
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
BRIEF RESEARCH REPORT article
Front. Vet. Sci. , 24 August 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.695466
This article is part of the Research Topic The Use of Nanoparticles in the Diagnosis and Therapy of Infectious Disease in Animals View all 6 articles
Porcine circovirus type 4 (PCV4), a recently reported circovirus, was first identified in pigs with clinical signs similar to porcine dermatitis nephropathy syndrome (PDNS), in Hunan province, China, in 2019. More knowledge regarding the assembly of capsid protein (Cap) into virus-like particles (VLPs), their structure and antigenic properties, are needed to provide new knowledge for diagnosis and further characterization of PCV4. In this study, high-level expression of PCV4 Cap was achieved in Escherichia coli with purified Cap self-assembling into VLPs (~20 nm) in vitro. Furthermore, these VLPs were internalized in vitro by PK15 and 3D4/21 cell lines. Significant structural differences between PCV4 and PCV2 capsids were demonstrated among loops (loop BC, CD, DE, EF, and GH), based on comparisons of 3D structures. In addition, five potential B cell epitopes identified in silico were mostly located in surface-exposed loops of PCV4 capsid. Cross-reaction between PCV4 and PCV2 or PCV3 conferred by humoral immune responses was deemed unlikely on the basis of ELISA and Western blotting for assessment of VLPs and using PCV4 or PCV2 VLPs. In conclusion, these studies provided new knowledge regarding PCV4 capsid surface patterns. It is noteworthy that the PCV4 VLPs prepared in our study have much potential for development of serological diagnostics for PCV4 and to further characterize this virus.
Porcine circoviruses (PCVs) are small, spherical, non-enveloped viruses composed of circular, single-stranded genomic DNA within an icosahedral capsid. These viruses belong to the genus Circovirus in the family Circoviridae. PCVs have high genetic variability, with four known genotypes: PCV1, PCV2, PCV3, and a novel PCV4 genotype. Although PCV1 is considered non-pathogenic, PCV2 is the causal agent of PCV2-associated diseases (PCVADs). PCV3, first identified in 2016 by metagenomic sequencing (1), is associated with porcine dermatitis nephropathy syndrome (PDNS), cardiac disease, reproductive failure, respiratory disease, etc. (1–3).
In 2019, a newly emerging PCV4 genotype was first identified in serum samples from pigs with respiratory and enteric signs as well as PDNS, in Hunan, China (4). PCV4 is suspected to be associated with these clinical signs, which also occur in pigs infected with PCV2 or PCV3 (1). Subsequently, PCV4 infections causing clinical signs in swine were reported in Henan, Shanxi, Jiangsu, Anhui, and Guangxi provinces, China (5–8), with PCV4-positive rates ranging from 5.1% (13/257) to 25.4% (16/63). Recently, PCV4 has also been detected on swine farms in Korea (9). As distribution, epidemiology, and pathogenicity (or putative disease association) of PCV4 have not been elucidated, there is an urgent need to characterize this virus.
The genome of PCV4 is ~1.7 kb long and contains two major open reading frames (ORFs). Whereas, ORF1 encodes replication protein (Rep) involved in virus replication, ORF2 encodes the capsid protein (Cap), the sole structural protein of this virus, capable of inducing neutralizing antibodies and representing a major target for vaccine design and serological diagnosis. In the absence of an efficient method of culturing PCV4, virus-like particles (VLPs) derived from genetic and protein engineering are considered a powerful model to investigate capsid assembly, tissue tropism, and pathogenesis and, in particular, develop effective serological diagnostics for PCV4 (to clarify its incidence). In this study, structure and antigenic properties of PCV4 Cap were analyzed, and expressed PCV4 Cap protein self-assembled into VLPs in an Escherichia coli expression system in vitro. These results should promote development of effective serological diagnostics for PCV4 and increase understanding of this virus.
A full-length PCV4 cap gene was codon optimized and synthesized by the GenScript Company (Nanjing, China) based on the PCV4-AHG-2019 genome sequence (GenBank accession number: MK986820). This gene was cloned into NdeI and BamHI restriction sites of the pET28a expression vector and the recombinant plasmid confirmed by DNA sequencing. To express recombinant protein, this plasmid was transformed into E. coli BL21 (DE3) competent cells (Biomed, Beijing, China) according to the instructions of the manufacturer. Subsequently, a single colony was selected and grown in LB medium (containing 50 μg/ml kanamycin) to an OD600 value reaching 0.6–0.8 before a final concentration of 0.5 mM IPTG was added to induce protein expression. After being induced, cells were grown at 25°C for 10 h, harvested, and soluble protein was purified by Ni-NTA affinity chromatography, as described (10). An E. coli BL21(DE3) cell pellet was placed into lysis buffer (100 mM NaH2PO4·2H2O, 50 mM imidazole, 10 mM Tris-HCl, 300 mM NaCl, and 100 mM KCl, pH 8.5) with 0.5 % Triton X-100, and 5 mM β-mercaptoethanol. Then, cells were disrupted by sonication (35% amplitude for 20 min), after which the homogenate was cleared by centrifugation (15,000 × g for 30 min). The supernatant was loaded on a prepacked HisTrap™ HP column (GE Healthcare Life Sciences, New York, NY, USA) according to the instructions of the manufacturer. The column was washed with wash buffer (50 mM NaH2PO4·2H2O, 50 mM imidazole, and 300 mM NaCl, pH 8.0), and PCV4 Cap was eluted with elution buffer (50 mM NaH2PO4·2H2O, 200 mM imidazole, and 300 mM NaCl, pH 8.0). Purified proteins were analyzed with SDS-PAGE and Western blot using mouse anti-6 × His tag antibody (1:3,000; Solarbio, Beijing, China) or mouse anti-PCV4 Cap serum (1:1,000).
Purified PCV4 Cap was dialyzed against phosphate-buffered saline (PBS) buffer (pH 7.4) for 48 h. The PCV4 VLPs were absorbed into carbon-coated copper grids for 10 min, stained with 1% phosphotungstic acid for 5 min, and imaged using TEM (CM100, Philips Electron Optics, Switzerland).
PCV2 VLPs were prepared as described (10). Female BALB/c mice (n = 4/group, 6 weeks old) were used to prepare anti-PCV4 and PCV2 Cap sera. Each mouse was initially immunized with 20 μg of PCV4 or PCV2 VLPs mixed with Freund's complete adjuvant (final volume, 0.2 ml). Booster immunizations using the same amount of VLPs mixed with Freund's incomplete adjuvant were done twice at 2-week intervals (weeks 2 and 4). Blood samples were collected 6 weeks after primary immunization, and sera were collected and stored at −20°C.
Two porcine cell lines (PK15 and 3D4/21) were used to detect cell entry of PCV4 VLPs. Cells cultured in 48-well plates were incubated with 2 μg of PCV4 VLPs in each well, with DMEM, in lieu of VLPs, added to cells as a control. After incubation for 1 h, cells were washed three times with PBST (containing 0.1% Tween 20). Subsequently, cell entry of PCV4 VLPs was detected with an indirect immunofluorescence assay (IFA), as described (11). Anti-PCV4 Cap mouse serum (1:1,000) was used as the primary antibody, and immunostained slides were examined with fluorescence microscopy (Olympus BX-51, Tokyo, Japan).
Sera of mice immunized with PCV2 and PCV4 VLPs were assessed using our established indirect ELISA, as described (10). Secondary antibody was horseradish peroxidase (HRP)-labeled goat anti-mouse IgG (Thermo Fisher Scientific, Shanghai, China). Optical densities were read at 450 nm.
All PCVs Cap sequences in the NCBI GenBank database (http://www.ncbi.nlm.nih.gov/) were retrieved. Amino acid sequences were aligned using the ESPript 3.0 online tool (http://espript.ibcp.fr/ESPript/ESPript/). The 3D structure homology modeling was reconstructed based on PCV2 Cap protein structure (Protein Data Bank (PDB) ID: 3R0R) in SWISS-MODEL (https://swissmodel.expasy.org/) and displayed with Pymol 1.8.0.3. Linear B cell epitopes of PCV4 Cap protein were predicted by BepiPred 2.0 online tool (http://www.iedb.org/) with a default threshold of 0.5. Antigenicity was evaluated with the Vaxijen v2.0 server online tool (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html).
The full length of the optimized cap gene was cloned into a pET28a expressed vector with a 6 × His tag fused at the NH2 terminus of the PCV4 Cap protein to aid purification. The PCV4 Cap was successfully expressed in bacteria. Furthermore, the soluble Cap was purified with Ni-NTA affinity chromatography. The purified Cap, analyzed by SDS-PAGE, had an apparent molecular weight of ~28 kDa (Figure 1A) and was recognized by anti-His tag antibody in a Western blot (Figure 1B).
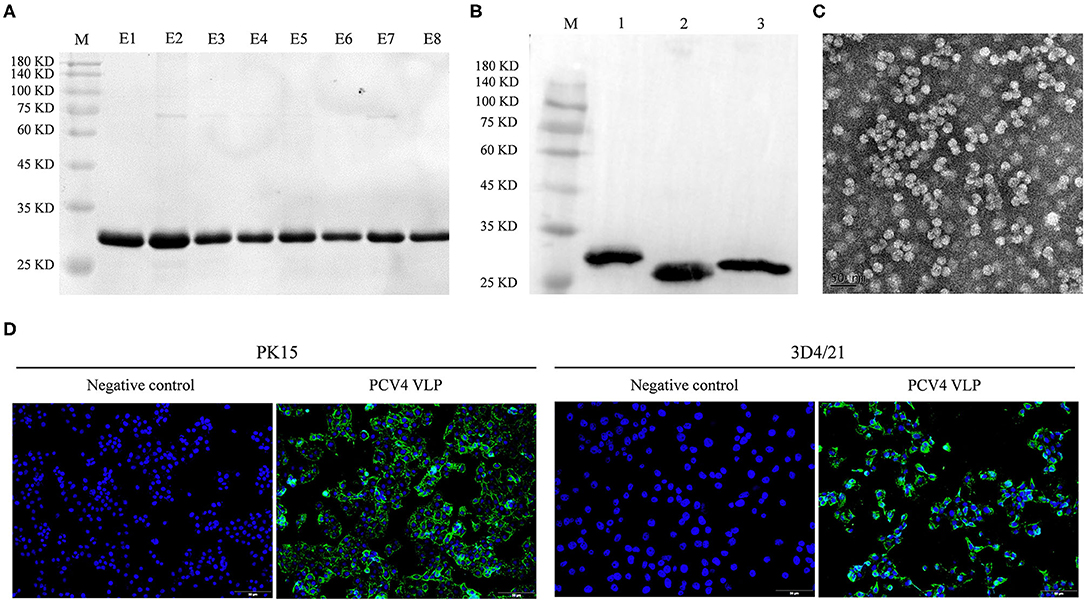
Figure 1. (A) Coomassie blue-stained SDS-PAGE of purified PCV4 Cap protein (lane M: protein marker; lanes E1–E8: purified PCV4 Cap protein). (B) Western blots of PCV Cap proteins using mouse anti-6 × His tag antibody (lane M: protein marker; lane 1: PCV4 Cap; lane 2: PCV3 Cap; and lane 3: PCV2 Cap). (C) Formation of PCV4 VLPs observed with TEM. (D) Entry of PCV4 VLPs into PK15 and 3D4/21 cell lines was confirmed by fluorescence microscopy. Green fluorescence represents PCV4 Cap in cells, whereas cell nuclei (blue) were stained by DAPI.
Purified PCV4 Cap protein was dialyzed against PBS buffer (pH 7.4) to facilitate in vitro assembly of VLPs. Spherical morphology (diameter, ~20 nm) was observed under TEM, confirming formation of PCV4 VLPs (Figure 1C). Therefore, the Cap self-assembled into VLPs in vitro.
Two porcine cell lines (PK15 and 3D4/21) were used to test internalization of PCV4 VLPs into cells in vitro. Based on IFA, PCV4 VLPs entered both PK15 and 3D4/21 cells (Figure 1D).
The PCV Cap has a canonical jelly roll fold; however, Cap sequence variations among various circovirus isolates have minimal effect on the jelly roll structure. Similar to PCV2 Cap structures, a viral jelly roll structure was also observed in the PCV4 Cap (Figure 2A), based on homology modeling using a crystal structure of PCV2 Cap as a template. Based on Cap sequence alignment, most variable residues between PCV2 and PCV4 Caps were located within the surface-exposed loops, especially the icosahedral five-fold axes composed of loops BC, DE, and HI (Figures 2A, 3A). Compared to the PCV2 Cap, loops BC, CD, DE, EF, and GH were distinctly different between PCV2 and PCV4 (Figure 2B).
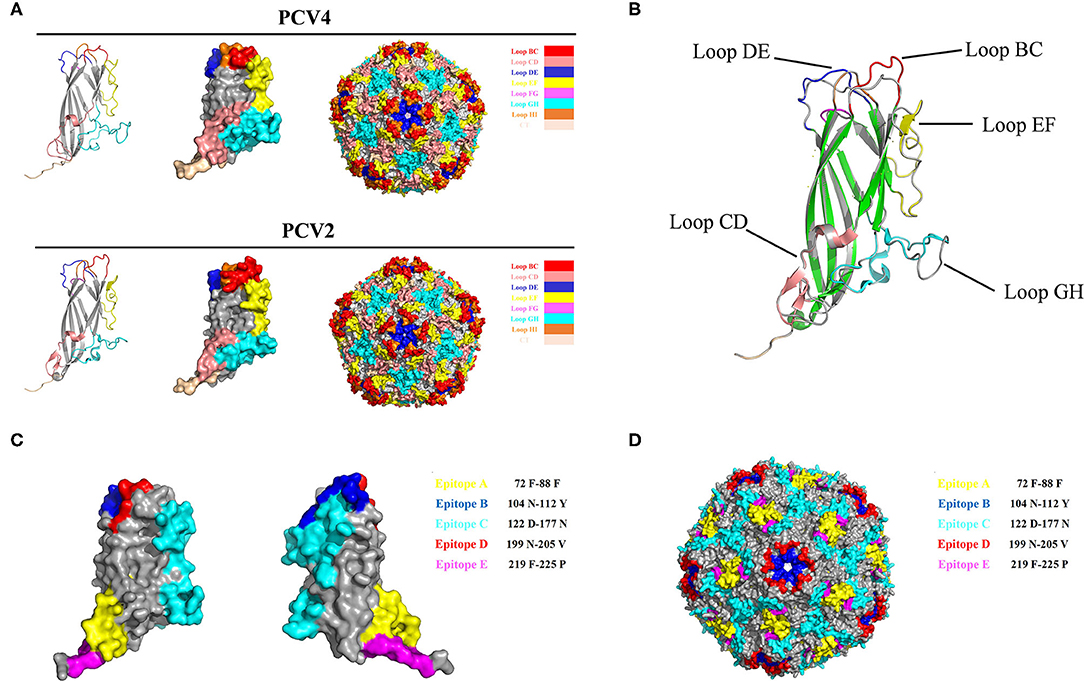
Figure 2. (A) 3D structure of PCV Caps and capsid. 3D Structure of the PCV4 Cap was simulated via homology modeling using the PCV2 Cap as a template (PDB ID: 3R0R). Loops were labeled with various colors. (B) 3D structure alignment of PCV4 and PCV2 Cap. Prediction of immune-epitopes of the PCV4 Cap (C) and capsid (D). The five predicted epitopes are indicated by various colors. (epitope A: yellow; epitope B: blue; epitope C: cyan; epitope D: red; and epitope E: magenta).
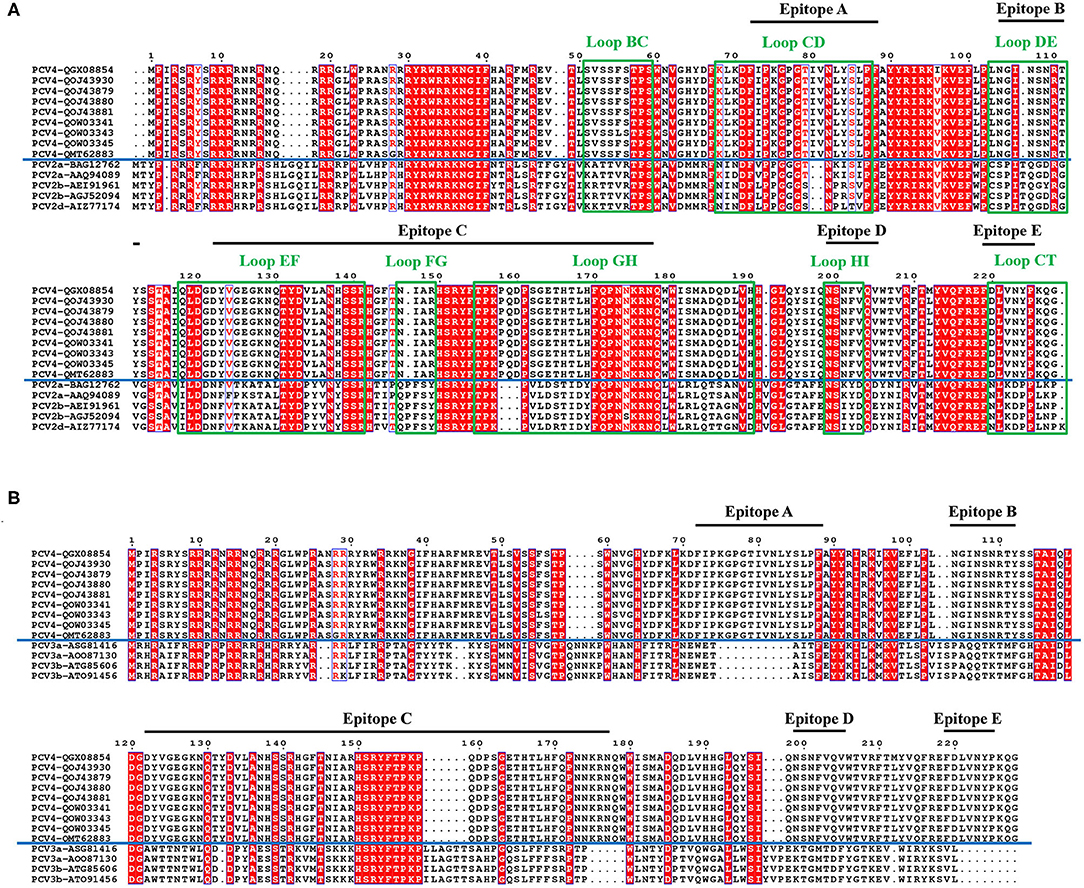
Figure 3. Sequence alignment of PCV Caps. Comparative amino acid sequence alignments between PCV4 and PCV2 Cap (A) or PCV4 and PCV3 Cap (B). All loops labeled with green rectangles and black lines represent predicted PCV4 potential linear B cell epitopes.
In the PCV4 Cap, five potential linear B cell epitopes with high antigenicity were predicted by BepiPred 2.0 based on default threshold, as indicated with various colors in the three-dimensional (3D) structure (Figure 2C). Notably, these five potential epitopes were all located in loops and mapped to the most exposed surface regions of the PCV4 capsid (Figures 2D, 3A). In-depth sequence analysis revealed the five B-cell epitopes of the PCV4 Cap shared less residue identity with either PCV2 or PCV3 Cap (Figure 3).
PCV2 VLPs with a diameter of ~20 nm were successfully obtained (Supplementary Figure 1). Cross-reactions of anti-PCV4 or anti-PCV2 IgG from mouse serum collected 6 weeks after the primary immunization with PCV4 or PCV2 VLPs, respectively, were determined by ELISA. Sera from PCV4-immunized mice did not react with PCV2 VLP, and vice versa, whereas sera from PCV2-immunized mice did not react with PCV4 VLP (Figures 4A,B; Supplementary Table 1). Therefore, PCV4 immunization induced genotype-specific antibody profiles in mice. Anti-PCV4 IgG had limited cross-reaction with PCV2; furthermore, antibodies elicited by the PCV2 Cap did not react with PCV4.
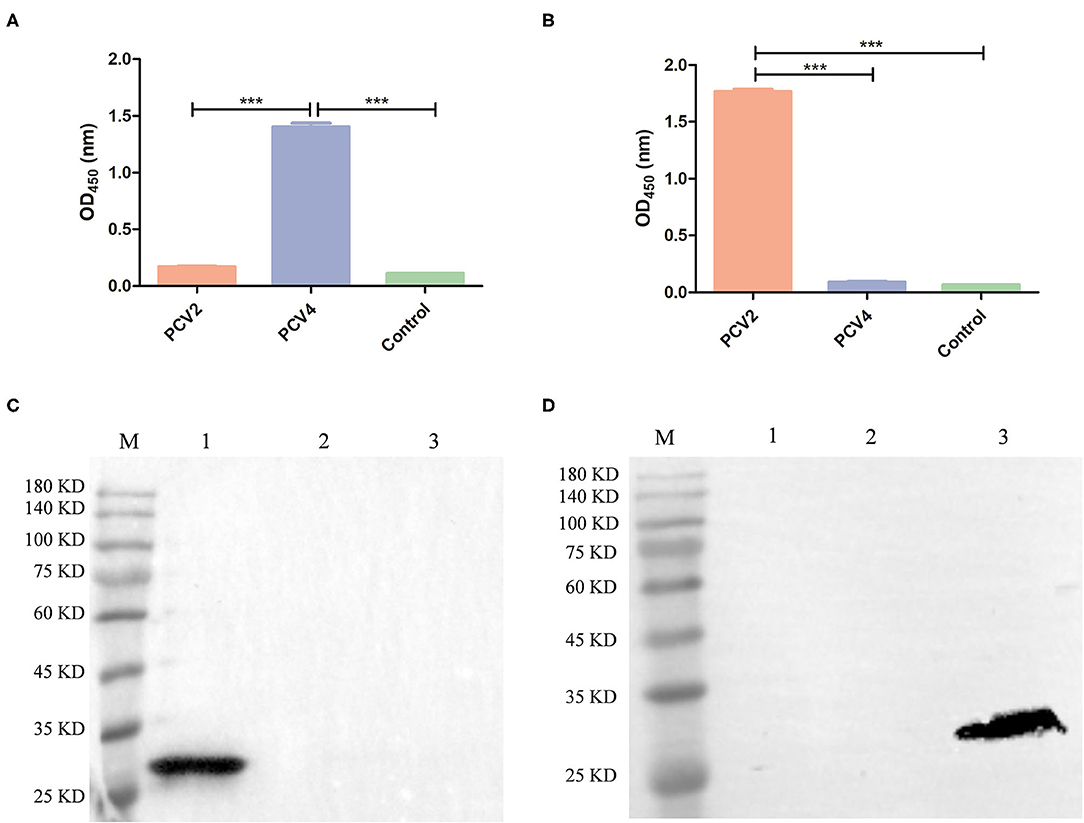
Figure 4. Cross-reaction between PCV4 and PCV2 by ELISA and Western blots. The reactivity of PCV4 VLP-immunized (A) or PCV2 VLP-immunized (B) mouse serum (n = 4) with PCV4 or PCV2 by ELISA. Reactivity of sera from PCV4 or PCV2 VLP-immunized mice, collected 6 weeks after initial immunization. Immunized mice (n = 2) received PBS as a control. Optical densities were read at 450 nm. ***p < 0.001. (C,D) Western blots of PCV Cap proteins using anti-PCV4 Cap serum or anti-PCV2 Cap serum, respectively (lane M: protein marker; lane 1: PCV4 Cap; lane 2: PCV3 Cap; and lane 3: PCV2 Cap).
Next, anti-PCV4 Cap serum was used for detecting cross-reactions with PCV2 or PCV3 Cap by Western blot, with anti-His tag antibody used as a positive control. The three PCV Caps positively reacted with anti-His tag antibody (Figure 1B). However, anti-PCV4 Cap serum recognized only the PCV4 Cap, but neither PCV2 nor PCV3 Cap (Figure 4C). In addition, anti-PCV2 Cap serum only reacted with PCV2 Cap, but neither PCV3 nor PCV4 Caps (Figure 4D). This lack of cross-reactions was attributed to the low residue identity among these Caps.
PCV4, a newly emerging porcine circovirus, has been identified in pigs with severe clinical disease, including PDNS-like clinical signs, diarrhea, and respiratory disease (4, 8). However, there is very limited information about its epidemiology, origin, evolution, pathogenesis, and transmission in swine herds. Notably, PCV4 has been detected as coinfections with PCV2, PCV3, or both (6–8). As PCV4 may be a serious risk for swine health, a better understanding of PCV4 pathogenesis, its epidemiology, and serological surveys to determine its prevalence is urgently needed.
VLPs are morphologically and immunogenically similar to their native viruses; furthermore, because of their ability to induce strong immune responses, they are widely used for novel vaccine design and serological diagnosis (12). Cap is the major structural protein of PCVs, and 60 Cap monomers self-assembled into VLPs in vitro (13, 14). The N-terminal nuclear location signal (NLS) domain of the Cap is abundant in arginine residues and contains several rare codons that limit gene expression in E. coli. In this study, a full-length PCV4 Cap protein via codon optimization was highly expressed in an inexpensive E. coli expression system. Purified Cap self-assembled into VLPs (~20 nm) in vitro with high yields. To characterize entry of PCV4 VLPs into cells, two porcine cell lines were used. As expected, PCV4 VLPs were capable of entering PK15 and 3D4/21 cells. Furthermore, PCV4 VLP-specific mouse serum recognized the PCV4 Cap prepared from the expression system. Therefore, PCV4 VLPs produced in E. coli were antigenic and well suited for future serological diagnostics and vaccine development.
We further analyzed the surface structure of PCV4 VLPs (capsid) and made comparisons to PCV2. Sequence alignment, together with 3D structure simulation, indicated substantial variation between PCV2 and PCV4 capsids in surface-exposed loop regions and surface patterns formed by various loops, although their whole structures were similar. In particular, the surface of the icosahedral five-fold axes constituted by loops BC, DE, and HI differed, as residues of these loopanalyzed the surface structures were highly divergent between PCV2 and PCV4 Caps (Figure 3A). In the PCV2 capsid, the structure of the five-fold axes was involved in conformational epitope formation. PCV2 neutralizing antibody bound to the residues around the surface of the five-fold symmetry axes, and this antibody recognized two adjacent capsid proteins as conformational epitopes (15). Diversity of the five-fold axes provided insights into antigenic differences between PCV2 and PCV4. Furthermore, these results highlighted differential structural and antigenic properties of PCV2 and PCV4 capsid proteins.
Interestingly, PCV4 Cap had high identity (70%) with the Cap of mink circovirus (4). However, it had low identities (~45 and 25%) with PCV2 and PCV3 Caps, respectively, whereas cross-reaction with PCV2 or PCV3 is unknown. To explore whether PCV4 cross-reacts with PCV2 or PCV3, anti-PCV4 Cap serum in BALB/c mice elicited by immunization with the PCV4 VLPs was used to detect cross-reaction with PCV2 or PCV3 Caps. Based on Western blot, only PCV4 Cap was recognized by anti-PCV4 Cap serum. Apparently, only PCV4 VLPs induced genotype-specific antibodies in mice. Moreover, the predicted PCV4 immune epitopes had very low identity to PCV2 or PCV3 Cap, which can further explain the absence of cross-reactions to the two Caps. Thus, cross-reaction between these Caps is unlikely, consistent with the result of the Western blot (Figure 4C). In parallel, anti-PCV2 Cap serum (produced by immunization with PCV2 VLPs) was used in Western blots to detect cross-reactions with PCV3 or PCV4 Cap; as expected, only the PCV2 Cap was specifically recognized by the anti-PCV2 Cap serum. Furthermore, using ELISA, we confirmed there was limited cross-reaction between PCV4 and PCV2. Therefore, we inferred that elicited antibody responses were unlikely to cross-protect between PCV4 and either PCV2 or PCV3.
We successfully expressed the PCV4 Cap and confirmed it was capable of self-assembling into VLPs in vitro with a diameter of ~20 nm. To our knowledge, this is the first report of PCV4 VLPs prepared from an E. coli expression system. The PCV4 VLPs produced in the present study had high antigenicity, with much potential for future serological diagnostics. Importantly, cross-reactions between PCV4 and either PCV2 or PCV3 were unlikely. Finally, these PCV4 VLPs are expected to be a valuable tool to study PCV4.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
The animal study was reviewed and approved by the Animal Ethics Committee of Hunan Agricultural University, Hunan, China.
NW and DW conceived and designed the experiments. DW and JM performed the generation, characterization of the PCV4 VLPs, designed, and carried out the immunoassays and bioinformatics analysis. BL and YZ prepared anti-PCV4 and PCV2 Cap serum from mouse. DW, YY, and NW wrote and revised the manuscript. All authors read and approved the final manuscript.
This research was funded by Hunan Provincial Natural Science Foundation of China, Grant Number 2018JJ2177; double first-class construction project of Hunan Agricultural University, Grant Number SYL2019048; and Post-graduate Research and Innovation Project of Hunan Province, Grant Number CX2018B394.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.695466/full#supplementary-material
1. Palinski R, Piñeyro P, Shang P, Yuan F, Guo R, Fang Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2017) 91:e01879–16. doi: 10.1128/JVI.01879-16
2. Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. (2016) 13:184. doi: 10.1186/s12985-016-0642-z
3. Jiang H, Wang D, Wang J, Zhu S, She R, Ren X, et al. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol. (2019) 93:e02045–18. doi: 10.1128/JVI.02045-18
4. Zhang HH, Hu WQ, Li JY, Liu TN, Zhou JY, Opriessnig T, et al. Novel circovirus species identified in farmed pigs designated as Porcine circovirus 4, Hunan province, China. Transbound Emerg Dis. (2020) 67:1057–61. doi: 10.1111/tbed.13446
5. Tian RB, Zhao Y, Cui JT, Zheng HH, Xu T, Hou CY, et al. Molecular detection and phylogenetic analysis of Porcine circovirus 4 in Henan and Shanxi Provinces of China. Transbound Emerg Dis. (2021) 68:276–82. doi: 10.1111/tbed.13714
6. Zhang D, Bai C, Ge K, Li Y, Gao W, Jiang S, et al. Establishment of an SYBR Green-based real-time PCR assay for porcine circovirus type 4 detection. J Virol Methods. (2020) 285:113963. doi: 10.1016/j.jviromet.2020.113963
7. Chen N, Xiao Y, Li X, Li S, Xie N, Yan X, et al. Development and application of a quadruplex real-time PCR assay for differential detection of porcine circoviruses (PCV1 to PCV4) in Jiangsu province of China from 2016 to 2020. Transbound Emerg Dis. (2021) 68:1615–24. doi: 10.1111/tbed.13833
8. Sun W, Du Q, Han Z, Bi J, Lan T, Wang W, Zheng M. Detection and genetic characterization of porcine circovirus 4 (PCV4) in Guangxi, China. Gene. (2021) 773:145384. doi: 10.1016/j.gene.2020.145384
9. Nguyen VG, Do HQ, Huynh TM, Park YH, Park BK, Chung HC. Molecular based detection, genetic characterization and phylogenetic analysis of porcine circovirus 4 from Korean domestic swine farms. Transbound Emerg Dis. (2021). doi: 10.1111/tbed.14017. [Epub ahead of print].
10. Zhang Y, Wang Z, Zhan Y, Gong Q, Yu W, Deng Z, et al. Generation of E. coli-derived virus-like particles of porcine circovirus type 2 and their use in an indirect IgG enzyme-linked immunosorbent assay. Arch Virol. (2016) 161:1485–91. doi: 10.1007/s00705-016-2816-9
11. Wang D, Zhang S, Zou Y, Yu W, Jiang Y, Zhan Y, et al. Structure-based design of porcine circovirus type 2 chimeric VLPs (cVLPs) displays foreign peptides on the capsid surface. Front Cell Infect Microbiol. (2018) 8:232. doi: 10.3389/fcimb.2018.00232
12. Yan D, Wei YQ, Guo HC, Sun SQ. The application of virus-like particles as vaccines and biological vehicles. Appl Microbiol Biotechnol. (2015) 99:10415–32. doi: 10.1007/s00253-015-7000-8
13. Khayat R, Brunn N, Speir JA, Hardham JM, Ankenbauer RG, Schneemann A, et al. The 2.3-angstrom structure of porcine circovirus 2. J Virol. (2011) 85:7856–62. doi: 10.1128/JVI.00737-11
14. Wang N, Zhan Y, Wang A, Zhang L, Khayat R, Yang Y. In silico analysis of surface structure variation of PCV2 capsid resulting from loop mutations of its capsid protein (Cap). J Gen Virol. (2016) 97:3331–44. doi: 10.1099/jgv.0.000634
Keywords: PCV4, Cap, VLPs, vaccine, serological diagnosis
Citation: Wang D, Mai J, Lei B, Zhang Y, Yang Y and Wang N (2021) Structure, Antigenic Properties, and Highly Efficient Assembly of PCV4 Capsid Protein. Front. Vet. Sci. 8:695466. doi: 10.3389/fvets.2021.695466
Received: 15 April 2021; Accepted: 28 July 2021;
Published: 24 August 2021.
Edited by:
Gerson Nakazato, State University of Londrina, BrazilReviewed by:
Kegong Tian, National Research Center for Veterinary Medicine, ChinaCopyright © 2021 Wang, Mai, Lei, Zhang, Yang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naidong Wang, bmFpZG9uZ3dhbmdAaHVuYXUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.