessed September 14, 2015).
42. Collins MD, Shah HN, Mitsuoka T. Reclassification of Bacteroides microfusus (Kaneuchi and Mitsuoka) in a new genus Rikenella, as Rikenella microfusus comb. nov. Syst Appl Microbiol. (1985) 6:79–81. doi: 10.1016/S0723-2020(85)80015-1
43. Su XL, Tian Q, Zhang J, Yuan XZ, Shi XS, Guo RB, et al. Acetobacteroides hydrogenigenes gen. nov., sp. nov., an anaerobic hydrogen-producing bacterium in the family Rikenellaceae isolated from a reed swamp. Int J Syst Evol Microbiol. (2014) 64:2986–91. doi: 10.1099/ijs.0.063917-0
44. Vicini JL, Brulla WJ, Davis CL, Bryant MP. Quin's oval and other microbiota in the rumens of molasses-fed sheep. Appl Environ Microbiol. (1987) 53:1273–6. doi: 10.1128/aem.53.6.1273-1276.1987
45. Seshadri R, Leahy SC, Attwood GT, Teh KH, Lambie SC, Cookson AL, et al. Cultivation and sequencing of rumen microbiome members from the Hungate1000 Collection. Nat Biotechnol. (2018) 36:359–67. doi: 10.1038/nbt.4110
46. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y. Induction of colonic regulatory t cells by indigenous clostridium species. Science. (2011) 331:337–41. doi: 10.1126/science.1198469
47. Mao SY, Huo WJ, Zhu WY. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model: Ruminal microbial communities and metabolite profiles of goats fed high-grain diet. Environ Microbiol. (2016) 18:525–41. doi: 10.1111/1462-2920.12724
48. Plaizier JC, Li SC, Danscher AM, Derakhshani H, Andersen PH, Khafipour E. Changes in microbiota in rumen digesta and feces due to a grain-based subacute ruminal acidosis (SARA) challenge. Microb Ecol. (2017) 74:485–95. doi: 10.1007/s00248-017-0940-z
49. Liu JH, Tian K, Sun Y, Wu Y, Chen JC, Zhang R. Effects of the acid–base treatment of corn on rumen fermentation and microbiota, inflammatory response and growth performance in beef cattle fed high-concentrate diet. Animal. (2020) 14:1876–84. doi: 10.1017/S1751731120000786
50. Ntaikou I, Gavala HN, Kornaros M, Lyberatos G. Hydrogen production from sugars and sweet sorghum biomass using ruminococcus albus. Int J Hydrogen Energy. (2008) 33:1153–63. doi: 10.1016/j.ijhydene.2007.10.053
51. Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, et al. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE. (2013) 8:e83424. doi: 10.1371/journal.pone.0083424
52. KEGG. Starch, and Sucrose Metabolism. (2017). Available online at: https://www.keggjp/keggbin/show_pathway?map00500 (accessed February 9, 2017).
53. KEGG. Pentose Phosphate Pathway. (2020). Available online at: https://www.kegg.jp/keggbin/show_pathway?map00030 (accessed October 6, 2020).
54. KEGG. Fructose, and Mannose Metabolism. (2020). Available online at: https://www.kegg.jp/kegg-bin/show_pathway?map00051 (accessed October 21, 2020).
55. KEGG. Glyoxylate, and Dicarboxylate Metabolism. (2020). Available online at: https://www.kegg.jp/kegg-bin/show_pathway?map00630 (accessed March 27, 2020).
Keywords: tannic acid treated corn, rumen fermentation characteristics, ruminal microbiota, high-throughput sequencing, rumen acidosis
Citation: Zhao FF, Zhang XZ, Zhang Y, Elmhadi M, Qin YY, Sun H, Zhang H, Wang MZ and Wang HR (2021) Tannic Acid-Steeped Corn Grain Modulates in vitro Ruminal Fermentation Pattern and Microbial Metabolic Pathways. Front. Vet. Sci. 8:698108. doi: 10.3389/fvets.2021.698108
Received: 22 April 2021; Accepted: 13 August 2021;
Published: 28 October 2021.
Edited by:
Amlan Kumar Patra, West Bengal University of Animal and Fishery Sciences (WBUAFS), IndiaReviewed by:
Arianna Buccioni, University of Florence, ItalyYosra Ahmed Soltan, Alexandria University, Egypt
Copyright © 2021 Zhao, Zhang, Zhang, Elmhadi, Qin, Sun, Zhang, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: H. R. Wang, aHJ3YW5nQHl6dS5lZHUuY24=
 F. F. Zhao
F. F. Zhao X. Z. Zhang1,2
X. Z. Zhang1,2 Mawda Elmhadi
Mawda Elmhadi