- Hunan Provincial Key Laboratory of Protein Engineering in Animal Vaccines, Laboratory of Functional Proteomics (LFP), Research Center of Reverse Vaccinology (RCRV), College of Veterinary Medicine, Hunan Agricultural University, Changsha, China
Porcine parvoviruses (PPVs) and porcine circoviruses (PCVs) infect pigs worldwide, with PPV1–7 and PCV2 infections common in pigs. Although PPV7 was only identified in 2016, co-infection of PPV7 and PCV2 is already common, and PPV7 may stimulate PCV2 replication. PCV3, a novel type of circovirus, is prevalent in pig populations worldwide and considered to cause reproductive disorders and dermatitis nephrotic syndrome. In recent studies, pigs were commonly infected with both PCV3 and PPV7. Our objective was to investigate the co-infections between PPV7 and PCV3 in samples from swine on farms in Hunan, China, and assess the potential impacts of PPV7 on PCV3 viremia. A total of 209 samples, known to be positive (105) or negative (104) for PCV3, were randomly selected from serum samples that were collected from commercial swine herds in seven regions from 2016 to 2018 in our previous studies; these samples were subjected to real-time PCR to detect PPV7. Of these samples, 23% (48/209) were positive for PPV7. Furthermore, the PPV7 positive rate was significantly higher in PCV3 positive serum (31.4%, 33/105) than in PCV3 negative serum (14.4%, 15/104). Another 62 PCV3 positive sow serum samples and 20 PCV3 positive aborted fetuses were selected from 2015 to 2016 in our other previous study. These samples were designated as being from farms with or without long-standing histories of reproductive failure (RF or non-RF), respectively, and they were also subjected to real-time PCR to detect PPV7 and to determine whether PPV7 affected PCV3 viremia. Among the 62 serum samples (39 PCV3 positive RF-serum and 23 PCV3 positive non-RF-serum), 45.1% (28/62) were positive for PPV7 and PCV3, and the PPV7 positive rate was significantly higher in PCV3 positive RF-serum (51.2%, 20/39) than in PCV3 positive non-RF-serum (34.8%, 8/23). In addition, there was a higher positive rate of PPV7 (55%, 11/20) in PCV3 positive aborted fetus samples. In addition, the copy number of PCV3 in PPV7 positive samples was significantly higher than that in PPV7 negative serum samples. Based on these findings, we concluded that PPV7 may stimulate PCV3 replication.
Introduction
Porcine parvoviruses (PPVs) have been prevalent in pigs globally, and PPV1 is considered as one of the main pathogens causing reproductive failure in pigs around the world (1). However, genotypes PPV2–PPV6 with pathogenic potential were also detected, e.g., by genome sequencing. Porcine parvovirus 7 (PPV7) was initially identified in 2016 by metagenomics sequencing of rectal swabs from healthy adult pigs in the United States and subsequently from pigs in Brazil, China, South Korea, Poland, and Sweden. In China, PPV7 is already prevalent in Guangxi, Hunan, Anhui, Fujian, Shandong, and Northeast China (2, 3), although the detailed information of its pathogenicity in pigs remains unavailable. Regarding novel PPVs, PPV4, PPV6, and PPV7 were detected in aborted fetuses, which implied that these viruses may cause reproductive failure (4–6). Moreover, detection of PPV7 in semen implies that this virus may cause reproductive dysfunction through vertical transmission (7).
PPV7 is a single-stranded DNA (ssDNA, ~4 kb), non-enveloped virus, with low homology with PPV1–6 (~30%). It belongs to the family Parvovirinae and is an emerging species of the genus Chapparvovirus. PPV7 can be isolated from healthy and sick pigs of all ages and was present in various tissues (liver, lung, lymph node, kidney, and spleen). Nucleotide mutation rates of NS1 and cap genes of PPV7 were higher than those of PPV1–4 (8), perhaps enabling PPV7 to adapt to various environmental conditions and posing a major threat to health security of pig herds.
PPV1–7 and porcine circovirus 2 (PCV2) co-infections are common in pigs. In recent studies, the level of PCV2 viremia was greater in serum samples that were positive for PPV1 and PPV7 than in those that were negative for PPVs (9, 10). Furthermore, there was a correlation between the Ct values of PPV7 and PCV2 (11). As a consequence, we inferred that, in addition to PPV1, PPV7 may potentially act as a co-factor infection by stimulating the replication of PCV2. PCV3, a novel type of circovirus discovered in 2016, is prevalent in many countries around the world and is regarded as causing reproductive disorders and dermatitis nephrotic syndrome, although the pathogenesis is not well established. It was reported that PCV3 positive samples have a high co-infection rate with PPV7 (12), although nothing is known about the impact of PPV7 on PCV3 viremia. In this study, we investigated co-infections between PPV7 and PCV3 in samples from swine on farms in Hunan, China, and assessed potential impacts of PPV7 on PCV3 viremia.
Materials and Methods
Serum and Aborted Fetuses
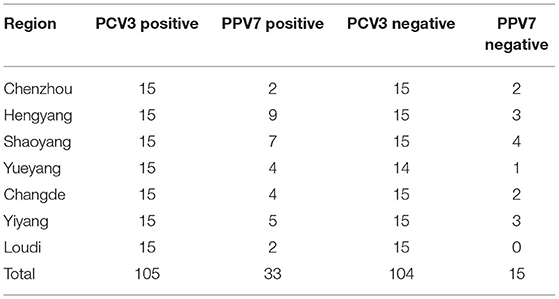
We previously detected PCV3 IgG antibodies in sow sera from commercial swine herds (n = 1038) in seven regions of Hunan Province, China using capsid protein-based indirect ELISA (13). Among them, a total of 209 serum samples (105 PCV3 positive and 104 PCV3 negative serum samples, Table 1), based on PCV3 detection by quantitative PCR (qPCR), as described (13, 14), were randomly selected and used to determine PPV7 prevalence in the present study. In other studies, we reported identification of PCV3 (using qPCR and ELISA, respectively) in sow sera (n = 190), which were selected from the farms (A–E) with or without reproductive failure (RF) in various regions in Hunan, China (14). In more detail (Table 2), 85 samples (with 39 PCV3 positive) were from sows that had aborted or had a history of reproductive failure (+RF), whereas the remaining 105 (with 23 PCV3 positive) were from healthy sows (from herds with no history of reproductive failure, –RF), among which copy numbers of PCV3 genome based on qPCR were determined and reported (13, 14). It was noteworthy that the PCV3 positive rate was significantly higher in sows with reproductive failure [+RF, 45.9% (39/85)] than in healthy sows [–RF, 21.9% (23/105)] (14). In addition, 60.6% (20/33) of aborted fetuses from Farms C and E were positive for PCV3 (13), based on qPCR assays (Table 2).
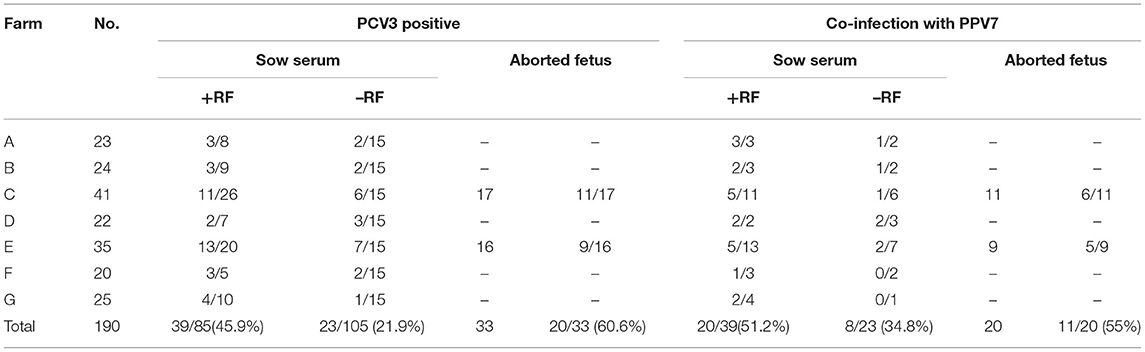
Table 2. Presence of PCV3 and PPV7 co-infections in serum of sows, with and without reproductive failure (RF), and in aborted fetuses.
As these important samples have already been tested for PCV3 and its viral load, they can also be used to detect co-infection with PPV7, facilitating an in-depth study of the co-infection of PCV3 and PPV7 and the interaction by co-infection to enhance or stimulate virus replication.
Real-Time PCR Assay for PCV3 and PPV7
qPCR for copy numbers of PCV3 genomic DNA with primers (QP3-F: YAGTGCTCCCCATTGAACGG and QP3-R: GCTCCAAGACGACCCTTATGC) in our previous report (13) was used to determine the copy number of PCV3 in the samples. In addition, a SYBR Green real-time PCR assay with primers (F1: GCGACCAGTCGAAAGTCTTC and R1: TTGGTGTTGCCCATTCTGTA) targeting a 165-bp region of PPV7, the conserved capsid gene for PPV7 detection, was done, as described (15). Based on results of real-time PCR, samples were deemed negative or positive for PCV3 and for PPV7.
In brief, we used a 20-μl reaction mixture containing 10 μl of AceQ qPCR SYBR Green Master Mix (Vazyme Biotech Co., Piscataway, NJ, USA), 0.4 μl PCV3 primer pairs or 0.6 μl PPV7 primer pairs (10 μM), 0.4 μl of 50 × ROX Reference Dye 1, 2 μl of DNA template, and 6.8 μl of RNase-free ddH2O. In addition, a pSP72 plasmid clone containing the full-length cap gene of PCV3 (pSP72-PCV3; GenBank accession number KY484769) or the full-length VP2 gene of PPV7 (pSP72-PPV7; GenBank accession number KU563733) and ddH2O were used as positive and negative controls, respectively. Copy numbers of viral genomic DNA extracted from samples were calculated based on a standard curve.
Statistical Analyses
All statistical analyses were performed using SPSS 21.0 software (SPSS Inc., Chicago, IL, USA) and GraphPad Prism version 8.0.0 for Windows (GraphPad Software, San Diego, CA, USA; www.graphpad.com). PCV3 and PPV7 serum categories were investigated using Fisher's exact test by pairwise comparisons. The one-way ANOVA was used for statistical comparison between PCV3 and PPV7 serum categories expressed as copy numbers. Pearson's correlations of copy numbers in PCV3 and PPV7 positive samples were determined. Statistical significance was set at p < 0.05, and confidence intervals were calculated.
Results
PPV7 Infections Occur Frequently in Pigs Affected With PCV3
The 209 samples (105 PCV3 positive and 104 PCV3 negative serum samples), derived from our previous study (13), were randomly selected from each region in Hunan, China, from 2016 to 2018. Among these 209 serum samples, 23% (48/209) were positive for PPV7. Of these, 31.4% (33/105) were positive for PPV7 in PCV3 positive serum samples (Table 1), whereas PPV7 was detected in 14.4% (15/104) of the randomly selected PCV3 negative samples (Table 1). The PPV7 positive rate was significantly higher (2.2 times) in PCV3 positive serum samples (31.4%) than in PCV3 negative serum samples (14.4%).
In this study, we also used sow sera and aborted fetuses that had been collected between 2015 and 2016 from seven sow farms with histories of long-standing reproductive problems (14). Among the 190 serum samples, there were 62 PCV3 positive and 128 PCV3 negative (14), whereas 24.7% (47/190) were positive for PPV7 (Table 2). The PPV7 positive rate was significantly higher (3.0 times) in PCV3 positive serum samples (28/62, 45.1%) than in PCV3 negative serum samples (19/128,14.8%).
The PPV7 detection rates in PCV3 positive serum samples with RF (+RF) were 51.2% (20/39), whereas they were only 34.8% (8/23) in PCV3 positive sera without RF (–RF). Furthermore, among 33 aborted fetuses from Farms C and E that had 20 PCV3 positive fetuses (14), 55% (11/20) were positive for both PPV7 and PCV3 (Table 2). In summary, the PPV7 positive rate was 1.5 times higher in PCV3 positive serum from sows with RF (+RF) vs. without RF (–RF); furthermore, there was a higher PPV7 prevalence (55%) in aborted fetus samples.
PCV3 Viremia Is Higher in PPV7 Positive Pigs
To evaluate impacts of PCV3 and PPV7 co-infections on their viremia, 190 sow serum samples (+RF and –RF) used in a previous report (14) were divided into the following groups: PCV3–PPV7 positive (n = 28), PCV3 positive–PPV7 negative (n = 34), and PCV3 negative–PPV7 positive (n = 19).
The copy number of PPV7 in PCV3 positive and negative serum samples was detected by real-time PCR; there was no significant difference in PPV7 between PCV3 positive (n = 28) and negative (n = 19) samples (Figure 1A). However, the copy number of PCV3 in PPV7 positive samples (n = 28, PCV3–PPV7 positive groups) was higher (p < 0.001) than that in PPV7 negative serum samples (n = 27, selected from 34 samples of PCV3 positive–PPV7 negative groups) (Figure 1B), and there was a very high correlation (p = 0.0002) in copy number between PCV3 and PPV7 from PCV3–PPV7 positive group samples (Figure 2). The linear correlation coefficient (r) between PPV7 and PCV3 copy numbers was 0.651. As the square of correlation (r2) score was 0.424, 42.4% of PCV3 copy number could be accounted for by PPV7 copy number.
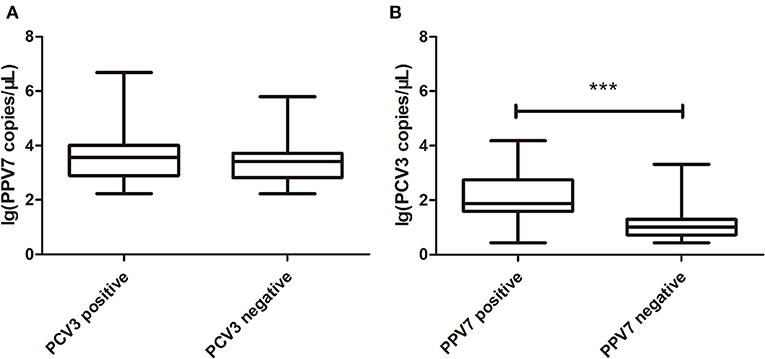
Figure 1. Boxplot comparison of real-time PCR copy number of PPV7 and PCV3. (A) Boxplot comparison of real-time PCR copy number of PPV7 in PCV3 positive (n = 28) and PCV3 negative (n = 19) serum samples. (B) Boxplot comparison of real-time PCR copy number of PCV3 in PPV7 positive (n = 28) and PPV7 negative (n = 27) serum samples. *** p < 0.001.
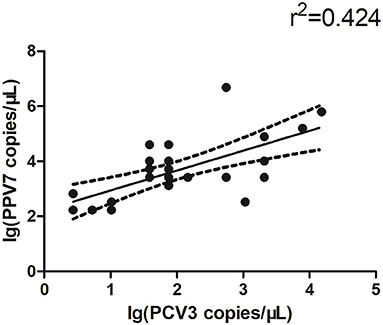
Figure 2. Scatterplots with trends for real-time PCR copy number for PPV7 and PCV3 positive samples (n = 28, p = 0.0002).
Discussion
PPV1 co-infects with PCV2 and PCV3, porcine reproductive and respiratory syndrome virus (PRRSV), pseudorabies virus (PRV), and classical swine fever virus (CSFV). The prevalence of PPV1–PCV2 co-infections is high, and PPV1 may trigger PCV2 associated disease (PCVAD) by supporting PCV2 replication, and increase PCVAD severity (e.g., pathological lesions in lymphoid tissues) (16). In addition, there are co-infections between novel PPVs and other well-known pathogens (e.g., PPVs, PCV2, PCV3, PRRSV, and TTSU) (3). Infections with PPV7 may become chronic, and PPV7 may contribute to virus persistence, with continuous excretion of virus in feces (17). In addition, fattening pigs without clinical symptoms had a high viral load (Ct <25), for PPV7 in their feces; therefore, variations in PPV7 viral loads may indicate various effects of PPV7 infection in pigs (17), or perhaps other conditions (e.g., co-infection) that made PPV7 pathogenic in pigs.
The prevalence of PPV7 ranged from 8.6 to 61.5% (2, 9, 11, 17–20). In our study of pigs from Hunan, China, the prevalence of PPV7 for both PCV3 negative and positive serum samples combined was 23% (48/209), and the prevalence in sow serum samples with or without RF was 24.7% (47/190). There was no basis to conclude that PPV7 contributed to all observed pathologic changes, as not all pathogens were consistently detected in diseased pigs and the prevalence of PPV7 in serum samples was higher than that in other tissues (18). Furthermore, none of the diseased pigs was only infected with PPV7. Therefore, it remains to be determined whether infection with PPV7 per se induces disease in pigs. It was also reported that the positive rate of PPV7 in PCV2 positive pig farms was significantly higher than that in negative farms (65.5 vs. 5.7%, respectively) (18). Moreover, the co-infection rate of PPV7 and PCV2 was high (17.4–59.5%) composed of 17.4% (67/385) and 59.5% (147/247) in Guangxi, 18.2% (29/159) in Poland, and 17.5% (21/120) in Anhui, respectively (9, 11, 19, 21). Therefore, it was speculated that PPV7 was an important cofactor of PCVAD (9). Although clinical symptoms and pathology of PPV7 remain unclear, it may act as a co-factor of disease caused by other porcine pathogens, or it may trigger disease development.
The co-infection rate of PCV3 and PPV7 was 9.1% (11/120) in samples from commercial farms with various clinical symptoms, including respiratory and gastrointestinal (19). In another report, in PCV3 positive samples, PCV3 had a high co-infection rate with both PPV6 (60.0%, 21/35) and PPV7 (74.3%, 26/35) (12). Based on these data, we inferred that there is a possible association between PCV3 and PPV7 infections. In our study, PCV3 also had a high co-infection rate with PPV7 [45.1% (28/62), 55% (11/20)]. Since both circovirus and parvovirus are ssDNA viruses, active proliferation of target cells is required for efficient viral replication. Virus-induced lymphocyte proliferation or immunosuppression can enhance the susceptibility to other virus replication and infection (22–25). For PCV2, its infection directly targets immune cells and causes immunosuppression (26–28), which leads to secondary or mixed infections with other pathogens. Furthermore, evidence that PPV-induced immune dysfunction could promote PCV2 replication (29) supports our notion that a co-infection of PPV7 and PCV3 could enhance the pathogenicity of the latter virus.
In this study, the PPV7 positive rate was statistically significantly higher in PCV3 positive versus PCV3 negative samples, suggesting that PCV3 may also cause immunosuppression, similar to PCV2, leading to secondary infection. Interestingly, co-infection with PPV7 and PCV3 in sow serum with RF (+RF) was significantly higher than that in sow serum without RF (–RF), and we also noted a higher PPV7 prevalence in aborted fetus samples. Furthermore, there were higher PCV3 viral loads in samples that were PPV7 positive compared with PPV7 negative. It has been suggested that PPV7 may stimulate the replication of PCV2 (11, 30). We speculated that PPV7 stimulated the replication of PCV3, thereby enhancing PCV3 viremia. Based on the present results and previous studies, we concluded that PPV7 may be an important co-factor triggering PCV2 and PCV3-associated diseases. Regardless, the pathogenesis of PPV7 infections, with or without PCV3 co-infection, needs to be further confirmed. More frequent multifactorial co-infection in clinical conditions contributes to a range of disease syndromes and is one of the most difficult problems in swine production, where next-generation sequencing (NGS) will gain a new insight into how co-factor infections interact to cause syndromes.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
We confirm that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. This study was approved by the Animal Ethics Committee of Hunan Agricultural University, Hunan, China.
Author Contributions
NW and JM conceived and designed the experiments. JM, DW, and YZ designed and carried out the PCV3 and PPV7 RT-PCR detection. JM, SZ, and CM conducted statistical analysis on the data. JM, AW, and NW contributed to writing and revision of the manuscript. All authors read and approved the final manuscript.
Funding
This study was supported by the Hunan Provincial Natural Science Foundation of China (No. 2018JJ2177) and Double First-class Construction Project of Hunan Agricultural University (No. SYL2019048).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Mengeling WL, Lager KM, Vorwald AC. The effect of porcine parvovirus and porcine reproductive and respiratory syndrome virus on porcine reproductive performance. Anim Reprod Sci. (2000) 60:199–210. doi: 10.1016/S0378-4320(00)00135-4
2. Palinski RM, Mitra N, Hause BM. Discovery of a novel Parvovirinae virus, porcine parvovirus 7, by metagenomic sequencing of porcine rectal swabs. Virus Genes. (2016) 52:564–7. doi: 10.1007/s11262-016-1322-1
3. Mai JH, Wang DL, Tan L, Wang ND. Advances in porcine parvovirus 7. Chin Vet Sci. (2021) 51:361–7. doi: 10.16656/j.issn.1673-4696.2021.0054
4. Cságola A, Lorincz M, Cadar D, Tombácz K, Biksi I, Tuboly T. Detection, prevalence and analysis of emerging porcine parvovirus infections. Arch Virol. (2012) 157:1003–10. doi: 10.1007/s00705-012-1257-3
5. Ni J, Qiao C, Han X, Han T, Kang W, Zi Z, et al. Identification and genomic characterization of a novel porcine parvovirus (PPV6) in China. Virol J. (2014) 11:203. doi: 10.1186/s12985-014-0203-2
6. Ouh IO, Park S, Lee JY, Song JY, Cho IS, Kim HR. First detection and genetic characterization of porcine parvovirus 7 from Korean domestic pig farms. J Vet Sci. (2018) 19:855–7. doi: 10.4142/jvs.2018.19.6.855
7. Chung HC, Nguyen VG, Park YH, Park KT, Park BK. PCR-based detection and genetic characterization of porcine parvoviruses in South Korea in 2018. BMC Vet Res. (2020) 16:113. doi: 10.1186/s12917-020-02329-z
8. Wang D, Mai J, Yang Y, Wang ND. Porcine parvovirus 7: evolutionary dynamics and identification of epitopes toward vaccine design. Viruses-Basel. (2020) 8:359. doi: 10.3390/vaccines8030359
9. Wang W, Cao L, Sun W, Xin J, Zheng M, Tian M, et al. Sequence and phylogenetic analysis of novel porcine parvovirus 7 isolates from pigs in Guangxi, China. PLoS ONE. (2019) 14:e0219560. doi: 10.1371/journal.pone.0219560
10. Opriessnig T, Xiao CT, Gerber PF, Halbur PG. Identification of recently described porcine parvoviruses in archived North American samples from 1996 and association with porcine circovirus associated disease. Vet Microbiol. (2014) 173:9–16. doi: 10.1016/j.vetmic.2014.06.024
11. Miłek D, Wozniak A, Podgórska K, Stadejek T. Do porcine parvoviruses 1 through 7 (PPV1-PPV7) have an impact on porcine circovirus type 2 (PCV2) viremia in pigs? Vet Microbiol. (2020) 242:108613. doi: 10.1016/j.vetmic.2020.108613
12. Ha Z, Xie CZ, Li JF, Wen SB, Zhang KL, Nan FL, et al. Molecular detection and genomic characterization of porcine circovirus 3 in pigs from Northeast China. BMC Vet Res. (2018) 14:321. doi: 10.1186/s12917-018-1634-6
13. Zhang S, Wang D, Jiang Y, Li Z, Zou Y, Li M, et al. Development and application of a baculovirus-expressed capsid protein-based indirect ELISA for detection of porcine circovirus 3 IgG antibodies. BMC Vet Res. (2019) 15:79. doi: 10.1186/s12917-019-1810-3
14. Zou Y, Zhang N, Zhang J, Zhang S, Jiang Y, Wang D, et al. Molecular detection and sequence analysis of porcine circovirus type 3 in sow sera from farms with prolonged histories of reproductive problems in Hunan, China. Arch Virol. (2018) 163:2841–7. doi: 10.1007/s00705-018-3914-7
15. Li YD, Yu ZD, Bai CX, Zhang D, Sun P, Peng ML, et al. Development of a SYBR Green I real-time PCR assay for detection of novel porcine parvovirus 7. Pol J Vet Sci. (2021) 24:43–9.
16. Opriessnig T, Fenaux M, Yu S, Evans RB, Cavanaugh D, Gallup JM, et al. Effect of porcine parvovirus vaccination on the development of PMWS in segregated early weaned pigs coinfected with type 2 porcine circovirus and porcine parvovirus. Vet Microbiol. (2004) 98:209–20. doi: 10.1016/j.vetmic.2003.11.006
17. Miłek D, Wozniak A, Podgórska K, Stadejek T. The detection and genetic diversity of novel porcine parvovirus 7 (PPV7) on Polish pig farms. Research in Vet Sci. (2018) 120:28–32. doi: 10.1016/j.rvsc.2018.08.004
18. Xing X, Zhou H, Tong L, Chen Y, Sun Y, Wang H, et al. First identification of porcine parvovirus 7 in China. Arch Virol. (2018) 163:209–13. doi: 10.1007/s00705-017-3585-9
19. Wang Y, Yang KK, Wang J, Wang XP, Zhao L, Sun P, et al. Detection and molecular characterization of novel porcine parvovirus 7 in Anhui province from Central-Eastern China. Infect Genet Evol. (2019) 71:31–5. doi: 10.1016/j.meegid.2019.03.004
20. Da Silva MS, Budaszewski RF, Weber MN, Cibulski SP, Paim WP, Mósena ACS, et al. Liver virome of healthy pigs reveals diverse small ssDNA viral genomes. Infect Genet Evol. (2020) 81:104203. doi: 10.1016/j.meegid.2020.104203
21. Cao L, Sun WC, Wang W, Tian MY, Guo DD, Liu YX, et al. Sequence and phylogenetic analysis of porcine parvovirus 7 isolates from pigs in Guangxi Province, China. Chin J Vet Sci. (2020) 40:457–62.
22. Saekhow P, Kishizuka S, Sano N, Mitsui H, Akasaki H, Mawatari T, et al. Coincidental detection of genomes of porcine parvoviruses and porcine circovirus type 2 infecting pigs in Japan. J Vet Med Sci. (2016) 77:1581–6. doi: 10.1292/jvms.15-0167
23. Saekhow P, Mawatari T, Ikeda H. Coexistence of multiple strains of porcine parvovirus 2 in pig farms. Microbiol Immunol. (2014) 58:382–7. doi: 10.1111/1348-0421.12159
24. Ellis J, Clark E, Haines D, West K, Krakowka S, Kennedy S, et al. Porcine circovirus-2 and concurrent infections in the field. Vet Microbiol. (2004) 98:159–63. doi: 10.1016/j.vetmic.2003.10.008
25. Darwich L, Mateu E. Immunology of porcine circovirus type 2 (PCV2). Adv Virus Res. (2012) 164:61–67. doi: 10.1016/j.virusres.2011.12.003
26. Darwich L, Segalés J, Mateu E. Pathogenesis of postweaning multisystemic wasting syndrome caused by Porcine circovirus 2: an immune riddle. Arch Virol. (2004) 149:857–74. doi: 10.1007/s00705-003-0280-9
27. Meng XJ. Porcine circovirus type 2 (PCV2): pathogenesis and interaction with the immune system. Annu Rev Anim Biosci. (2013) 1:43–64. doi: 10.1146/annurev-animal-031412-103720
28. Segalés J, Rosell C, Domingo M. Pathological findings associated with naturally acquired porcine circovirus type 2 associated disease. Vet Microbiol. (2004) 98:137–49. doi: 10.1016/j.vetmic.2003.10.006
29. Cadar D, Cságola A, Kiss T, Tuboly T. Capsid protein evolution and comparative phylogeny of novel porcine parvoviruses. Mol Phylogenet Evol. (2013) 66:243–53. doi: 10.1016/j.ympev.2012.09.030
Keywords: porcine parvovirus 7, porcine circovirus type 3, co-infections, viremia, RT-PCR
Citation: Mai J, Wang D, Zou Y, Zhang S, Meng C, Wang A and Wang N (2021) High Co-infection Status of Novel Porcine Parvovirus 7 With Porcine Circovirus 3 in Sows That Experienced Reproductive Failure. Front. Vet. Sci. 8:695553. doi: 10.3389/fvets.2021.695553
Received: 15 April 2021; Accepted: 25 June 2021;
Published: 29 July 2021.
Edited by:
Fabrizio Passamonti, University of Perugia, ItalyReviewed by:
Giovanni Franzo, University of Padua, ItalyPaula Louise Lagan Tregaskis, Agri-Food and Biosciences Institute (AFBI), United Kingdom
Copyright © 2021 Mai, Wang, Zou, Zhang, Meng, Wang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naidong Wang, bmFpZG9uZ3dhbmdAaHVuYXUuZWR1LmNu
†These authors have contributed equally to this work
 Jinhui Mai
Jinhui Mai Dongliang Wang
Dongliang Wang Yawen Zou
Yawen Zou Sujiao Zhang
Sujiao Zhang Aibing Wang
Aibing Wang Naidong Wang
Naidong Wang