- 1The Key Lab of Veterinary Biological Products, Henan University of Science and Technology, Luoyang, China
- 2Henan Provincial Open Laboratory of Key Disciplines in Environmental and Animal Products Safety, Henan University of Science and Technology, Luoyang, China
In this study, we compared the virulence of the most common serovars of Glaesserella parasuis in China, serovars 4, 5, 12, and 13 (36 strains in total) in BALB/c mice and piglets. In mice, the median lethal doses (LD50s) of the four serovars were roughly 9.80 × 107–4.60 × 109 CFU, 2.10 × 108–8.85 × 109 CFU, 4.81 × 107–7.01 × 109 CFU, and 1.75 × 108–8.45 × 108 CFU, respectively. Serovar 13 showed the strongest virulence, followed by serovar 4, serovar 12, and serovar 5, but a significant difference in virulence was only observed between serovars 5 and 13. The virulence of strains of the same serovars differed significantly in piglets. Virulent and attenuated strains were present in all serovars, but serovar 5 was the most virulent in piglets, followed by serovars 13, 4, and 12. A significant difference in virulence was observed between serovars 5 and 4 and between serovars 5 and 12. However, the virulence of serovars 5 and 13 did not differ significantly. This comprehensive analysis of G. parasuis virulence in mice and piglets demonstrated that: (1) the order of virulence of the four domestic epidemic serovars (from strongest to weakest) in piglets was serovars 5, 13, 4, and 12; (2) both virulent and attenuated strains were present in all serovars, so virulence did not necessarily correlate with serovar; (3) Although G. parasuis was fatal in BALB/c mice, its virulence is inconsistent with that in piglets, indicating that BALB/c mice are inadequate as an alternative model of G. parasuis infection.
Background
Glaesserella parasuis is a small Gram-negative bacillus belonging to the family Pasteurella, which causes polyserositis, arthritis, and sepsis in pigs, collectively called “Glässer's disease” (1). This is a common disease that seriously threatens the pig industry worldwide (2). In 1992, Kielstein and Rapp-Gabrielson developed a serological typing assay using an agar diffusion test that successfully categorizes G. parasuis into at least 15 serovars (3). Serovars 4 and 5 are most common, followed by serovars 13 and 14, and these have distinct spatial and temporal distributions (4–10). Kielstein and Rapp-Gabrielson also defined serovars 1, 5, 10, 12, 13, and 14 as virulent strains, serovars 2, 4, 8, and 15 as mesogenic strains, and serovars 3, 6, 7, 9, and 11 as attenuated strains (3). However, follow-up studies have shown that G. parasuis virulence does not necessarily correlate with the serovar (11). In recent years, the virulence-related genes of G. parasuis have been identified, but the pathogenic mechanisms of G. parasuis infection have not been demonstrated (12–16). Therefore, the determination of G. parasuis virulence still relies on infection tests in pigs.
Specific-pathogen-free (SPF) pigs (17, 18), Cesarean-derived, colostrum-deprived (CDCD) piglets (19), naturally farrowed, artificially reared (NFCD) pigs (20), and 9–10-week-old piglets negative for anti-G. parasuis antibodies (21, 22) have been successfully investigated. However, using piglets as a model of infection requires high standard laboratory conditions and expensive equipment. Alternative models have been explored with different strains of mice (23–26), but the BALB/c mouse is the only widely used alternative model. However, some previous studies using BALB/c mice as an alternative infection model have produced inconsistent and even contradictory results (26–29), and there has been no comparative study of the alternative BALB/c mouse model and the piglet model.
Experimental intraperitoneal infection model in BALB/c mice and piglets are used extensively in research on information for virulence and immunology of G. parasuis (22–26). In this study, BALB/c mice and 8–9-week-old G. parasuis seronegative piglets were used as the challenge models. The virulence of 36 common strains of G. parasuis in China was measured, and the reliability of the infection model in BALB/c mice was evaluated. Our results provide new data that extend our understanding of the potential harm posed by these G. parasuis strains, and should have utility in the diagnosis and prevention of Glässer's disease.
Methods
Strains, Media, and Reagents
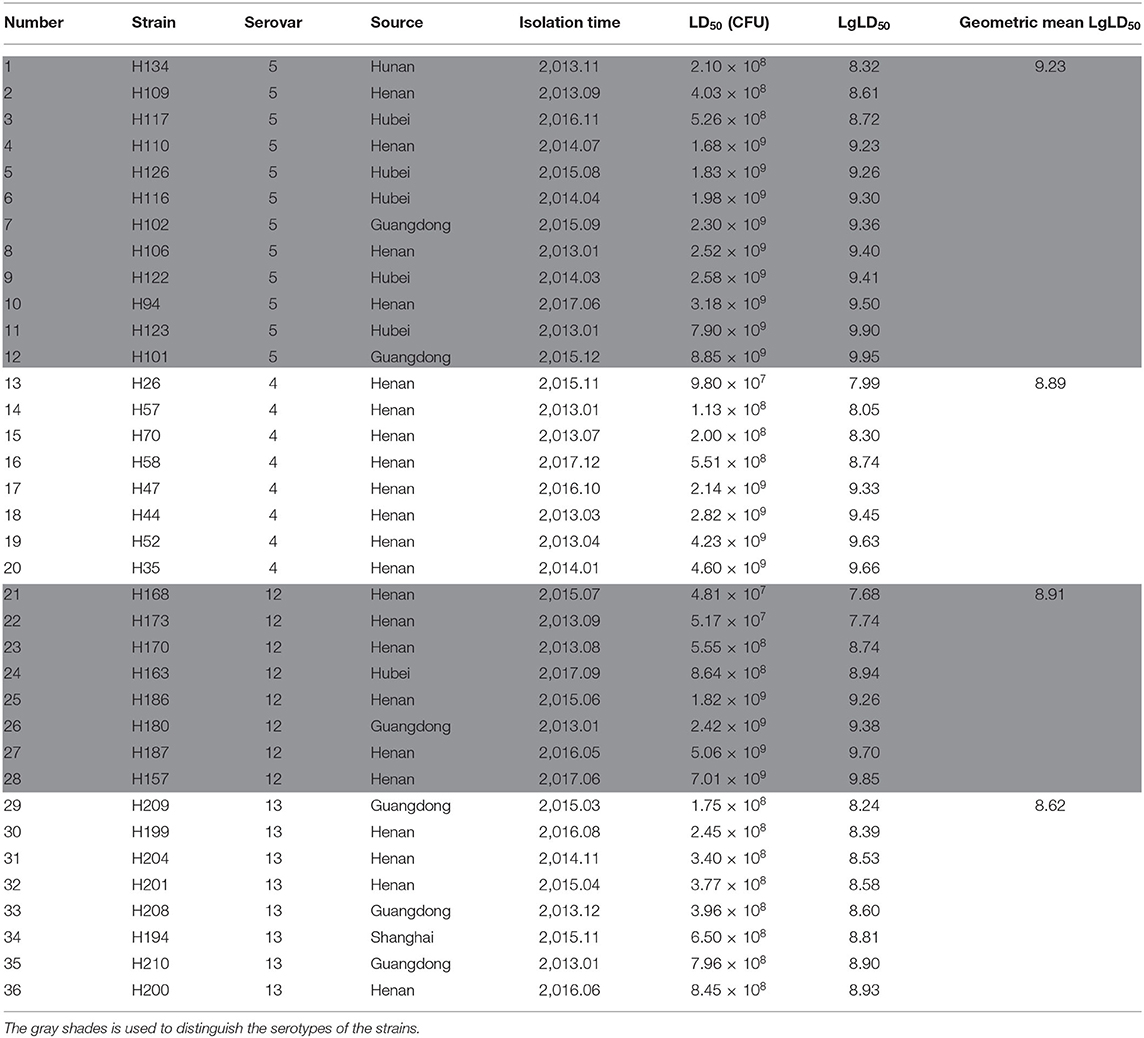
Thirty-six G. parasuis strains, including eight strains of serovar 4, 12 strains of serovar 5, eight strains of serovar 12, and eight strains of serovar 13, were isolated, identified, and stored at the Veterinary Laboratory of Bioengineering of Henan University of Science and Technology (Table 1). Tryptose Soya Agar (TSA) and Tryptose Soya Broth (TSB) media were purchased from Becton, Dickinson and Company (Sparks, MD, USA). Newborn calf serum was purchased from Zhejiang Tianhang Biotechnology Co. Ltd. Nicotinamide adenine dinucleotide (NAD) was purchased from Sigma-Aldrich.
Animals
Female SPF BALB/c mice (6–8 weeks old) were purchased from Charles River Co., Ltd, and 8–9-week-old piglets were purchased from farmers in Yichuan County, Henan Province, China. Nasal swabs were collected from all the piglets used in this study and shown to be G. parasuis-negative with PCR (30). The collected sera were shown to be G. parasuis-antibody negative with a microplate agglutination test (MAT) (31). All animal experiments were conducted in accordance with the guidelines and with the approval of the Animal Experiment Committee of Henan University of Science and Technology (No. 20190719016).
Mouse Virulence Test
TSA plates containing 10% newborn calf serum and 10 μg/mL NAD were inoculated with G. parasuis strain H134 of serovar 5 and incubated at 37°C for 24–48 h. Individual colonies were picked, purified, and cultured for 24–36 h. The cultured colonies were transferred to culture dishes containing 5–8 mL of phosphate-buffered saline (PBS; pH 7.2) with a sterile technique. The colonies were thoroughly suspended by pipetting. Three concentrations of the H134 bacterial suspension were prepared with two-fold dilution. When determining the virulence of strain H134, sixteen BALB/c mice were divided into four groups (four mice in each group). The original H134 bacterial suspension and the three dilutions were injected intraperitoneally into the mice of each group (0.2 mL per mouse). The H134 bacteria in the original suspension were counted to determine the actual numbers inoculated.
The remaining G. parasuis strains were cultured and used to challenge the mice in the same way. One PBS blank control was established for each batch. The incidence of disease and death were observed and recorded. The experiment was terminated 14 d post-challenge; survivors being killed with an intravenous overdose of sodium pentobarbital. Autopsies were performed immediately on the dead mice to record the numbers of lesions. Cardiovascular and pulmonary blood samples were collected and the bacteria in the samples identified. The median lethal dose (LD50) of G. parasuis in the mice was then calculated with the Reed-Muench method (32).
Piglet Virulence Test
G. parasuis strain H134 was cultured in medium plates as described above. Then a single colony of strain H134 was picked and cultured in TSB medium containing newborn calf serum (10%) and NAD (10 μg/mL), with shaking at 180 rpm at 37°C for 12–16 h. The culture solution was used as the stock solution and diluted 1:100, transferred to TSB medium, and cultured with shaking at 180 rpm at 37°C for 10–14 h. The H134 bacteria were harvested when their concentration was maximum, and the optical density at a wavelength of 600 nm (OD600) as well as growth curve of each strain were previously detailed (33) (Supplementary Figure 2). The cultured H134 bacterial solution (3 mL) was injected into the abdominal cavities of five piglets housed in isolation in pens with a concrete floor after they had been fasted for 4 h. The number of H134 bacteria in the original solution was counted to determine the actual number of bacteria inoculated (34).
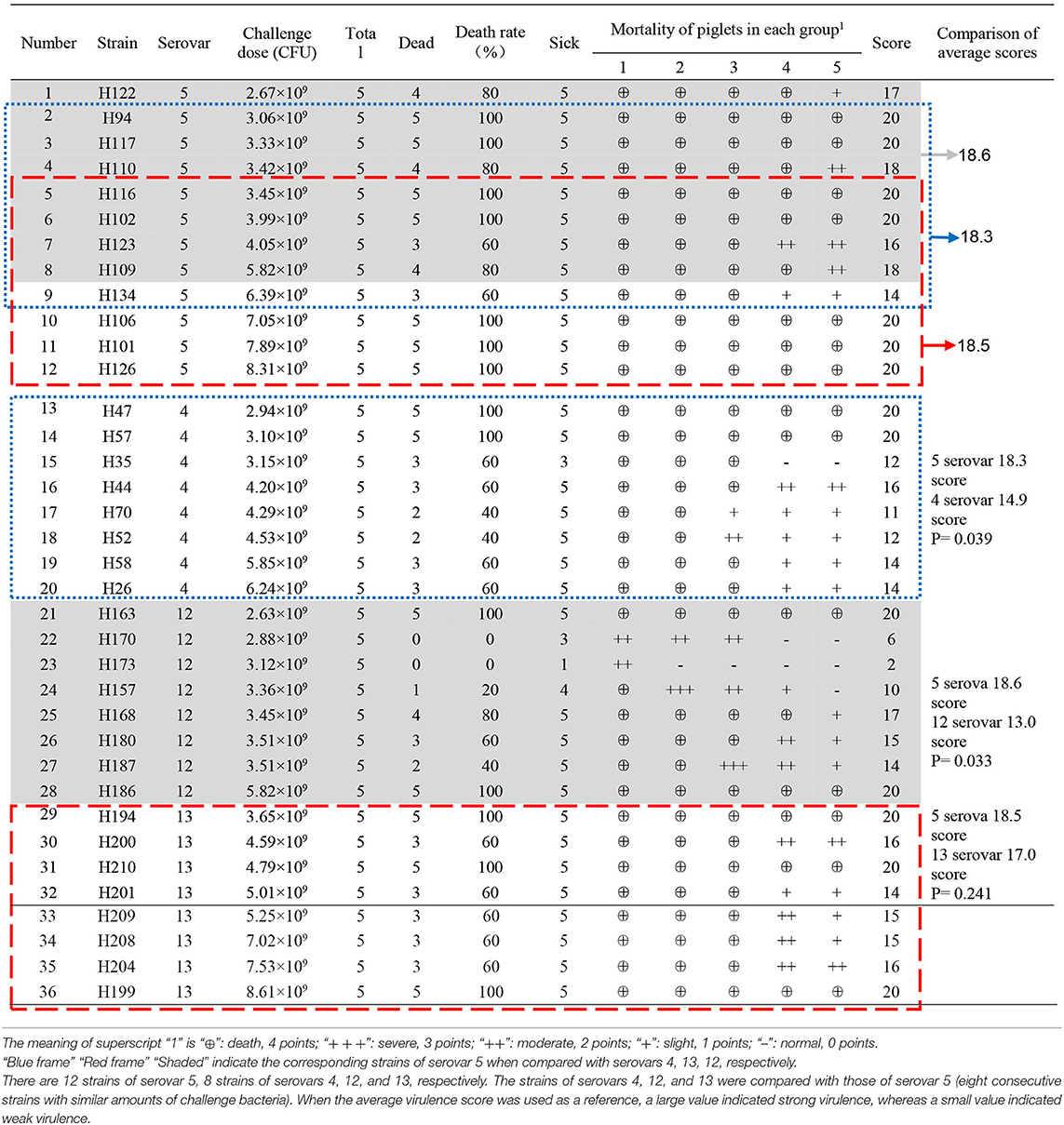
The remaining G. parasuis strains were cultured and used to challenge the pigelts in the same way. Thirty-six G. parasuis strains were tested in batches, and five piglets inoculated with sterile PBS were included in each batch as the controls. Disease onset and death were observed and recorded for 14 days. Autopsies were performed immediately on the deceased piglets, and bacteria were isolated from the heart blood and lungs and identified. After observation, all the survivors were killed with an intravenous overdose of sodium pentobarbital and dissected to check for lesions. The G. parasuis strains were quantitatively scored according to the morbidity and death of the piglets (Table 2).
Data Analysis
The logarithm of median lethal dose (LgLD50) value is used to indicate virulence (a higher value indicates weaker virulence). ANOVA was used to compare the LgLD50 values. P < 0.05 was deemed to indicate a significant difference.
The comprehensive virulence of various serovars in piglets was compared after normalization by sample size and the amount of bacteria used in the challenge. There were 12 strains of serotype 5 and 8 strains of serotype 4, serotype 5 and serotype 12 respectively. “Blue frame,” “Red frame,” and “Shaded” indicate the corresponding strains of serovar 5 when compared with serovars 4, 13, 12, respectively. The strains of serovars 4, 12, and 13 were compared with those of serovar 5 (eight consecutive strains with similar amounts of challenge bacteria; Table 2). When the average virulence score was used as a reference, a large value indicated strong virulence, whereas a small value indicated weak virulence. Student's t-test was used to evaluate the virulence scores of each type of strain. P < 0.05 indicated a significant difference.
SPSS 25.0 was used for all data analyses.
Results
Virulence of Epidemic Serovars in Mice
After the mice were infected with strains of four G. parasuis serovars, they displayed listlessness and disordered hair, and ceased or reduced their consumption of food. All the dead mice died within the first 3 days of challenge, whereas the surviving mice quickly recovered both their mental state and food intake. The mice in the blank control group showed no symptoms or deaths. G. parasuis was isolated from the blood samples collected from the heart and lungs of the dead mice during autopsy. All the mice infected with G. parasuis showed anorexia, psychiatric disorders, astasia, convulsion, cyanosis with visible mucous membrane. Hemorrhage and congestion were the main pathological features in the heart, liver, and other organs (Supplementary Figure 1).
The LD50 values of the strains were calculated with the Reed-Muench method, and the results (Table 1) showed that the LD50s of eight strains of G. parasuis of serovar 13 were between 1.75 × 108 and 8.45 × 108 CFU. The LD50 values of the three other serovars varied, and could be divided into two intervals. In the eight G. parasuis strains of serovar 4, the LD50s of four strains ranged between 9.80 × 107 and 5.51 × 108 CFU, whereas those of the other four strains ranged between 2.14 × 109 and 4.60 × 109 CFU. In the eight strains of serovar 12, the LD50s of four strains ranged between 4.81 × 107 and 8.64 × 108 CFU, whereas those of the other four strains ranged between 1.58 × 109 and 7.01 × 109 CFU. In the 12 G. parasuis strains of serovar 5, the LD50s of three strains ranged between 2.10 × 108 and 5.26 × 108 CFU, whereas those of the other nine strains ranged between 1.68 × 109 and 8.85 × 109 CFU.
The geometric means of the LgLD50 values were examined to analyze the virulence of each serovar in mice (35, 36) (Table 1). Serovar 13 was most virulent, followed by serovars 4, 12, and 5, which displayed the weakest virulence. However, ANOVA indicated that only the virulence of serovars 5 and 13 differed significantly (P < 0.05), and no significant differences were observed in the virulence of the other strains (P > 0.05; Figure 1).
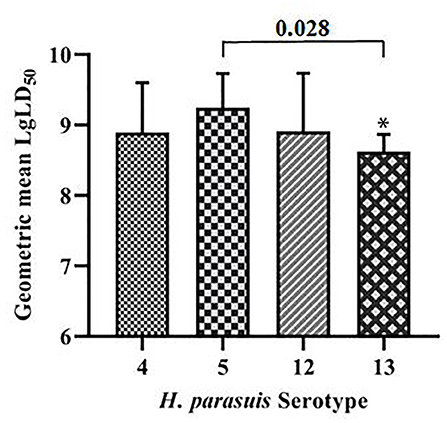
Figure 1. Comparative results of mouse virulence tests for the strains of each serovar. *P < 0.05 and **P < 0.01, compared with G. parasuis serovar 5.
Virulence of Epidemic Serovars in Piglets
None of the piglets in the control group manifested symptoms. However, symptoms typical of G. parasuis disease appeared in the piglets infected with each serovar strain, mainly manifesting as increased body temperature (>40.5°C), depression, loss or diminution of appetite, difficulty in breathing, skin cyanosis, joint swelling, and claudication. In a later stage, disordered hair, weight loss, lying on the ground, and death were observed. The dead piglets were immediately necropsied, and severe serous, fibrinous exudations and sepsis were observed. G. parasuis was isolated from the heart, blood, and lungs of the dead piglets (Figures 2, 3).
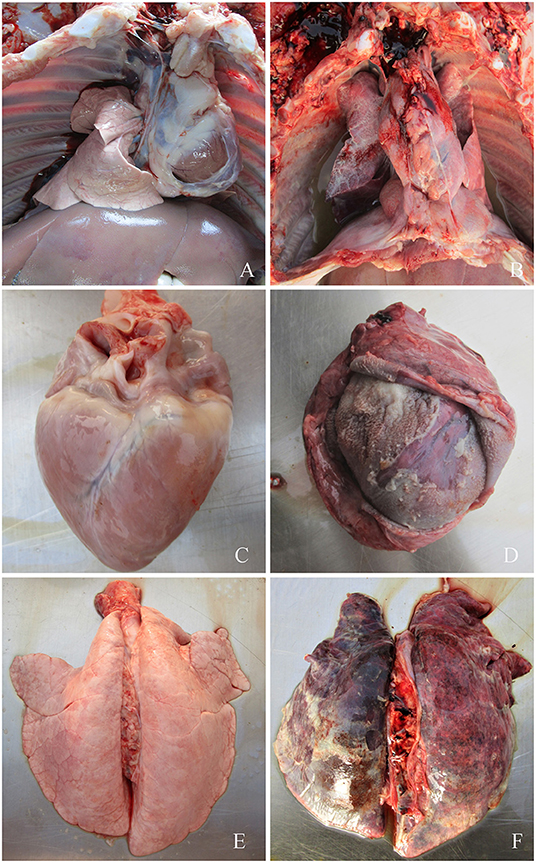
Figure 2. (A) Symptoms of piglets infected with G. parasuis. (A,C,E) control group; (B) pleural effusion; (D) pericardial thickening, typical “fluff heart”; (F) pulmonary congestion, bleeding, necrosis.
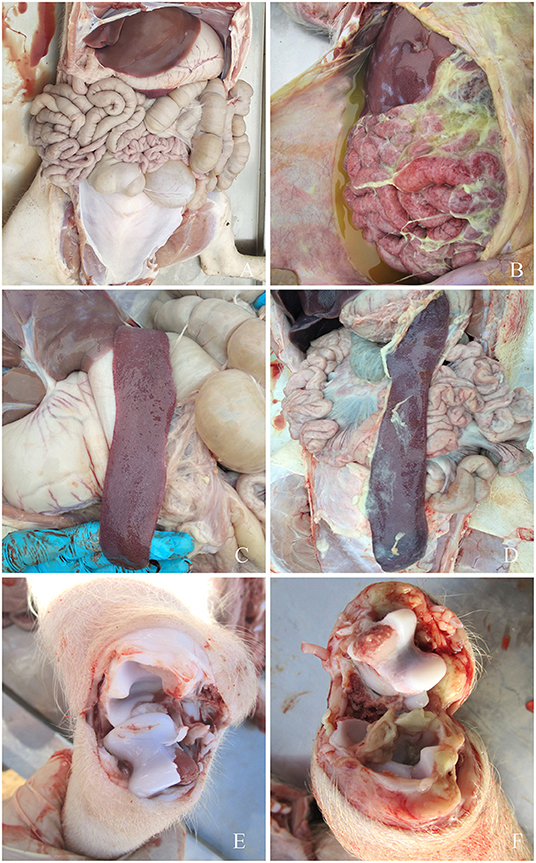
Figure 3. Symptoms of piglets infected with G. parasuis. (A,C,E) control group; (B) peritoneal fibrous exudate, intestinal wall congestion, adhesions; (D) spleen edge infarction, covered with fibrous pseudomembrane; (F) joints effusion.
The detailed results are shown in Table 2. Seven strains of G. parasuis serovar 5 caused the death of all piglets (7/12). The remaining five strains caused a death rate of 60% or above. Three strains of G. parasuis serovar 13 (3/8) caused death in all the piglets, and the remaining five strains resulted in a death rate of 60%. Two strains of G. parasuis serovar 4 caused the death of all piglets (2/8), four strains resulted in a death rate of 60%, and the remaining two strains caused a death rate of only 40%. Serovar 12 showed the weakest overall virulence (Table 2).
The comprehensive virulence of three serovars was compared with that of serovar 5. The comprehensive scores were calculated according to the severity of the onset of symptoms (Table 2) and the results were analyzed statistically. The comprehensive scores for serovar 5 and 4 were 18.3 and 14.9, respectively, which differed significantly (P = 0.039); the comprehensive scores for serovars 5 and 12 were 18.6 and 13.0, respectively, which differed significantly (P = 0.033); the comprehensive scores for serovars 5 and 13 were 18.5 and 17.0, respectively, which differed significantly (P = 0.241). These results showed that serovar 5 had the strongest comprehensive virulence, followed by serovar 13 and 4, and that serovar 12 had the weakest comprehensive virulence. Moreover, the comprehensive virulence of serovar 5 was significantly stronger than that of serovar 4 (P = 0.039) and 12 (P = 0.033; Figure 4).

Figure 4. Comparison of the comprehensive virulence of serovars 4, 12, and 13 with that of serovar 5 in piglets. *P < 0.05 and **P < 0.01, compared with G. parasuis serovars 5.
Comparison of G. parasuis Virulence in Mice and Piglets
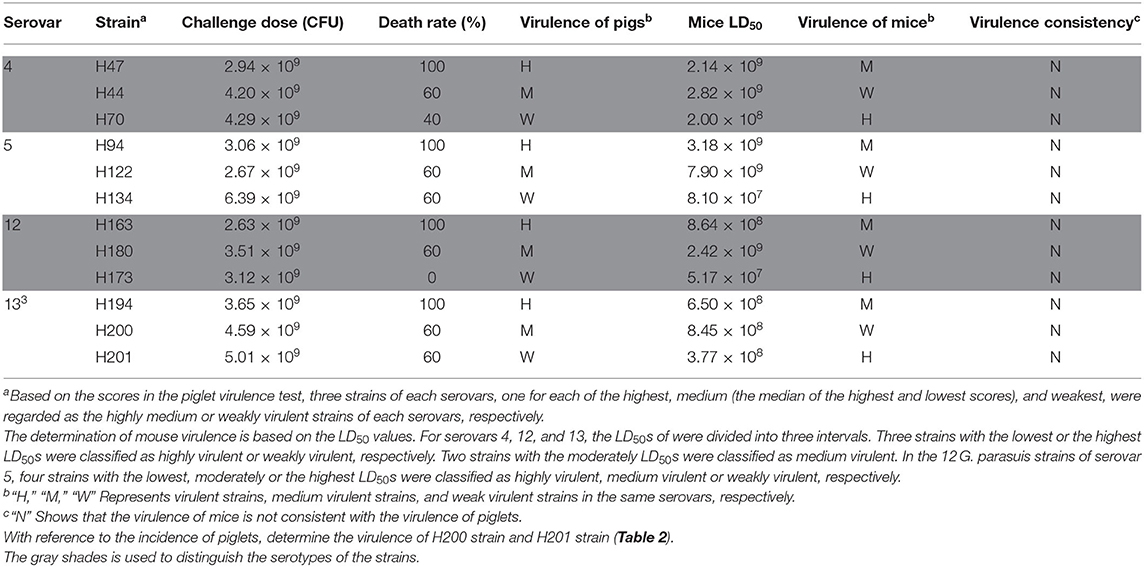
The virulence of the various G. parasuis serovars differed significantly in mice and piglets. Furthermore, the virulence of different strains of the same serovar also varied significantly. However, the comprehensive virulence of each serovar was inconsistent in mice and piglets. Serovar 13 showed the strongest comprehensive virulence in mice, followed by serovar 4, 12, and 5, and the comprehensive virulence of serovar 5 was significantly weaker than that of serovar 13 (P < 0.05). However, G. parasuis serovar 5 showed the strongest virulence in piglets, followed by serovar 13, 4, and 12. Furthermore, the comprehensive virulence of serovar 5 was significantly stronger than that of serovar 4 and 12 in piglets (P < 0.05). The virulence of 36 strains in mice and piglets was compared, and six strains were consistent. Furthermore, one strain showing either strong, medium, or weak virulence in piglets was selected from each serovar and their virulence (LD50) in mice examined to test the consistency of the results (Table 3). However, although G. parasuis could kill mice, the virulence of its various serovars was inconsistent with that in piglets.
Discussion
G. parasuis is a common respiratory bacterium with numerous serovars. It specific culture conditions, and in particular, strictly requires NAD (factor V) for growth (1). Even under the same culture conditions, the growth rates of different G. parasuis strains can vary (37). For example, G. parasuis could not be detected when it was placed in saline or PBS at 42°C for 1 h, at 37°C for 2 h, or at 25°C for 8 h. However, the number of live bacteria was only slightly reduced when it was stored at 5°C for 8 h (38). These observations imply that the actual challenge dose of each strain differed in the piglet virulence tests in that study; The minimum challenge dose was 2.63 × 109 CFU and the maximum dose was 8.61 × 109 CFU. In 2004, Oliveira reported that the intraperitoneal inoculation of piglets with 108-109 CFU of G. parasuis resulted in fibrinous polyserositis, arthritis, and meningitis, whereas piglets inoculated with 106–107 CFU showed no symptoms (39). The results of our study using 8–9-week-old G. parasuis-seronegative piglets challenged with 36 G. parasuis strains showed that the intraperitoneal injection of G. parasuis successfully caused Glässer's disease, and could be used to evaluate the virulence of different strains.
According to the Kielstein-Rapp-Gabrielson serotyping scheme, serovars 5, 12, and 13 are classified as strongly virulent, and serovar 4 is classified as moderately virulent (2). However, the piglet virulence test results showed that the comprehensive virulence of the four serovars followed the order: serovar 5 > 13 > 4 > 12. Although some strains of serovars 5 and 13 caused 100% death rates among the inoculated piglets, and were therefore strongly virulent strains (strains H94 and H194), some strains did not cause a 100% death rate in the inoculated pigs, and were therefore moderately virulent or attenuated strains (strains H134 and H201). All eight strains of serovar 4 caused the death of some inoculated piglets, exhibiting virulent strains (strains H47 and H57), but it could not be identified as a moderately virulent serovar in general. The results for serovar 12 were the most divergent among the strains, insofar as strain H163 was strongly virulent, whereas strains H170, H173, and H157 caused no deaths (although disease onset was observed) and therefore display attenuated virulence. These results are inconsistent with the serological classification, suggesting that there is no correlation between the serovar and virulence of G. parasuis. Aragon et al. also showed that some strains of virulent serovars 4 and 10 showed no pathogenicity to NFCD pigs, whereas in contrast, some strains of avirulent serovar 7 caused severe disease in piglets (40). Yu et al. studied 10 G. parasuis strains and demonstrated that strains of the same serovars showed different virulence in 4-week-old York piglets (25), indicating that in G. parasuis, serovar does not necessarily correlate with virulence. In fact, the relationship between serovar and virulence in G. parasuis is not well-established because the virulence of serovars was established using only reference strains (3, 17).
Strain classification of G. parasuis has been widely studied serotypically and genotypically, since differentiation of non-virulent strains from virulent strains is essential for diagnosis and control of the disease. Serotyping is the most commonly used subtyping method, and is traditionally considered to be associated with virulence, but increasing evidence indicated that serovar is a poor proxy for virulence (14, 40, 41). To resolve this problem, such as genome-wide association study (GWAS), enterobacterial repetitive intergenic consensus polymerase chain reaction (ERIC-PCR) (42), multiplex PCR (mPCR) (43) and multilocus variable number of tandem repeats analysis (MLVA) (44), have been used to differentiate G. parasuis strains and predicte virulence-associated genes. Although several studies have identified putative virulence-associated genes, their role in pathogenesis has not been demonstrated (45–48). It is probable that the virulence of G. parasuis strains may have varied due to vertical and/or horizontal transfer(s) of DNA in the past 20 years. Thus, it was not exact to decide the virulence only by the Kielsteine-Rappe-Gabrielson serotyping scheme. The animal challenge and gene virulence detection would be useful supplements to the Kielsteine-Rappe-Gabrielson serotyping scheme, which evaluates bacterial virulence of G. parasuis.
We used BALB/c mice as an alternative model of G. parasuis infection to test the virulence of the four most domestically common serovars of the bacterium. The results showed that the LD50s of serovar 13 were between 1.75 × 108 and 8.45 × 108 CFU, whereas the LD50s of the other three serovars varied more widely. These results are consistent with those of many scholars, who showed that the use of intraperitoneal injection of G. parasuis causes death in BALB/c mice (49, 50); However, these scholars didn't compare the virulence of these strains in piglets. We speculated that different infection routes would produce different virulence results. Gao et al. (51) studied various inoculation routes of G. parasuis in guinea pigs and demonstrated that intraperitoneal or intrapulmonary routes were more sensitive than intranasal, intramuscular, and subcutaneous routes (52, 53). In addition, considering the experimental operation, abdominal infection is simpler and easier to operate than other infection methods. To compare the virulence of G. parasuis in mice and piglets in this study, the intraperitoneal injection was used in mice and piglets. The results of our virulence tests in BALB/c mice were not consistent with those in piglets. The mouse virulence test results showed that serovar 13 displayed the strongest virulence, followed by serovars 4, 12, and 5, and that the virulence of serovar 5 was significantly weaker than that of serovar 13 (P < 0.05). However, the piglet virulence test results showed that serovar 5 was most virulent, followed by serovars 13, 4, and 12, and that the virulence of serovars 5 and 13 did not differ significantly (P > 0.05). Our results also showed that even the same strain of G. parasuis could display different virulence in mice and piglets. From an anatomical pathology perspective, we found that the BALB/c mice predominantly died from internal organ bleeding. Because pigs are a natural host of G. parasuis, the virulence test results for piglets better reflect the virulence of these common strains, our results also indicate that BALB/c mice are inadequate as an alternative model of G. parasuis infection.
Conclusion
In this study, the virulence of the commonest serovars (4, 5, 12, and 13) of G. parasuis in China (a total of 36 strains) was compared in BALB/c mice and piglets. Three conclusions can be drawn, from our results: (1) in piglets, the virulence of serovar 5 was the strongest, followed by that of serovars 13, 4, and 12; (2) both virulent and avirulent strains were present in all the serovars and there was no correlation between serovar and virulence; and (3) although G. parasuis caused death in BALB/c mice, the virulence test results of mice were not consistent with those of piglets, indicating that BALB/c mice are inadequate as an alternative model of G. parasuis infection.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
All procedures performed in studies involving animals were in accordance with the Animal Experiment Committee of Henan University of Science and Technology (No. 20180719009). The samples were collected and handled in accordance with the good animal practices required by the Animal Ethics Procedures and Guidelines of the People's Republic of China.
Author Contributions
ZZ designed the study. BQ performed experiments. BQ, FL, KC, WD, YW, YX, HW, and KD performed data analysis. BQ and ZZ wrote the draft and revised the manuscript. All authors have read and approved the manuscript.
Funding
This study was supported by grants from the National Natural Science Foundation of China (U1704117 and 32072899) and the Research and Development Foundation of Henan University of Science and Technology (2015ZDCXY04). The funding body had no role in the design of the study, collection, analysis, or interpretation of data or in the writing of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank International Science Editing Ltd. for editing the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.659244/full#supplementary-material
Supplementary Figure 1. Symptoms of mice infected with G. parasuis. (A–F) pleural effusion, peritoneal fibrous exudate; (G) liver hemorrhage, congestion and necrosis; (H) pulmonary congestion and hemorrhage; (I) heart congestion.
Supplementary Figure 2. The vitro growth curve for 36 isolates in 6–14 h. A single colony of G. parasuis was picked and cultured in TSB medium containing newborn calf serum (10%) and NAD (10 μg/mL), with shaking at 180 rpm at 37°C for 12–16 h. The culture solution was used as the stock solution and diluted 1:100, transferred to TSB medium, and cultured with shaking at 180 rpm at 37°C. From 6 to 14 h, the number of living bacteria in the bacterial liquid was recorded every 2 h.
Abbreviations
CDCD, Cesarean-derived, colostrum-deprived; NFCD, Naturally-arrowed, artificially-reared; NAD, Nicotinamide adenine dinucleotide; MAT, Microplate agglutination test; LD50, Median lethal dose; ANOVA, Analysis of variance; TSB, Tryptic soy broth; TSA, Tryptic soy agar; CFU, Colony–forming units.
References
1. Chen F. Veterinary Infectious Disease. 6th ed. Beijing: China Agriculture Ture Press. (2015). p. 212–269.
2. Ma L, Wang L, Chu Y, Li X, Cui Y, Chen S, et al. Characterization of Chinese Haemophilus parasuis isolates by traditional serotyping and molecular serotyping methods. PLoS ONE. (2016) 11:e0168903. doi: 10.1371/journal.pone.0168903
3. Kielstein P, Rapp-Gabrielson VJ. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J Clin Microbiol. (1992) 30:862–865. doi: 10.1128/jcm.30.4.862-865.1992
4. Tadjine M, Mittal KR, Bourdon S, Gottschalk M. Development of a new serological test for serotyping Haemophilus parasuis isolates and determination of their prevalence in North America. J Clin Microbiol. (2004) 42:839–40. doi: 10.1128/JCM.42.2.839-840.2004
5. Cai X, Chen H, Blackall PJ, Yin Z, Wang L, Liu Z, et al. Serological characterization of Haemophilus parasuis isolates from China. Vet Microbiol. (2005) 111:231–6. doi: 10.1016/j.vetmic.2005.07.007
6. Castilla KS, de Gobbi DDS, Moreno LZ, Paixão R, Coutinho TA, dos Santos JL, et al. Characterization of Haemophilus parasuis isolated from Brazilian swine through serotyping, AFLP and PFGE. Res Vet Sci. (2012) 92:366–71. doi: 10.1016/j.rvsc.2011.04.006
7. Luppi A, Bonilauri P, Dottori M, Iodice G, Gherpelli Y, Merialdi G, et al. Haemophilus parasuis serovars isolated from pathological samples in Northern Italy. Transbound Emerg Dis. (2013) 60:140–2. doi: 10.1111/j.1865-1682.2012.01326.x
8. Rúbies X, Kielstein P, Costa L, Riera P, Artigas C, Espuña E. Prevalence of Haemophilus parasuis serovars isolated in Spain from 1993 to 1997. Vet Microbiol. (1999) 66:245–8. doi: 10.1016/S0378-1135(99)00007-3
9. Morikoshi T, Kobayashi K, Kamino T, Owaki S, Hayashi S, Hirano S. Characterization of Haemophilus parasuis isolated in Japan. Nippon Juigaku Zasshi. (1990) 52:667–9. doi: 10.1292/jvms1939.52.667
10. Angen O, Svensmark B, Mittal KR. Serological characterization of Danish Haemophilus parasuis isolates. Vet Microbiol. (2004) 103:255–8. doi: 10.1016/j.vetmic.2004.07.013
11. Rapp-Gabrielson V, Kocus G, Clark J. Virulence of Different Serovars of Haemophilus parasuis for Cesarean-Derived, Colostrum-Deprived Pigs in Haemophilus, Actinobacillus, and Pasteurella. New York, NY: Plenum Press (1995) 204 p.
12. Metcalf DS, MacInnes JI. Differential expression of Haemophilus parasuis genes in response to iron restriction and cerebrospinal fluid. Can J Vet Res. (2007) 71:181–8.
13. Howell KJ, Weinert LA, Chaudhuri RR, Luan SL, Peters SE, Corander J, et al. The use of genome wide association methods to investigate pathogenicity, population structure and serovar in Haemophilus parasuis. BMC Genomics. (2014) 15:e1179. doi: 10.1186/1471-2164-15-1179
14. Brockmeier SL, Register KB, Kuehn JS, Nicholson TL, Loving CL, Bayles DO, et al. Virulence and draft genome sequence overview of multiple strains of the swine pathogen Haemophilus parasuis. PLoS ONE. (2014) 8:e103787. doi: 10.1371/journal.pone.0103787
15. Galofré-Milà N, Correa-Fiz F, Lacouture S, Gottschalk M, Strutzberg-Minder K, Bensaid A, et al. A robust PCR for the differentiation of potential virulent strains of Haemophilus parasuis. BMC Vet Res. (2017) 13:124. doi: 10.1186/s12917-017-1041-4
16. Li G, Niu H, Zhang Y, Li Y, Xie F, Langford PR, et al. Haemophilus parasuis cytolethal distending toxin induces cell cycle arrest and p53-dependent apoptosis. PLoS ONE. (2017) 12:e0177199. doi: 10.1371/journal.pone.0177199
17. Nielsen R. Pathogenicity and immunity studies of Haemophilus parasuis serotypes. Acta Vet Scand. (1993) 34:193–8. doi: 10.1186/BF03548209
18. Amano H, Shibata M, Kajio N, Morozumi T. Pathogenicity of Haemophilus parasuis serovars 4 and 5 in contact-exposed pigs. J Vet Med Sci. (1996) 58:559–61. doi: 10.1292/jvms.58.559
19. Vahle JL, Haynes JS, Andrews JJ. Experimental reproduction of Haemophilus parasuis infection in swine: clinical, bacteriologic, and morphologic findings. J Vet Diagnostic Investig. (1995) 7:476–80. doi: 10.1177/104063879500700409
20. Oliveira S, Galina L, Blanco I, Canals A, Pijoan C. Naturally-farrowed, artificially-reared pigs as an alternative model for experimental infection by Haemophilus parasuis. Can J Vet Res. (2003) 67:146–50.
21. Cai X. Isolation, Identification and Diagnosis of Haemophilus parasuis and Study on Inactivated Vaccine. Wuhan: Huazhong Agricultural University (2006).
22. Zhao Z, Liu H, Xue Y, Chen K, Liu Z, Xue Q, et al. Analysis of efficacy obtained with a trivalent inactivated Haemophilus parasuis serovars 4, 5, and 12 vaccine and commercial vaccines against Glässer's disease in piglets. Can J Vet Res. (2017) 81:22–7.
23. Morozumi T, Hiramune T, Kobayashi K. Glasser's disease in piglets produced by intraperitoneal inoculation with Haemophilus parasuis. Natl Inst Anim Health Q. (1981) 21:121–8.
24. Zhao G, Li X, Dai A, Yang S, Yang X. Isolation and identification of Haemophilus parasuis and the study of its virulence in the west of Fujian province. Heilong Anim Sci Vet Med. (2015) 10–13. doi: 10.13881/j.cnki.hljxmsy.2015.0651
25. Yu J, Zhang Y, Du Y, Li J, Huang B, Sun W, et al. The BALB/c mouse infection model for improving the Haemophilus parasuis serotyping scheme. Res Vet Sci. (2016) 109:166–8. doi: 10.1016/j.rvsc.2016.10.011
26. Gao P, Chu Y, Zhao P, He Y, Lu Z. Pathogenicity test of Haemophilus parasuis on some commonly used experimental animals. Acta Vet Zootech Sinica. (2009) 40:138–41. doi: 10.3321/j.issn:0366-6964.2009.01.024
27. He Y, Xu H, Ye F, Sun H, Wang H, Gong Y, et al. Study on the virulence of 15 standard isolates of Haemophilus parasuis in mice. Heilong Anim Sci Vet Med. (2011) 90–9. doi: 10.13881/j.cnki.hljxmsy.2011.17.060
28. Wang Z, Liu M J, Wang L, Shao G. Pathogenicity of Haemophilus parasuis to guinea pigs and mice. Chin J Vet Med. (2015) 51:96–8. doi: 10.3969/j.issn.0529-6005.2015.05.032
29. Tian H, Fu F, Li X, Chen X, Wang W, Lang K, et al. Identification of the immunogenic outer membrane protein A antigen of Haemophilus parasuis by a proteomics approach and passive immunization with monoclonal antibodies in mice. Clin Vaccine Immunol. (2011) 18:1695–701. doi: 10.1128/CVI.05223-11
30. Howell KJ, Peters SE, Wang J, Hernandez-Garcia J, Weinert LA, Luan SL, et al. Development of a multiplex PCR assay for rapid molecular serotyping of Haemophilus parasuis. J Clin Microbiol. (2015) 53:3812–21. doi: 10.1128/JCM.01991-15
31. Xue Q, Xue Y, Zhao Z, Chen K, Liu H, Wang L. First comparison of adjuvant for trivalent inactivated Haemophilus parasuis serovars 4, 5 and 12 vaccines against Glässer's disease. Vet Immunol Immunopathol. (2015) 168:153–8. doi: 10.1016/j.vetimm.2015.11.001
32. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol. (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
33. Ding W. Study on the pathogenic epidemiology and biological characteristics of Haemophilus parasuis. Luoyang: Henan University of Science and Technology (2019). Available online at: https://d.wanfangdata.com.cn/thesis/D01765747
34. Wang L, Zhao Z, Xue Y, Liu H, Deng W. Comparative study on pathogenicity of the most prevalent serovars of Haemophilus parasuis isolates in China. Chin J Prevent Vet Med. (2014) 34:1748–52.
35. Shi T, Li Y, Yu Y. Comparison of neurovirulence and full-length sequence of two rabies virus clones with different virulence. Chin J Zoonoses. (2018) 34:986–90. doi: 10.3969/j.issn.1002-2694.2018.00.181
36. Shi Q, Lan P, Huang D, Hua X, Jiang Y, Zhou J, et al. Diversity of virulence level phenotype of hypervirulent Klebsiella pneumoniae from different sequence type lineage. BMC Microbiol. (2018) 94:e1186. doi: 10.1186/s12866-018-1236-2
37. Xu H, Xue Y, Zhao Z, Chen K, Wang L, Deng W. Comparative study on biological characteristics of Haemophilus parasuis serotypes in China. Chin J Prevent Vet Med. (2014) 34:729–35.
38. Morozumi T, Hiramune T. Effect of temperature on the survival of Haemophilus parasuis in physiological saline. Natl Inst Anim Health Q. (1982) 22:90–1. doi: 10.1007/BF02282589
39. Oliveira S, Pijoan C. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol. (2004) 99:1–12. doi: 10.1016/j.vetmic.2003.12.001
40. Aragon V, Cerdà-Cuéllar M, Fraile L, Mombarg M, Nofrarías M, Olvera A, et al. Correlation between clinico-pathological outcome and typing of Haemophilus parasuis field strains. Vet Microbiol. (2010) 142:387–93. doi: 10.1016/j.vetmic.2009.10.025
41. Lawrence PK, Wiener BL, Kolander-Bremer T, Bey RF, Stine DL, Kittichotirat W, et al. Genome-wide association studies of virulent and avirulent Haemophilus parasuis serotype 4 strains. Genome Announce. (2014) 2:e00884–14. doi: 10.1128/genomeA.00884-14
42. Lin WH, Shi CH, Lin CF, Yang C, Lin C, Chiou M. Genotypic analyses and virulence characterization of Glaesserella parasuis isolates from Taiwan. Peer J. (2019) 7:e6960. doi: 10.7717/peerj.6960
43. Howell KJ, Weinert LA, Wang JH, Peters SE, Wang JH, Luan S, et al. “Pathotyping” multiplex PCR Assay for Haemophilus parasuis: a Tool for prediction of virulence. J Clin Microbiol. (2017) 14:2617–8. doi: 10.1128/JCM.02464-16
44. Boerlin P, Poljak Z, Gallant J, Chalmers G, Nicholson V, Soltes G, et al. Genetic deversity of Haemophilus parasuis from sick and healthy pigs. Vet Microbiol. (2013) 167:459–67. doi: 10.1016/j.vetmic.2013.07.028
45. Li JX, Xu LH, Su F, Yu B, Yuan X. Association between iscR-based phylogeny, serovars and potential virulence markers of Haemophilus parasuis. Peer J. (2019) 5:14:e6950. doi: 10.7717/peerj.6950
46. Chen H, Li C, Fang M, Zhu M, Li X, Zhou R, et al. Understanding Haemophilus parasuis infection in porcine spleen through a transcriptomics approach. BMC Genom. (2009) 10:64. doi: 10.1186/1471-2164-10-64
47. Pina S, Olvera A, Barcelo A, Bensaid A. Trimeric autotransporters of Haemophilus parasuis: generation of an extensive passenger domain repertoire specific for pathogenic strains. J Bacteriol. (2009) 191:576–87. doi: 10.1128/JB.00703-08
48. Sack M, Baltes N. Identification of novel potential virulence-associated factors in Haemophilus parasuis. Vet Microbiol. (2009) 136:382–6. doi: 10.1016/j.vetmic.2008.11.008
49. Li A, Xu C, Zhang J, Guo L, Zhang B, He Y, et al. Biological characteristics and pathogenicity of two Haemophilus parasuis strains in South China. China Anim Husb Vet Med. (2011) 38:179–82.
50. Lin B, Liu J, Li X, Yang X. Biological characteristics and pathogenicity of Haemophilus parasuis serotypes 4 and 5. J Longyan Univer. (2013) 31:61–4. doi: 10.3969/j.issn.1673-4629.2013.05.014
51. Gao P, Chu Y, Zhao P, He Y, Lu Z. Experimental infections of guinea pigs with Haemophilus parasuis and its tissue distribution. Chin J Anim Vet Sci. (2009) 40:1376–82.
52. Chen Q. Selection of Attenuated Haemophilus parasuis Strains Using Guinea Pigs Model. Wuhan: Huazhong Agricultural University (2016).
Keywords: Glaesserella parasuis, serovar, virulence, mouse, piglet, correlation
Citation: Qi B, Li F, Chen K, Ding W, Xue Y, Wang Y, Wang H, Ding K and Zhao Z (2021) Comparison of the Glaesserella parasuis Virulence in Mice and Piglets. Front. Vet. Sci. 8:659244. doi: 10.3389/fvets.2021.659244
Received: 27 January 2021; Accepted: 24 May 2021;
Published: 24 June 2021.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Jianmin Zhang, South China Agricultural University, ChinaYinsheng Qiu, Wuhan Polytechnic University, China
Chao-Nan Lin, National Pingtung University of Science and Technology, Taiwan
Copyright © 2021 Qi, Li, Chen, Ding, Xue, Wang, Wang, Ding and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhanqin Zhao, zhaozhanqin@126.com
 Baichuan Qi
Baichuan Qi Feiyue Li1
Feiyue Li1 Yang Wang
Yang Wang Hongwei Wang
Hongwei Wang