- 1Warnell School of Forestry and Natural Resources, University of Georgia, Athens, GA, United States
- 2Department of Population Health, Southeastern Cooperative Wildlife Disease Study, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
- 3Department of Population Health, Poultry Diagnostic and Research Center, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
- 4Department of Biological Sciences, Eastern Kentucky University, Richmond, KY, United States
- 5Department of Geography, University of Georgia, Athens, GA, United States
- 6Davee Center for Epidemiology and Endocrinology and the Urban Wildlife Institute, Lincoln Park Zoo, Chicago, IL, United States
- 7Athens Veterinary Diagnostic Laboratory, College of Veterinary Medicine, University of Georgia, Athens, GA, United States
- 8Enteric Diseases Laboratory Branch, Centers for Disease Control and Prevention, Atlanta, GA, United States
- 9Department of Environmental Health Science, College of Public Health, University of Georgia, Athens, GA, United States
Reptile-associated human salmonellosis cases have increased recently in the United States. It is not uncommon to find healthy chelonians shedding Salmonella enterica. The rate and frequency of bacterial shedding are not fully understood, and most studies have focused on captive vs. free-living chelonians and often in relation to an outbreak. Their ecology and significance as sentinels are important to understanding Salmonella transmission. In 2012–2013, Salmonella prevalence was determined for free-living aquatic turtles in man-made ponds in Clarke and Oconee Counties, in northern Georgia (USA) and the correlation between species, basking ecology, demographics (age/sex), season, or landcover with prevalence was assessed. The genetic relatedness between turtle and archived, human isolates, as well as, other archived animal and water isolates reported from this study area was examined. Salmonella was isolated from 45 of 194 turtles (23.2%, range 14–100%) across six species. Prevalence was higher in juveniles (36%) than adults (20%), higher in females (33%) than males (18%), and higher in bottom-dwelling species (31%; common and loggerhead musk turtles, common snapping turtles) than basking species (15%; sliders, painted turtles). Salmonella prevalence decreased as forest cover, canopy cover, and distance from roads increased. Prevalence was also higher in low-density, residential areas that have 20–49% impervious surface. A total of 9 different serovars of two subspecies were isolated including 3 S. enterica subsp. arizonae and 44 S. enterica subsp. enterica (two turtles had two serotypes isolated from each). Among the S. enterica serovars, Montevideo (n = 13) and Rubislaw (n = 11) were predominant. Salmonella serovars Muenchen, Newport, Mississippi, Inverness, Brazil, and Paratyphi B. var L(+) tartrate positive (Java) were also isolated. Importantly, 85% of the turtle isolates matched pulsed-field gel electrophoresis patterns of human isolates, including those reported from Georgia. Collectively, these results suggest that turtles accumulate Salmonella present in water bodies, and they may be effective sentinels of environmental contamination. Ultimately, the Salmonella prevalence rates in wild aquatic turtles, especially those strains shared with humans, highlight a significant public health concern.
Introduction
Salmonella enterica infections are a significant public health threat, responsible for over 93 million annual illnesses worldwide (1). In the United States alone, over 1 million cases of salmonellosis and 600 deaths are reported annually (2). Most cases of human salmonellosis are caused by food-borne Salmonella strains associated with contaminated meat, eggs, or produce. Produce has become a significant source of foodborne outbreaks associated with Salmonella (3–5), accounting for half the outbreaks and one quarter of the illnesses reported for the U.S. in 2016 alone (4). Water is central to growth and processing of fruits, vegetables, and nuts; and it is the most likely source of product contamination with Salmonella (6).
There has also been a significant number of human cases of salmonellosis linked to animal (4, 7–13) and environmental exposure (14–16) and a geographic disparity in reported cases of salmonellosis in the United States (17–19). Georgia has the highest annual Salmonella prevalence among the states; and within the state, the southern Coastal Plain has the highest incidence (20). There is a link between human cases and the Little River watershed in South Georgia, where 46% of Salmonella isolated from the Little River matched human isolates by pulsed-field gel electrophoresis (PFGE) (21). Understanding disease transmission in this region is difficult due to Salmonella strain diversity, its low abundance in water, and seasonal and weather-related fluctuations in its prevalence (14, 20, 21). Wildlife captured in the Little River watershed harbor some of the same Salmonella strains present in the river. However, only half of the raccoons and opossums sampled in this region possess the same Salmonella strains present in the CDC PulseNet database of human isolates (21). There are significant logistical challenges associated with sampling wildlife populations. Might an aquatic species prove a better sentinel for monitoring pathogenic Salmonella strains in watersheds or irrigation ponds?
For example, the American White Ibis (Eudocimus albus), an abundant aquatic bird, forms large nesting colonies in natural wetlands but have become habituated to living in agricultural and urban areas. Seventeen percent of ibis sampled in South Florida harbor Salmonella and 44% of these isolates match human isolates in the CDC-PulseNet database. Most notable was the spatial and temporal overlap in the isolation of these pathogenic strains with human cases of salmonellosis in South Florida. While this avian species is less likely to interact with humans and directly transmit Salmonella to people (11), it is a likely sentinel of environmental contamination (22). As a sentinel, the White Ibis is limited by its geographic distribution. Aquatic turtles, on the other hand, have a wide distribution across many different landscapes and habitat types, and often thrive in anthropogenic settings. Among wildlife, they are easy to capture, handle and sample.
Reptile-associated salmonellosis was a serious health problem in the 1960 and 1970s but was ameliorated, particularly in children, with public education and the 1975 federal ban of the sale of turtles <4 inches in carapacial length. In recent years, reptile-associated salmonellosis has increased again to comprise ~6% (74,000) of salmonellosis cases in the United States per year (24, 25). Most of these patients report contact with pet turtles (24, 26) and turtles have been responsible for several outbreaks of salmonellosis in the United States (8, 27, 28). Such outbreaks are typically associated with small turtles sold by street vendors and pet stores, despite the ban on their trade (29). Reptile-associated salmonellosis is most common in children (30, 31), and more likely to require hospitalization than other types of salmonellosis in other age groups (32). Understanding of turtle-associated salmonellosis primarily stems from epidemiological studies following outbreaks with reptile-associated serotypes (28) and surveys of captive turtles (33, 34). Several serotypes have been reported from turtles including the S. enterica serovars Muenchen, Typhimurium, Newport, Pomona, Litchfield and Paratyphi B. var L(+) tartrate positive (formerly Java) (8, 24–30). However, unique serovars are still being reported, e.g., Salmonella Agbeni (35), and there are many epidemiological gaps in understanding Salmonella carriage in turtles.
Salmonella enterica is routinely isolated from healthy, asymptomatic wild and pet chelonians (turtles and tortoises) and it is generally considered a normal component of their microbiota. The rate and frequency at which turtles shed Salmonella, and the conditions that may promote shedding, are not fully understood (36, 37). For commercial or pet turtles, hygiene, crowding, stress and other environmental factors may play a role in Salmonella shedding (38). This may, in part, explain the variability in past prevalence studies and may facilitate the role of turtles as Salmonella reservoirs for humans. Although some studies report low prevalence of Salmonella in free-ranging turtles, there is evidence that the prevalence can be higher in free-living turtles relative to captive turtles (39–42), likely influenced by species natural history (e.g., foraging behavior and habitat use), habitat quality, and other factors (e.g., landscape) that are largely unexplored. Several studies have investigated carriage of Salmonella enterica in free-living chelonians, with reported prevalence rates varying considerably depending on species, location and sampling methodology (40–45). To date, only a few studies have investigated the link between Salmonella prevalence in free-living turtles and human illness (7, 39, 46–48).
A better understanding of the public health risks of environmental exposure and the role of free-living turtles in transmission is especially important in regions where human salmonellosis is particularly high, such as the southeastern United States (19, 49). Of particular relevance, this region holds 10% of the world's aquatic turtle biodiversity (50). Aquatic turtles are ubiquitous throughout urban, suburban, and natural environments. In urban environments, they readily colonize ponds contaminated with runoff, often at high densities. Therefore, turtles are hypothesized to be good indicators of environmental contamination with Salmonella.
In 2012–2013, aquatic turtles from man-made ponds in north-central Georgia (Clarke and Oconee Counties) were surveyed for Salmonella. Salmonella prevalence by turtle species, basking ecology, demographics (age/sex), season, and landscape variables were investigated. Given the paucity of information regarding how various factors influence Salmonella prevalence in wild turtle populations, landscape variables related to water quality were examined to identify any associations with prevalence. Additionally, to better understand the role of wild turtles in non-foodborne human salmonellosis cases in Georgia, PFGE patterns were compared among Salmonella isolates recovered from wild turtles, archived animal, water, and human isolates.
Materials and Methods
Study Sites, Geographic Description, Animal Capture, and Sampling
Turtles were captured from April 2012 to June 2013 at eight small man-made ponds. Seven ponds were in Clarke County, Georgia (Algae Pond, Sisters Pond, Lake Chapman, UGA Golf Course, Milledge Pond, County Park, and Recreational Lake). Two of these ponds, Algae Pond and Sisters Pond, were located within the Whitehall Experimental Forest; the others were public or private ponds located on separate properties (Figure 1). The remaining pond was in Oconee County, Georgia at a private school. All water bodies were within the Oconee River watershed.
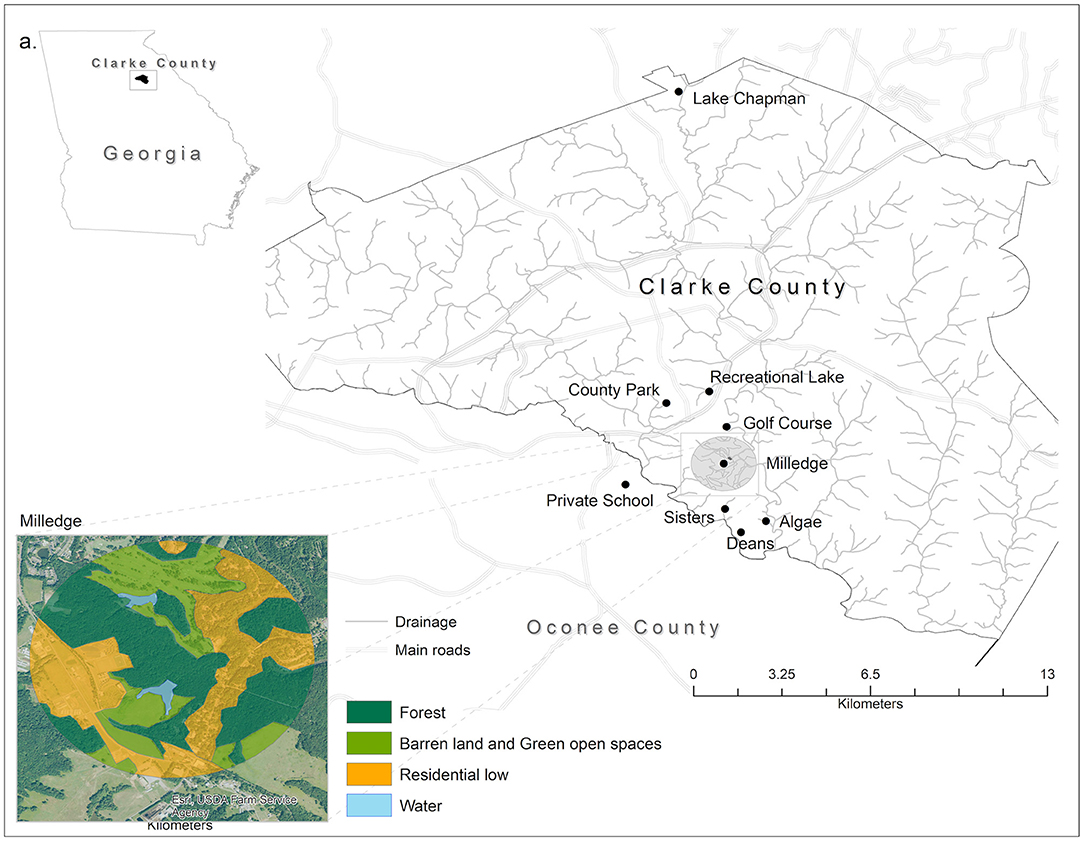
Figure 1. Map of the eight capture sites where turtles were captured for Salmonella testing. Seven sites were in Athens-Clarke County, Georgia (Algae Pond, Sisters Pond, Lake Chapman, UGA Golf Course, Milledge Pond, County Park, and Recreational Lake) and one was in Oconee County, Georgia (Private School). Inset shows and example of the land use categories surrounding each sample site. Land cover data are derived from the National Agriculture Imagery Program (NAIP, 2016) by the USDA's Farm Service Agency (FSA). Classes' denomination is based on the National Land Cover Classification. ArcGIS 10.5 licensed to Salisbury University, MD was used to extract the land cover features. All vector data used can be found freely available at TIGER from the U.S. Census Bureau Database.
Turtles were captured using standard hoop traps (Memphis Net & Twine, Inc., Memphis, TN), baited with oil-packed sardines or herring and placed such that turtles could surface to breathe. Within each pond, traps were positioned in locations that were predicted to be the most successful: e.g., in areas that were shaded or more densely vegetated, or near substrates suitable for turtle basking such as exposed logs or rocks. Traps were placed in these locations for 2–4 days at a time, checked daily, and rebaited after 2 or 3 days. Once in hand, turtles were identified to species, measured (utilizing standard morphometrics for chelonians), and weighed. The age and sex of each individual was determined by species-specific morphological characteristics as described by Buhlmann et al. (51). Turtles were individually held in clean plastic containers until they defecated or overnight (maximum time needed for all to defecate). Feces were collected with sterile plastic pipettes and ~1 g of feces was suspended in 10 ml of dulcitol selenite (Difco; Detroit, MI). Turtles were subsequently released at the capture site. All containers were cleaned with soap and water and disinfected with a 10% bleach solution before they were reused. All fecal samples in selenite were submitted on the same day of collection to the Athens Diagnostic Laboratory (Athens, GA) for culture. All animal capture and animal handling procedures were approved by the University of Georgia's Institutional Animal Care and Use Committee (AUP# A2010 10-186).
Salmonella Isolation and PFGE Molecular Characterization
Feces in selenite was incubated overnight at 42°C for Salmonella enrichment (52). A 10 μl loopful of the overnight enrichment was plated onto xylose lysine deoxycholate (XLD) and brilliant green (BG) plates (Remel Inc., Lenexa, KS) and incubated overnight at 37°C as previously described (10, 53). H2S-positive, black colonies were picked and subcultured onto blood agar plates (tryptic soy agar with 5% sheep blood). Final Salmonella confirmation was determined with the following tests on a single isolated colony: citrate, triple sugar iron (TSI), and motility-indole-ornithine media (Beckton and Dickson, Franklin Lakes, NJ); and a whole-cell agglutination test using Salmonella-specific poly A-I and Vi antiserum (Fisher Scientific, Pittsburgh, PA). Microbial identification as Salmonella was based on possessing all of the following criteria. Salmonella grows on TSI slant producing a red slant, yellow/black (H2S-production) butt, and gas. In addition, Salmonella is motile, citrate-positive, indole and ornithine negative and agglutinates with poly A-I/Vi antiserum (54). Samples were considered culture negative if no black or pink colonies were observed on XLD or BG sections, respectively. A delayed-secondary enrichment was done for samples that were culture negative after the primary enrichment and initial plating on XLD and BG. A 10 μl loopful of the secondary enrichment in selenite overnight was plated onto XLD and BG plates. Salmonella identification of suspect colonies was confirmed as previously stated. Isolates were forwarded to the National Veterinary Service Laboratory (NVSL) at Ames, Iowa, for definitive Salmonella serotyping.
At the time of sample submission, PFGEs were still the primary method utilized by the CDC to determine genetic relatedness among Salmonella isolates by comparison with human isolates in the CDC PulseNet USA national database. Agarose plugs and PFGE conditions were performed as previously described (53, 55–57). Electrophoresis was done using the CHEF DR II electrophoresis unit (Bio-Rad; Hercules, CA), with 0.5X Tris-borate-EDTA buffer (Sigma-Aldrich; St. Louis, MO); 6 V/cm with pulse times 2.25–63.85 s at 14°C for 15.5 h. A master database of Salmonella PFGE patterns in BioNumerics (Applied Maths; Austin, TX) contains over 1,000 PFGE entries for Salmonella isolated from water and various animal species (10, 11, 21). Comparisons were made between PFGE patterns in BioNumerics using Dice coefficient and unweighted pair group method of arithmetic averages (UPGMA) clustering. Clusters were based on a 75% similarity cut-off (21). Turtle isolates were also compared to archived isolates previously acquired from animal and water samples from the Oconee River watershed (21).
Landscape Data
All landscape data were public and freely available on government databases. The geographic coordinates of each pond were collected using a GPS hand device during the turtle captures. From each GPS location, a buffer of 1 km was established. Land cover data was extracted from the National Agriculture Imagery Program (NAIP-USDA) and was classified based on the National Land Cover Classification system (58), applied to the state level. The NAIP ortho-corrected imagery was used because the high spatial resolution of NAIP was suitable for a more precise land cover classification at 1 km around the ponds. NAIP imagery was classified based on the national land cover classification system (United States Geological Survey) (58). Within each 1 km buffer, the summarized land cover classes accounted for 100% of all classes surrounding that pond. For instance, the Low Intensity Urban areas were defined as 20–49% of impervious surfaces, which most commonly included single-family housing units (59). The land cover classifications utilized and the percentages of each class per pond are summarized on Supplementary Material. The National Road System data were used to measure the pond distances to the road types. Road types and human population were collected at the Topologically Integrated Geographic Encoding and Referencing database—TIGER, U.S. Census Bureau Database. ArcMap 10.6 (60) was used to extract the land cover, measure the distance from pond to features and measure the areas of each land cover class.
Statistical Analyses
Salmonella prevalence rates in turtles were calculated as the number of individual turtles with a Salmonella shedding status of positive divided by the total number of turtles captured and tested (all turtles captured regardless of Salmonella status). Salmonella prevalence rates were analyzed using generalized linear mixed effects models in R version 3.3.1 (61) with the lme4 package (62). The response variable, Salmonella prevalence, was modeled using a binomial error distribution, and all models included the pond from which turtles were sampled as a random effect. Likelihood ratio tests were used to test the significance of the following predictor variables for Salmonella prevalence rates: turtle species, turtle basking ecology (basking vs. non-basking species), pond area size, distance (meters) of pond to the closest highway or street (tested separately), and the percentages of (a) canopy cover over pond, (b) forest cover over pond, (c) low density residential land around each pond, (d) water, and green open space. All percentages were calculated at the 1 km landscape around the pond. The likelihood ratio test uses a chi-square distribution to determine the contribution of a single factor by comparing the fit of the model with and without the factor of interest (63). Chi-square analysis was used to examine differences in Salmonella prevalence between age and sex classes.
A candidate set of 11 single-factor and two-factor models were tested using an AICc model selection approach to understand which factors at the local and landscape scale influenced Salmonella prevalence rates in turtles (Table 1). The candidate set included a subset of the above listed predictor variables based on a priori hypotheses about the factors posited to influence Salmonella in turtles. The null model, including only the random effect of pond, was included in the candidate set.
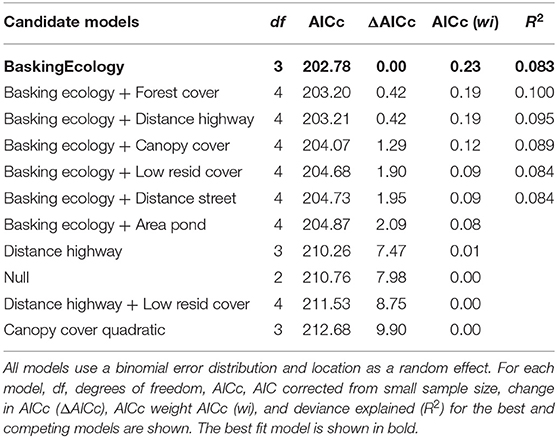
Table 1. Selection parameters of candidate generalized linear mixed models explaining variation in Salmonella prevalence rates of turtles.
Results
Fecal samples were collected from 194 individual wild turtles representing six species: common snapping turtle, Chelydra serpentina (CHSER, n = 20); common musk turtle, Sternotherus odoratus (STODO, n = 48); Eastern painted turtle, Chrysemys picta (CHPIC, n = 65); yellow-bellied slider, Trachemys scripta scripta (TRSCR, n = 50); spiny softshell turtle, Apalone spinifera (APSPI, n = 4); and loggerhead musk turtle, Sternotherus minor (STMIN, n = 7). In total, Salmonella was isolated from 45 of the sampled turtles (23.2%). Salmonella prevalence between species ranged from 14 to 100%: snapping (45 ± 0.25, 95%CI), common musk (22.9 ± 0.12%), painted (16.9 ± 0.09%), slider (14 ± 0.10%), softshell (100 ± 0.00%), and loggerhead musk (42.8 ± 0.50%; Figure 2; Chisq = 23.20; p < 0.001). Salmonella prevalence in softshell turtles was significantly higher than in painted, common musk, and sliders, and the prevalence in painted turtles was significantly lower than in softshell and snapping turtles (Figure 2).
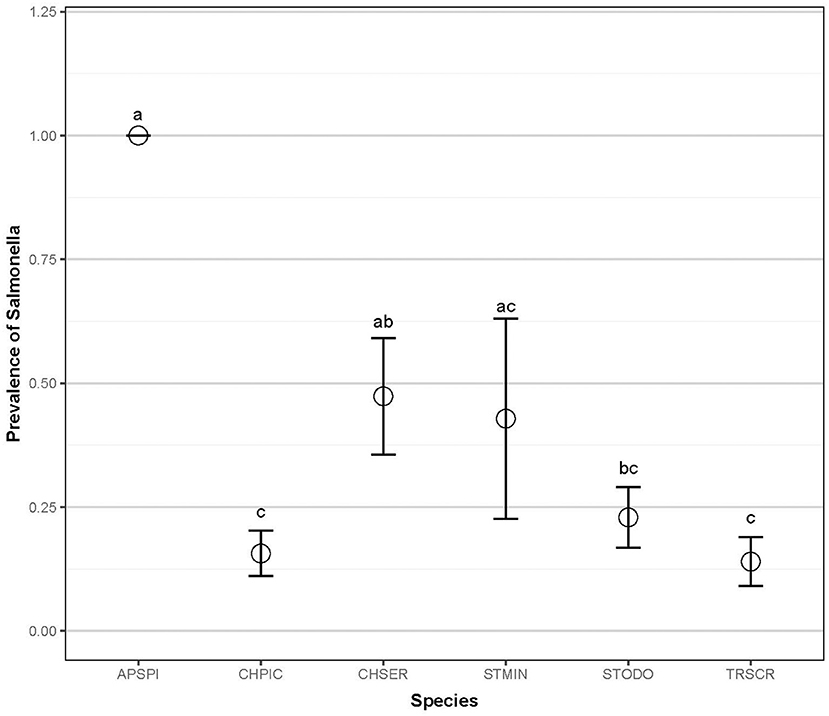
Figure 2. Salmonella prevalence of six aquatic turtle species (2012–2013). Letters indicate statistical significance. Common snapping turtle, Chelydra serpentina (CHSER, n = 20); common musk turtle, Sternotherus odoratus (STODO, n = 48); Eastern painted turtle, Chrysemys picta (CHPIC, n = 65); yellow-bellied slider, Trachemys scripta scripta (TRSCR, n = 50); Spiny softshell turtle, Apalone spinifera (APSPI, n = 4); and loggerhead musk turtle, Sternotherus minor (STMIN, n = 7).
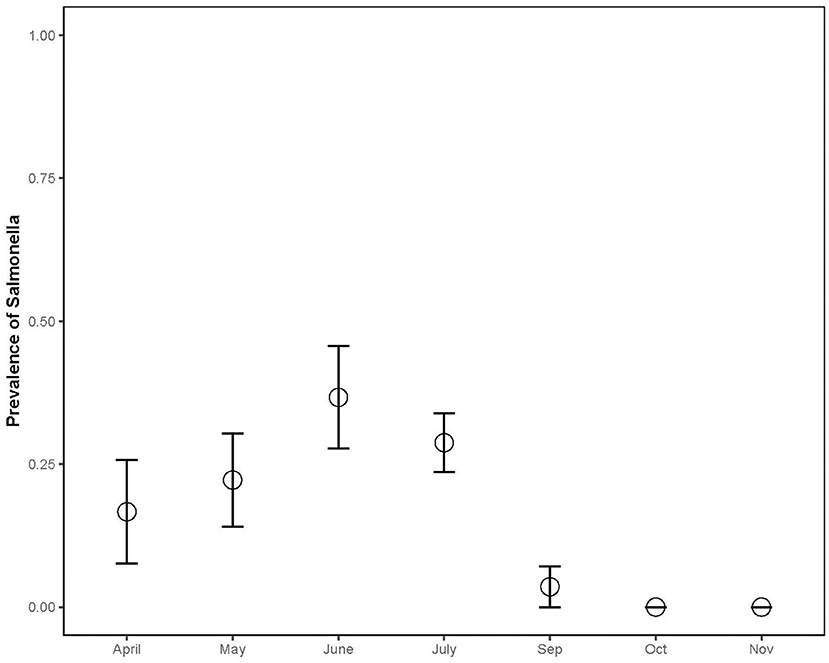
The prevalence in juveniles (n = 25, 36%) was significantly higher than in adults (n = 124, 20%) (Chisq = 5.884, p = 0.0153), and the prevalence in bottom-dwelling species (n = 78, 31%) (common and loggerhead musk and snapping turtles) was significantly higher than in basking species (n = 114, 15%; Chisq = 10.04; p = 0.001; Figure 3) (sliders and painted turtles). The prevalence for females (n = 63, 33%) was significantly higher than males (n = 93, 18%) (Chisq = 4.006, p = 0.045). The combined prevalence rates of Salmonella during the capture months for both sampling years were as follows: April (n = 20, 17%), May (n = 27, 22%), June (n = 29, 37%), July (n = 79, 29%), September (n = 28, 4%), October (n = 7, 0%), and November (n = 2, 0%) (Chisq = 17.83; p = 0.007; Figure 4). Post-hoc pairwise comparisons did not find significant differences between months, but prevalence did increase with increasing ambient temperatures, inferred from seasonal monthly data.
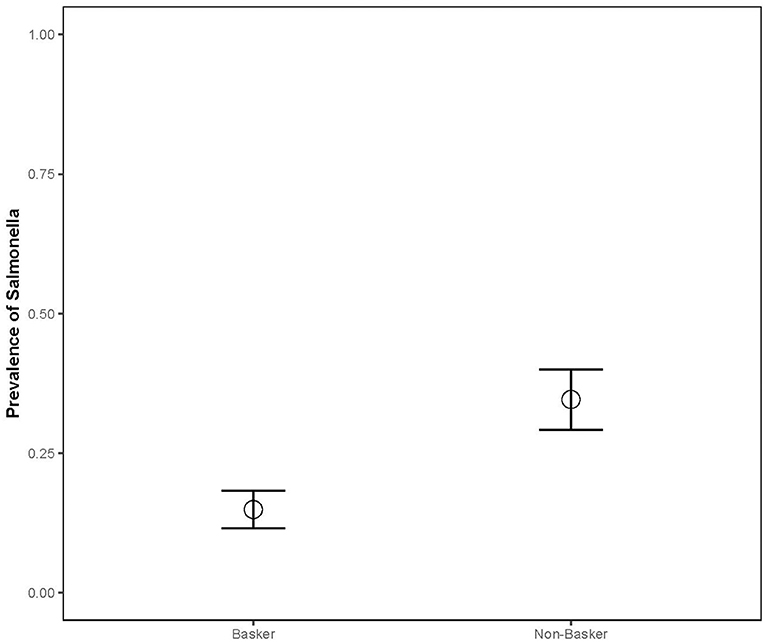
Figure 3. Salmonella prevalence of aquatic turtle species varied based on their basking behavior (2012–2013). CHPIC and TRSCR were classified as baskers, while CHSER, STODO, and STMIN were classified as non-baskers. APSPI were not included in basking analysis due to sample size and inconsistent basking behavior.
The best model explaining Salmonella prevalence rates in turtles (23% support) included only the single predictor of “basking ecology” (Table 1). The candidate set of models contained additional supported models that included basking class, plus a land cover variable. Two of these competing models had 19% support; one model included the effect of basking class and percent forest cover in the landscape, and the other included the effect of basking class and the distance of the pond from the closest highway. In both of these models, landscape variables had a negative effect on the prevalence of Salmonella in turtle populations, in other words, as forest cover increased in the landscape, there was a lower prevalence of Salmonella in the turtles, and the closer a pond was to the highway, the higher the prevalence of Salmonella. A third competing model with 12% support included the effect of basking class and the percent canopy cover over the pond sampled. As canopy cover increased over the pond, the prevalence of Salmonella in the turtle population decreased. Finally, two competing models had 9% support; one model included the effect of basking class and percent cover of low-density residential land in the landscape, and the other model included the effect of basking class and the distance of the pond to the closest street. As the percent cover of low-density residential area in the landscape increased, the prevalence of Salmonella in turtle populations increased. In addition, the distance to the closest street had a negative effect on Salmonella prevalence (i.e., the closer the pond was to the street, the higher the Salmonella prevalence).
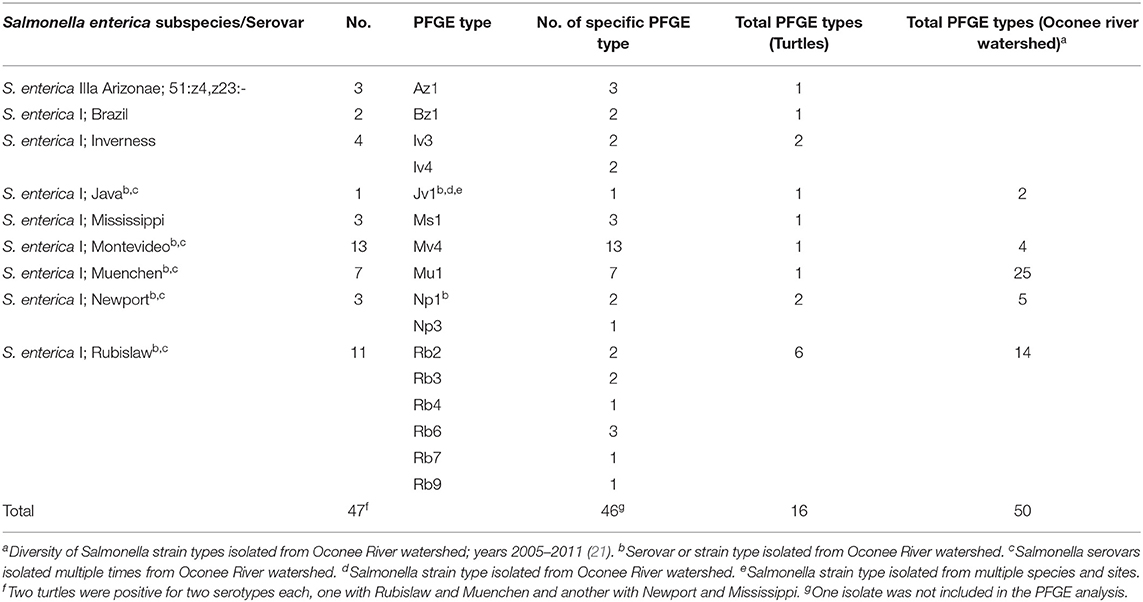
A total of nine different serovars of two subspecies were isolated from turtles including 3 S. enterica subsp. arizonae and 44 S. enterica subsp. enterica (Table 2). Two turtles had two serotypes isolated from each individual, one with Rubislaw and Muenchen and another with Newport and Mississippi. Among the S. enterica serovars, two (Montevideo (n = 13) and Rubislaw (n = 11) were predominant, and fewer numbers of serovars Muenchen, Newport, Mississippi, Inverness, Brazil, and Paratyphi B. var L(+) tartrate positive (Java) were isolated (Table 2). Salmonella enterica IIIa Arizonae, a subspecies commonly isolated from reptiles (64, 65), was only isolated from three turtles (6.7% of the total turtles positive for Salmonella). Five of the nine Salmonella serovars isolated from turtles were previously isolated from wildlife and water in the Oconee River watershed (Table 2) (21). PFGE patterns of Salmonella isolates clustered into 16 types indicating genetic relatedness. These clusters were unique for each serovar, with the exception of S. Inverness which consisted of two distinct PFGE clusters, E and G (Figure 5). Salmonella Rubislaw had the greatest diversity of PFGE types (Table 2). Of the 16 PFGE types found in turtles, five were previously found in animal and water samples from the Oconee watershed. The diversity in PFGE patterns was lower in S. enterica isolated from turtles compared to the same Salmonella serovars in the aforementioned samples from the Oconee River watershed (21, 66): 16 vs. 50, respectively (Table 2). Two Salmonella strain types, Muenchen (Mu1) and Montevideo (Mv4), were the most common PFGE types identified in turtle isolates: 13.3 and 28.9%, respectively.
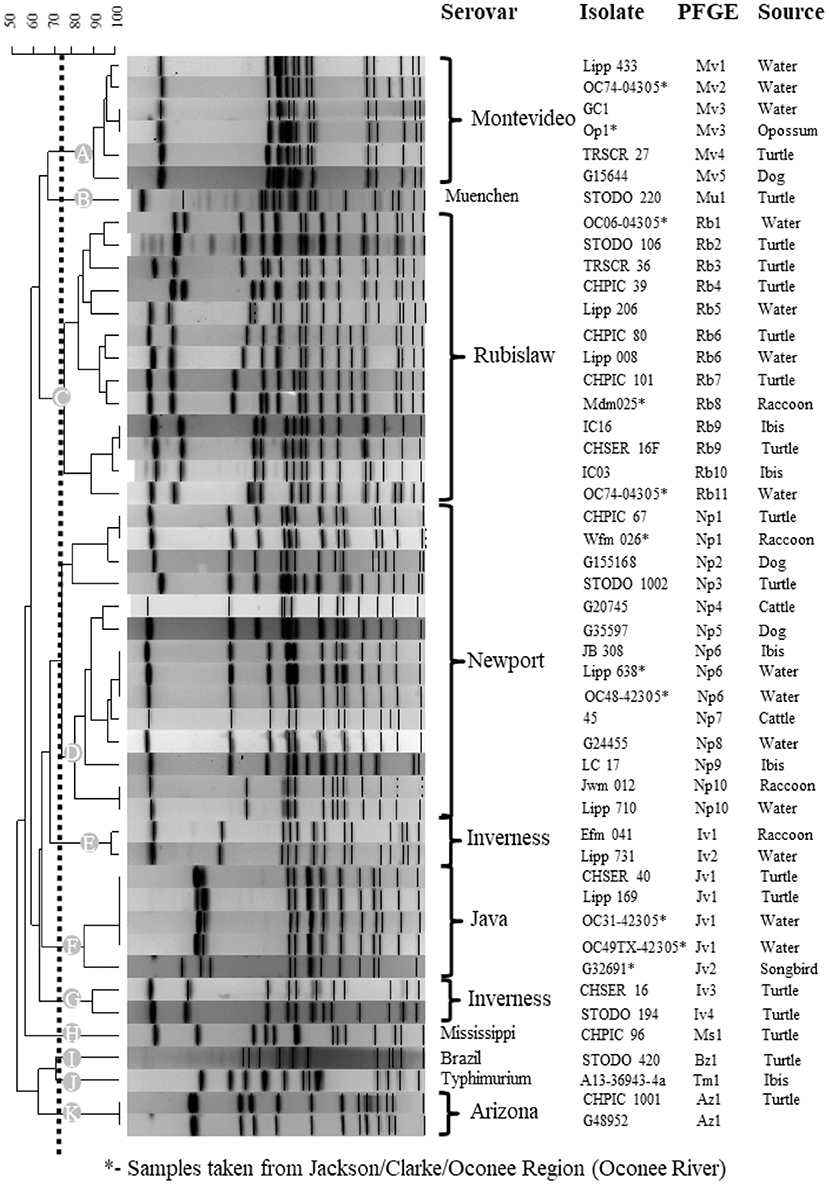
Figure 5. Genetic relatedness of Salmonella enterica isolated from turtles, other animal species and water collected from the Oconee River watershed and Athens-Clarke Georgia. Pulsed gel-electrophoresis (PFGE) was used to determine genetic relatedness among turtle isolates and archived animal and water isolates. A subset of similar or matching Salmonella PFGE patterns (>75%) are presented for the over 1,000 PFGE entries in the BioNumerics database. Level of similarity was calculated by the band-based Dice coefficient. Clustering of samples was performed using the unweighted pair-group method with arithmetic averaging (UPGMA) to generate this dendrogram. Dotted line illustrates the 75% cutoff used to identify similar and matching PFGE patterns and the 11 clusters (A-K) identified in this analysis. * indicates PFGE patterns for Salmonella isolates collected from the Oconee River watershed which runs through Athens-Clarke county, Georgia, and neighboring Jackson and Oconee counties.
The majority of turtle isolates (40/46; 86.9%) had PFGE patterns that matched human cases in the CDC PulseNet USA database (Table 3). Fifty-one percent of the turtle isolates that matched human PFGE patterns (n = 20) in the PulseNet database were outbreak-related strains. Two-thirds of the turtle isolates had PFGE patterns that matched temporally or spatially with human cases reported from Georgia (Table 3). Salmonella strain types Jv1 (Java) and Ms1 (Mississippi) were also reported among human cases in Athens, Georgia (2009, 2016–2018).
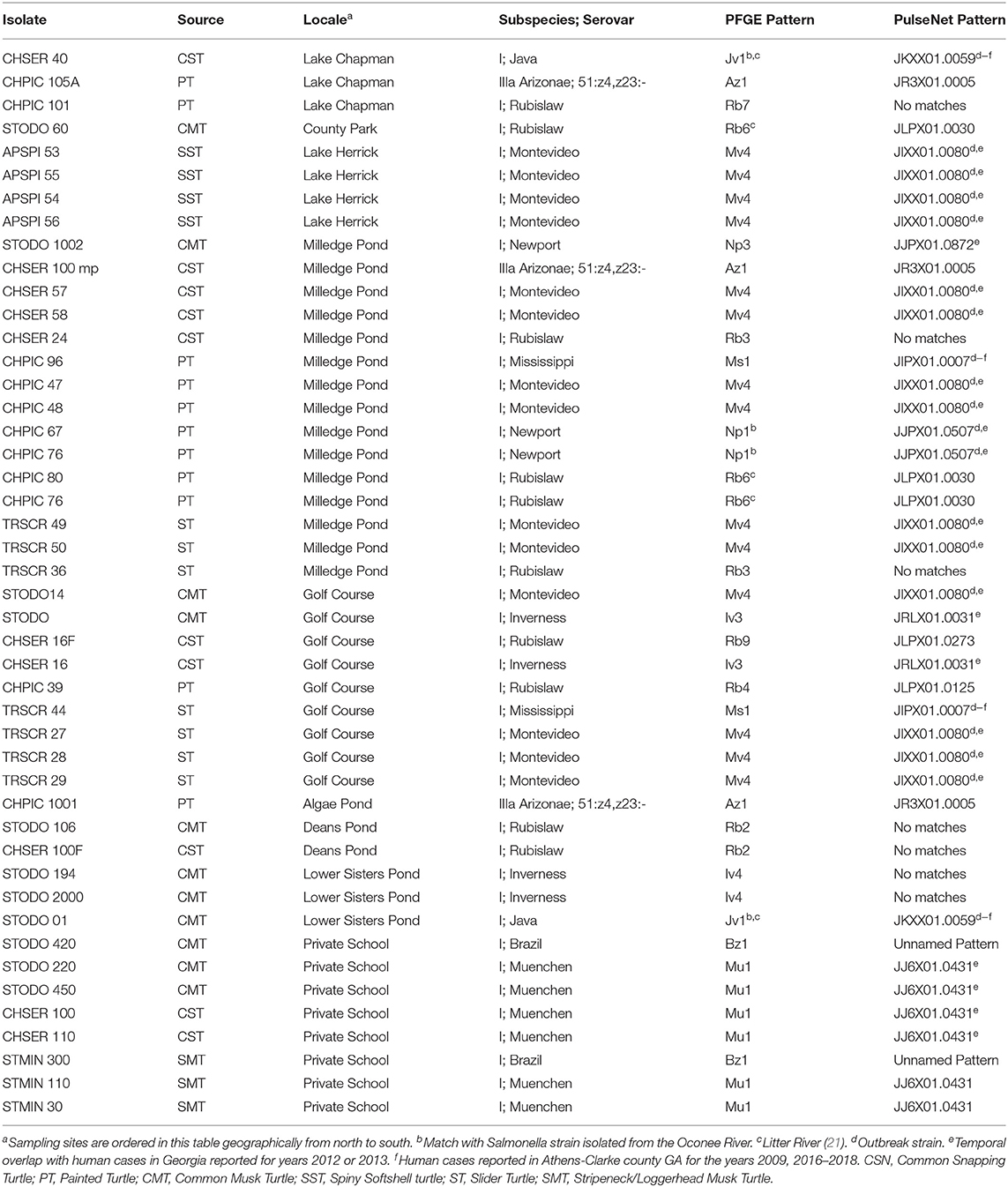
Table 3. Matching Pulsed Field Gel Electrophoresis (PFGE) Patterns between Salmonella enterica isolated from turtles and humans.
Discussion
Wild turtles in the United States are often presumed to harbor a high prevalence of Salmonella because most of the published information regarding Salmonella prevalence in turtles comes from studies focused primarily on pet turtles or from epidemiological investigations following an outbreak, which often involve commercial breeding facilities (28, 67–69). Compared to studies in other countries (7, 34, 39, 40, 43, 45, 47, 70–73), there is a paucity of information about Salmonella prevalence in wild turtles, in the United States (41, 42, 44, 74). This study attempts to fill that knowledge gap and demonstrated a wide range of prevalence of Salmonella among free-living turtle species (14–100%). In general, these findings were actually consistent with other studies that reported moderate prevalence in wild aquatic turtles (44, 45, 70, 74). Variation in Salmonella prevalence reported in past studies could be attributed to differences in sample collection (39, 42) or culture methodology (23, 75–77). Depending on sample type, the use and type of Salmonella enrichment media results in significant differences in isolation efficacy (75–77) and secondary enrichment can significantly increase Salmonella isolation (23). Variations may also be related to species-specific susceptibility to Salmonella infection (39), habitat type (39), geographic location (20, 78, 79), and/or degree of anthropogenic influence (e.g., sewage or agricultural runoff) (80–82) on water bodies.
There were significant differences in Salmonella prevalence by turtle species. Painted turtles and sliders had significantly lower prevalence rates compared to snapping turtles, which may be attributable to undetermined ecological differences. Although all of the spiny softshell turtles were colonized by Salmonella (100%), these results should be interpreted with caution because the sample size of this species was very small (n = 4) and all four turtles came from the same pond (Lake Herrick). Interestingly, bottom-dwelling species (musk and snapping turtles) had a higher prevalence than basking species. Gaertner et al. (44) also found a higher prevalence of Salmonella in cloacal swabs of non-basking turtles [12/19 (63%), musk and snapping turtles)] compared with basking turtles [8/30 (27%), red-eared sliders and Texas river cooters (Pseudemys texana)]. Research is needed to determine whether this is due to exposure (e.g., Salmonella settling in pond detritus) or the effects of higher temperatures on turtles' immune system function, as behavioral basking is associated with increased immune system activity in ectotherms (83). A statistically-significantly higher prevalence of Salmonella infection occurred in juveniles than adults. This follows the general pattern in other animals of Salmonella infection in juveniles vs. adults, due to immature immune systems or gastrointestinal microflora (37). In addition, juvenile turtles may have a higher probability of Salmonella exposure through their more omnivorous diet, may spend more time hiding in detritus, or may be being more easily stressed than adults, which may increase susceptibility or shedding. As expected, there was a trend toward increasing Salmonella prevalence with higher ambient temperature, inferred from seasonal monthly data. Higher temperatures create a favorable environment for Salmonella, and previous studies have shown that Salmonella isolations from water bodies increased during summer months due to enhanced environmental persistence and replication of the bacteria, as well as increased storm events that flush more bacteria into river systems or stir up sediment (20).
Lastly, although the most plausible model found in model selection included only the effect of basking class, other models in the candidate set were found to be competing models, with delta AIC of <2. Competing models represent other plausible explanations for the dataset. These four competing models all included the effect of basking class, but each also included a landscape variable, meaning that these variables also likely explain some of the variation in Salmonella prevalence. For example, one competing model included the effect of basking class and percent of forest cover in the landscape, and a second included the effect of basking class and the canopy cover above the pond. A third and fourth competing model included the effect of basking class with the distance from streets and highways. In general, the prevalence of Salmonella decreased as the percent of forest and canopy cover and the distance from streets and highways increased and Salmonella prevalence increased as the percent of low-density residential areas increased. These results are consistent with the general theory that anthropogenic modifications to the landscape affect Salmonella contamination of water bodies (84, 85).
The nine serovars isolated from the majority of turtles included those commonly reported in reptiles (e.g., S. Java and S. Arizonae) (86), but they also included other types not historically reported commonly in turtles (e.g., S. Montevideo, S. Newport). The isolation of these atypical serovars suggests that turtles could be colonized with S. enterica serotypes that are concurrently present in the water body, possibly because of anthropogenic influences (20, 21, 78, 79). Zoomorphic variables (vicinity of poultry or cattle farms, animal manure application to pastureland, etc.) might also factor into environmental contamination of the turtles' habitat (78). While our sampling sites were in the mostly suburban area of Athens, GA, poultry farms occur along the Oconee River at the northern and western part of this watershed. Application of poultry manures to pastureland is also a common practice and runoff from these fields could find its way to this watershed (87). Many of these turtle isolates appear to be pathogenic for humans, given that 86% of these isolates matched PulseNet PFGE patterns of isolates from humans. In addition, a significantly higher proportion of turtle Salmonella isolates matched PFGE patterns of human salmonellosis cases, compared to the proportion of isolates from other animals and water bodies in the Oconee River watershed that matched human salmonellosis cases (84.8 vs. 50%, mostly comprised of mesomammals [e.g., raccoons (Procyon lotor) and Virginia opossums (Didelphis virginianus)] utilizing water bodies (21). While temporally there is considerable diversity in Salmonella serovar and strain types present in the Oconee River watershed, there are specific serovars and strain types that are repeatedly detected (21, 66). One serovar, S. Rubislaw, is of particular significance; this serovar is increasing in frequency in humans in Georgia and across the Southeastern United States (49, 88). Despite its strain type diversity, there was a significantly higher proportion of S. Rubislaw from turtles that matched human PulseNet PFGE patterns than Rubislaw isolated from other animals and water in the Oconee River watershed (21). While many of the Salmonella PFGE patterns in turtle isolates matched PulseNet patterns associated with outbreaks, temporal and spatial overlap between Salmonella isolation from turtles and humans was not as strong as similar studies (7, 10, 11, 21). It is likely turtles have acquired Salmonella from a human source (e.g., wastewater). A large number of aging septic systems and sewer lines may contribute to surface water contamination, including where turtles are found (89, 90).
The higher prevalence in turtles of PFGE patterns associated with human clinical disease may be because turtles inherently have more contact with water than mesomammals and birds (21), and sampling water requires collecting and filtering large amounts of water for testing. Turtles may accumulate Salmonella from their environment at higher levels that are easier to detect than in water alone. This accumulation could have important practical applications for better estimating bacterial contamination in the environment. For example, Salmonella bacteria in irrigation ponds have been identified as one likely source of contamination of produce (91); yet, the generally low detection rate of Salmonella in these aquatic environments (92) poses an epidemiological challenge for identifying the environmental sources of produce-associated outbreaks. These results suggest that sampling turtles in these possibly source environments might be more efficient than sampling the water bodies themselves. Lastly, whole genome sequencing has very recently replaced PFGE for PulseNet comparisons of isolates and is recommended for future studies.
In conclusion, there is significant overlap in the S. enterica serovars and strains that are associated with both wild turtles and human populations in Georgia. Whether the turtles are the source of human cases or just a sentinel of environmental contamination is currently unknown. Reptile-associated salmonellosis remains a public health concern (4, 7, 8, 28, 35, 93, 94). The current investigation by the CDC of an ongoing outbreak of salmonellosis linking human infections with contact with infected wild birds or contaminated feeders should serve as a reminder of the significance of understanding salmonellae dynamics in wildlife (95). There is high variability in the rates of S. enterica shedding among turtles but to better understand the epidemiology of Salmonella in turtles in the United States, a large-scale, nationwide study investigating the Salmonella prevalence of healthy, asymptomatic free-living turtles is needed. Of note, the data in this study were acquired in 2012–2013 so contemporary studies are needed to determine if the epidemiologic patterns observed are consistent. However, since this study was conducted, there have been no additional studies on Salmonella in turtles in Georgia or the Southeastern United States, so these data provide the most recent, but historic prospective on prevalence and epidemiology factors related to turtle infections which can guide future studies. Despite the age of these data, we suggest that turtles could be excellent indicators for levels of Salmonella contamination in those water bodies because turtles spend most of their lives in often poor-quality ponds, acquire Salmonella from those water bodies, and are relatively easy to attract and capture with baited traps.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by University of Georgia's Institutional Animal Care and Use Committee (AUP# A2010 10-186).
Author Contributions
SH, MY, and JM contributed to study design, collected data, and wrote manuscript. VP conducted statistical analyses and contributed to the manuscript. JM, KH, PG-S, and AP analyzed and interpreted data and contributed to the manuscript. JH, KJ, and TK collected data. SS, PG-S, and KH analyzed, interpreted data, and contributed to manuscript revisions. KJ and TK analyzed and interpreted data. JM, EL, MM, and SC contributed to manuscript revisions. EL analyzed data, contributed funds, and contributed to the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was partially funded by a National Institutes of Health grant (1R15AI089565-01), the Young Scholars Program, the Georgia Veterinary Scholars Program, and by UGA's REU Population Biology of Infectious Diseases which was funded by the National Science Foundation (1659683). JM was supported by USDA HATCH grant VA-160130. Additional funding was provided by state and federal supporters of SCWDS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the following students for field assistance: Madeleine Mullen, Scarlet Sumner, Candance Cooper, and Kayla Garrett.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.674973/full#supplementary-material
References
1. Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, O'Brien SJ, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. (2010) 50:882–9. doi: 10.1086/650733
2. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States–major pathogens. Emerg Infect Dis. (2011) 17:7–15. doi: 10.3201/eid1701.P11101
3. Jackson BR, Griffin PM, Cole D, Walsh KA, Chai SJ. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998-2008. Emerg Infect Dis. (2013) 19:1239–44. doi: 10.3201/eid1908.121511
4. Marshall KE, Nguyen TA, Ablan M, Nichols MC, Robyn MP, Sundararaman P, et al. Investigations of possible multistate outbreaks of Salmonella, shiga toxin-producing Escherichia coli, and Listeria monocytogenes infections-United States, 2016. MMWR Surveill Summ. (2020) 69:1–14. doi: 10.15585/mmwr.ss6906a1
5. Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, et al. Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998-2008. Emerg Infect Dis. (2013) 19:407–15. doi: 10.3201/eid1903.111866
6. Markland SM, Ingram D, Kniel KE, Sharma M. Water for agriculture: the convergence of sustainability and safety. Microbiol Spectr. (2017) 5. doi: 10.1128/microbiolspec.PFS-0014-2016
7. Collins J, Simpson KMJ, Bell G, Durrheim DN, Hill-Cawthorne GA, Hope K, et al. A One Health investigation of Salmonella enterica serovar Wangata in north-eastern New South Wales, Australia, 2016-2017. Epidemiol Infect. (2019) 147:e150. doi: 10.1017/S0950268819000475
8. Bosch S, Tauxe RV, Behravesh CB. Turtle-associated salmonellosis, United States, 2006-2014. Emerg Infect Dis. (2016) 22:1149–55. doi: 10.3201/eid2207.150685
9. Compton JA, Baney JA, Donaldson SC, Houser BA, San Julian GJ, Yahner RH, et al. Salmonella infections in the common raccoon (Procyon lotor) in western Pennsylvania. J Clin Microbiol. (2008) 46:3084–6. doi: 10.1128/JCM.00685-08
10. Hernandez SM, Keel K, Sanchez S, Trees E, Gerner-Smidt P, Adams JK, et al. Epidemiology of a Salmonella enterica subsp. enterica serovar Typhimurium strain associated with a songbird outbreak. Appl Environ Microbiol. (2012) 78:7290–8. doi: 10.1128/AEM.01408-12
11. Hernandez SM, Welch CN, Peters VE, Lipp EK, Curry S, Yabsley MJ, et al. Urbanized White Ibises (Eudocimus albus) as carriers of Salmonella enterica of significance to public health and wildlife. PLoS ONE. (2016) 11:e0164402. doi: 10.1371/journal.pone.0164402
13. Conrad CC, Stanford K, Narvaez-Bravo C, Callaway T, McAllister T. Farm fairs and petting zoos: a review of animal contact as a source of zoonotic enteric disease. Foodborne Pathog Dis. (2017) 14:59–73. doi: 10.1089/fpd.2016.2185
14. Lee D, Chang HH, Sarnat SE, Levy K. Precipitation and salmonellosis incidence in Georgia, USA: interactions between extreme rainfall events and antecedent rainfall conditions. Environ Health Perspect. (2019) 127:97005. doi: 10.1289/EHP4621
15. Williams S, Patel M, Markey P, Muller R, Benedict S, Ross I, et al. Salmonella in the tropical household environment–everyday, everywhere. J Infect. (2015) 71:642–8. doi: 10.1016/j.jinf.2015.09.011
16. MacDonald E, White R, Mexia R, Bruun T, Kapperud G, Brandal LT, et al. The role of domestic reservoirs in domestically acquired Salmonella infections in Norway: epidemiology of salmonellosis, 2000-2015, and results of a national prospective case-control study, 2010-2012. Epidemiol Infect. (2018) 147:1–8. doi: 10.1017/S0950268818002911
17. Castronovo DA, Chui KK, Naumova EN. Dynamic maps: a visual-analytic methodology for exploring spatio-temporal disease patterns. Environ Health. (2009) 8:61. doi: 10.1186/1476-069X-8-61
18. Boore AL, Hoekstra RM, Iwamoto M, Fields PI, Bishop RD, Swerdlow DL. Salmonella enterica infections in the United States and assessment of coefficients of variation: a novel approach to identify epidemiologic characteristics of individual serotypes, 1996-2011. PLoS ONE. (2015) 10:e0145416. doi: 10.1371/journal.pone.0145416
19. Centers for Disease Control and Prevention. An atlas of Salmonella in the United States (1968-2011) Available online at: https://www.cdc.gov/salmonella/pdf/salmonella-atlas-508c.pdf.
20. Haley BJ, Cole DJ, Lipp EK. Distribution, diversity, and seasonality of waterborne salmonellae in a rural watershed. Appl Environ Microbiol. (2009) 75:1248–55. doi: 10.1128/AEM.01648-08
21. Maurer JJ, Martin G, Hernandez S, Cheng Y, Gerner-Smidt P, Hise KB, et al. Diversity and persistence of Salmonella enterica strains in rural landscapes in the southeastern United States. PLoS ONE. (2015) 10:e0128937. doi: 10.1371/journal.pone.0128937
22. Becker DJ, Teitelbaum CS, Murray MH, Curry SE, Welch CN, Ellison T, et al. Assessing the contributions of intraspecific and environmental sources of infection in urban wildlife: Salmonella enterica and white ibis as a case study. J R Soc Interface. (2018) 15:20180654. doi: 10.1098/rsif.2018.0654
23. Rigby CE, Pettit JR. Delayed secondary enrichment for the isolation of salmonellae from broiler chickens and their environment. Appl Environ Microbiol. (1980) 40:783–6. doi: 10.1128/aem.40.4.783-786.1980
24. Mermin J, Hutwagner L, Vugia D, Shallow S, Daily P, Bender J, et al. Reptiles, amphibians, and human Salmonella infection: a population-based, case-control study. Clin Infect Dis. (2004) 38(Suppl. 3):S253–61. doi: 10.1086/381594
25. Harris JR, Bergmire-Sweat D, Schlegel JH, Winpisinger KA, Klos RF, Perry C, et al. Multistate outbreak of Salmonella infections associated with small turtle exposure, 2007-2008. Pediatrics. (2009) 124:1388–94. doi: 10.1542/peds.2009-0272
26. Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. (1997) 35:2786–90. doi: 10.1128/jcm.35.11.2786-2790.1997
27. Harris JR, Neil KP, Behravesh CB, Sotir MJ, Angulo FJ. Recent multistate outbreaks of human Salmonella infections acquired from turtles: a continuing public health challenge. Clin Infect Dis. (2010) 50:554–9. doi: 10.1086/649932
28. Gambino-Shirley K, Stevenson L, Concepcion-Acevedo J, Trees E, Wagner D, Whitlock L, et al. Flea market finds and global exports: Four multistate outbreaks of human Salmonella infections linked to small turtles, United States-2015. Zoonoses Public Health. (2018) 65:560–8. doi: 10.1111/zph.12466
29. Cohen ML, Potter M, Pollard R, Feldman RA. Turtle-associated salmonellosis in the United States. effect of public health action, 1970 to 1976. JAMA. (1980) 243:1247–9. doi: 10.1001/jama.243.12.1247
30. Centers for Disease C Prevention. Multistate outbreak of human Salmonella infections associated with exposure to turtles–United States, 2007-2008. MMWR Morb Mortal Wkly Rep. (2008) 57:69–72.
31. Mermin J, Hoar B, Angulo FJ. Iguanas and Salmonella marina infection in children: a reflection of the increasing incidence of reptile-associated salmonellosis in the United States. Pediatrics. (1997) 99:399–402. doi: 10.1542/peds.99.3.399
32. Ackman DM, Drabkin P, Birkhead G, Cieslak P. Reptile-associated salmonellosis in New York State. Pediatr Infect Dis J. (1995) 14:955–9. doi: 10.1097/00006454-199511000-00006
33. Marin C, Vega S, Marco-Jimenez F. Tiny turtles purchased at pet stores are a potential high risk for Salmonella human infection in the Valencian region, eastern Spain. Vector Borne Zoonotic Dis. (2016) 16:455–60. doi: 10.1089/vbz.2016.1950
34. McWhorter A, Owens J, Valcanis M, Olds L, Myers C, Smith I, et al. In vitro invasiveness and antimicrobial resistance of Salmonella enterica subspecies isolated from wild and captive reptiles. Zoonoses Public Health. (2021) 68:402–12. doi: 10.1111/zph.12820
35. Koski L, Stevenson L, Huffman J, Robbins A, Latash J, Omoregie E, et al. Notes from the field: an outbreak of Salmonella sgbeni Infections linked to turtle exposure - United States, 2017. MMWR Morb Mortal Wkly Rep. (2018) 67:1350. doi: 10.15585/mmwr.mm6748a5
36. Jacobson ER. Infectious Disease and Pathology of Reptiles. Color Atlas and Text. Boca Raton, FL: CRC Press Taylor Francis Group (2007).
37. Mitchell MA, Shane SM. Salmonella in reptiles. Sem Avian Exotic Pet Med. (2001) 10:25–35. doi: 10.1053/saep.2001.19798
38. Shane SM, Gilbert R, Harrington KS. Salmonella colonization in commercial pet turtles (Pseudemys scripta elegans). Epidemiol Infect. (1990) 105:307–16. doi: 10.1017/S0950268800047907
39. Gay N, Le Hello S, Weill FX, de Thoisy B, Berger F. Salmonella serotypes in reptiles and humans, French Guiana. Vet Microbiol. (2014) 170:167–71. doi: 10.1016/j.vetmic.2014.01.024
40. Hidalgo-Vila J, Diaz-Paniagua C, Perez-Santigosa N, de Frutos-Escobar C, Herrero-Herrero A. Salmonella in free-living exotic and native turtles and in pet exotic turtles from SW Spain. Res Vet Sci. (2008) 85:449–52. doi: 10.1016/j.rvsc.2008.01.011
41. Richards JM, Brown JD, Kelly TR, Fountain AL, Sleeman JM. Absence of detectable Salmonella cloacal shedding in free-living reptiles on admission to the wildlife center of Virginia. J Zoo Wildl Med. (2004) 35:562–3. doi: 10.1638/03-070
42. Saelinger CA, Lewbart GA, Christian LS, Lemons CL. Prevalence of Salmonella spp in cloacal, fecal, and gastrointestinal mucosal samples from wild North American turtles. J Am Vet Med Assoc. (2006) 229:266–8. doi: 10.2460/javma.229.2.266
43. Marin C, Ingresa-Capaccioni S, Gonzalez-Bodi S, Marco-Jimenez F, Vega S. Free-living turtles are a reservoir for Salmonella but not for Campylobacter. PLoS ONE. (2013) 8:e72350. doi: 10.1371/journal.pone.0072350
44. Gaertner JP, Hahn D, Rose FL, Forstner MR. Detection of salmonellae in different turtle species within a headwater spring ecosystem. J Wildl Dis. (2008) 44:519–26. doi: 10.7589/0090-3558-44.2.519
45. Hidalgo-Vila J, Diaz-Paniagua C, de Frutos-Escobar C, Jimenez-Martinez C, Perez-Santigosa N. Salmonella in free living terrestrial and aquatic turtles. Vet Microbiol. (2007) 119:311–5. doi: 10.1016/j.vetmic.2006.08.012
46. Gruszynski K, Pao S, Kim C, Toney D, Wright K, Ross PG, et al. Evaluating wildlife as a potential source of Salmonella serotype Newport (JJPX01.0061) contamination for tomatoes on the eastern shore of Virginia. Zoonoses Public Health. (2014) 61:202–7. doi: 10.1111/zph.12061
47. Gong S, Wang F, Shi H, Zhou P, Ge Y, Hua L, et al. Highly pathogenic Salmonella Pomona was first isolated from the exotic red-eared slider (Trachemys scripta elegans) in the wild in China: Implications for public health. Sci Total Environ. (2014) 468–469:28–30. doi: 10.1016/j.scitotenv.2013.08.025
48. Zhang J, Kuang D, Wang F, Meng J, Jin H, Yang X, et al. Turtles as a possible reservoir of nontyphoidal Salmonella in Shanghai, China. Foodborne Pathog Dis. (2016) 13:428–33. doi: 10.1089/fpd.2015.2107
49. Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Report. (2016). Available online at: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf.
50. Lydeard C, Mayden RL. A Diverse and endangered aquatic ecosystem of the southeast United States. Conserv Biol. (1995) 9:800–5. doi: 10.1046/j.1523-1739.1995.09040800.x
51. Buhlmann K, Tuberville T, Gibbons W. Turtles of the Southeast. Athens: University of Georgia Press (2008).
52. WHO Global Foodborne Infections Network. Laboratory Protocol: Isolation of Salmonella and Shigella From Faecal Specimens (2010). Available online at: https://antimicrobialresistance.dk/CustomerData/Files/Folders/6-pdf-protocols/62_gfn-stool-culture-nov2010.pdf.
53. Hunter SB, Vauterin P, Lambert-Fair MA, Van Duyne MS, Kubota K, Graves L, et al. Establishment of a universal size standard strain for use with the PulseNet standardized pulsed-field gel electrophoresis protocols: converting the national databases to the new size standard. J Clin Microbiol. (2005) 43:1045–50. doi: 10.1128/JCM.43.3.1045-1050.2005
54. Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, Richter SS, et al. Manual of Clinical Microbiology, 11th Edn. Washington DC : American Society of Microbiology (2015).
55. Koort JMK, Lukinmaa S, Rantala M, Unkila E, Siitonen A. Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J Clin Microbiol. (2002) 40:3497–8. doi: 10.1128/JCM.40.9.3497-3498.2002
56. Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157 : H7 Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. (2006) 3:59–67. doi: 10.1089/fpd.2006.3.59
57. Zamperini K, Soni V, Waltman D, Sanchez S, Theriault EC, Bray J, et al. Molecular characterization reveals Salmonella enterica serovar 4,(5),12:i:- from poultry is a variant Typhimurium serovar. Avian Dis. (2007) 51:958–64. doi: 10.1637/7944-021507-REGR.1
58. Anderson JR, Hardy EE, Roach JT, Witmer RE. “A land use and land cover classification system for use with remote sensor data,” in Geological Survey Professional Paper. (1976).
59. Irwin EG, Bockstael NE. The evolution of urban sprawl: Evidence of spatial heterogeneity and increasing land fragmentation. Proc Natl Acad Sci USA. (2007) 104:20672–7 doi: 10.1073/pnas.0705527105
61. R Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing. Available online at: http://www.R-project.org/.
62. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. (2015) 67:48. doi: 10.18637/jss.v067.i01
63. Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, et al. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol Evol. (2009) 24:127–35. doi: 10.1016/j.tree.2008.10.008
64. Lukac M, Pedersen K, Prukner-Radovcic E. Prevalence of Salmonella in captive reptiles from Croatia. J Zoo Wildl Med. (2015) 46:234–40. doi: 10.1638/2014-0098R1.1
65. Bauwens L, Vercammen F, Bertrand S, Collard JM, De Ceuster S. Isolation of Salmonella from environmental samples collected in the reptile department of Antwerp Zoo using different selective methods. J Appl Microbiol. (2006) 101:284–9. doi: 10.1111/j.1365-2672.2006.02977.x
66. Meinersmann RJ, Berrang ME, Jackson CR, Fedorka-Cray P, Ladely S, Little E, et al. Salmonella, Campylobacter and Enterococcus spp.: their antimicrobial resistance profiles and their spatial relationships in a synoptic study of the upper Oconee River basin. Microbial Ecol. (2008) 55:444–52. doi: 10.1007/s00248-007-9290-6
67. Back DS, Shin GW, Wendt M, Heo GJ. Prevalence of Salmonella spp. in pet turtles and their environment. Lab Anim Res. (2016) 32:166–70. doi: 10.5625/lar.2016.32.3.166
68. Kuroki T, Ishihara T, Nakajima N, Furukawa I, Une Y. Prevalence of Salmonella enterica subspecies enterica in red-eared sliders Trachemys scripta elegans retailed in pet shops in Japan. Jpn J Infect Dis. (2019) 72:38–43. doi: 10.7883/yoken.JJID.2018.140
69. Marenzoni ML, Zicavo A, Veronesi F, Morganti G, Scuota S, Coletti M, et al. Microbiological and parasitological investigation on chelonians reared in Italian facilities. Vet Ital. (2015) 51:173–8. doi: 10.12834/VetIt.7.21.3
70. Nowakiewicz A, Ziolkowska G, Zieba P, Dziedzic BM, Gnat S, Wojcik M, et al. Aerobic bacterial microbiota isolated from the cloaca of the European pond turtle (Emys orbicularis) in Poland. J Wildl Dis. (2015) 51:255–9. doi: 10.7589/2013-07-157
71. Edwards JJ, Amadi VA, Soto E, Jay-Russel MT, Aminabadi P, Kenelty K, et al. Prevalence and phenotypic characterization of Salmonella enterica isolates from three species of wild marine turtles in Grenada, West Indies. Vet World. (2021) 14:222–9. doi: 10.14202/vetworld.2021.222-229
72. Lecis R, Paglietti B, Rubino S, Are BM, Muzzeddu M, Berlinguer F, et al. Detection and characterization of Mycoplasma spp. and Salmonella spp. in free-living European tortoises (Testudo hermanni, Testudo graeca, and Testudo marginata). J Wildl Dis. (2011) 47:717–24. doi: 10.7589/0090-3558-47.3.717
73. Hidalgo-Vila J, Díaz-Paniagua C, Ruiz X, Portheault A, El Mouden H, Slimani T, et al. Salmonella species in free-living spur-thighed tortoises (Testudo graeca) in central western Morocco. Vet Rec. (2008) 162:218–19. doi: 10.1136/vr.162.7.218
74. Hahn D, Gaertner J, Forstner MR, Rose FL. High-resolution analysis of salmonellae from turtles within a headwater spring ecosystem. FEMS Microbiol Ecol. (2007) 60:148–55. doi: 10.1111/j.1574-6941.2007.00275.x
75. Hammack TS, Amaguana RM, June GA, Sherrod PS, Andrews WH. Relative effectiveness of selenite cystine broth, tetrathionate broth, and Rappaport-Vassiliadis medium for the recovery of Salmonella spp. from foods with a low microbial load. J Food Prot. (1999) 62:16–21. doi: 10.4315/0362-028X-62.1.16
76. Aspinall ST, Hindle MA, Hutchinson DN. Improved isolation of salmonellae from faeces using a semisolid Rappaport-Vassiliadis medium. Eur J Clin Microbiol Infect Dis. (1992) 11:936–9. doi: 10.1007/BF01962379
77. Corrente M, Madio A, Friedrich KG, Greco G, Desario C, Tagliabue S, et al. Isolation of Salmonella strains from reptile faeces and comparison of different culture media. J Appl Microbiol. (2004) 96:709–15. doi: 10.1111/j.1365-2672.2004.02186.x
78. Thomas JL, Slawson RM, Taylor WD. Salmonella serotype diversity and seasonality in urban and rural streams. J Appl Microbiol. (2013) 114:907–22. doi: 10.1111/jam.12079
79. Walters SP, Gonzalez-Escalona N, Son I, Melka DC, Sassoubre LM, Boehm AB. Salmonella enterica diversity in central Californian coastal waterways. Appl Environ Microbiol. (2013) 79:4199–209. doi: 10.1128/AEM.00930-13
80. Harris CS, Tertuliano M, Rajeev S, Vellidis G, Levy K. Impact of storm runoff on Salmonella and Escherichia coli prevalence in irrigation ponds of fresh produce farms in southern Georgia. J Appl Microbiol. (2018) 124:910–21. doi: 10.1111/jam.13689
81. Weller D, Belias A, Green H, Roof S, Wiedmann M. Landscape, Water quality, and weather factors associated with an increased likelihood of foodborne pathogen contamination of New York streams used to source water for produce production. Front Sustain Food Syst. (2020) 3:124. doi: 10.3389/fsufs.2019.00124
82. Bardsley CA, Weller DL, Ingram DT, Chen Y, Oryang D, Rideout SL, et al. Strain, soil-type, irrigation regimen, and poultry litter influence Salmonella survival and die-off in agricultural soils. Front Microbiol. (2021) 12:590303. doi: 10.3389/fmicb.2021.590303
83. Zimmerman LM, Vogel LA, Bowden RM. Understanding the vertebrate immune system: insights from the reptilian perspective. J Exp Biol. (2010) 213:661–71. doi: 10.1242/jeb.038315
84. Luo Z, Gu G, Ginn A, Giurcanu MC, Adams P, Vellidis G, et al. Distribution and characterization of Salmonella enterica isolates from irrigation ponds in the southeastern United States. Appl Environ Microbiol. (2015) 81:4376–87. doi: 10.1128/AEM.04086-14
85. Levantesi C, Bonadonna L, Briancesco R, Grohmann E, Toze S, Tandoi V. Salmonella in surface and drinking water: occurrence and water-mediated transmission. Food Res Int. (2012) 45:587–602. doi: 10.1016/j.foodres.2011.06.037
86. Warwick C, Lambiris AJ, Westwood D, Steedman C. Reptile-related salmonellosis. J R Soc Med. (2001) 94:124–26. doi: 10.1177/014107680109400306
87. Vereen E Jr, Lowrance RR, Jenkins MB, Adams P, Rajeev S, et al. Landscape and seasonal factors influence Salmonella and Campylobacter prevalence in a rural mixed use watershed. Water Res. (2013) 47:6075–85. doi: 10.1016/j.watres.2013.07.028
88. Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Report, 2013. (2016). Available online at: https://www.cdc.gov/nationalsurveillance/pdfs/salmonella-annual-report-2013-508c.pdf.
89. Capps KA, Bateman McDonald JM, Gaur N, Parsons R. Assessing the socio-environmental risk of onsite wastewater treatment systems to inform management decisions. Environ Sci Technol. (2020) 54:14843–53. doi: 10.1021/acs.est.0c03909
90. Carroll GD, Rasmussen TC. Identifying Sewage Leaks in Urban Environments: Examples From Athens, GA. Available online at: https://smartech.gatech.edu/bitstream/handle/1853/47154/CarrollD%20paper%20April%2014.pdf?sequence=1&isAllowed.
91. Liu H, Whitehouse CA, Li B. Presence and persistence of Salmonella in water: the impact on microbial quality of water and food safety. Front Public Health. (2018) 6:159. doi: 10.3389/fpubh.2018.00159
92. Topalcengiz Z, Strawn LK, Danyluk MD. Microbial quality of agricultural water in Central Florida. PLoS ONE. (2017) 12:e0174889. doi: 10.1371/journal.pone.0174889
93. Walters MS, Simmons L, Anderson TC, DeMent J, Van Zile K, Matthias LP, et al. Outbreaks of salmonellosis from small turtles. Pediatrics. (2016) 137:e20151735. doi: 10.1542/peds.2015-1735
94. Sodagari HR, Habib I, Shahabi MP, Dybing NA, Wang P, Bruce M. A review of the public health challenges of Salmonella and turtles. Vet Sci. (2020) 7:56. doi: 10.3390/vetsci7020056
95. Centers for Disease Control and Prevention. Salmonella Outbreak Linked to Wild Songbirds. Available online at: https://www.cdc.gov/salmonella/typhimurium-04-21/index.html.
Keywords: chelonia, turtle, Salmonella, Salmonella enterica, reptile-associated salmonellosis
Citation: Hernandez SM, Maurer JJ, Yabsley MJ, Peters VE, Presotto A, Murray MH, Curry S, Sanchez S, Gerner-Smidt P, Hise K, Huang J, Johnson K, Kwan T and Lipp EK (2021) Free-Living Aquatic Turtles as Sentinels of Salmonella spp. for Water Bodies. Front. Vet. Sci. 8:674973. doi: 10.3389/fvets.2021.674973
Received: 02 March 2021; Accepted: 25 June 2021;
Published: 22 July 2021.
Edited by:
Armanda Bastos, University of Pretoria, South AfricaReviewed by:
Narayan Paudyal, Nepal Agricultural Research Council, NepalSamantha Wisely, University of Florida, United States
Carla Grattarola, Liguria e Valle d'Aosta, Italy
Copyright © 2021 Hernandez, Maurer, Yabsley, Peters, Presotto, Murray, Curry, Sanchez, Gerner-Smidt, Hise, Huang, Johnson, Kwan and Lipp. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sonia M. Hernandez, c2hlcm56QHVnYS5lZHU=
†Present address: John Maurer, Department of Animal and Poultry Sciences, Virginia Tech, Blacksburg, VA, United States
Andrea Presotto, Department of Geography and Geosciences, Salisbury University, Salisbury, MD, United States
 Sonia M. Hernandez
Sonia M. Hernandez John J. Maurer
John J. Maurer Michael J. Yabsley
Michael J. Yabsley Valerie E. Peters4
Valerie E. Peters4 Maureen H. Murray
Maureen H. Murray Susan Sanchez
Susan Sanchez Peter Gerner-Smidt
Peter Gerner-Smidt Kelley Hise
Kelley Hise Erin K. Lipp
Erin K. Lipp