
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 16 July 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.692521
This article is part of the Research Topic Lawsonia intracellularis : A Problem Well Understood is a Problem Half Solved View all 7 articles
 Fernando L. Leite1
Fernando L. Leite1 Brittanie Winfield2
Brittanie Winfield2 Elizabeth A. Miller2
Elizabeth A. Miller2 Bonnie P. Weber2
Bonnie P. Weber2 Timothy J. Johnson2
Timothy J. Johnson2 Fred Sylvia1
Fred Sylvia1 Erika Vasquez3
Erika Vasquez3 Fabio Vannucci3
Fabio Vannucci3 Dana Beckler4
Dana Beckler4 Richard E. Isaacson2*
Richard E. Isaacson2*Porcine proliferative enteropathy remains one of the most prevalent diseases in swine herds worldwide. This disease is caused by Lawsonia intracellularis, an intracellular bacterial pathogen that primarily colonizes the ileum. In this study, we evaluated changes to the microbiome of the ileal mucosa, ileal digesta, cecal digesta, and feces subsequent to challenge with L. intracellularis and to an oral live vaccine against L. intracellularis. Given that gut homogenates have been used since 1931 to study this disease, we also characterized the microbial composition of a gut homogenate from swine infected with L. intracellularis that was used as challenge material. The L. intracellularis challenge led to a dysbiosis of the microbiome of both the small and large intestine marked by an increase of pathobionts including Collinsella, Campylobacter, Chlamydia, and Fusobacterium. This microbiome response could play a role in favoring L. intracellularis colonization and disease as well as potentially predisposing to other diseases. Vaccination altered both small and large intestine microbiome community structure and led to a significant 3.03 log10 reduction in the amount of L. intracellularis shed by the challenged pigs. Vaccination also led to a significant decrease in the abundance of Collinsella, Fusobacterium, and Campylobacter among other microbial changes compared with non-vaccinated and challenged animals. These results indicate that L. intracellularis infection is associated with broad changes to microbiome composition in both the large and small intestine, many of which can be mitigated by vaccination.
Lawsonia intracellularis is a Gram-negative intracellular bacterial pathogen and the causative agent of porcine proliferative enteropathy (PPE). Infection with L. intracellularis has been estimated to occur in more than 90% of swine farms worldwide (1, 2). L. intracellularis infects intestinal epithelial cells, preferentially those of the terminal ileum, and are present in cells in the crypts, causing hyperplasia leading to thickened intestinal tissue. Lesions in swine are also marked by a loss of mucin-producing goblet cells and a decrease of villus height (1, 3, 4). There are two clinical forms of PPE: porcine intestinal adenomatosis (PIA) and porcine hemorrhagic enteropathy (PHE). PHE occurs primarily in older animals, is hard to reproduce experimentally, and is associated with death and hemorrhagic diarrhea. PIA occurs in growing pigs and can lead to significant loss in production performance, diarrhea, and intestinal lesions (1, 2).
The gut microbiome is known to influence health as well as production performance of swine (5). It is also known that the composition of the microbiome varies between different portions of the intestinal tract and that the ileum is the preferential site of L. intracellularis infection (2, 5, 6). Research from our group has found that L. intracellularis can significantly alter the gut microbiome, with one of the likely consequences being the promotion of Salmonella enterica infection in swine (7, 8).
This study had the objective of extending the characterization of the microbiome response to L. intracellularis infection by evaluating microbial changes at different stages of disease, in different portions of the intestinal tract and in association with diagnostic measurements, in an attempt to better understand the underlying microbial community interactions occurring with disease. Additionally, the response to an oral live vaccine against L. intracellularis was investigated to determine if alterations to the microbiome of animals that were immunized were correlated with a less severe disease.
The animal protocol used was approved by the Boehringer Ingelheim Animal Health USA Inc., Institutional Animal Care and Use Committee (protocol number 100896); and all experiments were performed in accordance with relevant guidelines and regulations. All pigs used in this study were obtained from a sow herd with no previous history of clinical signs from L. intracellularis infection. After being weaned, at 3 weeks of age, pigs were randomly allocated to three different treatments groups: Control (non-infected, non-vaccinated control group), Law (challenged with L. intracellularis), and LawVac (vaccinated and challenged with L. intracellularis). The LawVac group was vaccinated with the oral live vaccine (Enterisol® Ileitis, Boehringer Ingelheim Animal Health USA Inc., Duluth, GA, USA) after 5 days of acclimation. To investigate the impact of the vaccine alone on the microbiome, a group of six random pigs in the Control and LawVac groups were necropsied at 21 days post-vaccination [0 days post-infection (dpi)]. At 6 weeks of age, 3 weeks post-vaccination, the Law and LawVac groups were challenged with a gut homogenate containing 4 × 107 L. intracellularis organisms. To evaluate the microbiome response at different stages of infection, six pigs were randomly selected from each treatment group (Control, Law, and LawVac) and euthanized at 7, 21, and 28 dpi. To minimize the exposure of the Control group to L. intracellularis, this treatment group was housed in a separate barn from the challenged and vaccinated groups. Separation of animals within the same room has been found to be insufficient to prevent transmission of L. intracellularis (9). To further prevent chances of cross contamination among groups, strict biosecurity measures were followed. This included not exposing the Control group to any equipment or attire used in caring for Lawsonia-challenged animals. Additionally, empty pens were kept between the Law and LawVac groups to minimize the chances of contact and transmission of microorganisms between these treatment groups. All pigs were held in slatted floor pens at a density appropriate for their size. All pigs received the same feed, which was formulated by a nutritionist and did not contain antibiotics. Each treatment group comprised three pens with eight animals each, except the Law group, which had three pens with six animals each. This was because the Law treatment group was not different from the Control group at time of challenge; thus, no animals were euthanized at this time point from this treatment group. Two animals died of mulberry heart disease during the study, and samples were not collected for microbiome characterization from these animals (one pig in the Control and one pig in the LawVac treatment groups).
Since the microbiome is known to differ along the different sections of the intestinal tract of pigs (6, 10), we collected samples to represent both the large and small intestine to better characterize the intestinal microbiome. Considering that L. intracellularis preferentially infects epithelial cells located in the terminal ileum, we also collected mucosal scrapings from this segment as previously described, with the addition of RNAlater (Thermo Fisher Scientific, Waltham, MA, USA) (4). In addition to ileum scrapings, digesta of the terminal ileum (luminal contents) and digesta from the cecum and feces were also collected during necropsy. These samples were frozen at −80°C soon after collection. Fecal samples were also collected from all animals 5 days after allocation to treatment (−23 dpi), as well as on 0, 7, 21, and 28 dpi.
To characterize the infection status and seroconversion of animals, blood samples were collected on all animals 5 days after allocation to treatment (−23 dpi), as well as on 0, 7, 21, and 28 dpi. Samples were tested by the immune peroxidase monolayer test at the University of Minnesota Veterinary Diagnostic Laboratory (11). The entire intestinal tract was examined for macroscopic lesions characteristic of PPE. Gross lesions were evaluated with the following rubric: 0 = no gross lesion; 1 = mild edema and hyperemia of mucosa or serosa; 2 = edema, hyperemia, and reticulated serosa and mucosa (thickening); 3 = edema, hyperemia, reticulated serosa and mucosa, and gross thickening of the mucosa; and 4 = severe thickening mucosal hemorrhaging or necrosis. A portion of the terminal ileum was collected in formalin for microscopic evaluation to determine the presence or absence of lesions. Immunohistochemistry (IHC) was used to assess the presence of L. intracellularis-specific antigens in the tissues as previously described (12). IHC was evaluated on a 5-point scale, as follows: 0 = absence of Lawsonia antigen; 1 = 0–25% of crypts with antigen; 2 = 25–50% of crypts with antigen; 3 = 50–75% of crypts with antigen; and 4 = 75–100% of crypts with antigen.
DNA was extracted from 250 mg of each sample using the QIAGEN DNeasy PowerSoil HTP 96 Kit (QIAGEN, Germantown, MD, USA) following the manufacturer's protocol. The resulting DNA was stored at −80°C. The V4 region of the 16S rRNA gene was amplified from DNA samples and sequenced on the Illumina, Inc. (San Diego, CA, USA) MiSeq platform (2 × 300 bp, v3 chemistry) at the University of Minnesota Genomics Center (Minneapolis, MN, USA), as previously described (13).
Raw reads were processed using the DADA2 pipeline (v2.0) (14) in R (v3.6.1) (15). Forward and reverse reads were trimmed to 200 and 160 bp, respectively, with all other parameters set to default values. Taxonomy was assigned using the SILVA small subunit rRNA sequence database (v132) (16). The resulting amplicon sequence variant (ASV) count table was filtered to remove samples with <2,500 total reads. ASVs were removed if they were classified as Archaea, Eukarya, mitochondria, chloroplasts, or unknown; had <10 reads; and/or occurred in only one sample.
For alpha diversity analyses, cecal digesta, feces, ileal mucosa, and ileal digesta samples were rarefied to 12,027, 8,156, 2,447, and 12,223 reads, respectively. ASV richness and the Shannon diversity index were calculated with the R package, phyloseq (v1.28.0) (17). To assess differences in alpha diversity between the treatment groups, non-parametric Wilcoxon rank-sum tests were performed, and p-values were adjusted for multiple testing using the Benjamini–Hochberg procedure.
Prior to performing beta diversity analyses and differential abundance testing, cumulative sum scaling (CSS) was used to normalize the unrarefied data using the phyloseq phyloseq_transform_css function. The Bray–Curtis dissimilarities and weighted UniFrac distances were then calculated using the phyloseq distance function. Differences in microbial composition between the treatment groups and dpi were visualized with principal coordinate analysis (PCoA) plots with multivariate t-distribution ellipses. Significant differences in composition between groups were assessed by permutational multivariate analysis of variance (PERMANOVA) tests with 999 permutations with the adonis function from the R package, vegan (v2.5.5) (18). The betadisper function, for PERMDISP analysis, was then used to confirm that the PERMANOVA results were not simply artifacts of heterogeneous dispersion between groups. Differential abundance testing of genus-level CSS-normalized counts between treatment groups (Law vs. Control and Law vs. LawVac) was conducted with zero-inflated Gaussian mixture models using the fitZIG function from the R package, metagenomSeq (v1.26.3) (19) with default parameters. Separate models were fit for each comparison (Law vs. Control and Law vs. LawVac) at each dpi time point (7, 21, and 28 dpi) in each intestinal segment (cecal digesta, feces, ileal mucosa, and ileal digesta). Genera occurring in only one sample were removed. Model coefficients and associated statistics, including the log2-fold changes in abundance, were viewed with the function MRcoefs. p-Values were adjusted for multiple comparisons using the Benjamini–Hochberg procedure. An alpha of 0.05 was used as the significance threshold in all statistical tests.
The preparation of gut homogenate challenge material followed procedures previously described (20). Briefly, mucosal scrapings from the ileum of pigs with confirmed L. intracellularis infection and gross PPE lesions were homogenized and diluted in sucrose potassium glutamate diluent and Dulbecco's Modified Eagle Medium (DMEM) prior to intragastric inoculation of animals.
Quantification of L. intracellularis present in fecal samples, cecal digesta, and gut homogenate was performed by qPCR (21). Briefly, extraction of fecal samples was performed using the MagMAX Pathogen RNA/DNA Kit (Thermo Fisher). qPCR was performed using the Thermo Scientific VetMAX™ L. intracellularis PCR Kit on a Roche Diagnostics (Basel, Switzerland) 480 LightCycler per manufacturer's protocol. A standard curve was included in each run for the quantification of L. intracellularis. Statistical differences in fecal shedding of L. intracellularis were assessed by Wilcoxon's rank-sum test.
With the objective of gaining a broad understanding of the bacterial and viral taxa of the gut homogenate inoculum material, shotgun metagenomic analysis was performed. Metagenomic characterization of the gut homogenate challenge material was performed at Newport Laboratories (Worthington, MN, USA). DNA and RNA extraction was performed using MagMax Core Nucleic Acid Purification Kit (Thermo Fisher). cDNA was synthesized from RNA with the Maxima H Minus Double-Stranded cDNA Synthesis Kit (Thermo Fisher). Library preparation was performed with the Nextera XT kit (Illumina), and sequencing was performed on MiSeq sequencer with 2 × 300 bp V3 chemistry (Illumina). Sequence read files were uploaded on to Illumina BaseSpace for analysis. Scaffolds were created using Illumina BaseSpace application SPAdes Genome Assembler. Metagenomic analysis was performed using Illumina BaseSpace application Kraken (v1.0.0) (22). The generated summary report was filtered for virus and bacterial species detected.
Shotgun metagenomic analysis of the gut homogenate challenge material revealed the presence of 66 different bacterial species among 164,123 sequencing reads. The most abundant was L. intracellularis, which represented approximately 0.40% of total reads and 34.9% of bacterial reads (Supplementary Table 1). Other bacteria present included Campylobacter fetus, Campylobacter jejuni, Campylobacter concisus, Chlamydia trachomatis, Bacteroides fragilis, and Fusobacterium nucleatum. At a much lower abundance, sequences matching eight different viruses were also detected in the gut homogenate and included porcine type C oncovirus (Supplementary Table 2).
To better understand how the composition of the gut microbiome changed during infection with L. intracellularis, sequential necropsies were performed at 0, 7, 21, and 28 days post-challenge. First, shedding of L. intracellularis was measured at the various time points. Fecal shedding was evaluated at 0 and 7 dpi among 18 pigs per group and at 21 and 28 dpi among 12 and six pigs per group, respectively. Shedding of L. intracellularis was not detected in the feces of pigs in the Control group and the group challenged with L. intracellularis at 0 dpi. In the challenged (“Law”) group, shedding increased from 4.3 log10 organisms per gram at 7 dpi to 7.87 log10 organisms per gram at 21 dpi and then decreased to 6.69 log10 organisms per gram at 28 dpi (Figure 1). Pigs in the vaccinated and challenged (“LawVac”) group had on average 0.60 log10 L. intracellularis/gram in feces at 0 dpi. Since the pigs at 0 dpi had not been challenged yet, detectable L. intracellularis likely was from the live oral vaccine strain. At 21 dpi, the LawVac group shed on average 6.59 log10 organisms per gram and at 28 dpi, 3.66 log10, the latter representing a 3.03 log10 reduction in the shedding of L. intracellularis (p = 0.008) as compared with non-vaccinated Law pigs (Figure 1). Both Law and LawVac groups had positive IHC staining of L. intracellularis at 7, 21, and 28 dpi. The LawVac group did not have positive IHC staining at 0 dpi and had less severe microscopic lesions at 28 dpi. At this time point, the LawVac group had a median lesion score of 0.5 with three of six (50%) pigs with positive IHC staining, while the Law group had a median lesion score of 2 with five of six pigs (83%) with positive IHC staining. While both Law and LawVac groups had macroscopic lesions at 21 dpi, only the Law group had macroscopic lesions characteristic of L. intracellularis infection at 28 dpi with two of six (33%) pigs having a lesion score of 2 and 4, respectively. No macroscopic lesions were found in the LawVac group at this time point. No macroscopic lesions were found at 0 or 7 dpi. At 3 weeks post-vaccination, only one of the 24 (4.2%) LawVac animals had serum antibodies against L. intracellularis. This proportion increased to 23.5% (4/17), 90.9% (10/11), and 83.3% (5/6) at 7, 21, and 28 dpi, respectively. All Law animals were seronegative at 0 dpi, and seroconversion increased to 33.3% (6/18), 63.6% (7/11), and 100% (6/6) of pigs with serum antibodies against L. intracellularis at 7, 21, and 28 dpi, respectively. Control animals remained negative for all diagnostic measures of L. intracellularis infection throughout the trial.
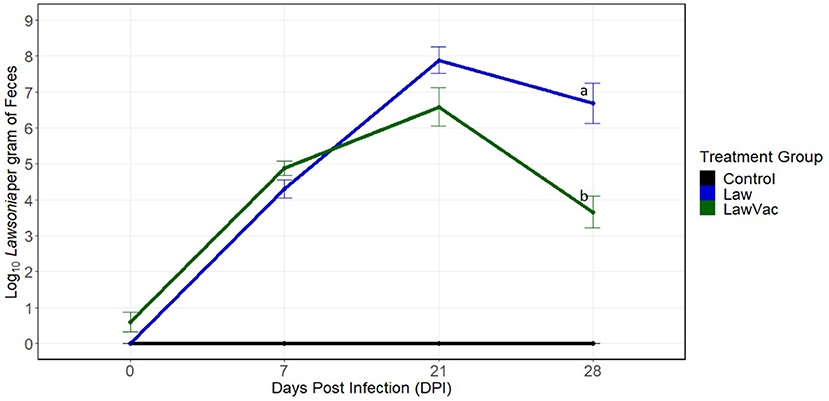
Figure 1. Quantification of Lawsonia intracellularis shedding in fecal samples over time, measured by qPCR. Mean and standard error of the mean at 0 days post-infection (n = 18 pigs per treatment group), 7 days post-infection (n = 18 pigs per treatment group), 21 days post-infection (n = 12 pigs per treatment group), and at 28 days post-infection (n = 6 pigs per treatment group). Different letters indicate statistical significance [Wilcoxon rank-sum test (p < 0.05)].
Sequencing of the 16S rRNA gene for microbiome analysis generated mean reads per sample of 23,599 ± 5,059, 21,645 ± 4,831, 16,639 ± 6,742, and 21,938 ± 3,748 for the cecal digesta, feces, ileal mucosa, and ileal digesta, respectively. To determine if changes in microbiome community composition were altered by infection with L. intracellularis, a beta diversity analysis was performed among the challenged and non-challenged Law and Control groups utilizing the Bray–Curtis dissimilarity index. A clear effect of challenge on community structure was observed. Figure 2 shows a clear segregation between Law and Control treatments that becomes more evident as infection progresses. At the site of L. intracellularis infection, the terminal ileal mucosa, no significant differences were observed at 7 dpi (PERMANOVA: p = 0.43, R2 = 0.082; Supplementary Table 3), while a clear change in community structure was observed at 21 dpi (p = 0.0010, R2 = 0.21) and 28 dpi (p = 0.0070, R2 = 0.28; Figure 2A). Among ileal digesta, a significant difference was observed at 7 dpi (p = 0.046, R2 = 0.13), but similar to ileal mucosa, differences also increased as infection progressed at 21 dpi (p = 0.011, R2 = 0.31) and 28 dpi (p = 0.0040, R2 = 0.380; Figure 2B). Among fecal samples (Figure 2D), while at 0 dpi, a significant difference among groups was detected (p = 0.0010, R2 = 0.078); this difference was not present at −23 dpi (p = 0.36, R2 = 0.025). Treatment group explained a drastically increased proportion of microbial community variation as infection progressed, with PERMANOVA R2-values of 0.095, 0.31, and 0.30 at 7, 21, and 28 dpi, respectively (all p ≤ 0.005). Finally, a similar trend to other communities was observed in the cecal digesta, with no significant differences observed at 7 dpi (p = 0.13, R2 = 0.12) and an increase in the distinction between treatment group microbial communities as infection progressed at 21 dpi (p = 0.0050, R2 = 0.31) and 28 dpi (p = 0.0040, R2 = 0.41; Figure 2C). All tests for homogeneity of variance between sample groups were non-significant, providing support for true differences in composition between dpi. Similar results were also observed when analyses were conducted using weighted UniFrac distance instead of Bray–Curtis dissimilarity (Supplementary Figure 1 and Supplementary Table 3). Together, these findings indicate broad changes to community composition due to L. intracellularis challenge, which progress during infection in both the small and large intestine.
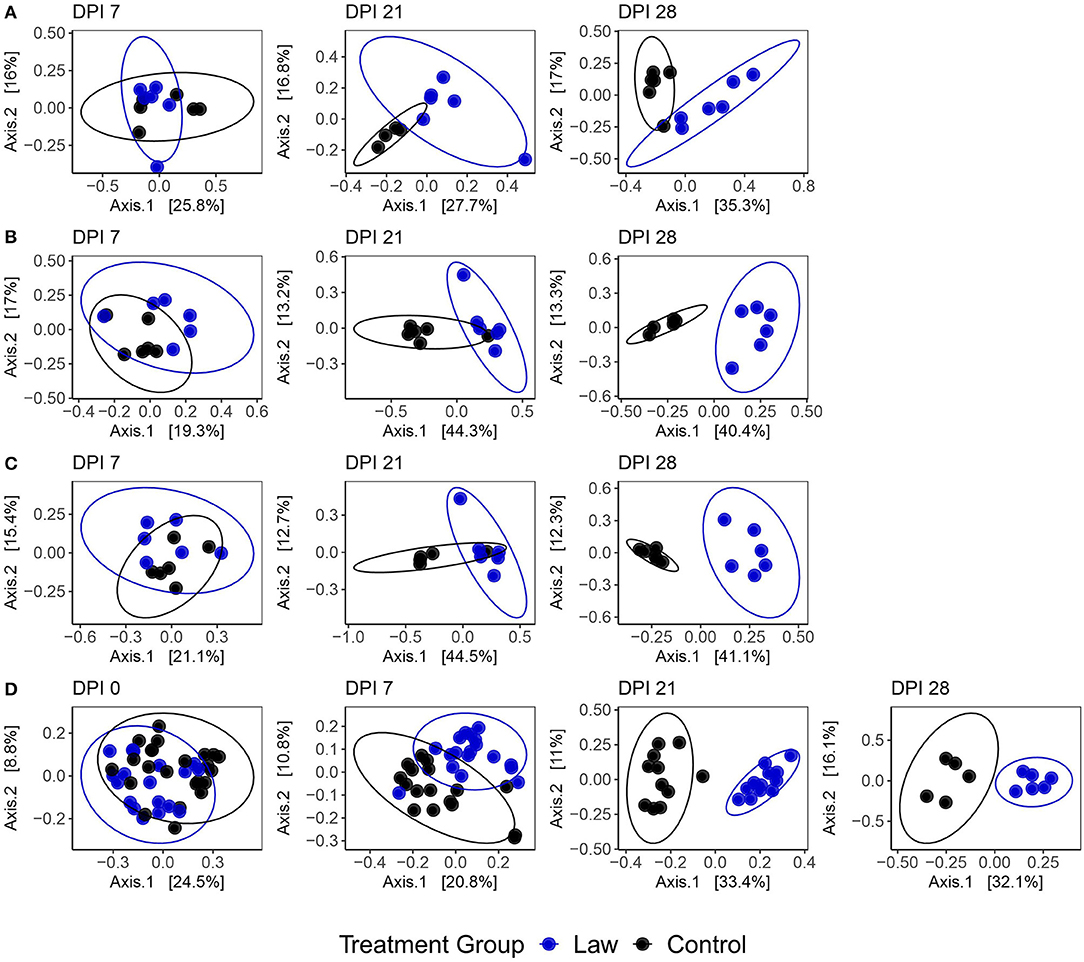
Figure 2. Principal coordinate analysis plots using the Bray–Curtis dissimilarity of samples from (A) ileal mucosa; (B) ileal digesta; (C) cecal digesta; and (D) feces among Law and Control groups at different days post-infection (dpi). Ellipses represent the 95% confidence level of the multivariate Student's t-distribution.
Changes to alpha diversity were not as evident as those found in beta diversity. Among fecal samples, Shannon's diversity index was significantly higher in the Law group compared with the Control group at 0 and 7 dpi (Supplementary Figure 2A). For the number of ASVs, the Law group was higher than the Control group at −23 dpi, but this difference was not subsequently observed at 0 dpi through 28 dpi (Supplementary Figure 2B). No significant differences in the Shannon diversity index or number of ASVs were found in ileal mucosa, ileal digesta, and cecal digesta between the Law and Control groups (data not shown).
As expected, microbial composition differed between the intestinal segments sampled (Supplementary Figure 3). The Lawsonia genus was detected by 16S rRNA gene analysis and found to be significantly differentially abundant between the Control and Law groups. Following a similar trend to that observed by qPCR quantification, the greatest differential abundance of Lawsonia was found at 21 dpi, at which point Lawsonia was the most differentially abundant genus among the Control and Law groups in both feces and ileal mucosa samples (Figures 3, 4).
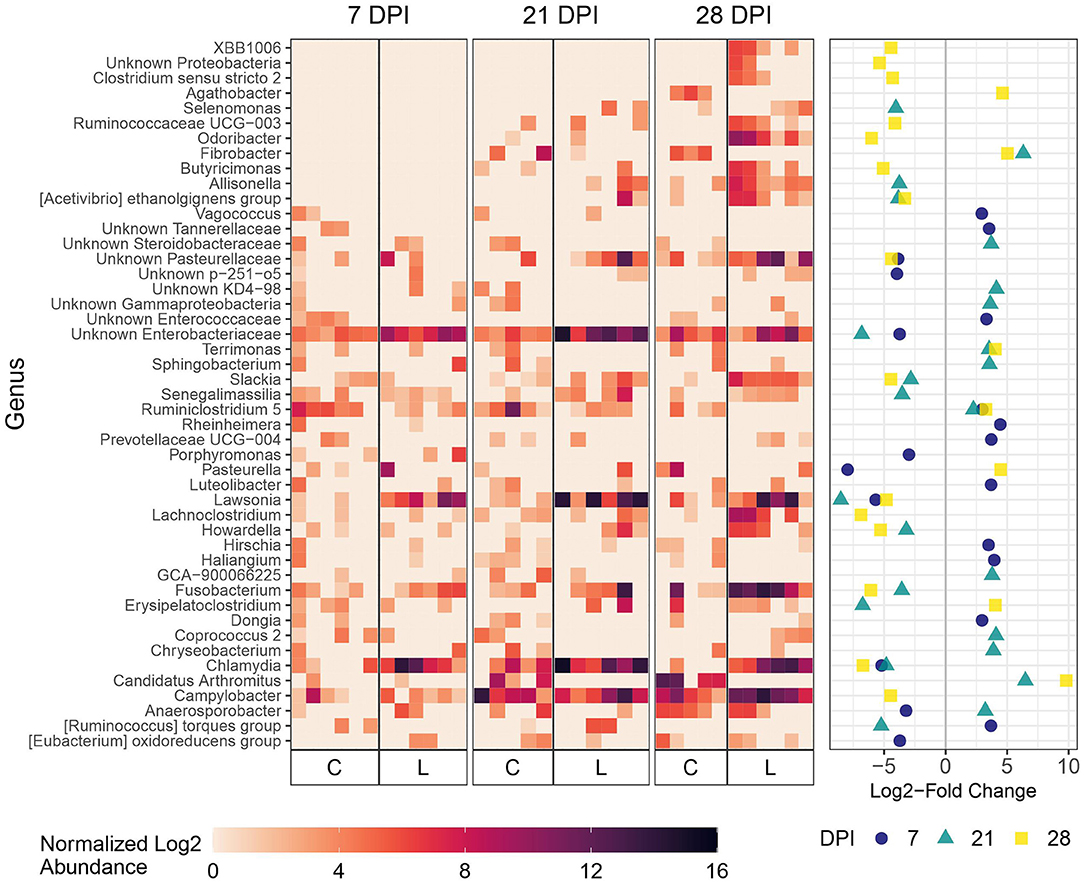
Figure 3. Differential abundance of microbial genera in ileal mucosa between the Law (L) and Control (C) groups at 7, 21, and 28 dpi. The 20 genera with the largest significant log2-fold change at each dpi are included. The heatmap shows the cumulative sum scaling (CSS)-normalized log2 abundance of each genera per sample, with the significant log2-fold changes by dpi plotted on the right panel. Note that a genus only has to be one of the top 20 significantly differentially abundant at 1 dpi for it to be included in the figure. A positive log2-fold change indicates greater abundance in the Control group, and a negative log2-fold change indicates greater abundance in the Law group.
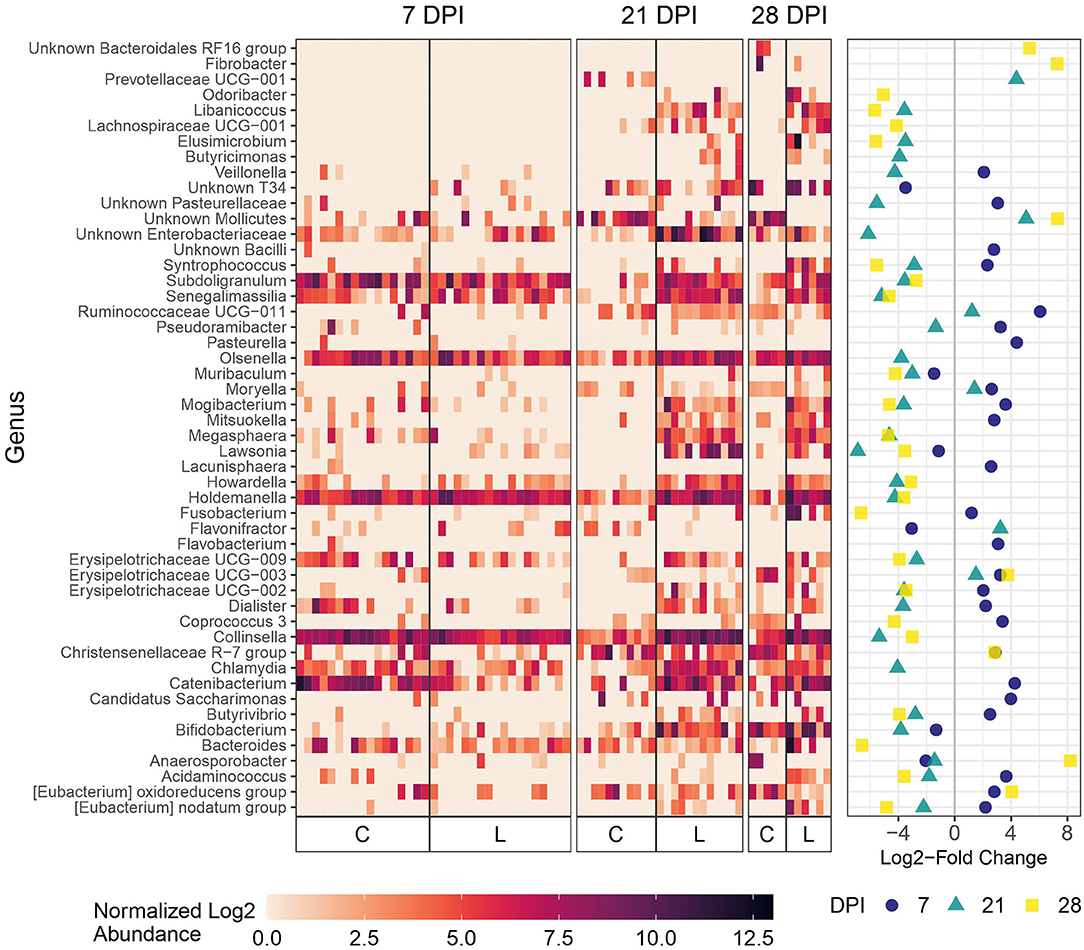
Figure 4. Differential abundance of microbial genera in feces between the Law (L) and Control (C) groups at 7, 21, and 28 dpi. The 20 genera with the largest significant log2-fold change at each dpi are included. The heatmap shows the cumulative sum scaling (CSS)-normalized log2 abundance of each genera per sample, with the significant log2-fold changes by dpi plotted on the right panel. Note that a genus only has to be one of the top 20 significantly differentially abundant at 1 dpi for it to be included in the figure. A positive log2-fold change indicates greater abundance in the Control group, and a negative log2-fold change indicates greater abundance in the Law group.
At 21 dpi, Lawsonia was significantly more abundant in the ileal mucosa of pigs in the Law group compared with Control (Figure 3). Chlamydia, Fusobacterium, and unknown members of the family Enterobacteriaceae were also significantly more abundant (Figure 3). Several genera were more abundant in Control animals at this time point compared with Law pigs, including Candidatus Arthromitus, Fibrobacter, and Ruminiclostridium 5 (Figure 3). At 7 dpi, Lawsonia, Chlamydia, and unknown Enterobacteriaceae were also more abundant in the Law group (Figure 3). At 28 dpi, along with Lawsonia and Chlamydia, Campylobacter along with more genera were more abundant in the Law group (Figure 3).
In the ileal digesta, only two genera were differentially abundant between Law and Control pigs at 7 dpi: Slackia and Catenibacterium both being more abundant in Control animals (p < 0.01). Interestingly, at both 21 and 28 dpi, Slackia was more abundant in Law than Control pigs (p < 0.01; Supplementary Figure 4). At 21 dpi, Campylobacter was more abundant in the Law than Control pigs, along with Lawsonia and Collinsella, among others. At 28 dpi, again Collinsella was among the most differentially abundant genus in Law pigs compared with Control pigs along with Lawsonia (Supplementary Figure 4).
Similar to ileal digesta, in cecal digesta, only few genera were differentially abundant between the Law and Control groups at the initial stage of infection at 7 dpi. Again, Slackia along with Catenibacterium was differentially abundant and more abundant in Control than Law pigs (p < 0.05) at this time point. At 21 dpi, many more differentially abundant bacteria were found; among those most abundant in Law pigs were Lawsonia, Chlamydia, Collinsella, and Fusobacterium (Supplementary Figure 5). At 28 dpi, even more bacteria were differentially abundant; in addition to those seen at 21 dpi, Bacteroides among others were more abundant in the Law group than the Control group (Supplementary Figure 5).
At 21 days dpi when L. intracellularis was at the highest levels in feces as measured by qPCR, differential abundance was detected for Lawsonia and several other bacteria including Chlamydia, Collinsella, and unknown Enterobacteriaceae (Figure 4) in challenged animals compared with non-challenged Control animals. At 28 dpi, the two most differentially abundant genera with the greatest abundance in the Law group compared with the Control group were Fusobacterium and Bacteroides, while some of the same bacteria differentially abundant at 21 dpi were found at 28 dpi including Collinsella. At the initial stage of infection (7 dpi), Lawsonia was already significantly differentially abundant among groups in feces (Figure 4). Similar to ileal and cecal digesta, Catenibacterium was more abundant in Control animals at this time point (Figure 4).
The differential abundances of Chlamydia and Collinsella between treatment groups are shown in Figure 5. In all time points post-infection, Chlamydia was significantly more abundant in the ileal mucosa of the Law group, while only at 21 dpi was it significantly greater in abundance in ileal digesta and feces (Figure 5A). For Collinsella, only at 21 dpi it was found to be significantly more abundant in ileal mucosa, while at both 21 and 28 dpi, it was significantly more abundant in feces and ileal digesta of Law pigs (Figure 5B). This further highlights the differential abundance observed between Law and Control animals in the different intestinal segments and time points of infection.
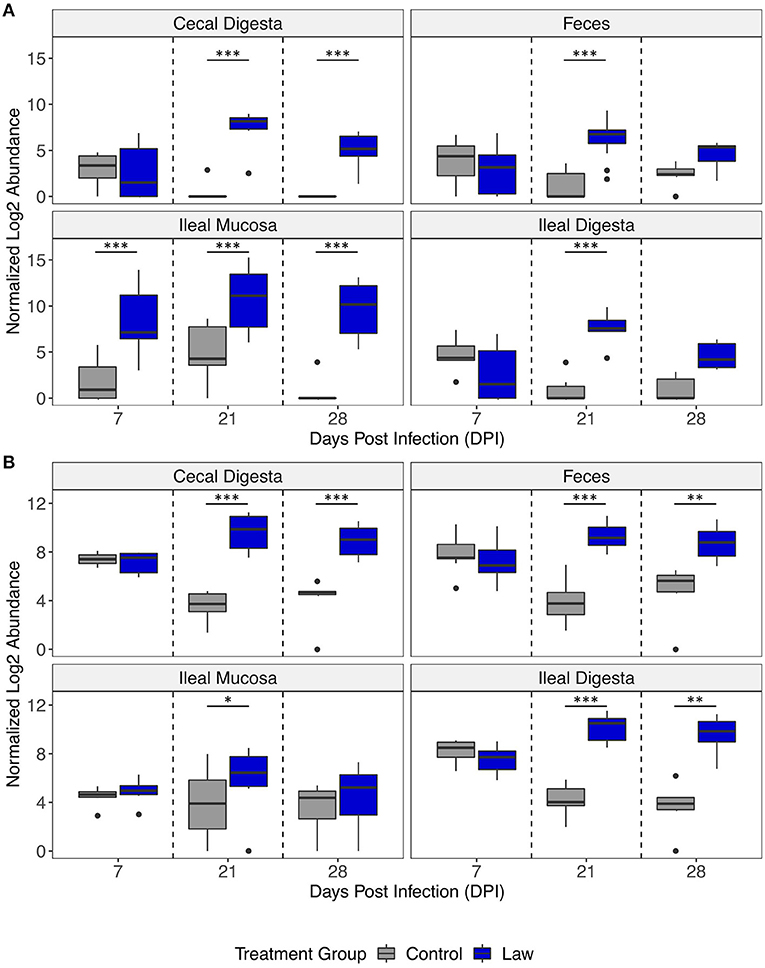
Figure 5. Differential abundances of the genera (A) Chlamydia and (B) Collinsella between Law and Control treatment groups in different intestinal segments over days post-infection. Asterisks indicate significant differences between treatment groups. *p < 0.05; **p < 0.01; and ***p < 0.001.
An oral live vaccine has been used to control L. intracellularis infection since 2001, and while its effect on the microbiome has been previously described (8), we wanted to extend these findings to investigate different intestinal segments and time points as well as focus on its impact to L. intracellularis challenge. At 3 weeks post-vaccination (0 dpi), a significant difference in community structure was found between LawVac and Law pigs in feces (Bray–Curtis dissimilarity PERMANOVA: p = 0.042, R2 = 0.041; Supplementary Table 4), while no differences in community structure were found between groups prior to vaccination at −23 dpi (p = 0.41, R2 = 0.025). Vaccination had a significant effect (p < 0.05) on the community structure of ileal mucosa, ileal digesta, and cecal digesta relative to non-vaccinated Control pigs at 0 dpi (data not shown). Investigating composition further, at 0 dpi, there were 73 significantly differentially abundant genera between LawVac and Law pigs in feces (Supplementary Table 5).
During infection, at 7, 21, and 28 dpi, fecal community structure was also significantly different between the LawVac and Law groups. An increase in composition dissimilarity between groups was observed with the progression of infection from 7 dpi (p = 0.023, R2 = 0.054) to 28 dpi (p = 0.061, R2 = 0.14; Supplementary Figure 6D). Among cecal digesta, a significant difference in community structure was only observed at 28 dpi (p = 0.024, R2 = 0.16; Supplementary Figure 6C). While no significant differences in community structure were noted in ileal digesta between the Law and LawVac groups, they did appear as distinct groups at 28 dpi (p = 0.067, R2 = 0.14; Supplementary Figure 6B). Among ileal mucosa, a significant difference in community structure was observed at both 7 dpi (p = 0.024, R2 = 0.17) and 28 dpi (p = 0.0070, R2 = 0.20) time points between the LawVac and Law groups (Supplementary Figure 6A). Again, there was no significant heterogeneous dispersion between sample groups and similar ordination plots; and PERMANOVAs were observed when analyses were conducted using weighted UniFrac distance instead of Bray–Curtis dissimilarity (Supplementary Figure 7 and Supplementary Table 4).
When comparing all treatment groups together, it was also observed that as infection progressed, so did the fecal microbial community differences among groups. While at 0 and 7 dpi, there was no clear visual distinction between treatments, at 21 dpi, both groups challenged with L. intracellularis were noticeably different from Control pigs (p = 0.0010, R2 = 0.32); and at 28 dpi, each group had a distinct community structure (p = 0.001, R2 = 0.30; Figure 6).
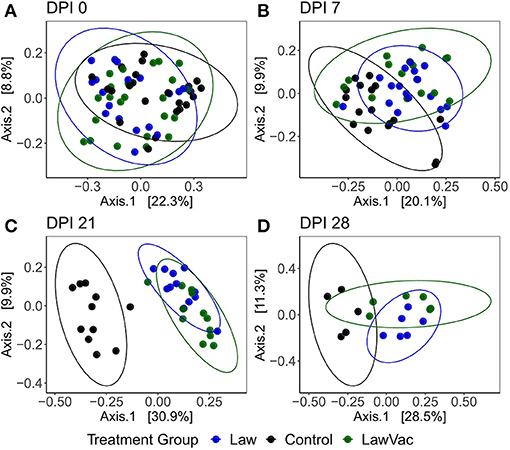
Figure 6. Principal coordinate analysis plots using the Bray–Curtis dissimilarity of fecal microbiota among Control, Law, and LawVac groups at (A) 0 days post-infection; (B) 7 days post-challenge; (C) 21 days post-infection; and (D) 28 days post-infection. Ellipses represent the 95% confidence level of the multivariate Student's t-distribution.
We then focused on the 28 dpi time point since this was the time point when vaccinated animals exhibited a significant 3.03 log10 reduction in the shedding of L. intracellularis (p = 0.008; Figure 1). Among fecal microbiota, only two genera were more abundant in the LawVac group in comparison with the Law group: Ruminococcaceae UCG-008 and Unknown Mollicutes (Figure 7A). Lawsonia, Fusobacterium, and Bacteroides among other genera were significantly more abundant in the Law group compared with the LawVac group at this time point (Figure 7A). In ileal mucosa samples, Fusobacterium and Bacteroides were the two most significantly abundant genera in the ileal mucosa of non-vaccinated pigs along with Campylobacter (Figure 7B). Only eight genera were differentially abundant in ileal digesta. While unknown Mollicutes were significantly more abundant in the vaccinated group, Bacteroides and Fusobacterium were significantly more abundant in the Law group (Supplementary Figure 8). Among cecal digesta, Lawsonia along with Fusobacterium, Campylobacter, and Bacteroides among others were significantly more abundant in non-vaccinated challenged pigs (Supplementary Figure 9).

Figure 7. Differential abundances of microbial genera between the Law and LawVac groups at 28 dpi in (A) fecal samples and (B) ileal mucosa samples. The 20 genera with the largest significant log2-fold change are included for ileal mucosa samples. The heatmap shows the cumulative sum scaling (CSS)-normalized log2 abundance of each genera per sample, with the significant log2-fold changes plotted on the right panel. A positive log2-fold change indicates greater abundance in the LawVac group, and a negative log2-fold change indicates greater abundance in the Law group.
This study characterized the porcine intestinal microbiome response of pigs to a L. intracellularis gut homogenate challenge and vaccination with an oral live vaccine. In the first description of PPE by Biester and Schwarte (23), it was noted that disease could be reproduced by feeding healthy swine intestinal contents and scrapings from affected pigs. Since then, gut homogenates, which comprise homogenate intestinal material from pigs with gross PPE lesions, have been described and used extensively to study L. intracellularis infection (4, 20, 24, 25). In this study, we used shotgun metagenomic sequencing to investigate the bacterial and viral composition of L. intracellularis gut homogenate challenge material. While L. intracellularis was the predominant bacterium found in the gut homogenate, other bacteria were detected along with some viruses. The relative abundance of L. intracellularis among other bacteria found by shotgun sequencing was similar to that found in a previous report by 16S rRNA gene sequencing, with relative abundances of 34.9% and 37.5%, respectively (26).
When analyzing the challenge dose of L. intracellularis provided to pigs (7.60 log10 L. intracellularis organisms), it was clear that infection and proliferation of L. intracellularis occurred, as Lawsonia-infected animals shed on average 7.87 log10 organisms/gram at 21 dpi. Along with detection by quantitative real-time PCR, 16S rRNA gene analysis also confirmed the presence of Lawsonia, which was significantly differentially abundant between Control non-infected and Law infected groups in feces, ileal mucosa, ileal digesta, and cecal digesta. Typical macroscopic lesions and positive IHC staining for the presence of L. intracellularis in terminal ileum tissue were also observed in these animals.
Past studies have found an intriguing relationship between L. intracellularis infection and gut microbiome composition. This study is not the first description of bacteria other than Lawsonia being enriched in the intestine of pigs challenged with a gut homogenate. McOrist et al. (24) studied gnotobiotic pigs challenged with a gut homogenate, and similar to the present study, they also found that species of Campylobacter and Bacteroides were present in the homogenate challenge material and enriched in the challenged pigs that developed PPE lesions. While we identified a low number of viruses in the gut homogenate material, McOrist et al. (24) did note that gut homogenate material was not fully infective after being more extensively filtered, suggesting that bacteria were the primary cause of disease. This indicates that underlying microbial interactions occur during infection and could contribute to development of lesions and disease.
Previous work from our research group has found that L. intracellularis challenge using a pure culture in conventionally raised specific pathogen free pigs does lead to a significant change in the microbial community structure sampled in cecal mucosa, colon mucosa, and feces of pigs (7, 8). In the present study, we also observed that L. intracellularis challenge with the gut homogenate led to a very clear and dramatic change in microbial community structure not only of the cecum and feces but also of the ileal mucosa and digesta. The fact that the differences in community structure found in Law challenged pigs increased as the infection progressed is further evidence of the interaction of L. intracellularis with the microbiome. While the evidence supports the hypothesis that L. intracellularis is the cause of disease, it is not possible to rule out the interaction of one or more additional bacteria required for disease.
When evaluating the differential abundance of various microbes due to L. intracellularis challenge, 16S rRNA gene analysis was used to confirm that Lawsonia was present in the terminal ileal mucosa, the site known to be the preferential niche of infection (1, 2). At all time points post infection, in addition to Lawsonia, Chlamydia and unknown members of the family Enterobacteriaceae also were more abundant in the ileal mucosa of pigs in the Law group. Chlamydia is a Gram-negative intracellular bacteria that can replicate in mucosal epithelial cells, including those of the gastrointestinal tract, and can cause enteritis (27). Chlamydia was also detected in the gut homogenate challenge material, indicating it was present in the pigs with PPE lesions used to make the challenge material and might have been the source of Chlamydia in the pigs in the Law group.
Among the bacteria significantly less abundant in the ileal mucosa of challenged animals were Ruminiclostridium 5, Fibrobacter, and Candidatus Arthromitus. The latter is a candidate genus of segmented filamentous bacteria, which are known to bind to the surface of the ileal epithelium and have the capacity of modulating immune response (28). In ileal digesta and cecal digesta, bacteria of the Ruminiclostridium genus were also less abundant in the challenged pigs. Interestingly, Ruminiclostridium abundance in fecal samples has recently been shown to be significantly and inversely correlated to Mycoplasma hyopneumoniae abundance in infected swine and are known to produce butyrate (29). Both L. intracellularis and M. hyopneumoniae are prevalent bacterial pathogens known to cause significant production losses, including during co-infection even when pigs are not exhibiting clinical signs (30).
At 28 dpi when Law pigs still had macroscopic lesions, Fusobacterium and Bacteroides among other bacteria were more abundant in the ileal mucosa of pigs in the Law group. Fusobacterium and Bacteroides, including F. nucleatum and B. fragilis, were also found in the gut homogenate material. Fusobacterium and F. nucleatum abundance in colonic mucosa have been found to be significantly more abundant in patients with adenoma and colorectal cancer, and these bacteria have the capacity to induce severe TNF-mediated inflammation (31, 32). Interestingly, gene expression pathways observed in intestinal tissue of pigs with PPE lesions have been found to be associated with a cellular proliferation and inflammatory signature resembling those of cancer and other inflammatory diseases of the intestinal tract (4, 33). PPE of course is not a cancer, as lesions do resolve. Similar to F. nucleatum, Bacteroides, and B. fragilis also have been implicated in colorectal cancer, in which certain strains have been found to be capable of promoting cellular proliferation by stimulation of Wnt signaling. The latter is a signaling pathway that has also been described to be altered in pigs with PPE (4, 34–36). Increased abundance of Bacteroides has also been found in the fecal microbiome of pigs challenged with a pure culture of L. intracellularis and to be negatively associated with weight gain in swine (8, 10). Considering one of the hallmarks of PPE lesions is the loss of goblet cells and the protective intestinal mucin layer (1, 37), it is likely that bacteria other than L. intracellularis could contribute to the development of lesions as they would have increased access to the epithelium. Additionally, mucin is known to harbor antimicrobial peptides, which, if altered during infection, could also contribute to dysbiosis (38, 39).
In this study, we also evaluated the response of pigs to an oral live vaccine against L. intracellularis. We observed that this vaccine led to a 3.03 log10 reduction in the shedding of L. intracellularis at 28 dpi, when LawVac pigs shed on average 3.66 log10 organisms per gram. L. intracellularis shedding level post-challenge, at levels above 6 log10 in particular, have a significant positive correlation to intestinal lesion severity and a significant negative correlation to average daily weight gain (25). When evaluating differential abundance at the time point when vaccinated animals shed significantly less L. intracellularis, several of the bacteria that were found to be more abundant in infected animals were decreased in the vaccinated group. These included Fusobacterium, Campylobacter, and Bacteroides. We have previously found that vaccination against L. intracellularis led to a decrease in the abundance of Collinsella and Prevotella in pigs co-infected with L. intracellularis and S. enterica serovar Typhimurium. Although not among the genera with the greatest differential abundant fold change, vaccinated pigs did have a significant (p < 0.05) decrease in abundance of Collinsella in cecal digesta and of Prevotella 2 in ileal mucosa (data not shown). Again, similar to previous findings, several of the sequences of Collinsella were closely related those of Collinsella aerofaciens and sequences of Prevotella to Prevotella copri. These have been considered pathobiont bacteria, as they have the capacity to induce inflammatory responses, which could favor Salmonella infection (40, 41). The decrease in abundance of Collinsella and Prevotella by vaccination was also previously associated with a significant 2.12 log10 reduction in the shedding of S. Typhimurium (8). This suggests that one of the consequences of L. intracellularis infection on microbiome composition could be increased susceptibility to other pathogens, an effect that can be minimized by vaccination. These observations suggest that oral vaccination not only can mitigate traditional measures of disease, such as reduction of lesions and shedding, but also can prevent some of the negative effects of pathogen challenge over microbiome composition, being a tool to protect this important community. The resulting microbiome could also be involved in reducing susceptibility to other pathogens, decreasing the shedding of L. intracellularis, decreasing PPE lesions, and improving production performance, which have been shown in studies evaluating this vaccine (8, 42–45).
It is remarkable that a vaccine that only contains antigens to one bacterium (L. intracellularis) also altered the abundance of other bacterial members of the gut microbiome. Perhaps this indicates that L. intracellularis is the primary causative agent of PPE and driver of dysbiosis, which when controlled with an effective mucosal immune response allows for minimizing its effects on microbial community composition. It is also possible but unclear if the changes observed in the gut microbiome by the oral vaccine alone or prior to challenge had any effect in the protection conferred to L. intracellularis.
Dysbiosis has been broadly defined as any change to the composition of resident commensal communities relative to the community found in healthy individuals. Dysbiosis can be further categorized into to three types: (i) loss of beneficial microbial organisms, (ii) expansion of pathobionts, and (iii) loss of overall microbial diversity (46). Pathobionts are bacteria that reside in the host and, although not considered primary pathogens, have the capacity to induce and contribute to disease (40, 46). This study suggests that L. intracellularis challenge induces dysbiosis not only due to changes to the composition of community commensal bacteria of the small and large intestine but also by an increase of pathobionts such as Campylobacter, Chlamydia, Fusobacterium, Bacteroides, and Collinsella. These effects may be mitigated by vaccination, which in this study not only significantly decreased the abundance of L. intracellularis but also led to a different community structure and a decreased abundance of several pathobionts induced by challenge.
Our findings support the strong association of L. intracellularis with other bacteria present in the swine gut that could contribute to the development and severity of disease, many of which were found to be altered by vaccination in this study. Future studies should focus on investigating microbiome changes induced by L. intracellularis in natural field conditions as well as utilizing gnotobiotic models with different defined microbial communities to allow for more conclusions on the exact contribution of organisms other than L. intracellularis in the establishment and severity of PPE.
The sequencing datasets generated for this study can be found in the NCBI BioProject repository under ID PRJNA728719, https://www.ncbi.nlm.nih.gov/bioproject/728719.
The animal study was reviewed and approved by the Boehringer Ingelheim Animal Health USA Inc., Institutional Animal Care and Use Committee and all experiments were performed in accordance with relevant guidelines and regulations.
FL and RI conceived the study. BW, EM, BPW, and TJ conducted microbiome analysis. FS conducted metagenomic analysis of gut homogenate. EV, DB, and FV conducted pathological measurements and collected samples. FL, RI, and TJ wrote the manuscript. All authors have read and critically reviewed the manuscript.
This work was funded by USDA-AFRI grant 1005423 as well as Boehringer Ingelheim Animal Health USA Inc.
This study was partially funded by Boehringer Ingelheim Animal Health USA Inc., Duluth, GA. FL and FS are employees of Boehringer Ingelheim Animal Health USA Inc. DB is employed by Gut Bugs Inc.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We wish to thank Kristin Parker and Karenzha Schemmer for their help in conducting the shotgun metagenomic analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.692521/full#supplementary-material
1. Lawson GHK, Gebhart CJ. Proliferative enteropathy. J Comp Pathol. (2000) 122:77–100. doi: 10.1053/jcpa.1999.0347
2. Vannucci F, McOrist S, Gebhart CJ. Proliferative enteropathy. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, Zhang J, editors. Diseases of Swine. Ames: Wiley (2019). p. 898–912.
3. Rowland AC, Lawson GHK, Maxwel A. Intestinal adenomatosis in the pig : occurrence of a bacterium in affected cells pepsin inhibitory activity amongst activation peptides of pepsinogen. Nature. (1973) 243:1973.
4. Leite FL, Abrahante JE, Vasquez E, Vannucci F, Gebhart CJ, Winkelman N, et al. A cell proliferation and inflammatory signature is induced by Lawsonia intracellularis infection in swine. MBio. (2019) 10:1–14. doi: 10.1101/384230
5. Isaacson R, Kim HB. The intestinal microbiome of the pig. Anim Health Res Rev. (2012) 13:100–9. doi: 10.1017/S1466252312000084
6. Holman DB, Brunelle BW, Trachsel J, Allen HK. Meta-analysis to define a core microbiota in the swine gut. mSystems. (2017) 2:e00004-17. doi: 10.1128/msystems.00004-17
7. Borewicz KA, Kim HB, Singer RS, Gebhart CJ, Sreevatsan S, Johnson T, et al. Changes in the porcine intestinal microbiome in response to infection with Salmonella enterica and Lawsonia intracellularis. PLoS ONE. (2015) 10:e0139106. doi: 10.1371/journal.pone.0139106
8. Leite FLL, Singer RS, Ward T, Gebhart CJ, Isaacson RE. Vaccination against Lawsonia intracellularis decreases shedding of salmonella enterica serovar typhimurium in co-infected pigs and alters the gut microbiome. Sci Rep. (2018) 8:2857. doi: 10.1038/s41598-018-21255-7
9. Jordan DM, Knitted JP, Schwartz KJ, Roof MB, Hoffman LJ. A Lawsonia intracellularis transmission study using a pure culture inoculated seeder-pig sentinel model. Vet Microbiol. (2004) 104:83–90. doi: 10.1016/j.vetmic.2004.09.004
10. De Rodas B, Youmans BP, Danzeisen JL, Tran H, Johnson TJ. Microbiome profiling of commercial pigs from farrow to finish. J Anim Sci. (2018) 96:1778–94. doi: 10.1093/jas/sky109
11. Guedes RMC, Gebhart CJ, Deen J, Winkelman NL. Validation of an immunoperoxidase monolayer assay as a serologic test for porcine proliferative enteropathy. J Vet Diagnostic Investig. (2002) 14:528–30. doi: 10.1177/104063870201400618
12. Guedes RMC, Gebhart CJ, Winkelman NL, Mackie-Nuss RAC, Marsteller TA, Deen J. Comparison of different methods for diagnosis of porcine proliferative enteropathy. Can J Vet Res. (2002) 66:99–107.
13. Gohl DM, Vangay P, Garbe J, MacLean A, Hauge A, Becker A, et al. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat Biotechnol. (2016) 34:942–9. doi: 10.1038/nbt.3601
14. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. (2016) 13:581–3. doi: 10.1038/nmeth.3869
15. R: A Language Environment for Statistical Computing. R Core Team (2019). Available online at: http://www.r-project.org/ (accessed June 15, 2021).
16. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:590–6. doi: 10.1093/nar/gks1219
17. McMurdie PJ, Holmes S. Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. (2013) 8:e61217. doi: 10.1371/journal.pone.0061217
18. Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, et al. Vegan: Community Ecology Package. R package version 2.5–7 (2020).
19. Paulson JN, Colin Stine O, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. (2013) 10:1200–2. doi: 10.1038/nmeth.2658
20. Roberts L, Rowland AC LG. Experimental reproduction of porcine intestinal adenomatosis and necrotic enteritis. Vet Rec. (1977) 100:12–13. doi: 10.1136/vr.100.1.12
21. Pusteria N, Mapes S, Rejmanek D, Gebhart C. Detection of Lawsonia intracellularis by real-time PCR in the feces of free-living animals from equine farms with documented occurrence of equine proliferative enteropathy. J Wildl Dis. (2008) 44:992–8. doi: 10.7589/0090-3558-44.4.992
22. Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. (2014) 15:R46. doi: 10.1186/gb-2014-15-3-r46
24. McOrist S, Lawson GH. Reproduction of proliferative enteritis in gnotobiotic pigs. Res Vet Sci. (1989) 46:27–33. doi: 10.1016/s0034-5288(18)31112-3
25. Collins AM, Barchia IM. The critical threshold of Lawsonia intracellularis in pig faeces that causes reduced average daily weight gains in experimentally challenged pigs. Vet Microbiol. (2014) 168:455–8. doi: 10.1016/j.vetmic.2013.12.003
26. Muwonge A, Karuppannan AK, Opriessnig T. Probiotics mediated gut microbiota diversity shifts are associated with reduction in histopathology and shedding of Lawsonia intracellularis. Anim Microbiome. (2021) 3:22. doi: 10.1186/s42523-021-00084-6
27. Schautteet K, Vanrompay D. Chlamydiaceae infections in pig. Vet Res. (2011) 42:29. doi: 10.1186/1297-9716-42-29
28. Hedblom GA, Reiland HA, Sylte MJ, Johnson TJ, Baumler DJ. Segmented filamentous bacteria - metabolism meets immunity. Front Microbiol. (2018) 9:1991. doi: 10.3389/fmicb.2018.01991
29. Valeris-Chacin R, Sponheim A, Fano E, Isaacson R, Singer RS, Nerem J, et al. Relationships among fecal, air, oral, and tracheal microbial communities in pigs in a respiratory infection disease model. Microorganisms. (2021) 9:252. doi: 10.3390/microorganisms9020252
30. Helm ET, Curry SM, Schwartz KJ, Lonergan SM, Gabler NK. Mycoplasma hyopneumoniae– Lawsonia intracellularis dual challenge modulates intestinal integrity and function. J Anim Sci. (2019) 97:2376–84. doi: 10.1093/jas/skz112
31. McCoy AN, Araújo-Pérez F, Azcárate-Peril A, Yeh JJ, Sandler RS, Keku TO. Fusobacterium is associated with colorectal adenomas. PLoS ONE. (2013) 8: e53653. doi: 10.1371/journal.pone.0053653
32. Brennan CA, Garrett WS. Fusobacterium nucleatum — symbiont, opportunist and oncobacterium. Nat Rev Microbiol. (2019) 17:156–66. doi: 10.1038/s41579-018-0129-6
33. Smith SH, Wilson AD, Van Ettinger I, Macintyre N, Archibald AL, Ait-Ali T. Down-regulation of mechanisms involved in cell transport and maintenance of mucosal integrity in pigs infected with Lawsonia intracellularis. Vet Res. (2014) 45:55. doi: 10.1186/1297-9716-45-55
34. Huan YW, Bengtsson RJ, MacIntyre N, Guthrie J, Finlayson H, Smith SH, et al. Lawsonia intracellularis exploits β-catenin/Wnt and Notch signalling pathways during infection of intestinal crypt to alter cell homeostasis and promote cell proliferation. PLoS ONE. (2017) 12:e0173782. doi: 10.1371/journal.pone.0173782
35. Helm ET, Burrough ER, Leite FL, Gabler NK. Lawsonia intracellularis infected enterocytes lack sucrase-isomaltase which contributes to reduced pig digestive capacity. Vet Res. (2021) 52:90. doi: 10.1186/s13567-021-00958-2
36. Vacante M, Ciuni R, Basile F, Biondi A. Gut microbiota and colorectal cancer development: a closer look to the adenoma-carcinoma sequence. Biomedicines. (2020) 8:1–19. doi: 10.3390/biomedicines8110489
37. Bengtsson RJ, MacIntyre N, Guthrie J, Wilson AD, Finlayson H, Matika O, et al. Lawsonia intracellularis infection of intestinal crypt cells is associated with specific depletion of secreted MUC2 in goblet cells. Vet Immunol Immunopathol. (2015) 168:61–7. doi: 10.1016/j.vetimm.2015.08.005
38. Mowat AM, Agace WW. Regional specialization within the intestinal immune system. Nat Rev Immunol. (2014) 14:667–85. doi: 10.1038/nri3738
39. Fang J, Wang H, Zhou Y, Zhang H, Zhou H, Zhang X. Slimy partners: the mucus barrier and gut microbiome in ulcerative colitis. Exp Mol Med. (2021) 53:772–87. doi: 10.1038/s12276-021-00617-8
40. Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. (2011) 23:473–80. doi: 10.1016/j.coi.2011.07.010
41. Gomez-Arango LF, Barrett HL, Wilkinson SA, Callaway LK, McIntyre HD, Morrison M, et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes. (2018) 9:189–201. doi: 10.1080/19490976.2017.1406584
42. Kroll JJ, Roof MB, McOrist S. Evaluation of protective immunity in pigs following oral administration of an avirulent live vaccine of Lawsonia intracellularis. Am J Vet Res. (2004) 65:559–65. doi: 10.2460/ajvr.2004.65.559
43. McOrist S, Smits RJ. Field evaluation of an oral attenuated Lawsonia intracellularis vaccine for porcine proliferative enteropathy (ileitis). Vet Rec. (2007) 161:26–8. doi: 10.1136/vr.161.1.26
44. Park S, Lee J-B, Kim K-J, Oh Y-S, Kim M-O, Oh Y-R, et al. Efficacy of a commercial live attenuated Lawsonia intracellularis vaccine in a large scale field trial in Korea. Clin Exp Vaccine Res. (2013) 2:135. doi: 10.7774/cevr.2013.2.2.135
45. Peiponen KS, Tirkkonen BT, Junnila JJT, Heinonen ML. Effect of a live attenuated vaccine against Lawsonia intracellularis in weaned and finishing pig settings in Finland. Acta Vet Scand. (2018) 60:1–7. doi: 10.1186/s13028-018-0374-8
Keywords: Lawsonia intracellularis, oral vaccine, microbiome, swine, pathobiont
Citation: Leite FL, Winfield B, Miller EA, Weber BP, Johnson TJ, Sylvia F, Vasquez E, Vannucci F, Beckler D and Isaacson RE (2021) Oral Vaccination Reduces the Effects of Lawsonia intracellularis Challenge on the Swine Small and Large Intestine Microbiome. Front. Vet. Sci. 8:692521. doi: 10.3389/fvets.2021.692521
Received: 08 April 2021; Accepted: 17 June 2021;
Published: 16 July 2021.
Edited by:
Mariano Enrique Fernández-Miyakawa, Instituto Nacional de Tecnología Agropecuaria (Argentina), ArgentinaReviewed by:
Juan M. Diaz Carrasco, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaCopyright © 2021 Leite, Winfield, Miller, Weber, Johnson, Sylvia, Vasquez, Vannucci, Beckler and Isaacson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard E. Isaacson, aXNhYWMwMTVAdW1uLmVkdQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.