
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Vet. Sci. , 15 June 2021
Sec. Animal Reproduction - Theriogenology
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.680539
This article is part of the Research Topic Molecular Biomarkers in Animal Reproduction View all 11 articles
Embryonic losses constitute a major burden for reproductive efficiency of farm animals. Pregnancy losses in ungulate species, which include cattle, pigs, sheep and goats, majorly occur during the second week of gestation, when the embryo experiences a series of cell differentiation, proliferation, and migration processes encompassed under the term conceptus elongation. Conceptus elongation takes place following blastocyst hatching and involves a massive proliferation of the extraembryonic membranes trophoblast and hypoblast, and the formation of flat embryonic disc derived from the epiblast, which ultimately gastrulates generating the three germ layers. This process occurs prior to implantation and it is exclusive from ungulates, as embryos from other mammalian species such as rodents or humans implant right after hatching. The critical differences in embryo development between ungulates and mice, the most studied mammalian model, have precluded the identification of the genes governing lineage differentiation in livestock species. Furthermore, conceptus elongation has not been recapitulated in vitro, hindering the study of these cellular events. Luckily, recent advances on transcriptomics, genome modification and post-hatching in vitro culture are shedding light into this largely unknown developmental window, uncovering possible molecular markers to determine embryo quality. In this review, we summarize the events occurring during ungulate pre-implantation development, highlighting recent findings which reveal that several dogmas in Developmental Biology established by knock-out murine models do not hold true for other mammals, including humans and farm animals. The developmental failures associated to in vitro produced embryos in farm animals are also discussed together with Developmental Biology tools to assess embryo quality, including molecular markers to assess proper lineage commitment and a post-hatching in vitro culture system able to directly determine developmental potential circumventing the need of experimental animals.
Optimal reproductive performance in farm animals relies on the proper accomplishment of the different biological processes leading to delivery. Starting from the ovulation of a competent oocyte, conception requires a successful fertilization to produce a zygote, which marks the onset of preimplantation development. During preimplantation development, complex cell proliferation, differentiation and migration processes must be finely controlled to ensure embryo viability and subsequent embryo implantation. Following embryo implantation, a fetus will be developed, ultimately resulting in a newborn. When global reproductive failures in farm ungulates are dissected into these steps, preimplantation development (i.e., the period compromised between fertilization and implantation) clearly stands out as the most problematic. For instance, in the case of cattle, embryonic losses prior to day 16 (D16) post-insemination can rise up to 50% in high yielding dairy cows (1), whereas in pigs it has been estimated that one into five embryos dies before implantation (2).
Preimplantation development in ungulates can be divided into two periods. The first period spans from fertilization to blastocyst hatching, i.e., the release of the embryo from a glycoprotein protective shell termed zona pellucida. This pre-hatching period is common to all mammals and constitutes the whole preimplantation period in rodents and humans, where blastocysts implant right after hatching. In contrast, ungulates exhibit a second preimplantation period termed conceptus elongation. During conceptus elongation the ungulate blastocyst must undergo dramatic morphological changes that, in the case of cattle, convert a ~150 μm D7 blastocyst into a ~30 cm long D21 conceptus around implantation (3, 4). Reproductive failures occurring during preimplantation development can be originated in any of these periods, but developmental collapse during conceptus elongation is the main responsible for global reproductive failures in ungulates (5). To illustrate the magnitude of this problem in cattle farms, it has been estimated that one third of the viable D6 blastocysts fail to elongate and maintain pregnancy by D28 (6), and embryo mortality during early conceptus elongation (D7–D14) oscillates between 26 and 34% (4). In this perspective, the study of the cell differentiation, proliferation, and migration processes occurring during preimplantation development is crucial to understand conceptus collapse and, thereby, finding suitable markers to assess proper lineages development is key to improve reproductive efficiency in livestock ungulates.
The molecular basis of the developmental processes occurring during the first weeks of pregnancy in ungulates is only partially understood, mainly due to two technical limitations: the lack of an in vitro system able to recapitulate conceptus elongation, and the difficulties for performing loss-of-function studies in these species. Luckily, recent advances in in vitro culture of post-hatching blastocysts in cattle (7) and sheep (own unpublished data), together with the development of CRISPR-Cas9 technology to perform loss of function studies in livestock species are set to boost our knowledge on molecular markers for assessing proper embryo development.
In this review, we discuss the differences between ungulate embryo development and that of rodents and humans, highlighting the molecular markers involved in the first lineages differentiation events occurring in ungulates. We also revise different studies that have reported impaired lineages development in in vitro-produced embryos, and provide insights into the potential of lineages markers and post-hatching embryo culture systems to assess embryo quality in farm animals.
Segregation of the first cell lineages in the embryo is critical for proper pregnancy establishment and fetus development. Unfortunately, comprehensive understanding of this process is only available in mice, which became a classical model in Developmental Biology due to its low maintenance cost, fast life cycle and, particularly, due to the well-developed techniques for genome modification in this species. Although the first stages of early embryo development are broadly conserved in mammals, increasing amount of research using novel technologies such as single cell transcriptomics or generation of knock-out embryos by CRISPR-Cas9 are revealing important differences in gene regulatory networks between rodent and non-rodent species.
During the first cell divisions, the embryo relies on maternal transcripts and proteins until the embryonic genome is activated between the 2- (rodents) and 4/8-cell stages (lagomorphs, ungulates, and primates) (8). At these early stages of development, blastomeres are morphologically indistinguishable, but from the 8-cell stage in the mouse, cells located in the outside of the embryo undergo a process of polarization that will influence their fate, biasing outer polar cells toward trophectoderm (TE) and inner apolar cells toward inner cell mass (ICM) (9–11) (Figure 1). The formation of an apical domain in the outer cells triggers a transcriptional network involving Hippo/YAP signaling and the activation of Tead4 in the mouse, which leads to the downregulation of the pluripotency factor Sox2 and the upregulation of Cdx2 from the morula stage (12–15). Activation of CDX2 downregulates Oct4 (14, 16) and activates the expression of other TE markers such as Gata3, Eomes or Elf5 (17), allowing the emergence of the first cell lineages during the formation of the blastocyst: TE, which will mediate implantation, and ICM that will form the embryo proper.
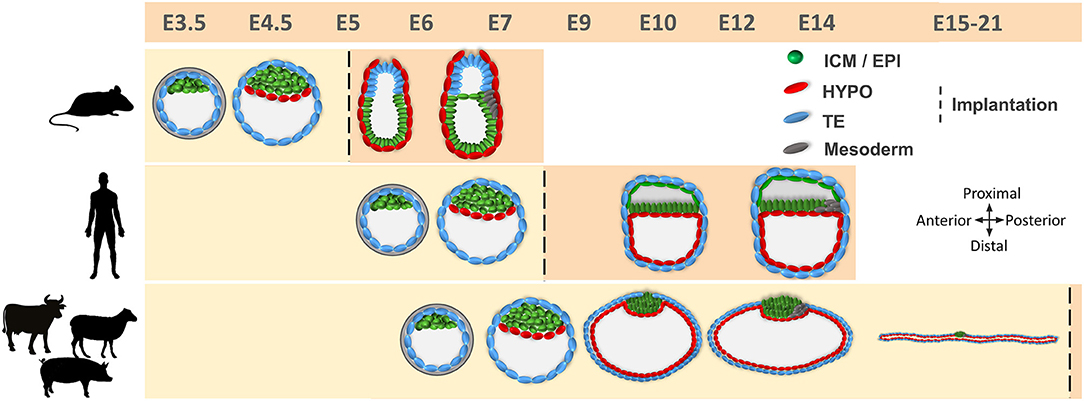
Figure 1. Comparative developmental timeline between mice, humans, and farm ungulates. Different cell lineages are indicated. Embryo development in humans and farm ungulates is delayed compared to mice. Implantation takes place after blastocyst hatching in mice and humans, while in farm ungulates it occurs following conceptus elongation. Mouse epiblast develops in a cup shape after the blastocyst stage, while in humans and farm ungulates it forms a flat embryonic disc. E, embryonic day; ICM, inner cell mass; EPI, epiblast; HYPO, hypoblast; TE, trophectoderm.
These transcription factors exhibit different temporal expression profiles and roles in cell differentiation events in non-rodent mammals (Table 1). TE-specific genes CDX2, GATA3, or TEAD4 are also expressed in the TE of bovine (18–20), porcine (21, 22), and human (20, 23, 24) blastocysts, but remarkable differences in timing of expression and function have been observed between ungulates and rodents. For instance, CDX2 protein expression before the blastocyst stage is restricted to some scattered cells in pig and cattle, contrasting to the ubiquitous expression in mice (14, 25–27), and both CDX2 and OCT4 are expressed in the TE until late blastocyst stages in humans, pigs and cattle (25, 26, 28, 29), in contrast to the mutually exclusive expression observed in mouse blastocysts (14). In the same line, whereas CDX2 represses Oct4 expression in the TE of murine blastocysts (16), CDX2 downregulation does not affect OCT4 expression in bovine embryos (25, 30), and bovine OCT4 promoter lacks an essential region -CR4- necessary for repression of OCT4 in mouse TE (25). Recent KO experiments have also highlighted a different function of OCT4 in rodents vs. ungulates and humans. Oct4 KO mouse embryos develop to blastocyst, although hypoblast formation—explained in the next section—is impaired, but the absence of OCT4 protein in OCT4 KO morulae has been observed to impair blastocyst formation and reduce CDX2 expression in both cattle (31) and humans (32, 33).
Other transcription factors involved in TE vs. ICM differentiation in mouse also seem to play different roles in ungulate development. TEAD4, the upstream regulator of TE genes such as CDX2 and EOMES, does not appear to be essential for TE specification in ungulates, as its downregulation does not impair blastocyst formation in cattle (34, 35), whereas Tead4 ablation in mice completely abolishes blastocoel formation (36, 37). Furthermore, transcription factors downstream TEAD4 such as EOMES and ELF5 are still not expressed at the blastocyst stage in cattle (18, 30, 38), pigs (21) and humans (24). Although gene ablation experiments in ungulates are required to faithfully elucidate the role of these genes, these evidences strongly suggest that the transcriptional network required for TE specification in ungulates is considerably different to that of mice. In this perspective, early TE specification could have emerged as an evolutionary mechanism in rodents to allow implantation at an earlier stage than in other species. Fortunately, recent improvements in genome edition techniques in ungulates (39, 40) currently allow the exploration of the molecular machinery involved in ICM/TE specification in farm animals. Genome editing constitutes also an unvaluable tool to study other reproductive processes (41). For instance, the generation of aromatase-null porcine conceptuses has uncovered that intrinsic estrogen conceptus production is not required for early maternal recognition of pregnancy or implantation in pig, in striking contrast to previous beliefs (42).
After the differentiation of the TE, the second cell fate decision takes place in the ICM and determines the emergence of the pluripotent epiblast and the extraembryonic hypoblast (Figure 1). In mice, hypoblast markers are sequentially expressed, starting with GATA6 at the 8-cell, PDGFRα at the 16-cell, SOX17 at the 32-cell, and GATA4 at the 64-cell stages (43, 44). Later on, the cells forming the early ICM (E3.5) co-express both epiblast (OCT4, SOX2, and NANOG) and hypoblast (GATA6, SOX17, and PDGFRA) proteins, but by E4.5 these cells will show mutually exclusive expression for both markers (43, 45). Although the detailed temporal expression pattern of these markers is not available for most domestic mammals, there are broad similarities in the gene regulatory network controlling the second lineage specification within mammals (46). For example, PDGFRα and SOX17 are also co-expressed with epiblast markers in the bipotent ICM cells and become restricted to hypoblast cells in late blastocysts in human, pig and cattle embryos (18, 21, 24). However, important differences have been also reported between rodent and non-rodent species (Table 1). GATA6, together with the epiblast marker NANOG, are expressed in all cells of the mouse morula and their mutual repression in the late ICM is essential for epiblast vs. hypoblast specification (43, 47). However, this might not be a strict requirement in all mammals, since GATA6 is expressed in all cell lineages of the blastocyst in primates, pigs and cattle, only becoming restricted to the hypoblast at later developmental stages (18, 21, 24, 48–50), and NANOG is not expressed in the human, pig and cattle morula (21, 26, 51, 52). Once specified, hypoblast cells reorganize to form an epithelium lying in contact with the blastocoel cavity (43) and migrate to cover the inner embryo surface in primates and ungulates (7) (Figure 1).
A characteristic aspect of ungulate development is TE fate. After blastocyst hatching, the TE can be classified into mural TE, which covers the blastocoel cavity, and polar TE, covering the ICM. While in rodents and primates, the polar TE forms the extraembryonic ectoderm (ExE), contributing to implantation and becoming part of the placenta, in lagomorphs (53) and ungulates (3) this function is accomplished by the mural TE. The polar TE, also known as Rauber's layer (RL), is removed through an apoptotic mechanism (54) around D9–D11 in pigs (55), D10–D12 in horses (56), D11–D12 in sheep (own unpublished observations) and by day 14 in cattle (54), directly exposing the epiblast to the uterine histotroph (Figure 1). Shortly after the disappearance of the RL, the extraembryonic membranes (EEMs, composed by mural TE and hypoblast) undergo extensive proliferation. As a consequence of this proliferation, the embryo is termed conceptus (EEMs + embryo proper) and it progresses from spherical through ovoid, tubular and filamentous stages reaching a length of ~30 cm in cattle (3) and ~100 cm in pigs (57) by the time of implantation, which starts about D14 in pigs (58), D15 in the sheep (59), and D19 in cattle (60). In other non-ungulate domestic species, such as rabbits and horses, the blastocyst also experiences a massive growth of EEMs before implantation, reaching up to 20 mm in horses and 5 mm in rabbits, but it remains spherical (8).
Besides the massive proliferation of EEMs, major developmental events take place in the epiblast before implantation: an anterioposterior axis is established that will outline the body plan, and the three germ layers become specified, together with the germline (61–63). Before the RL disintegrates, the epiblast forms a small cavity that will be opened once the RL disappears, unfolding the epiblast (64, 65). This contrasts with murine and human development, where the epiblast cavitates to form the amniotic cavity (66–68) (Figure 1). Roughly concomitant to RL disintegration, the ungulate epiblast develops into a clearly identifiable circular light structure: the embryonic disc (ED), where epiblast cells develop tight junctions and form a basal lamina toward the hypoblast (63). When the ED is fully formed, expression of core pluripotency markers SOX2, OCT4, and NANOG is restricted to the epiblast (21, 25, 69). During the next days, the ED will acquire an oval shape and a higher density at the posterior edge, associated with the ingression of the first cells into the primitive streak and the beginning of gastrulation. Some cells at the posterior part of the epiblast start to express the mesoderm marker BRACHYURY (T) and to downregulate SOX2 before the primitive streak is morphologically visible (59, 69–72). These T-positive cells will be the first cells to egress into the space between the epiblast and the hypoblast and will form the mesoderm, which quickly migrates to cover the whole embryonic disc. At the same time, more epiblast cells continue to egress through the primitive streak to form the endoderm, which lies on the dorsal hypoblast, while the mesoderm forms a mesenchyme between the epiblast and the endoderm. Epiblast cells that do not pass through the streak will form the ectoderm (73, 74). The primitive streak will be extended in an anterior direction, being the anterioposterior axis of the embryo proper aligned with the proliferation of the extra-embryonic membranes (i.e., conceptus elongation axis) (75).
Our knowledge of the genes and signaling pathways controlling gastrulation in mammals is mainly derived from the mouse embryo, in which gastrulation occurs following implantation. Key genes involved in gastrulation such as BRACHYURY, EOMES, BMP4, NODAL, CER1, or FOXA2 seem to play conserved roles in ungulates, although with some differences in their location and temporal expression (3, 59, 63, 65, 71, 76, 77).
The developmental defects ultimately leading to embryo mortality during conceptus elongation have been difficult to explore, given the challenges for obtaining elongating embryos in vivo. However, several in vitro evidences and in vivo observations point to the development of the epiblast as the most vulnerable process. In vitro evidences show that the requirements for trophectoderm and hypoblast development are less restrictive than those required for epiblast survival. Primary bovine trophectoderm cell cultures can be established using relatively simple media supplemented with 10 % serum (78, 79), whereas conditions required for truly pluripotent epiblast cell culture in farm animals remain to be captured (80). In the same line, early in vitro culture systems designed for the development of post-hatching ungulate embryos were successful in achieving trophectoderm proliferation and some degree of hypoblast migration, but the epiblast degenerated (81–83). To attain epiblast survival in vitro, we required a way more complex medium (termed N2B27) containing aminoacids, lipids, vitamins, hormones, and growth factors not present in previous systems (7). Yet, under our system epiblast survival is observed in 55–60% of the in vitro produced embryos, whereas trophoblast and hypoblast proliferation is found in all surviving structures.
Failure in epiblast development has been also observed in vivo. Embryo transfer of in vitro produced (IVP) embryos often results in lower pregnancy rates compared to their in vivo counterparts, as it will be discussed below, and failures in epiblast development seem to be the main responsible for such developmental arrest. IVP-derived bovine conceptuses have been reported to exhibit smaller EDs than their in vivo counterparts (84), and multiple studies have reported a remarkably high percentage (23–65%) of IVP-derived conceptuses lacking EDs (4, 85–90), as reviewed by Ealy et al. (91). Impaired ED development has also been observed in ovine and bovine embryos produced by somatic cell nuclear transfer (SCNT), a technology that induces pleiotropic effects over different lineages (92, 93). Interestingly, embryos lacking an ED were also observed after SCNT at a very high rate, ranging from 20 to 58% (Table 2) (89, 94, 96–99), and failures in a mechanism that populates inner cells based on asymmetric divisions of outer cells have been proposed to be responsible for developmental arrest in SCNT rabbit embryos (95). Moreover, transcriptional alterations in embryonic lineages of SCNT embryos were 10–20-fold more abundant in the epiblast than in extraembryonic lineages, both in cattle and mice (89, 98, 101, 102). Accordingly, some authors have observed that normal elongation in SCNT cattle embryos was more frequent (46/50 embryos) than normal ED formation and gastrulation (38/50) (98).
In vitro embryo production enables a myriad of applications in livestock species, ranging from boosting the number of embryos obtained from females of high genetic merit to overcoming infertility problems associated to heat stress (103–105). According to the International Embryo Transfer Society (IETS), while the number of in vivo derived embryos transferred seems to have stabilized in cattle, the use of in vitro-produced (IVP) embryos is currently increasing (742,908 embryos transferred worldwide in 2018) (106). However, despite all efforts performed to optimize assisted reproductive technologies (ARTs), embryo production systems are still not fully efficient and important differences have been reported between in vitro and in vivo embryos (107). Many studies have shown that pregnancy rates after transfer of an in vitro embryo are between 10 and 40% lower than with embryos generated by artificial insemination or by Multiple Ovulation Embryo Transfer (MOET) (6, 108–111). As previously mentioned, IVP embryos often show compromised development of embryonic lineages—particularly the epiblast—following embryo transfer, and it has been estimated that 80% of pregnancy failures following embryo transfer of IVP embryos occur before day 40 of pregnancy (91). Unfortunately, these rates have not improved in the last decades.
In order to improve in vitro embryo production techniques, it is essential to assess embryo quality, i.e., the odds of post-transfer survival, to determine which modifications of current protocols are beneficial for subsequent embryo survival. Arguably, the best embryo quality assessment would be the analysis of embryo development following embryo transfer, but this test holds two major drawbacks (1) it is expensive, time-consuming and requires the use of experimental animals and (2) it is inherently bound to intrinsic variabilities in uterine receptivity between females (112, 113). Morphological evaluation (114), widely used both in humans and farm animals to select embryos before transfer due to its non-invasive nature, is certainly useful, as pregnancy rates are higher when better-quality grade embryos are transferred (115–118). However, embryo grade is a subjective criterion; it does not always reflect competence to establish pregnancy (119), and it does not necessarily infer proper development of embryonic lineages. For instance, early mouse mutant embryos lacking a specific cell lineage cannot be visually distinguished from their wildtype counterparts, although they hardly progress beyond implantation (37, 120–123). In this perspective, the analysis of the development of specific lineages provides deeper insights of embryo quality.
Successful development of the first cell lineages is essential for implantation and further development to term (124). The most commonly used method to analyse the first lineage differentiation (i.e., ICM vs. TE) in blastocysts from livestock species has been differential cell staining, a technique based on selective permeabilization of the outer blastocyst cells which will be subsequently stained with propidium iodide (125, 126). Unfortunately, this technique only provides information on cell location, but fails to assess if those cells are properly committed to TE or ICM, as the expression of specific lineage markers is not determined. Recent studies performing single-cell transcriptomics in farms animals have enabled the identification of lineage markers. The first study using this technology in mouse pre-implantation embryos (127) was soon followed by other reports in human (24, 48), monkey (49), cow (18, 19, 128) and pig (21), which revealed relevant differences between rodents and non-rodents mammals. In bovine, two studies based on single cell qPCR analysis of IVP morulae and expanded blastocysts observed that while some classical hypoblast markers in the mouse (GATA6, GSC, and HNF4A) were not specific to this lineage in bovine, SOX17, GATA4, and PDGFRA were largely specific (18, 19). Furthermore, the core pluripotency markers NANOG, SOX2, and OCT4 were detected in epiblast cells, although NANOG, FGF4, and TDGF1 were deemed as the most epiblast-specific, and trophectoderm cells exclusively expressed CDX2, GATA2, GATA3, KRT8, PECAM1, or DAB2 (18, 19). In the pig, scRNAseq of in vivo-derived morulae, early and late blastocysts, and spherical embryos, revealed that although NANOG, SOX2, and OCT4 are expressed in epiblast cells, SOX2 is the most specific epiblast marker, followed by NANOG (21). GATA2 and GATA3 were reliable trophoblast markers, while CDX2 was barely expressed in early blastocysts, and TEAD4 was expressed in all cell lineages. Finally, hypoblast cells were characterized by specific expression of SOX17, PDGFRA, GATA4, or NID2 (21).
Expression of different lineage-specific markers can be analyzed at the protein level through embryo immunostaining (Figure 2). Although most commercial antibodies are designed to react with mouse and human proteins, some of them can also be used to label pig, sheep and cow embryos (Figure 3 and Table 3). Antibodies against the core pluripotency markers OCT4, NANOG, and SOX2 have been regularly used to label ICM and epiblast cells. Particular caution must be paid when analyzing OCT4, as this protein is expressed in all cell lineages in bovine and porcine blastocysts (25, 26, 130, 133), being its expression restricted to the epiblast only at later stages [E11 in bovine (25)]. NANOG protein has been specifically detected in ICM cells in bovine (26, 51, 131, 134) and porcine blastocysts (21). However, SOX2 seems to be the most specific epiblast marker in pig (21, 55, 130, 135), sheep (Figure 2) and bovine embryos (7, 30, 31, 131, 134). Regarding TE markers, CDX2 remains the most commonly used marker, being TE-specific in pig (26, 130), sheep (Figure 2) and cow blastocysts (7, 25, 26). In our experience (unpublished observations, Figure 2) GATA3 is also a reliable TE marker in porcine, ovine and bovine embryos. Finally, regarding to hypoblast, SOX17 specifically labels hypoblast cells from the blastocyst up to the elongated conceptus in bovine (7), ovine (Figure 2) and porcine embryos (21, 69). Other classical hypoblast markers in mice include GATA6, ubiquitously expressed in early blastocysts and becoming restricted to hypoblast cells in late bovine (26, 51), ovine (Figure 2) and porcine blastocysts (130), and GATA4, ubiquitously expressed in bovine late blastocysts and becoming specifically restricted to hypoblast cells in post-hatching E10.5 embryos (51, 129), and in some cells close to the epiblast in late pig blastocysts (132).
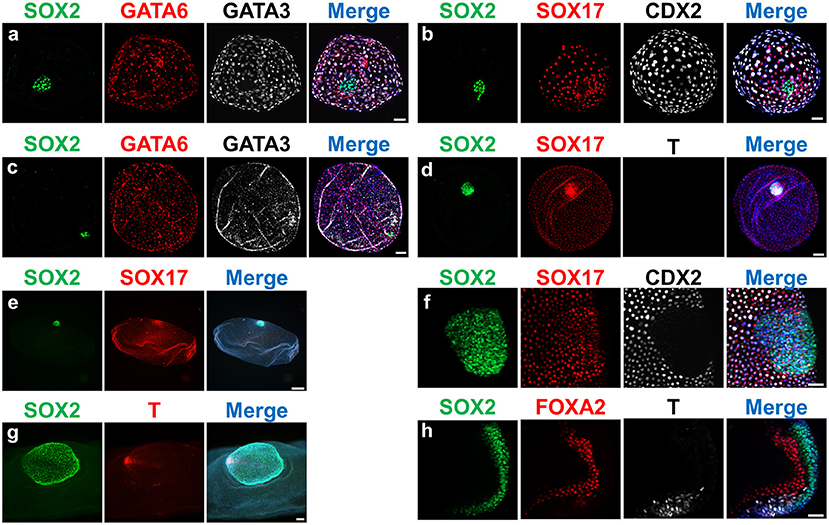
Figure 2. Expression of lineage-specific markers in ovine embryos at different developmental stages. Fluorescence images of embryos stained for SOX2 (epiblast); SOX17/GATA6/FOXA2 (hypoblast/endoderm); CDX2/GATA3 (trophectoderm); T (mesoderm). Nuclei were counterstained with DAPI (merge). (a,b) D9 Hatched blastocysts; (c,d): D11 Spherical embryos; (e) D13 Tubular embryo; (f) D13 ED without Rauber's layer; (g) gastrulating ED; (h) section of a gastrulating ED. Scale bars = 50 μm for (a,b,f,h); 100 μm for (c,d,g); 500 μm for panel (e).
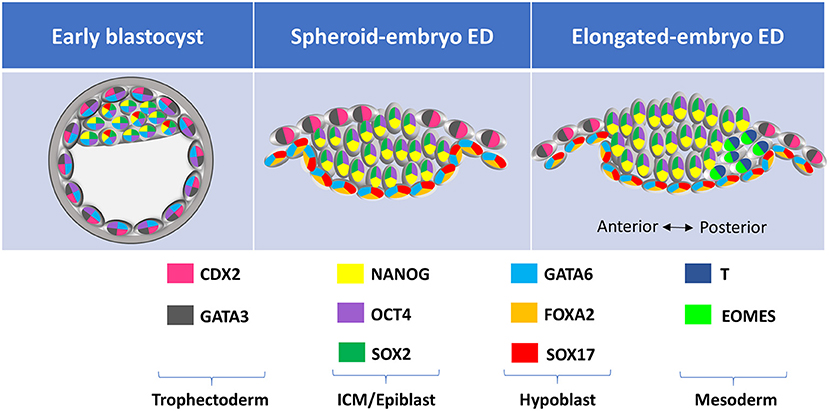
Figure 3. Lineage-specific markers expressed at different developmental stages in farm ungulate embryos. ED, embryonic disc; ICM, inner cell mass; TE, trophectoderm.
Similarly to the conventional quality assessment in blastocysts, the developmental analysis of elongated conceptuses has been traditionally based on morphology, generally limited to conceptus length (136–139). Conceptus length constitutes a good proxy for the development of extra-embryonic membranes but such development may not be coupled to the development of the epiblast, the most sensitive lineage. In other words, the “bigger is better” concept routinely applied to assess conceptus development may be wrong, as an structure only composed by EEMs (i.e., lacking an ED) will not develop any fetus. In this regard, verifying the presence and proper development of the ED is essential to determine conceptus quality. To this aim, gastrulation markers have still not received as much attention as early lineages differentiation markers, since conceptuses at gastrulation stages are less accessible for experimental studies. However, BRACHYURY protein has been first observed in nascent mesoderm cells in the posterior epiblast of ovoid pig embryos (69, 71), and both BRACHYURY and EOMES are located in the posterior part of the ED at the same time that SOX2 expression is restricted to the anterior part in elongating sheep and cow embryos (Figure 2). Finally, SOX17 and FOXA2 are expressed in the migrated hypoblast and in the definitive endoderm in pig (71), sheep and cow elongating embryos (own unpublished observations, Figure 2).
Direct assessment of the developmental potential of IVP embryos beyond the blastocyst stage (i.e., during the most vulnerable period) has been traditionally hampered by the requirement of time and resource consuming in vivo experiments involving embryo transfer and posterior recovery (4, 89, 100, 138), as no in vitro system able to support embryo development beyond blastocyst hatching was available for any farm animal. In the last years, an increasing interest has been placed on developing post-blastocyst in vitro culture systems to better investigate embryo development and mortality during this developmental window in different mammalian species. Human embryos have been cultured in the absence of maternal tissues up to D13 (140, 141) in a system that also allows mouse post-blastocyst culture up to egg cylinders (142). This system was further improved to allow development of human embryos up to gastrulating stages, and deep embryo characterization by single-cell transcriptome and methylome mapping (143, 144).
In farm animals, hatched blastocysts attach to the bottom of the culture dish or grow in rounded form until they collapse under normal pre-hatching embryo culture conditions. Although explanted EDs were cultured in vitro in rabbits (145, 146), a livestock species where gastrulation also occurs in a flat embryonic disc, limited success was achieved in ungulates until recently. Pioneer studies established an in vitro post-hatching development (PHD) system based on agarose gel tunnels and serum- and glucose-enriched medium that achieved some expansion of the trophectoderm and certain proliferation of hypoblast cells in bovine embryos up to D15, but hypoblast migration was incomplete and epiblast cells were unable to survive (81–83). In order to promote the development of the epiblast—the most stringent cell lineage—we have developed a system based on N2B27 medium, a defined medium composed by Neurobasal and DMEM/F12 media, and N2 and B27 supplements which was initially designed to culture neurons (147), and it was employed later for embryonic stem cells derivation and culture (148). Under this system, bovine (7) and ovine (own unpublished data) blastocysts develop beyond hatching, attaining complete hypoblast migration and epiblast survival and development into an early ED.
This pioneer system paves the way for future research focused on improving the conditions of in vitro embryo production or associated techniques such as embryo freezing or vitrification, as it allows direct embryo quality assessment without the need of experimental animals. The potential roles on lineages development of specific metabolites, hormones, or growth factors whose levels are altered in detrimental conditions for embryo survival, such as negative energy balance in post-partum dairy cows (90), can also be analyzed in this system.
Being the most vulnerable period for embryo survival in ungulate species, conceptus elongation still constitutes a black box in Developmental Biology. Luckily, recent findings based on transcriptome analysis and gene ablation have started to shed light into the cell differentiation, proliferation and migration processes governing conceptus elongation, which in some cases differ greatly from those occurring in mice, the most studied mammalian model. The molecular markers of lineage differentiation in ungulates unveiled by these experiments are extremely useful to assess proper lineage development in in vitro produced embryos, where epiblast development has been highlighted as the major obstacle to attain a successful pregnancy. Besides, recent advances on in vitro culture have moved forward the limits of in vitro embryo development, providing a system to directly evaluate the developmental potential of IVP embryos during the most sensitive period for developmental failure. Such system provides a direct embryo quality assessment for testing diverse modifications in the IVP protocol or in associated techniques such as vitrification or freezing, aimed to improve pregnancy rates following embryo transfer.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Research on conceptus elongation carried out by the authors and the publication costs of this review were funded by the projects StG-757886-ELONGAN from the European Research Council and AGL2017-58739-R from the Spanish Ministry of Economy and Competitiveness (MINECO). Images shown on Figure 2 were taken on an equipment funded by the project ECQ2018-005184-P from MINECO. PR-I is funded by a Ramón y Cajal Contract from MINECO (RYC2018-025666-I).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the slaughterhouses Transformación Ganadera de Leganés SA, Matadero Madrid-Norte SA, and Matadero Mondejano SL (especially to Reyes Prieto Cabañas) for gently providing the ovaries required for research on ungulate lineage differentiation markers.
1. Dunne LD, Diskin MG, Sreenan JM. Embryo and foetal loss in beef heifers between day 14 of gestation and full term. Anim Reprod Sci. (2000) 58:39–44. doi: 10.1016/S0378-4320(99)00088-3
2. Bennett GL, Leymaster KA. Integration of ovulation rate, potential embryonic viability and uterine capacity into a model of litter size in swine. J Anim Sci. (1989) 67:1230–41. doi: 10.2527/jas1989.6751230x
3. Maddox-Hyttel P, Alexopoulos NI, Vajta G, Lewis I, Rogers P, Cann L, et al. Immunohistochemical and ultrastructural characterization of the initial post-hatching development of bovine embryos. Reproduction. (2003) 125:607–23. doi: 10.1530/reprod/125.4.607
4. Berg DK, van Leeuwen J, Beaumont S, Berg M, Pfeffer PL. Embryo loss in cattle between Days 7 and 16 of pregnancy. Theriogenology. (2010) 73:250–60. doi: 10.1016/j.theriogenology.2009.09.005
5. Diskin MG, Morris DG. Embryonic and early foetal losses in cattle and other ruminants. Reprod Domest Anim. (2008) 43(Suppl 2):260–7. doi: 10.1111/j.1439-0531.2008.01171.x
6. Santos JE, Thatcher WW, Chebel RC, Cerri RL, Galvao KN. The effect of embryonic death rates in cattle on the efficacy of estrus synchronization programs. Anim Reprod Sci. (2004) 82–83:513–35. doi: 10.1016/j.anireprosci.2004.04.015
7. Ramos-Ibeas P, Lamas-Toranzo I, Martinez-Moro A, de Frutos C, Quiroga AC, Zurita E, et al. Embryonic disc formation following post-hatching bovine embryo development in vitro. Reproduction. (2020) 160:579–89. doi: 10.1530/REP-20-0243
8. Piliszek A, Madeja ZE. Pre-implantation development of domestic animals. Curr Top Dev Biol. (2018) 128:267–94. doi: 10.1016/bs.ctdb.2017.11.005
9. Korotkevich E, Niwayama R, Courtois A, Friese S, Berger N, Buchholz F, et al. The apical domain is required and sufficient for the first lineage segregation in the mouse embryo. Dev Cell. (2017) 40:235–47.e7. doi: 10.1016/j.devcel.2017.01.006
10. Maître JL, Turlier H, Illukkumbura R, Eismann B, Niwayama R, Nédélec F, et al. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature. (2016) 536:344–8. doi: 10.1038/nature18958
11. Artus J, Hue I, Acloque H. Preimplantation development in ungulates: a ‘menage a quatre' scenario. Reproduction. (2020) 159:R151–R72. doi: 10.1530/REP-19-0348
12. Wicklow E, Blij S, Frum T, Hirate Y, Lang RA, Sasaki H, et al. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet. (2014) 10:e1004618. doi: 10.1371/journal.pgen.1004618
13. Ralston A, Cox BJ, Nishioka N, Sasaki H, Chea E, Rugg-Gunn P, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development. (2010) 137:395–403. doi: 10.1242/dev.038828
14. Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. (2005) 132:2093–102. doi: 10.1242/dev.01801
15. Hirate Y, Hirahara S, Inoue K, Suzuki A, Alarcon VB, Akimoto K, et al. Polarity-dependent distribution of angiomotin localizes Hippo signaling in preimplantation embryos. Curr Biol. (2013) 23:1181–94. doi: 10.1016/j.cub.2013.05.014
16. Niwa H, Toyooka Y, Shimosato D, Strumpf D, Takahashi K, Yagi R, et al. Interaction between Oct3/4 and Cdx2 determines trophectoderm differentiation. Cell. (2005) 123:917–29. doi: 10.1016/j.cell.2005.08.040
17. Ng RK, Dean W, Dawson C, Lucifero D, Madeja Z, Reik W, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol. (2008) 10:1280–90. doi: 10.1038/ncb1786
18. Wei Q, Zhong L, Zhang S, Mu H, Xiang J, Yue L, et al. Bovine lineage specification revealed by single-cell gene expression analysis from zygote to blastocyst. Biol Reprod. (2017) 97:5–17. doi: 10.1093/biolre/iox071
19. Negron-Perez VM, Zhang Y, Hansen PJ. Single-cell gene expression of the bovine blastocyst. Reproduction. (2017) 154:627–44. doi: 10.1530/REP-17-0345
20. Gerri C, McCarthy A, Alanis-Lobato G, Demtschenko A, Bruneau A, Loubersac S, et al. Initiation of a conserved trophectoderm program in human, cow and mouse embryos. Nature. (2020) 587:443–7. doi: 10.1038/s41586-020-2759-x
21. Ramos-Ibeas P, Sang F, Zhu Q, Tang WWC, Withey S, Klisch D, et al. Pluripotency and X chromosome dynamics revealed in pig pre-gastrulating embryos by single cell analysis. Nat Commun. (2019) 10:500. doi: 10.1038/s41467-019-08387-8
22. Kong Q, Yang X, Zhang H, Liu S, Zhao J, Zhang J, et al. Lineage specification and pluripotency revealed by transcriptome analysis from oocyte to blastocyst in pig. FASEB J. (2020) 34:691–705. doi: 10.1096/fj.201901818RR
23. Niakan KK, Eggan K. Analysis of human embryos from zygote to blastocyst reveals distinct gene expression patterns relative to the mouse. Dev Biol. (2013) 375:54–64. doi: 10.1016/j.ydbio.2012.12.008
24. Blakeley P, Fogarty NM, del Valle I, Wamaitha SE, Hu TX, Elder K, et al. Defining the three cell lineages of the human blastocyst by single-cell RNA-seq. Development. (2015) 142:3151–65. doi: 10.1242/dev.123547
25. Berg DK, Smith CS, Pearton DJ, Wells DN, Broadhurst R, Donnison M, et al. Trophectoderm lineage determination in cattle. Dev Cell. (2011) 20:244–55. doi: 10.1016/j.devcel.2011.01.003
26. Kuijk EW, Du Puy L, Van Tol HT, Oei CH, Haagsman HP, Colenbrander B, et al. Differences in early lineage segregation between mammals. Dev Dyn. (2008) 237:918–27. doi: 10.1002/dvdy.21480
27. Bou G, Liu S, Sun M, Zhu J, Xue B, Guo J, et al. CDX2 is essential for cell proliferation and polarity in porcine blastocysts. Development. (2017) 144:1296–306. doi: 10.1242/dev.141085
28. Cauffman G, Van de Velde H, Liebaers I, Van Steirteghem A. Oct-4 mRNA and protein expression during human preimplantation development. Mol Hum Reprod. (2005) 11:173–81. doi: 10.1093/molehr/gah155
29. Kirchhof N, Carnwath JW, Lemme E, Anastassiadis K, Schöler H, Niemann H. Expression pattern of Oct-4 in preimplantation embryos of different species. Biol Reprod. (2000) 63:1698–705. doi: 10.1095/biolreprod63.6.1698
30. Goissis MD, Cibelli JB. Functional characterization of CDX2 during bovine preimplantation development in vitro. Mol Reprod Dev. (2014) 81:962–70. doi: 10.1002/mrd.22415
31. Daigneault BW, Rajput S, Smith GW, Ross PJ. Embryonic POU5F1 is required for expanded bovine blastocyst formation. Sci Rep. (2018) 8:7753. doi: 10.1038/s41598-018-25964-x
32. Fogarty NME, McCarthy A, Snijders KE, Powell BE, Kubikova N, Blakeley P, et al. Genome editing reveals a role for OCT4 in human embryogenesis. Nature. (2017) 550:67–73. doi: 10.1038/nature24033
33. Stamatiadis P, Boel A, Cosemans G, Popovic M, Bekaert B, Guggilla R, et al. Comparative analysis of mouse and human preimplantation development following POU5F1 CRISPR/Cas9 targeting reveals interspecies differences. Hum Reprod. (2021) 36:1242–52. doi: 10.1093/humrep/deab027
34. Sakurai N, Takahashi K, Emura N, Hashizume T, Sawai K. Effects of downregulating TEAD4 transcripts by RNA interference on early development of bovine embryos. J Reprod Dev. (2017) 63:135–42. doi: 10.1262/jrd.2016-130
35. Akizawa H, Kobayashi K, Bai H, Takahashi M, Kagawa S, Nagatomo H, et al. Reciprocal regulation of TEAD4 and CCN2 for the trophectoderm development of the bovine blastocyst. Reproduction. (2018) 155:563–71. doi: 10.1530/REP-18-0043
36. Nishioka N, Yamamoto S, Kiyonari H, Sato H, Sawada A, Ota M, et al. Tead4 is required for specification of trophectoderm in pre-implantation mouse embryos. Mech Dev. (2008) 125:270–83. doi: 10.1016/j.mod.2007.11.002
37. Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, DePamphilis ML, et al. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. (2007) 134:3827–36. doi: 10.1242/dev.010223
38. Degrelle SA, Campion E, Cabau C, Piumi F, Reinaud P, Richard C, et al. Molecular evidence for a critical period in mural trophoblast development in bovine blastocysts. Dev Biol. (2005) 288:448–60. doi: 10.1016/j.ydbio.2005.09.043
39. Lamas-Toranzo I, Galiano-Cogolludo B, Cornudella-Ardiaca F, Cobos-Figueroa J, Ousinde O, Bermejo-Alvarez P. Strategies to reduce genetic mosaicism following CRISPR-mediated genome edition in bovine embryos. Sci Rep. (2019) 9:14900. doi: 10.1038/s41598-019-51366-8
40. Lamas-Toranzo I, Martinez-Moro A, E OC, Millan-Blanca G, Sanchez JM, Lonergan P, et al. RS-1 enhances CRISPR-mediated targeted knock-in in bovine embryos. Mol Reprod Dev. (2020) 87:542–9. doi: 10.1002/mrd.23341
41. Lamas-Toranzo I, Fonseca Balvis N, Querejeta-Fernandez A, Izquierdo-Rico MJ, Gonzalez-Brusi L, Lorenzo PL, et al. ZP4 confers structural properties to the zona pellucida essential for embryo development. Elife. (2019) 8:e48904. doi: 10.7554/eLife.48904
42. Meyer AE, Pfeiffer CA, Brooks KE, Spate LD, Benne JA, Cecil R, et al. New perspective on conceptus estrogens in maternal recognition and pregnancy establishment in the pigdagger. Biol Reprod. (2019) 101:148–61. doi: 10.1093/biolre/ioz058
43. Plusa B, Piliszek A, Frankenberg S, Artus J, Hadjantonakis AK. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. (2008) 135:3081–91. doi: 10.1242/dev.021519
44. Artus J, Piliszek A, Hadjantonakis AK. The primitive endoderm lineage of the mouse blastocyst: sequential transcription factor activation and regulation of differentiation by Sox17. Dev Biol. (2011) 350:393–404. doi: 10.1016/j.ydbio.2010.12.007
45. Chazaud C, Yamanaka Y. Lineage specification in the mouse preimplantation embryo. Development. (2016) 143:1063–74. doi: 10.1242/dev.128314
46. Frankenberg SR, de Barros FR, Rossant J, Renfree MB. The mammalian blastocyst. Wiley Interdiscip Rev Dev Biol. (2016) 5:210–32. doi: 10.1002/wdev.220
47. Bessonnard S, De Mot L, Gonze D, Barriol M, Dennis C, Goldbeter A, et al. Gata6, Nanog and Erk signaling control cell fate in the inner cell mass through a tristable regulatory network. Development. (2014) 141:3637–48. doi: 10.1242/dev.109678
48. Petropoulos S, Edsgard D, Reinius B, Deng Q, Panula SP, Codeluppi S, et al. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell. (2016) 165:1012–26. doi: 10.1016/j.cell.2016.03.023
49. Nakamura T, Okamoto I, Sasaki K, Yabuta Y, Iwatani C, Tsuchiya H, et al. A developmental coordinate of pluripotency among mice, monkeys and humans. Nature. (2016) 537:57–62. doi: 10.1038/nature19096
50. Stirparo GG, Boroviak T, Guo G, Nichols J, Smith A, Bertone P. Integrated analysis of single-cell embryo data yields a unified transcriptome signature for the human pre-implantation epiblast. Development. (2018) 145:dev158501. doi: 10.1242/dev.158501
51. Kuijk EW, van Tol LT, Van de Velde H, Wubbolts R, Welling M, Geijsen N, et al. The roles of FGF and MAP kinase signaling in the segregation of the epiblast and hypoblast cell lineages in bovine and human embryos. Development. (2012) 139:871–82. doi: 10.1242/dev.071688
52. Cauffman G, De Rycke M, Sermon K, Liebaers I, Van de Velde H. Markers that define stemness in ESC are unable to identify the totipotent cells in human preimplantation embryos. Hum Reprod. (2009) 24:63–70. doi: 10.1093/humrep/den351
53. Williams BS, Biggers JD. Polar trophoblast (Rauber's layer) of the rabbit blastocyst. Anat Rec. (1990) 227:211–22. doi: 10.1002/ar.1092270210
54. van Leeuwen J, Rawson P, Berg DK, Wells DN, Pfeffer PL. On the enigmatic disappearance of Rauber's layer. Proc Natl Acad Sci U S A. (2020) 117:16409–17. doi: 10.1073/pnas.2002008117
55. Sun R, Lei L, Liu S, Xue B, Wang J, Shen J, et al. Morphological changes and germ layer formation in the porcine embryos from days 7-13 of development. Zygote. (2015) 23:266–76. doi: 10.1017/S0967199413000531
56. Enders AC, Lantz KC, Liu IK, Schlafke S. Loss of polar trophoblast during differentiation of the blastocyst of the horse. J Reprod Fertil. (1988) 83:447–60. doi: 10.1530/jrf.0.0830447
57. Geisert RD, Brookbank JW, Roberts RM, Bazer FW. Establishment of pregnancy in the pig: II. Cellular remodeling of the porcine blastocyst during elongation on day 12 of pregnancy. Biol Reprod. (1982) 27:941–55. doi: 10.1095/biolreprod27.4.941
58. Waclawik A, Kaczmarek MM, Blitek A, Kaczynski P, Ziecik AJ. Embryo-maternal dialogue during pregnancy establishment and implantation in the pig. Mol Reprod Dev. (2017) 84:842–55. doi: 10.1002/mrd.22835
59. Guillomot M, Turbe A, Hue I, Renard JP. Staging of ovine embryos and expression of the T-box genes Brachyury and Eomesodermin around gastrulation. Reproduction. (2004) 127:491–501. doi: 10.1530/rep.1.00057
60. Bazer FW, Spencer TE, Johnson GA, Burghardt RC, Wu G. Comparative aspects of implantation. Reproduction. (2009) 138:195–209. doi: 10.1530/REP-09-0158
61. Johnson AD, Alberio R. Primordial germ cells: the first cell lineage or the last cells standing? Development. (2015) 142:2730–9. doi: 10.1242/dev.113993
62. Gao Y, Hyttel P, Hall VJ. Dynamic changes in epigenetic marks and gene expression during porcine epiblast specification. Cell Reprogram. (2011) 13:345–60. doi: 10.1089/cell.2010.0110
63. Oestrup O, Hall V, Petkov SG, Wolf XA, Hyldig S, Hyttel P. From zygote to implantation: morphological and molecular dynamics during embryo development in the pig. Reprod Domest Anim. (2009) 44(Suppl. 3):39–49. doi: 10.1111/j.1439-0531.2009.01482.x
64. Hall VJ, Jacobsen JV, Rasmussen MA, Hyttel P. Ultrastructural and molecular distinctions between the porcine inner cell mass and epiblast reveal unique pluripotent cell states. Dev Dyn. (2010) 239:2911–20. doi: 10.1002/dvdy.22424
65. van Leeuwen J, Berg DK, Pfeffer PL. Morphological and gene expression changes in cattle embryos from hatched blastocyst to early gastrulation stages after transfer of in vitro produced embryos. PLoS One. (2015) 10:e0129787. doi: 10.1371/journal.pone.0129787
66. Hertig AT, Rock J, Adams EC. A description of 34 human ova within the first 17 days of development. J Anat. (1956) 98:435–93. doi: 10.1002/aja.1000980306
67. Luckett WP. Origin and differentiation of the yolk sac and extraembryonic mesoderm in presomite human and rhesus monkey embryos. J Anat. (1978) 152:59–97. doi: 10.1002/aja.1001520106
68. Bedzhov I, Zernicka-Goetz M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell. (2014) 156:1032–44. doi: 10.1016/j.cell.2014.01.023
69. Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, et al. Principles of early human development and germ cell program from conserved model systems. Nature. (2017) 546:416–20. doi: 10.1038/nature22812
70. Gaivão MM, Rambags BP, Stout TA. Gastrulation and the establishment of the three germ layers in the early horse conceptus. Theriogenology. (2014) 82:354–65. doi: 10.1016/j.theriogenology.2014.04.018
71. Wolf XA, Serup P, Hyttel P. Three-dimensional immunohistochemical characterization of lineage commitment by localization of T and FOXA2 in porcine peri-implantation embryos. Dev Dyn. (2011) 240:890–7. doi: 10.1002/dvdy.22602
72. Idkowiak J, Weisheit G, Plitzner J, Viebahn C. Hypoblast controls mesoderm generation and axial patterning in the gastrulating rabbit embryo. Dev Genes Evol. (2004) 214:591–605. doi: 10.1007/s00427-004-0436-y
73. Alev C, Wu Y, Kasukawa T, Jakt LM, Ueda HR, Sheng G. Transcriptomic landscape of the primitive streak. Development. (2010) 137:2863–74. doi: 10.1242/dev.053462
74. Tam PP, Beddington RS. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development. (1987) 99:109–26. doi: 10.1242/dev.99.1.109
75. Eakin GS, Behringer RR. Diversity of germ layer and axis formation among mammals. Semin Cell Dev Biol. (2004) 15:619–29. doi: 10.1016/j.semcdb.2004.04.008
76. Hue I, Renard JP, Viebahn C. Brachyury is expressed in gastrulating bovine embryos well ahead of implantation. Dev Genes Evol. (2001) 211:157–9. doi: 10.1007/s004270100138
77. Blomberg Le A, Garrett WM, Guillomot M, Miles JR, Sonstegard TS, Van Tassell CP, et al. Transcriptome profiling of the tubular porcine conceptus identifies the differential regulation of growth and developmentally associated genes. Mol Reprod Dev. (2006) 73:1491–502. doi: 10.1002/mrd.20503
78. Talbot NC, Caperna TJ, Edwards JL, Garrett W, Wells KD, Ealy AD. Bovine blastocyst-derived trophectoderm and endoderm cell cultures: interferon tau and transferrin expression as respective in vitro markers. Biol Reprod. (2000) 62:235–47. doi: 10.1095/biolreprod62.2.235
79. Ramos-Ibeas P, Calle A, Pericuesta E, Laguna-Barraza R, Moros-Mora R, Lopera-Vasquez R, et al. An efficient system to establish biopsy-derived trophoblastic cell lines from bovine embryos. Biol Reprod. (2014) 91:15. doi: 10.1095/biolreprod.114.118430
80. Ramos-Ibeas P, Nichols J, Alberio R. States and origins of mammalian embryonic pluripotency in vivo and in a dish. Curr Top Dev Biol. (2018) 128:151–79. doi: 10.1016/bs.ctdb.2017.11.002
81. Brandao DO, Maddox-Hyttel P, Lovendahl P, Rumpf R, Stringfellow D, Callesen H. Post hatching development: a novel system for extended in vitro culture of bovine embryos. Biol Reprod. (2004) 71:2048–55. doi: 10.1095/biolreprod.103.025916
82. Vajta G, Alexopoulos NI, Callesen H. Rapid growth and elongation of bovine blastocysts in vitro in a three-dimensional gel system. Theriogenology. (2004) 62:1253–63. doi: 10.1016/j.theriogenology.2004.01.007
83. Machado GM, Ferreira AR, Pivato I, Fidelis A, Spricigo JF, Paulini F, et al. Post-hatching development of in vitro bovine embryos from day 7 to 14 in vivo versus in vitro. Mol Reprod Dev. (2013) 80:936–47. doi: 10.1002/mrd.22230
84. Bertolini M, Beam SW, Shim H, Bertolini LR, Moyer AL, Famula TR, et al. Growth, development, and gene expression by in vivo- and in vitro-produced day 7 and 16 bovine embryos. Mol Reprod Dev. (2002) 63:318–28. doi: 10.1002/mrd.90015
85. Loureiro B, Block J, Favoreto MG, Carambula S, Pennington KA, Ealy AD, et al. Consequences of conceptus exposure to colony-stimulating factor 2 on survival, elongation, interferon-τ secretion, and gene expression. Reproduction. (2011) 141:617–24. doi: 10.1530/REP-10-0511
86. Block J, Fischer-Brown AE, Rodina TM, Ealy AD, Hansen PJ. The effect of in vitro treatment of bovine embryos with IGF-1 on subsequent development in utero to Day 14 of gestation. Theriogenology. (2007) 68:153–61. doi: 10.1016/j.theriogenology.2007.04.045
87. Fischer-Brown AE, Lindsey BR, Ireland FA, Northey DL, Monson RL, Clark SG, et al. Embryonic disc development and subsequent viability of cattle embryos following culture in two media under two oxygen concentrations. Reprod Fertil Dev. (2004) 16:787–93. doi: 10.1071/RD04026
88. Bertolini M, Mason JB, Beam SW, Carneiro GF, Sween ML, Kominek DJ, et al. Morphology and morphometry of in vivo- and in vitro-produced bovine concepti from early pregnancy to term and association with high birth weights. Theriogenology. (2002) 58:973–94. doi: 10.1016/S0093-691X(02)00935-4
89. Alexopoulos NI, Maddox-Hyttel P, Tveden-Nyborg P, D'Cruz NT, Tecirlioglu TR, Cooney MA, et al. Developmental disparity between in vitro-produced and somatic cell nuclear transfer bovine days 14 and 21 embryos: implications for embryonic loss. Reproduction. (2008) 136:433–45. doi: 10.1530/REP-07-0392
90. Desmet KLJ, Marei WFA, Richard C, Sprangers K, Beemster GTS, Meysman P, et al. Oocyte maturation under lipotoxic conditions induces carryover transcriptomic and functional alterations during post-hatching development of good-quality blastocysts: novel insights from a bovine embryo-transfer model. Hum Reprod. (2020) 35:293–307. doi: 10.1093/humrep/dez248
91. Ealy AD, Wooldridge LK, McCoski SR. BOARD INVITED REVIEW: post-transfer consequences of in vitro-produced embryos in cattle. J Anim Sci. (2019) 97:2555–68. doi: 10.1093/jas/skz116
92. Chavatte-Palmer P, Camous S, Jammes H, Le Cleac'h N, Guillomot M, Lee RS. Review: Placental perturbations induce the developmental abnormalities often observed in bovine somatic cell nuclear transfer. Placenta. (2012) 33(Suppl.):S99–S104. doi: 10.1016/j.placenta.2011.09.012
93. Hall V, Hinrichs K, Lazzari G, Betts DH, Hyttel P. Early embryonic development, assisted reproductive technologies, and pluripotent stem cell biology in domestic mammals. Vet J. (2013) 197:128–42. doi: 10.1016/j.tvjl.2013.05.026
94. Tveden-Nyborg P, Peura TT, Hartwich KM, Walker SK, Maddox-Hyttel P. Morphological characterization of pre- and peri-implantation in vitro cultured, somatic cell nuclear transfer and in vivo derived ovine embryos. Reproduction. (2005) 130:681–94. doi: 10.1530/rep.1.00850
95. Fabreges D, Daniel N, Duranthon V, Peyrieras N. Control of the proportion of inner cells by asymmetric divisions and the ensuing resilience of cloned rabbit embryos. Development. (2018) 145:dev152041. doi: 10.1242/dev.152041
96. Ideta A, Urakawa M, Aoyagi Y, Saeki K. Early development in utero of bovine nuclear transfer embryos using early G1 and G0 phase cells. Cloning Stem Cells. (2007) 9:571–80. doi: 10.1089/clo.2007.0017
97. Rodríguez-Alvarez L, Sharbati J, Sharbati S, Cox JF, Einspanier R, Castro FO. Differential gene expression in bovine elongated (Day 17) embryos produced by somatic cell nucleus transfer and in vitro fertilization. Theriogenology. (2010) 74:45–59. doi: 10.1016/j.theriogenology.2009.12.018
98. Degrelle SA, Jaffrezic F, Campion E, Le Cao KA, Le Bourhis D, Richard C, et al. Uncoupled embryonic and extra-embryonic tissues compromise blastocyst development after somatic cell nuclear transfer. PLoS One. (2012) 7:e38309. doi: 10.1371/journal.pone.0038309
99. Smith CS, Berg DK, Berg M, Pfeffer PL. Nuclear transfer-specific defects are not apparent during the second week of embryogenesis in cattle. Cell Reprogram. (2010) 12:699–707. doi: 10.1089/cell.2010.0040
100. van Leeuwen J, Berg DK, Smith CS, Wells DN, Pfeffer PL. Specific epiblast loss and hypoblast impairment in cattle embryos sensitized to survival signalling by ubiquitous overexpression of the proapoptotic gene BAD. PLoS One. (2014) 9:e96843. doi: 10.1371/journal.pone.0096843
101. Oda M, Shiota K, Tanaka S. Trophoblast cell lineage in cloned mouse embryos. Dev Growth Differ. (2010) 52:285–91. doi: 10.1111/j.1440-169X.2010.01173.x
102. Hirasawa R, Matoba S, Inoue K, Ogura A. Somatic donor cell type correlates with embryonic, but not extra-embryonic, gene expression in postimplantation cloned embryos. PLoS One. (2013) 8:e76422. doi: 10.1371/journal.pone.0076422
103. Ealy AD, Drost M, Hansen PJ. Developmental changes in embryonic resistance to adverse effects of maternal heat stress in cows. J Dairy Sci. (1993) 76:2899–905. doi: 10.3168/jds.S0022-0302(93)77629-8
104. Looney CR, Lindsey BR, Gonseth CL, Johnson DL. COMMERCIAL ASPECTS OF OOCYTE RETRIEVAL AND IN-VITRO FERTILIZATION (IVF) FOR EMBRYO PRODUCTION IN PROBLEM COWS. Theriogenology. (1994) 41:67–72. doi: 10.1016/S0093-691X(05)80050-0
105. Block J, Bonilla L, Hansen PJ. Efficacy of in vitro embryo transfer in lactating dairy cows using fresh or vitrified embryos produced in a novel embryo culture medium. J Dairy Sci. (2010) 93:5234–42. doi: 10.3168/jds.2010-3443
107. Ramos-Ibeas P, Heras S, Gómez-Redondo I, Planells B, Fernández-González R, Pericuesta E, et al. Embryo responses to stress induced by assisted reproductive technologies. Mol Reprod Dev. (2019) 86:1292–306. doi: 10.1002/mrd.23119
108. Lucy MC. Reproductive loss in high-producing dairy cattle: where will it end? J Dairy Sci. (2001) 84:1277–93. doi: 10.3168/jds.S0022-0302(01)70158-0
109. Wiltbank MC, Baez GM, Garcia-Guerra A, Toledo MZ, Monteiro PL, Melo LF, et al. Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology. (2016) 86:239–53. doi: 10.1016/j.theriogenology.2016.04.037
110. Gatea AO, Smith MF, Pohler KG, Egen T, Pereira MHC, Vasconselos JLM, et al. The ability to predict pregnancy loss in cattle with ELISAs that detect pregnancy associated glycoproteins is antibody dependent. Theriogenology. (2018) 108:269–76. doi: 10.1016/j.theriogenology.2017.12.021
111. Pohler KG, Green JA, Geary TW, Peres RF, Pereira MH, Vasconcelos JL, et al. Predicting embryo presence and viability. Adv Anat Embryol Cell Biol. (2015) 216:253–70. doi: 10.1007/978-3-319-15856-3_13
112. Estrada-Cortes E, Ortiz WG, Chebel RC, Jannaman EA, Moss JI, de Castro FC, et al. Embryo and cow factors affecting pregnancy per embryo transfer for multiple-service, lactating Holstein recipients. Transl Anim Sci. (2019) 3:60–5. doi: 10.1093/tas/txz009
113. Geary TW, Burns GW, Moraes JG, Moss JI, Denicol AC, Dobbs KB, et al. Identification of beef heifers with superior uterine capacity for pregnancy. Biol Reprod. (2016) 95:47. doi: 10.1095/biolreprod.116.141390
114. Stringfellow D, Givens M. Manual of the International Embryo Transfer Society (IETS), 4th Edn. Champaign: IETS (2010).
115. Farin PW, Slenning BD, Britt JH. Estimates of pregnancy outcomes based on selection of bovine embryos produced in vivo or in vitro. Theriogenology. (1999) 52:659–70. doi: 10.1016/S0093-691X(99)00160-0
116. Hasler JF, Henderson WB, Hurtgen PJ, Jin ZQ, McCauley AD, Mower SA, et al. PRODUCTION, FREEZING AND TRANSFER OF BOVINE IVF EMBRYOS AND SUBSEQUENT CALVING RESULTS. Theriogenology. (1995) 43:141–52. doi: 10.1016/0093-691X(94)00020-U
117. Hasler JF. Factors affecting frozen and fresh embryo transfer pregnancy rates in cattle. Theriogenology. (2001) 56:1401–15. doi: 10.1016/S0093-691X(01)00643-4
118. Chebel RC, Demétrio DG, Metzger J. Factors affecting success of embryo collection and transfer in large dairy herds. Theriogenology. (2008) 69:98–106. doi: 10.1016/j.theriogenology.2007.09.008
119. Hansen PJ. The incompletely fulfilled promise of embryo transfer in cattle-why aren't pregnancy rates greater and what can we do about it? J Anim Sci. (2020) 98:skaa288. doi: 10.1093/jas/skaa288
120. Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. (1998) 95:379–91. doi: 10.1016/S0092-8674(00)81769-9
121. Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. (2003) 113:631–42. doi: 10.1016/S0092-8674(03)00393-3
122. Chazaud C, Yamanaka Y, Pawson T, Rossant J. Early lineage segregation between epiblast and primitive endoderm in mouse blastocysts through the Grb2-MAPK pathway. Dev Cell. (2006) 10:615–24. doi: 10.1016/j.devcel.2006.02.020
123. Kang M, Piliszek A, Artus J, Hadjantonakis AK. FGF4 is required for lineage restriction and salt-and-pepper distribution of primitive endoderm factors but not their initial expression in the mouse. Development. (2013) 140:267–79. doi: 10.1242/dev.084996
124. Piliszek A, Grabarek JB, Frankenberg SR, Plusa B. Cell fate in animal and human blastocysts and the determination of viability. Mol Hum Reprod. (2016) 22:681–90. doi: 10.1093/molehr/gaw002
125. Fouladi-Nashta AA, Alberio R, Kafi M, Nicholas B, Campbell KH, Webb R. Differential staining combined with TUNEL labelling to detect apoptosis in preimplantation bovine embryos. Reprod Biomed Online. (2005) 10:497–502. doi: 10.1016/S1472-6483(10)60827-9
126. Thouas GA, Korfiatis NA, French AJ, Jones GM, Trounson AO. Simplified technique for differential staining of inner cell mass and trophectoderm cells of mouse and bovine blastocysts. Reprod Biomed Online. (2001) 3:25–9. doi: 10.1016/S1472-6483(10)61960-8
127. Deng Q, Ramskold D, Reinius B, Sandberg R. Single-cell RNA-seq reveals dynamic, random monoallelic gene expression in mammalian cells. Science. (2014) 343:193–6. doi: 10.1126/science.1245316
128. Lavagi I, Krebs S, Simmet K, Beck A, Zakhartchenko V, Wolf E, et al. Single-cell RNA sequencing reveals developmental heterogeneity of blastomeres during major genome activation in bovine embryos. Sci Rep. (2018) 8:4071. doi: 10.1038/s41598-018-22248-2
129. du Puy L, Lopes SM, Haagsman HP, Roelen BA. Analysis of co-expression of OCT4, NANOG and SOX2 in pluripotent cells of the porcine embryo, in vivo and in vitro. Theriogenology. (2011) 75:513–26. doi: 10.1016/j.theriogenology.2010.09.019
130. Cao S, Han J, Wu J, Li Q, Liu S, Zhang W, et al. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genomics. (2014) 15:4. doi: 10.1186/1471-2164-15-4
131. Meng F, Forrester-Gauntlett B, Turner P, Henderson H, Oback B. Signal inhibition reveals JAK/STAT3 pathway as critical for bovine inner cell mass development. Biol Reprod. (2015) 93:132. doi: 10.1095/biolreprod.115.134254
132. Rodriguez A, Allegrucci C, Alberio R. Modulation of pluripotency in the porcine embryo and iPS cells. PLoS One. (2012) 7:e49079. doi: 10.1371/journal.pone.0049079
133. van Eijk MJ, van Rooijen MA, Modina S, Scesi L, Folkers G, van Tol HT, et al. Molecular cloning, genetic mapping, and developmental expression of bovine POU5F1. Biol Reprod. (1999) 60:1093–103. doi: 10.1095/biolreprod60.5.1093
134. Khan DR, Dube D, Gall L, Peynot N, Ruffini S, Laffont L, et al. Expression of pluripotency master regulators during two key developmental transitions: EGA and early lineage specification in the bovine embryo. PLoS One. (2012) 7:e34110. doi: 10.1371/journal.pone.0034110
135. Liu S, Bou G, Sun R, Guo S, Xue B, Wei R, et al. Sox2 is the faithful marker for pluripotency in pig: evidence from embryonic studies. Dev Dyn. (2015) 244:619–27. doi: 10.1002/dvdy.24248
136. Lonergan P, Woods A, Fair T, Carter F, Rizos D, Ward F, et al. Effect of embryo source and recipient progesterone environment on embryo development in cattle. Reprod Fertil Dev. (2007) 19:861–8. doi: 10.1071/RD07089
137. Barnwell CV, Farin PW, Whisnant CS, Alexander JE, Farin CE. Maternal serum progesterone concentration and early conceptus development of bovine embryos produced in vivo or in vitro. Domest Anim Endocrinol. (2015) 52:75–81. doi: 10.1016/j.domaniend.2015.03.004
138. Clemente M, de La Fuente J, Fair T, Al Naib A, Gutierrez-Adan A, Roche JF, et al. Progesterone and conceptus elongation in cattle: a direct effect on the embryo or an indirect effect via the endometrium? Reproduction. (2009) 138:507–17. doi: 10.1530/REP-09-0152
139. Barnwell CV, Farin PW, Ashwell CM, Farmer WT, Galphin SP Jr., et al. Differences in mRNA populations of short and long bovine conceptuses on Day 15 of gestation. Mol Reprod Dev. (2016) 83:424–41. doi: 10.1002/mrd.22640
140. Deglincerti A, Croft GF, Pietila LN, Zernicka-Goetz M, Siggia ED, Brivanlou AH. Self-organization of the in vitro attached human embryo. Nature. (2016) 533:251–4. doi: 10.1038/nature17948
141. Shahbazi MN, Jedrusik A, Vuoristo S, Recher G, Hupalowska A, Bolton V, et al. Self-organization of the human embryo in the absence of maternal tissues. Nat Cell Biol. (2016) 18:700–8. doi: 10.1038/ncb3347
142. Bedzhov I, Leung CY, Bialecka M, Zernicka-Goetz M. In vitro culture of mouse blastocysts beyond the implantation stages. Nat Protoc. (2014) 9:2732–9. doi: 10.1038/nprot.2014.186
143. Xiang L, Yin Y, Zheng Y, Ma Y, Li Y, Zhao Z, et al. A developmental landscape of 3D-cultured human pre-gastrulation embryos. Nature. (2020) 577:537–42. doi: 10.1038/s41586-019-1875-y
144. Zhou F, Wang R, Yuan P, Ren Y, Mao Y, Li R, et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature. (2019) 572:660–4. doi: 10.1038/s41586-019-1500-0
145. Stankova V, Tsikolia N, Viebahn C. Rho kinase activity controls directional cell movements during primitive streak formation in the rabbit embryo. Development. (2015) 142:92–8. doi: 10.1242/dev.111583
146. Puschel B, Bitzer E, Viebahn C. Live rabbit embryo culture. Cold Spring Harb Protoc. (2010) 2010:pdb prot5352. doi: 10.1101/pdb.prot5352
147. Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. (1993) 35:567–76. doi: 10.1002/jnr.490350513
Keywords: embryo quality, embryo transfer, developmental biology, lineage markers, viability, conceptus elongation, assisted reproductive technologies, post-hatching embryo culture
Citation: Pérez-Gómez A, González-Brusi L, Bermejo-Álvarez P and Ramos-Ibeas P (2021) Lineage Differentiation Markers as a Proxy for Embryo Viability in Farm Ungulates. Front. Vet. Sci. 8:680539. doi: 10.3389/fvets.2021.680539
Received: 14 March 2021; Accepted: 24 May 2021;
Published: 15 June 2021.
Edited by:
Cristina Alicia Martinez, Linköping University, SwedenReviewed by:
Deqiang Miao, Washington State University, United StatesCopyright © 2021 Pérez-Gómez, González-Brusi, Bermejo-Álvarez and Ramos-Ibeas. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pablo Bermejo-Álvarez, YmVybWVqby5wYWJsb0BpbmlhLmVz; Priscila Ramos-Ibeas, cmFtb3MucHJpc2NpbGFAaW5pYS5lcw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.