
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci., 15 July 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.647462
This article is part of the Research TopicNewcastle Disease in Low- and Middle-Income Countries: Challenges and Strategies for ControlView all 5 articles
 Shimaa M. G. Mansour1*†
Shimaa M. G. Mansour1*† Reham M. ElBakrey2†
Reham M. ElBakrey2† Fakry F. Mohamed1†
Fakry F. Mohamed1† Esraa E. Hamouda2
Esraa E. Hamouda2 Mona S. Abdallah3
Mona S. Abdallah3 Ahmed R. Elbestawy4
Ahmed R. Elbestawy4 Mahmoud M. Ismail5
Mahmoud M. Ismail5 Hanan M. F. Abdien3
Hanan M. F. Abdien3 Amal A. M. Eid2*
Amal A. M. Eid2*Avian orthoavulavirus 1, formerly known as avian paramyxovirus type-1 (APMV-1), infects more than 250 different species of birds. It causes a broad range of clinical diseases and results in devastating economic impact due to high morbidity and mortality in addition to trade restrictions. The ease of spread has allowed the virus to disseminate worldwide with subjective virulence, which depends on the virus strain and host species. The emergence of new virulent genotypes among global epizootics, including those from Egypt, illustrates the time-to-time genomic alterations that lead to simultaneous evolution of distinct APMV-1 genotypes at different geographic locations across the world. In Egypt, the Newcastle disease was firstly reported in 1947 and continued to occur, despite rigorous prophylactic vaccination, and remained a potential threat to commercial and backyard poultry production. Since 2005, many researchers have investigated the nature of APMV-1 in different outbreaks, as they found several APMV-1 genotypes circulating among various species. The unique intermingling of migratory, free-living, and domesticated birds besides the availability of frequently mobile wild birds in Egypt may facilitate the evolution power of APMV-1 in Egypt. Pigeons and waterfowls are of interest due to their inclusion in Egyptian poultry industry and their ability to spread the infection to other birds either by presence of different genotypes (as in pigeons) or by harboring a clinically silent disease (as in waterfowl). This review details (i) the genetic and pathobiologic features of APMV-1 infections in Egypt, (ii) the epidemiologic and evolutionary events in different avian species, and (iii) the vaccine applications and challenges in Egypt.
Newcastle disease (ND) is a highly contagious notifiable viral disease with significant clinical impact and heavy economic losses to the poultry industry worldwide (1). It is on the list A of OIE and ranked as the second-highest endemic disease in many countries (2). In developing or developed countries where chickens are raised in small household or commercial sectors, ND has a significant economic impact on poultry sector due to high mortality rates (up to 100%), decreased productivity, and disease prevention and control expenses as well as trade restrictions (1).
Faulty vaccination programs and incorrect administration, transport, and storage of vaccine, as well as concurrent infection, may play a role in maximizing the losses and impact of ND field challenge. Heterogeneous genotype cross-protection is controversial. Repeated ND virus (NDV) infections even in vaccinated birds could be attributed to improper vaccination and immune suppression along with viral mutation (3). The molecular epidemiology and sequence analysis of NDV in Egypt are important to determine the current situation and available control measures (4). In Egypt, NDV was firstly identified in 1947 (5) by virus isolation in embryonated chicken eggs (ECEs) and then identification using hemagglutination inhibition (HI) assay. Despite extensive vaccination programs against ND in both commercial and backyard poultry flocks across Egypt, many outbreaks have occurred since then, resulting in huge economic losses. This raises the inquiries about the genetic diversity of the indigenous strains as well as the feasibility of commercial vaccines on protection against circulating NDVs (6). Besides, mixed infections in birds with NDV, and other viral infections such as infectious bronchitis virus (IBV), avian influenza viruses (AIVs) (H5Nx or H9N2), or avian reovirus, usually aggravate losses (4, 7–10).
The urgent and long-awaited questions had affirmative answers: Do different genotypes change the keys to deal with prevention of ND and change the strategies of making effective vaccines in protecting against NDV? Are traditional vaccines on their way to extinction? For better answers, there is a need to analyze the outcome of available studies to achieve the most of the divine gift that concerns natural lentogenic and avirulent strains for protection against circulating velogenic NDVs. This review provides a historical overview of NDV status in Egypt, concerning the pathobiology and epidemiology of NDV and timely genetic and evolutionary insights in the virus genome, along with the background and rationale of vaccine strategies and challenges, aiming to develop insights toward solving the NDV endemic situation in Egypt.
The avian orthoavulavirus 1, also known as NDV, is an enveloped, negative-sense, single-stranded RNA virus that belongs to the family Paramyxoviridae under the order Mononegavirales (11). The viral genome is around 15,200 bp in length and encodes for six different protein, namely, nucleocapsid protein (NP), phosphoprotein (P), matrix (M) protein, large RNA polymerase (L), fusion (F) protein, and hemagglutinin–neuraminidase (HN). The F and HN surface glycoproteins are involved in the antigenicity and pathogenicity of NDV (12). Two other proteins (V and W) could also be coded through P protein mRNA editing (13).
Based on the phylogenetic analysis of F gene sequences, the NDV can be divided into two classes (I and II): class I usually includes avirulent viruses, and their natural reservoir is aquatic wild birds (14), whereas class II consists of at least 20 genotypes (I–XXI, as genotype XV that contains only recombinant sequences was excluded from the final analyses) and includes both avirulent and virulent strains (15–17). According to its pathogenicity, NDV is categorized into five pathotypes: asymptomatic enteric, lentogenic, mesogenic, neurotropic velogenic, and viscerotropic velogenic (18). One major determinant of NDV virulence is the F protein cleavage site (19). The lentogenic strains have a monobasic amino acid (aa) motif 112GR/K-Q-GR↓L117, while velogenic and mesogenic strains have a multi-basic aa motif, 112R/G/K-R-Q/KK/R-R↓F117 (20).
Pigeon paramyxovirus type-1 (PPMV-1) is an antigenic variant of Avian orthoavulavirus 1 and is known to infect pigeons, doves, wild birds, and domestic poultry. It induces nervous manifestations similar to the nervous form of ND, with few evident respiratory signs (21, 22). PPMV-1 can be differentiated from classical NDV by HI test, monoclonal antibodies, and restriction enzyme analysis of F gene. Phylogenetically, PPMV-1 isolates are classified as a distinct sub-genotype within genotype VI of class II (sub-genotype VIb) (23). Four major panzootics of ND have been reported in different avian species (24). The third outbreak, mostly affecting pigeons and doves, was caused by PPMV-1, which originated in the Middle East (Iraq) in the late 1970s and then spread rapidly to Europe (25, 26). Now, PPMV-1 is endemic in domestic and feral pigeons in many areas of the world including the USA and Europe (27). Despite the control measures, PPMV-1 remained enzootic in several countries, including Egypt, causing economic losses (7, 22).
The ND affects a wide range of domestic and wild birds, with greatly varying pathogenicity, spanning from peracute disease (with up to 100% mortality) to asymptomatic disease. Such variability makes it challenging to sort ND as a single clinicopathological entity. Over 250 species of birds are susceptible to NDV infection. It is known that PPMV-1 attacks mainly pigeons and less frequently chickens. In addition, there are cases of virus infection in birds kept in captivity and wild birds, including pheasants, partridges, falcons, swans, cockatoos, blackbirds, and budgerigars (28–32).
In this review, we represent a nationwide prospective of NDV in Egypt, including the ecology and prevalence in different avian species (Figure 1), vaccine approaches, and molecular aspects of previously isolated NDV strains, which shall hopefully reveal the potential causes of NDV emergence and dissemination in Egypt.
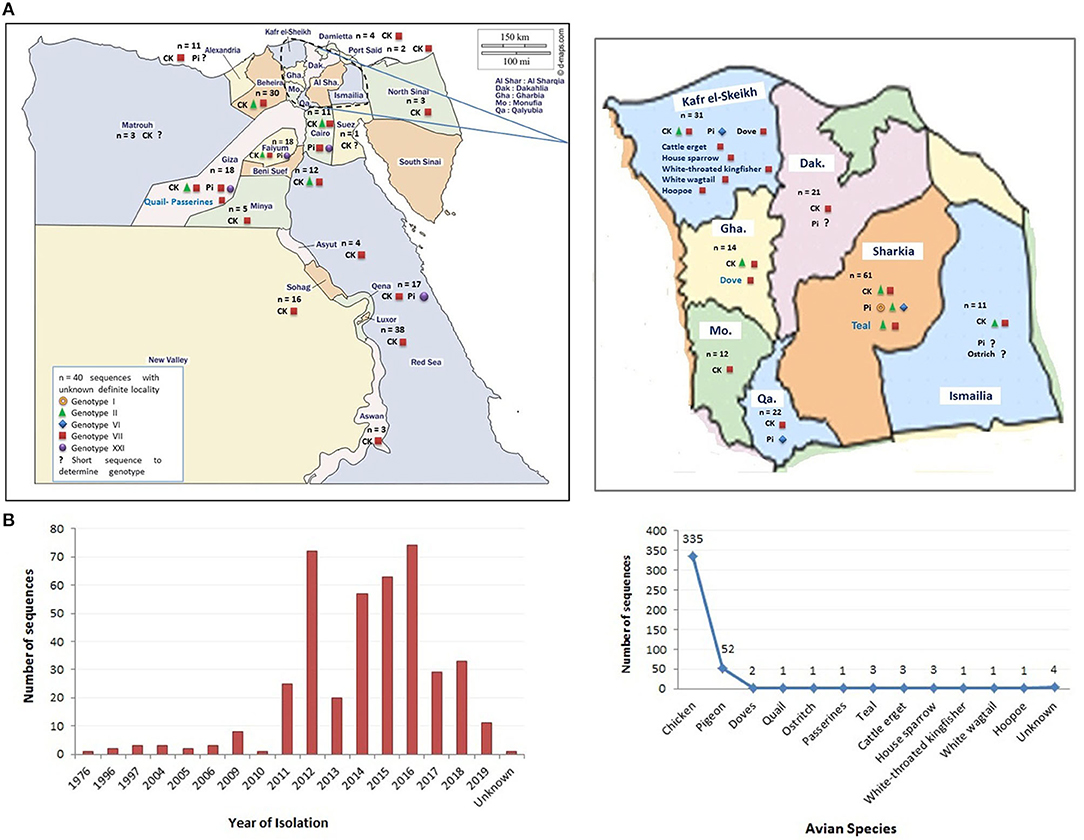
Figure 1. The distribution of Newcastle disease virus (NDV) strains (sequences) in Egypt. (A) The geographical detection of NDV in different Egyptian provinces (data obtained from GenBank in early December 2020). (B) The grouping of detected NDV strains according to year of detection and species of origin. Color codes are indicated in the figures.
Chickens are the most commonly affected avian species. Clinically, five NDV pathotypes were categorized. Additionally, in vivo testing in embryos or chickens/pigeons can be done to determine strain pathogenicity, including mean death time (MDT), intravenous (IV) pathogenicity index (IVPI), and intracerebral pathogenicity index (ICPI). The MDT is the time to death, measured in hours, after inoculation of ECEs. The MDT for the NDV strains, <60, 60–90, or >90 h, was considered velogenic, mesogenic, or lentogenic strains, respectively (2, 33). The IVPI involves scoring illness (0 = normal; 1 = sick; 2 = paralyzed or nervous signs; and 3 = death) after IV inoculation of 6-week-old chickens. The velogenic strains have an IVPI score of 2–3, mesogenic 0–0.5, and lentogenic of zero (34). Now, the definitive in vivo assessment of NDV virulence is based on the ICPI test. It is considered as the most sensitive and widely used test for measuring the virulence in ten 1-day-old chicks (2, 35). The ICPI involves scoring sick or dead (0 = normal; 1 = sick; and 2 = dead). Scores with an ICPI of 1.5–2 is considered velogenic, 1–1.5 mesogenic, and 0–0.5 lentogenic. ICPI score of ≥0.7 is considered “notifiable” to the OIE (2). Particular drawbacks of pathogenicity indices are in the interpretation of pathotype results. A previous study (36) reported 10 NDV isolates from pigeons to have ICPI values (1.2–1.45) and IVPI values (0–1.3), classifying the isolates as virulent. However, MDT was low (98 h) in lentogenic strains. Indeed, not all virulent strains have an MDT <60 h. The in vivo tests on NDV isolates from species other than chickens can present some problems and may not produce accurate interpretations until passaged in chickens or ECEs (37). The course of the NDV infection can vary widely depending on the virulence of the virus. Several studies were conducted in Egypt on the in vivo pathogenicity of NDV field isolates (Table 1). Earlier, in a surveillance among more than 100 chicken flocks at Sharkia Province in late 1980s, 26 NDVs were isolated, velogenic (n = 15), mesogenic (n = 9), and lentogenic (n = 2) isolates, from vaccinated birds, besides one mesogenic from egg shell and other lentogenic from drinking water. The velogenic NDVs showed IVPI (1.5–1.8) in chickens; however, higher figures of ICPI (2.3–3.7) were recorded in pigeon squabs (38). The values of pathogenicity indices of Egyptian NDV isolates (Table 1) revealed that the majority of NDV strains were velogenic. The MDT (36–60 h), IVPI values of 1.5–3, and ICPI scores (1.6–2.0) are indicative for velogenic nature NDV isolates. However, for mesogenic strains, the MDT and ICPI were >60 h and 0.5–1.5, respectively. Besides, 96–108 h and 0.38–0.44 for MDT and ICPI, respectively, indicate the lentogenic nature of strains. The aforesaid findings are consistent with the sequence analysis of F protein cleavage site. In contrast, Nagy et al. (51) characterized 13 NDV isolates from vaccinated chicken flocks during 2014–2017; two of them, Ck/ME3/Eg/16 and Ck/ME5/Eg/16, had the 112GRQGRL117 cleavage motif characteristic to lentogenic strains, although they showed MDT <36–48 h and high ICPI (1.89–2.00) congruent for velogenic pathotype. Similarly, Naguib et al. (53) recorded that the sequence of isolate R1973/11 represents a cleavage site of lentogenic viruses; however, it had an ICPI of 1.88, clearly categorizing the isolate as velogenic. The applied molecular pathotyping by RT-qPCR using the primers and probes specific for avirulent and virulent pathotypes revealed that two Egyptian NDV isolates during 2011 were positive for lentogenic and virulent pathotypes, indicating mixed infection.
In Egypt, NDV infection results in variable mortalities and clinical findings. The clinical signs observed in commercial broiler chickens were severe depression, green diarrhea, paresis, and death within 48–72 h after the onset of the disease. Other signs including severe conjunctivitis, facial swelling, and birds standing dull with drooping wings were recorded in many studies. Besides, a drop in egg production reached 50% in layer flocks. Necropsies revealed congestion of the meningeal blood vessels and signs of septicemia in the form of congested subcutaneous (SC) blood vessels; congestion of the liver, spleen, and lungs; and gallbladder enlargement. Tracheitis and airsacculitis were seen in the respiratory tract. Hemorrhages on the tips of the proventriculus gland, greenish mucous content in the gastrointestinal tract, elliptical raised ulcers in the intestine, and enlarged hemorrhagic cecal tonsils were also observed (40, 110, 111). Variable mortalities (10–100%) were recorded in backyard and commercial vaccinated and non-vaccinated broiler poultry flocks (9, 49, 111–113). The clinical signs were respiratory distress and elevated mortality with nervous manifestation and deaths occurring with 24–48 h after the onset of clinical signs (48).
National efforts to update the knowledge about NDV/AIV prevalence and an active surveillance undertaken on 195 broilers and layers farms from 18 Egyptian provinces resulted in 41/195 (21%) positive for matrix gene of NDV and 24/195 (12%) positive for virulent NDV (44). During 2014–2015, the NDV genotype VII was reported with a percentage of 37.8% (114). There was a similar incidence of 37.5% in chicken flocks of 10- to 240-day-olds located in different districts of Sharkia province (115). Lower prevalence (12.5–16.2%) was recorded (49, 52, 116). A higher incidence of NDV with a percentage of 57.5% was detected during 2012–2015 (46) and 45.46% during 2019 (117). The highest incidence of NDV with a percentage of 86.2% was recorded in commercial chicken flocks during 2012–2015 (48). Moreover, Moharam et al. (9) screened 26 chicken flocks (backyard and commercial) during 2015–2016. Small-scale holders including farms keeping layers (n = 9), broilers (n = 4), or Balady chickens (n = 3) situated in provinces Beheira and Monufia of the Nile delta region. Commercial broiler farms were located in the provinces of Giza (n = 9) and Monufia (n = 1). Although all flocks were ND vaccinated, a virus was in both holders with a percentage of 84.6%. However, virulent NDV was detected in 30.76%. A recent study was conducted on 120 poultry flocks from 10 Egyptian provinces in the Egyptian Delta region during 2015–2019. The highest prevalence of virulent NDV was reported in broiler flocks (41.1%; 37/90), followed by layer flocks (38.4% 5/13). Baladi and Sasso chickens represented a confirmed NDV of 71.4% (5/7) and 33.3% (1/3), respectively (10).
Regarding the prevalence of NDV according to localities, several studies were conducted to investigate the geographic prevalence of NDV in different provinces in Egypt. The overall incidence of NDV in Egyptian provinces during 2012–2019 (Figure 2) ranged from 8.3% in Luxor to 100% in many regions: Port Said, Damietta, Gharbia, Menofia, Qalubia, and Minya (44, 46, 47, 52). The incidence of NDV in Sharkia ranged from 14% (52) to 50% (10). No NDV was detected in some provinces: Qaluibia, Menofia, Matrouh, Qena (52), Giza (10), and Aswan (44). The variations mentioned above in the prevalence of NDV along the Egyptian provinces could be attributed to (i) individual concept of NDV surveillance either in domesticated or wild, free-living, and migratory birds; (ii) difference of testing procedures and sample size; (iii) variable distribution of poultry farm population and levels of biosafety and biosecurity; (iv) absence of efficient and reliable surveillance systems that are needed to document the disease status of a population at a given time; and (v) different factors like the disease awareness of persons reporting suspect cases. In this framework, and according the OIE, any national surveillance scheme for an animal/avian infection may be constructed on two different surveillance approaches: active as the regular periodic samples' collection by veterinary health authorities and passive surveillance, which is distinct from active surveillance, as birds are only tested if they show clinical signs, and then they are detected and reported to the national authorities (118). Besides mandatory standard parameters of ND prevalence, studies must be approved and applied.
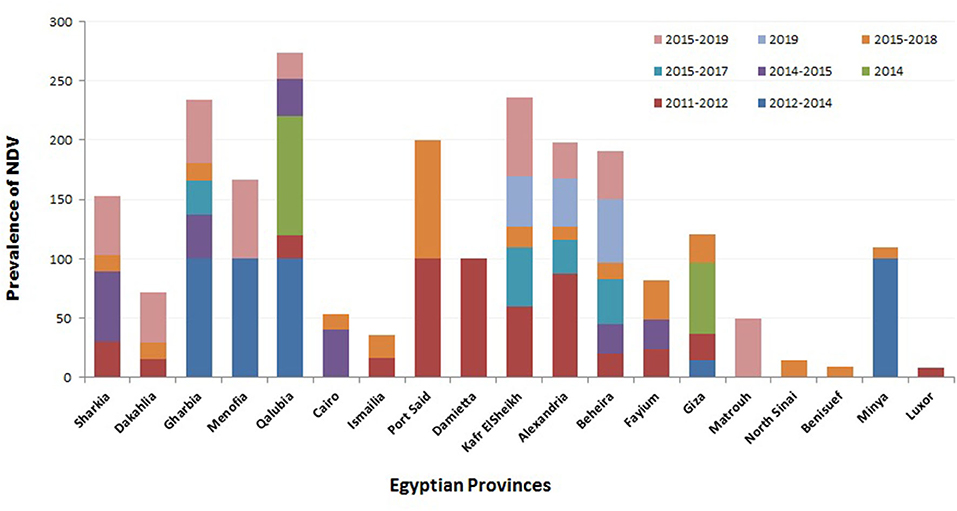
Figure 2. Summary of Newcastle disease virus (NDV) prevalence studies in Egypt according to geographical (Egyptian province) and time (year of detection) panels.
Columbiformes birds including pigeons and doves can be infected with NDV; however, the disease in pigeons is mainly caused by pigeon-specific viruses, e.g., PPMV-1. Pigeons and doves had been implicated as amplification or reservoir hosts, as they were frequently infected with virulent strains of NDV (119, 120). Since 1981, the clinical pictures in pigeons were consistent with ND, and then the virus infection was serologically confirmed in diseased pigeons along the Nile delta provinces in 1984 (26). Compulsory vaccination of racing pigeons is a local act in some countries. However, control of NDV in wild pigeons is almost impossible (121). Thus, pigeons are considered threatening carriers for the poultry industry (122). Pigeons in Egypt are not regularly vaccinated. As described by Mansour et al. (123), Elgendy et al. (124), and Rohaim et al. (54), pigeons suffered from NDV and AIV infections. In previous records of pigeons, the NDV infection caused a variable range of mortalities and morbidities (125, 126). Abou Hashem (127) did isolate 24 antigenically similar PPMV-1 viruses from pigeons in the Egyptian provinces of Sharkia (n = 14) and Dakalia (n = 10). Experimental IV or intraocular (IO) inoculation of pigeons revealed diarrhea and nervous signs [3–11 days post-infection (dpi)] followed by 100% death. The virus was transferred by contact with birds and caused mortality of 60–80%. The prevalence of PPMV-1 in Dakhlia province showed 18/100 pigeons suffering from diarrhea and nervous signs. When pigeons were infected experimentally with field viruses, the re-isolation of PPMV-1 was up to 13 days pi; and using HI, the relationship between PPMV-1 and the LaSota strain was confirmed (128). Moreover, Amer et al. (129) explored the virulence and transmissibility of NDV experimentally in pigeons, as 5% of the infected birds showed greenish diarrhea and 95% had positive virus isolation, with a detection rate of 70% in contact pigeons. In Kafr El-Sheikh province during 2014, wild pigeons showed dullness, lethargy, and neurological signs in addition to petechial hemorrhages in the heart and brain, congested lungs and liver, and enlargement of spleen upon necropsy. Molecularly, the detected virus (Pigeon/Egypt/VRLCU/2014) belonged to genotype VI, a well-known lineage representing PPMV-1 in pigeons. Biologically, as shown in Table 1, the virus had ICPI and MDT of 1.2 and 86 h, respectively (54). In 2014, a pooled brain sample from three diseased free-living pigeons (torticollis and whitish green diarrhea) in Desouk, Kafr El-Shiekh province, Egypt, was identified by HI and partial F-gene sequencing as genotype VI (130). One study focused on the outbreaks of influenza and paramyxovirus co-infections among clinically diseased pigeons (7), where mortalities varied from 10 to 92.5% in both single and mixed virus infections, with an indicative clinical picture of PPMV-1 and/or AIV, as pigeons showed tremors (83.3%), torticollis (17.7%), wing and leg paralysis (25%), and greenish diarrhea. Also, respiratory signs were observed only in few naturally infected pigeons. NDV was detected in 67.8% of diseased pigeons, mostly in pigeons aged ≤ 1 month. Mesogenic NDVs are rarely traced in prevalence along the history of ND. Nevertheless, Hamouda et al. (131) utilized restriction fragment length polymorphism (RFLP) to identify nine strains (mesogenic/lentogenic PPMV-1); 27 (mesogenic/lentogenic NDV) in the Sharkia province, in addition to one velogenic strain for each NDV and PPMV-1, were also detected. In another study, brain samples were collected from 12 pigeon farms with severe neurological symptoms and greenish diarrhea and showed a mortality rate of 3.3–38.5%. The ICPI values were 1.41–1.51, and MDT was 64–69 h (Table 1). However, IVPI in chickens was zero, indicative of moderate virulence (mesogenic nature) in chicken. Phylogenetically, all tested viruses were in genotype VI (56). Viruses of genotype VI had also been circulating in apparently healthy pigeons (132, 133), and in the study of Sabra et al. (55), it was confirmed that at least sub-genotype VIg is probably maintained in healthy captive pigeons in Egypt, with an ICPI value of 1.31 (Pigeon/Egypt/Giza/11/2015).
To follow the transmission dynamics of avian avulavirus (velogenic viscerotropic ND-genotype VII), intranasally (IN) or intramuscularly (IM) infected 8-week-old non-vaccinated native Egyptian Balady pigeons were kept in contact with non-vaccinated commercial Arbor Acres broiler chickens (4 weeks of age). The IM-infected birds had 100% mortality for chickens and 53.3% for pigeons, whereas the mortalities of IN-infected birds were 70 and 6.6% for chickens and pigeons, respectively. The viral shedding was higher in the oropharynx compared with the cloaca for both IN- and IM-infected pigeons. The IN-infected pigeons continued shedding the virus from the oropharynx at 4–16 dpi, while IM-infected pigeons had no oropharyngeal shedding at 11 dpi. Contact chickens had typical ND clinical picture, with mortalities of 40–60% and with higher virus shedding titers upon contact with IM-infected pigeons compared with IN-infected ones (134). It is worth mentioning that PPMV-1 strains could be isolated from the intestinal tract of infected pigeons suffering from greenish diarrhea. Those infected pigeons enhance the spread of the disease due to potential exposure of other birds to contaminated food with the pigeon fecal material (121, 135). Doves are common birds that come into contact with chickens in Egypt, on either open rural farms or live bird markets (LBMs), transmitting NDV to other bird species. When doves were inoculated with NDV using several routes, they were highly susceptible and showed nervous manifestations and congested organs. They can also shed the virus and transmit it to contact-susceptible chickens. The velogenic viscerotropic NDV strain had been detected in cloacal swabs (15/140) from free-flying doves in different localities in Egypt (136). During 2014–2015, NDV was detected in doves and characterized as genotype VII from samples collected from Gharbia (NDV/Dove/Bassioun/Egypt/MS2/2014KR082486) and Kafr El-Sheikh (NDV/dove/Desouk/Egypt/MS5/2015KT006286).
Taken together, pigeons and/or doves play significant roles in introduction, maintenance, transmission, epidemiology, and distribution of emergent NDV viruses in Egypt. It is of high value to track pigeons, at least those in close vicinity to poultry farms. The disease caused by NDVs of pigeon origin varies according to several factors, including host, environmental, and co-infection scenarios as well as lack of hygienic precautions.
Quails had been introduced to the commercial poultry sector in Egypt mainly for food consumption and were considered carriers and/or susceptible hosts to NDV (58, 137, 138). In Egypt, El-Zanaty and Abd El-Motelib (139) isolated the viscerotropic velogenic ND from quails of the Assiut province. In the Suez Canal University, a farm of 5,000 quails had 1.6% mortality; preliminary diagnosis suggested ND. Diseased birds, 3 weeks old, showed mild respiratory and nervous signs. Dead ones showed focal hemorrhagic lesions in the respiratory system and hemorrhagic spots on the liver, spleen, kidneys, and heart, without any obvious lesions in the digestive tract. Seroconversion confirmed NDV antibodies (1:32–1:256). The NDV was successfully isolated with a percentage of 75%. Hemagglutination assay (HA) titers ranged from 1:8 to 1:2,048, while MDT/minimal lethal dose (MLD) of three strains was 70, 80, and 75 h (mesogenic). The experimental ND infection resulted in 25% deaths after 1 week (58). Aly (140) investigated PPMV-1 isolates experimentally in quails and revealed that it could cause mild infection with 5% mortality in quails, but contact pigeons displayed greenish diarrhea and nervous signs (25%) followed by deaths (20%).
Japanese quails in Egypt were also susceptible to infection with NDV genotype VII, where the virus caused 33% mortality in quails and 100% mortality in chickens, with a typical ND picture, which was more severe in chickens compared with quails (141). The low death rates accompanied with nervous involvement and different shedding patterns are shared observations in partially resistant birds such as pigeons (142), cormorants (143), and ducks (144) upon infection with virulent NDV viruses. Also, vaccination protected quails against NDV infection (145, 146).
Migratory and non-migratory free-flying wild birds can play significant roles in NDV potentiation, transmission, and spread. It is believed that most wild birds got NDV as a direct result of spillover from domestic poultry species (24). Few exceptions are noticed, as NDVs are endemic in migratory birds, such as PPMV-1, which had taken wild pigeons as a reservoir/adaptation host, which lead to (i) emergence of highly virulent forms of ND in other avian species but not pigeons, (ii) global dissemination of such viruses, and (iii) increasing threats to the commercial poultry sector (23).
In 1976, 9/386 NDV isolates were identified from cloacal swabs of migratory birds in Northern Egypt (Bahig, Burg El-Arab, and Ikingi Mariut) (147). The MDT in chicken embryos was 59.2–77.6 h. But the pathogenicity indices in pigeons were 0.14–1.98 and 1.95–3.02 for IM and IC applications, respectively. Intracloacal application showed that all isolates are lethal to susceptible chickens, which suffered from provoked neurological signs and intestinal lesions (velogenic features of NDV) (57). Most recently, Rohaim et al. (148) isolated and identified vaccinal APMV-1 (5/297 oral and cloacal swabs) during a survey of apparently healthy wild birds in eight Egyptian provinces during early 2014 to late 2015. The APMV-1 isolate from teal (NDV/Teal/VRLCU-EG/2015) had an MDT of 96 h and an ICPI of 0.4375, harbored the GRQGRL motif at its F protein cleavage site, and belonged phylogenetically to genotype II (100% identity with the LaSota vaccinal strain), which collectively indicate its lentogenic nature and highlight the potential reverse spillover of NDV live vaccines from domestic poultry to wild birds (148). Furthermore, El Naggar et al. (59) also characterized NDV (4/112) of wild bird origin in Egypt. Teal (n = 2), quail (n = 1), and cattle egret (n = 1) tested positive for NDV, whereas house sparrow samples were negative. The ICPI and MDT ranged at 1.6–1.83 and 63.2–65 h, respectively (Table 1), suggestive of the velogenic potential of the four isolates. They were molecularly clustered into VII genotype and had the RRQKRF polybasic motif at the F protein cleavage site. Under experimental conditions, the previously mentioned four isolates did cause a pantropic infection in chickens, and the LaSota-vaccinated one failed to survive the disease (59).
Waterfowls, including ducks and geese, are less susceptible to NDV infection (24), as many NDV strains of different virulence had been isolated from either diseased or clinically healthy ducks, which raise the question of whether ducks/geese are only natural reservoirs or susceptible host to NDV (60, 61). Thus, the interest on natural infection of those birds with NDV has greatly increased (62–65). Prolonged viral shedding of waterfowl increased the risks of NDV of waterfowl origin (66). In Egypt, IM inoculation of Muscovy ducks with NDV genotype VII leads to only 5% mortality accompanied with higher and prolonged cloacal shedding compared with tracheal one. In contrast, IN inoculation did not cause deaths in ducks but elevated the tracheal shedding. The contact chicken had severe symptoms with very high mortality rates, emphasizing the fact that ducks are effective carriers of NDV (67). Besides, a virulent NDV of genotype VII was identified in 2/6 duck farms during 2017–2018 (68). Unexpectedly, NDV was also seen in co-infection reports, including two duck farms, but consistent with worldwide NDV reports from ducks (1, 64, 69). The role of ducks as a carrier of virulent NDV in Egypt remains to be investigated (68).
Mixed (co)-infection is defined as simultaneous infection of the same host with two different pathogens in the same time frame, a common event that is frequently occurring among birds, especially in intensive rearing systems and/or in developing countries. Several factors can control the outcome of co-infection, which could be either synergistic or antagonistic. This includes the interaction time, host immunity, and environmental conditions as well as biological activities of infectious agents. Viral–bacterial or viral–viral co-infection is highly common under poor biosecurity levels, which are frequently observed in Egypt. On the other side, viral interference is defined as a phenomenon in which initial virus infection prevents secondary homologous or heterologous virus by (i) blocking cellular receptors, (ii) competition on metabolic products required for viral replication, and (iii) initial sensitization of the host through virus-induced immune responses. Viral interference could interfere with proper diagnosis, as it could lead to undetectable/very low virus titers and atypical pathognomonic lesions (70, 71).
In Egyptian commercial poultry flocks, the disease outbreaks have increased during the last decade with high moralities and variable clinical pictures, especially respiratory signs. Lentogenic/velogenic NDV and high-pathogenicity AIV/low-pathogenicity AIV (LPAIV/HPAIV) are frequently reported from poultry, particularly in endemic areas, including Egypt. Both viruses share the same primary replication sites in the upper respiratory tracts of birds. Accordingly, both viruses could enhance or worsen the outcome of infection. Besides, co-infections with IBV were reported and represented significant alterations in the clinical picture, severity, and mortality rates (4, 72). Notably, a triple avian influenza subtypes (H5N1, H9N2, and H5N8) co-infection was also detected (68).
Variable NDV co-infection with other viruses was recorded (Figure 3). In Sharkia province, concurrent infection of NDV/H9 was detected in 5/50 flocks (10%) through 2012–2013 (73), 1/42 flocks (2.4%) during 2012–2014 (74), and 2/7 samples (28.6%) during 2013–2018 (75). Thirty percent of mixed infection of NDV/H5 was recorded in 2014–2015 (115).
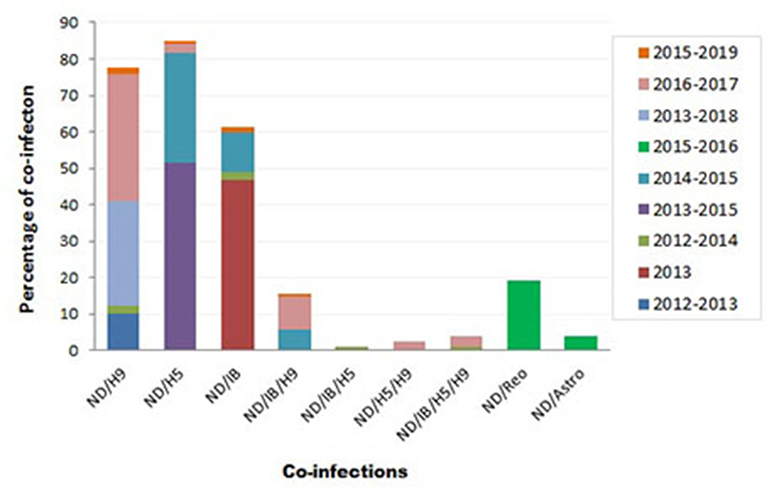
Figure 3. Summary of Newcastle disease virus (NDV) co-infection records with different infectious agents in Egypt.
During 2012–2014, mixed infection of NDV with IB (2.3%), NDV/IB/H5 (1.2%), and NDV/IB/H5/H9 (1.2%) was recorded (4). Sooner, increased rates of NDV/IB co-infection (10.8%) and NDV/IB/H9 (5.8%) were reported during 2014–2015 (116). Interestingly, Zaher and Girh (76) reported higher incidence of NDV/IBV co-infection with a percentage of 46.67%. In broiler and layer chickens, the co-infection was 48.1% (38/79) and 45% (32/71), respectively. In pigeons, NDV/H5 co-infection (51.6%) was recorded during 2013–2015. The co-infections appeared to predominate especially with foreign breeds of pigeons (63.6%), between 1 month and 1 year of age (55%), in summer (71.4%) and 60% in the closed rearing system (7).
During 2016–2017, respiratory viral pathogens were screened among poultry flocks (n = 50), commercial broilers (n = 39), commercial layers (n = 11), quails (n = 4), Bluebird (Sialis, n = 1), and Greenfinch (Chloris chloris, n = 1). Results revealed that the incidence of single ND infection was 33.3%. Concurrent infection of NDV/H9N2 was reported with a rate of 7.7% in commercial broilers and 27.3% in layers. Other mixed infections, i.e., NDV/H5N1, ND/H5N1/H9N2, and ND/IB/H5N1/H9N2, were also observed (2.6% each). Mixed ND/IB/H9N2 infection was observed in three separate farms (9.1% each) (8).
At the period of 2017–2018, poultry flocks (n = 39) showed severe respiratory signs: 32 chicken flocks (19 broiler, seven native broiler, four commercial layer, and two breeder farms), six duck farms, and one flock of outbred turkeys. Out of 39 farms, nine samples were positive for vNDV. In chicken farms, various virus combinations with NDV were detected: H5N1/H5N8/H9N2/IBV/NDV or H5N1/H5N8/H9N2/NDV or H5N8/H9N2/IBV/NDV, and H5N1/H9N2/IBV/NDV or H5N8/H9N2/NDV. Two duck farms revealed mixed infection of H5N1/H5N8/H9N2/NDV (68). More recently, the co-infection rate of NDV/H9N2 (2/120; 1.6%), NDV/H5N1 (1/120; 0.8%), NDV/IBV (2/120; 1.6%), and NDV/IBV/H9N2 (1/120; 0.8%) was recorded in 120 commercial farms or backyard houses in 10 Egyptian provinces during 2015–2019 (10).
Moreover, other mixed infections of NDV with avian astrovirus (3.8%) or avian reovirus (19.2%) were also identified (9). Earlier, in surveillance among more than 100 chicken flocks at Sharkia province in late 1980, mixed NDV with IB, reo, or pox viruses was recorded among examined flocks (77).
A total of 408 Egyptian NDV F protein strains were obtained from the GenBank to be included in our analysis (335 from chicken, 52 from pigeon, 17 from other avian species, and four of unknown origin). The last ones were considered to be of chicken origin. The official molecular NDV reports started in Egypt as early as 2005 (41). However, the National Center for Biotechnology Information (NCBI) database had other Egyptian NDV strains that are assumingly from 1976, mid-1990s, and early 2000s. Consistent with the second NDV outbreak in Egypt (VII), poultry researcher focused more on NDV starting from 2011, with maximum detection in 2012 and 2014–2016 (Figures 1A,B). All recorded sequences belonged to NDV class II.
However, only 136 had a complete F protein gene, as also most of the sequences came from NDV strains of chicken origin (n = 117), followed by pigeon (n = 14), and finally few sequences from teal (n = 3), quail (n = 1), and cattle egret (n = 1). A pilot phylogenetic tree was constructed based on the recommendations of Dimitrov et al. (17), from which these criteria were mainly included: (i) alignment using complete F protein nucleotide sequences, (ii) construction using maximum likelihood method using the general time-reversible (GTR) model with Gamma distribution (G), (iii) application of the model with 1,000 bootstrap replicates and values of ≥70 were indicated above the tree branches, and (iv) involvement of independent strains with no obvious epidemiologic link.
Chicken (broiler, layer, and breeder) is hugely involved in the poultry industry of Egypt, with an investment of nearly LE18 billion at that time of recording (78). Large industrial farms follow a strict vaccination program with intensive biosecurity. However, backyard/family farms do not usually vaccinate their birds and either treat their diseased ones symptomatically or submit them for slaughter. Such a discrepancy in NDV control strategies and weak governmental imposition of field regulations contribute significantly in disseminating the NDV. In this regard, following the molecular pattern of NDV in chicken is of great value.
The NDV F sequences from chicken origin are grouped under the VII.1.1 or II genotypes. Genotype VII was reported from different parts of the world and was dominantly circulating in Egypt since 2011 (79) and was found across almost the whole country (Figures 1A,B). Previously, complete F strains were classified as VIIb (9) or VIId (6, 50). Recent literature started to follow the new classification system and predominantly described VII.1.1 from chickens (10, 51, 80). Some partial F sequences were identified as VIIj (81), or generally as VII (79, 111), and most surprisingly as genotype VI (39) or I (accession number KR535623). However, studied sequences were too short to be considered for conclusive genotypic taxonomy. Since the VII.1.1 included anyhow previously characterized members of VIIb, VIId, VIIe, VIIj, and VII1, we could assume that all previously speculated NDV VII strains from chicken are actually one genetic linage of viruses or one genotype known as VII.1.1, which is responsible for the second epizootic outbreak/wave of NDV in Egypt, and still endemic in Egypt since then (Figures 4A, 5B). Meanwhile, NDV genotype II from chicken was initially described in 2005 (41) and 2006 (42), when it was responsible of the first reported NDV outbreak in Egypt. Subsequently, it was displaced by genotype VII. Nevertheless, genotype II is still existing but to a lesser extent (51) (Figures 4A, 5A). Pigeons are brought in Egypt for several reasons, including passion, gaming, investment, and meat production. They are mostly raised in spindly wooden columns that look like a medieval siege tower. Unfortunately, pigeons are a high potential source of NDV spread at least to commercial/backyard poultry sector due to their migratory behavior, free-living system of rearing, close contact with other birds in or out LBMs, and reluctance of owners to vaccination. Despite being a carrier reservoir for NDV (134), NDV strains of chickens were more pathogenic and transmissible to chickens compared with those from pigeons (82). The NDV surveillance system in the pigeon is quite weak and does not reflect the actual situation.
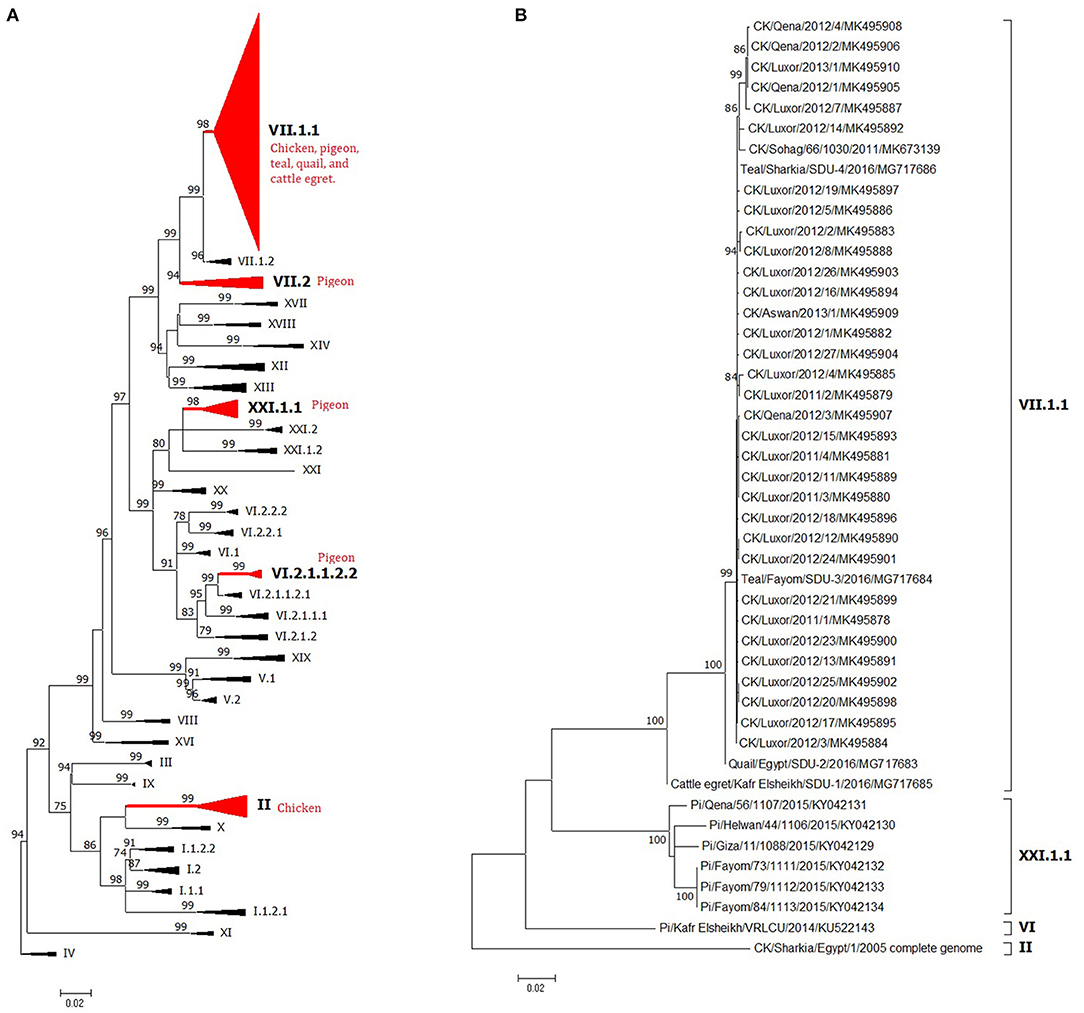
Figure 4. The phylogeny of Egyptian Newcastle disease viruses (NDVs) (A) full F protein gene or (B) full HN protein gene of different Egyptian isolates. The maximum likelihood (ML) tree was built based on the general time-reversible model with a discrete gamma distribution at 1,000 bootstrapping. All genotypes are in Newcastle disease virus (NDV) class II and were assigned with Roman numerals, as suggested by Dimitrov et al. (17). The taxons of F protein gene tree were compressed for better presentation. Sequences from Egypt are labeled with a red circle.
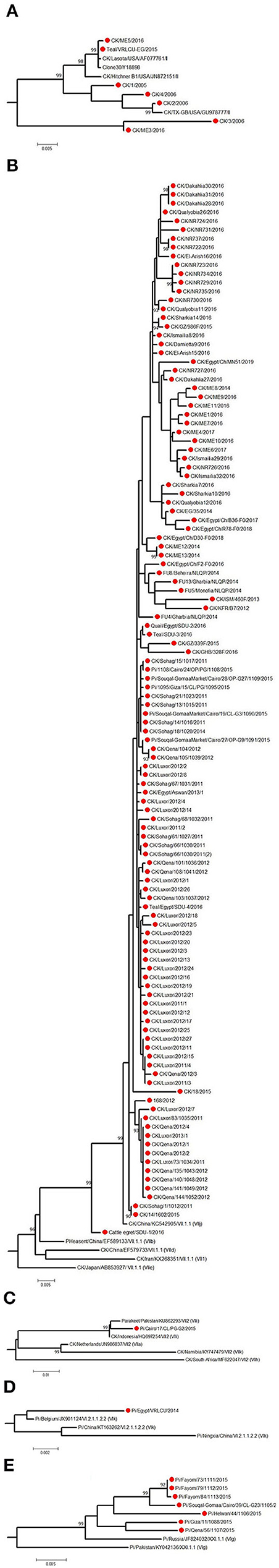
Figure 5. The detailed full F protein gene-based phylogenetic tree of Egyptian Newcastle disease viruses (NDVs), representing various detected strains from different bird species, which belonged to different lineages, as (A) genotype II, (B) genotype VII.1.1, (C) genotype VII.2, (D) genotype VI.2.1.1.2.2, and (E) XXI.1.1. The construction of the tree was done by using the maximum likelihood (ML) method, general time-reversible model, and bootstrap values of 1,000 (17). Sequences from Egypt are labeled with a red circle.
The low number of NDV/pigeon paramyxoviruses detected in pigeons reflected a high level of genetic diversity. At first, Rohaim et al. (54) reported the PPMV-1 from a single case (one diseased pigeon; KU522142) in 2014 and was classified as VI (sub-genotype VIb). Consequently, two investigations in clinically affected pigeons recorded partial F protein sequences of PPMV-1. Mansour et al. (7) found three different NDV genotypes during 2013–2015 (Ia, II, and VI), where the VI sequences belonged to VIb.2, a newly emerged cluster that was described mainly in Europe, while KU522142 clustered with VIb.1/re, another classical pigeon NDV lineage from Europe. The same genotype (VIb.2) was also found in 2016 and was renamed as VIg (56). Complete F protein gene sequences of VIg were also defined in apparently healthy pigeons in 2015 (55). Based on the classification system of our study, complete F protein sequences from pigeons clustered into four groups: the newly proposed genotype XXI (XXI.1.1; n = 7), VI (n = 1), VII.1.1 (n = 5), and VII.2 (n = 1). The XXI.1.1 separated basically VIg strains from the VI genotype. The sequence, KU522142, remained in VI and assigned in the sub-genotype VI.2.1.1.2.2, while VII.1.1 and VII.2 were seen also in 2015 (83) and described in Figures 4A, 5B–E. Other strains that had a partial F protein gene (identified as genotypes I and II plus unidentified ones) need further confirmation/analysis following complete F protein gene sequencing.
Other birds, including wild ones, are considered as another introducing, disseminating, and maintaining factor of NDV in Egypt due to their migration or involvement in LBMs. They are also involved directly or indirectly in the wild–domestic–human interfaces, imposing the importance of continuous surveillance of pathogens. All five complete F protein gene sequences from quail (n = 1), Cattle egret (n = 1), and teal (n = 3) belonged to genotype VII.1.1 (59) except one sequence from teal (148), which was in genotype II (Figures 4A, 5A,B). Another 12 NDV partial F protein sequences from passerines (n = 1), dove (n = 2), house sparrow (n = 3), cattle egret (n = 2), white wagtail (n = 1), white-throated kingfisher (n = 1), and hoopoe (n = 1) were found in genotype VII, while ostrich (n = 1) was placed in genotype II.
For the HN protein, a total of 46 complete gene sequences were collected and involved in the phylogenetic tree (Figure 4B) to collect some evolutionary criteria for an important NDV surface protein in Egypt. Both Egyptian HN and F protein followed the same phylogenetic topology. They were found in genotypes VII.1.1 (strains from chicken mainly, teal n = 2, quail n = 1, and cattle egret n = 1), II (one sequence from chicken), and VI or XXI.1.1 (strains of pigeon origin).
The F protein cleavage site (112-117) is considered a major determinant factor that plays a role in virus pathogenicity/virulence (19). However, other factors, for example, the HN stem region and globular head, are also involved (12). All isolates of Egyptian NDVs had an F protein of 552aa length. The deduced aa analysis showed the relative stability of F protein cleavage site in the Egyptian NDV isolates overtime, which tend to be genotype-specific, not species-specific. NDV isolates in VII.1.1 and VII.2 (from chicken, pigeon, teal, quail, or cattle egret) had the velogenic motif RRQKRF except one chicken isolate that had the RRKKRF motif.
Genotype II strains had either the velogenic motif RRQKRF (in chicken and pigeon) or the lentogenic motif GRQGRL (in chicken and teal). Pigeon NDV strains in XXI.1.1 had the velogenic KRQKRF motif, while pigeon strains in genotype VI and I had the velogenic RRQKRF and lentogenic GKQGRL ones, respectively. Two strains from pigeon (MF614961) and chicken (MK604215) also had the lentogenic motifs GRQGRL and GKQGRL, respectively (unidentified genotype; not in Supplementary Tables 1, 2). The previously mentioned 12 NDV partial F protein sequences from wild birds had all RRQKRF residues at their cleavage site, except the ostrich isolate, which had the RNQGRL motif. As previously reported, aa changes in the cleavage site affected the fusion efficiency of the F protein (84), gradual dominance of virulent strains in quasispecies (85), and virulence of the virus (ICPI decreased in Q114R mutant viruses) as reported by Samal et al. (86). However, the clinical pathogenicity of the virus under natural conditions is usually influenced by other viral, host, and environmental factors. Interestingly, Nagy et al. (51) reported high pathogenicity indices in lentogenic motif harboring NDV isolates of chicken origin, emphasizing the importance of following the OIE recommendations in determining virulence regarding NDV of field origin.
The F protein signal peptide (1–31) was highly variable among the Egyptian strains, consistent with the findings of Orabi et al. (6). Other regions, including the fusion peptide (117–142); heptad repeats a, b, and c (HRa 143–185, HRb 268–299, and HRc 471–500); transmembrane (TM) domain (501–522); and cytoplasmic (CT) tail (523–553) were subjected to aa analysis, where several mutations were reported, which might affect the folding and fusion activities of the protein as described in the fusion peptide (87, 88) or the HRa, b, and c (89). The HRa is also supposed to include a potential antigenic epitope (90, 149–163); however, the Egyptian NDV isolates had only few reports of aa substitutions in this epitope (V168I and D170N). The TM domain affects the structural confirmation of the F protein, F–HN protein interaction, and fusion activity (91). Also, mutant CT domain modulates the F protein biological characters, virulence, and pathogenicity (92). D170N was reported in VII.2 and VII.1.1 (one pigeon and four chicken strains, respectively), which is a neutralizing epitope (93). Also, residues D479 and S486 are critical for the fusogenic activity (94) (Supplementary Tables 1, 2).
The HN protein of NDV is a surface glycoprotein that mediates several functions, including (i) attaching of NDV to cellular sialic acid receptors (95), (ii) promoting F protein fusion activity (96), (iii) facilitating the NDV budding by its neuraminidase (NA)-mediated receptor cleavage (97), and (iv) determining NDV tropism and virulence (98). Structurally, the NDV HN protein consists of CT tail, N-terminal TM domain, a stalk region, and a C-terminal globular head. The HN stalk forms an interaction with the F protein through a stretch of amino acids (aa positions 74–110) that forms two conserved heptad repeats (HRA and HRB) (99).
The HN protein length of Egyptian NDV isolates was 572aa except the parent NDV isolate (II) from 2005 (FJ939313), which was slightly longer (578aa). The NDV HN length affects the replication and biological properties of the virus as extended HN showed increased HA titer and receptor binding but impaired NA, fusion, and replication abilities. However, the virulence of virus was not changed (100). TM and stalk domains of the Egyptian NDV strains were highly conserved among genotypes (for example, VII.1.1, XXI.1, VI, and II), with few substitution mutations, which may affect the structure and activity of the HN protein (99, 101).
Major epitopes in the C-terminal head of HN were also investigated. As it is basically involved in antibody recognition (102), a single aa change in that major linear epitope 345PDKQDYQIR353 (1/1–4) allows the escape of its corresponding antigenic variant from neutralizing monoclonal antibodies, which was reported at least at position 347 (103, 104). It might explain the high virulence of chicken VII.1.1 isolates in chickens compared with others. Other antigenic epitopes within the HN protein were compared as shown in Supplementary Tables 3, 4.
Amino acid residues involved in receptor recognition (95), HAD ability, NA activity, fusion activity (105), interaction with F protein (106), head–stalk linker region (107), and predicted B-cell epitope (108) were highly conversed with few exceptions. Due to the intensive unplanned vaccination in Egypt, selection/vaccination pressure could explain the occurrence of such aa substitutions in the epitopes of Egyptian NDV HN overtime, which in turn may affect, to different levels, the vaccine efficacy, induced immunity, and virus shedding in birds as a result of antigenic variability (108). HN protein is a determinant viral factor for thermostability, as mutant NDV viruses (S315P and I369V) were more thermostable and possessed more HA titers and NA fusion activity (109).
Besides good biosecurity regimens, the control of ND principally accounted for mandatory preventive vaccination of flocks and hygienic culling of infected birds. In Egypt, many NDV commercial traditional/classical genotype II vaccines are used in the Egyptian poultry field such as (i) live seed virus vaccine strains of LaSota, Hitchner, VG/GA, clone 30, PHYLMV, and others or (ii) inactivated (killed) virus vaccines, mainly the LaSota one (153). Recently, several recombinant and novel inactivated (Genotype VII; GVII) vaccines were introduced gradually to cope with the continuous evolution and spread of velogenic NDV-GVII (154, 155). ND is endemic in Egypt, and there is an enormous pressure from the field circulation of diverse genotypes II, VI, and VII; massive poultry production; and direct and/or indirect contact with free-living and migratory birds, which generally represent a significant challenge to poultry holders. Currently applied vaccination strategies are relatively effective in preventing severe illness and death of infected birds but may fail to prevent infection or shedding of the virus. In Egypt, the main goal of scientists and poultry producers is to minimize the economic impact of NDV infections. Accordingly, innovative vaccination strategies were applied to potentially increase protection and reduce viral shedding, and presumably the spreading and the transmission of the virus. Here, we reviewed all studies related to development of ND vaccine technology and strategies. The extent to which heterologous vaccines can protect against different genetic variants of NDV is still controversial. The applied preventive programs in Egyptian provinces include live (genotype II) and inactivated vaccines (genotypes II/VII). Proper vaccines and the design of efficient vaccination programs shall give the best protection against clinical disease and prevent/reduce either mortality or virus shedding in vaccinated flocks.
Over the past 25 years, many vaccination practices were tested experimentally in Egypt (Supplementary Table 5). Some studies were designated to evaluate different vaccines and/or vaccine regimens applied in Egypt. Upon evaluation, the ND live virus vaccine protected from clinical disease (90%) and deaths (80%) in case of homologous challenge with GII of VG/GA live strain (77) and HB1 with LaSota live strains (156) but only 75% protection rate from clinical disease and deaths as reported by Lebdah et al. (157). These protection percentages were raised up to 100% upon inclusion of inactivated vaccines with HB1 and LaSota live vaccines (156, 157). However, the results of Mohamed et al. (158) were drastic when they used DNA vaccine containing F and HN antigens in one dose (30–40%); then they improved the protection to 90–100% by introducing two or three vaccine doses at 1 week apart.
In most broiler chicken farms, it is recommended to administer several doses of live attenuated vaccines that should be primed as early as possible (159, 160). This could begin directly after hatching (at 1 day old) with HB1, followed by a booster dose of LaSota or Clone 30 vaccine two times at 2 weeks apart. As a result of insufficient protection of live vaccines in some intensive rearing localities, inactivated vaccines are usually included at the seventh or 14th day of age to maximize the protection. Day 1 recombinant vaccine application is still of limited use (153, 161).
After the involvement of new velogenic genotype VII of NDV (79), many studies were performed to evaluate the existing regimens to protect against the circulating field viruses, develop vaccine preparation, and update virus vaccine seeds (155, 158, 162–165). Three schemes of vaccination were proposed; the first included a heterologous vaccine regimen (massive genotype II vaccines) against challenge velogenic NDV genotype VII. The use of multiple live vaccines (genotype II; HB1, LaSota, and clone 30) induced lower protection that ranged from 72 to 93.4% (159, 166). In addition, programs included both inactivated and live genotype II-based vaccines; 100% of birds were completely protected from mortality upon heterologous virus challenge with genotype VII (45, 167). However, Nemr (168) and Shahin et al. (160) recorded 93.66 and 96% protection, respectively. On the other hand, trials to use only one shot of inactivated genotype II vaccine was contradictory, as Sediek et al. (169) recorded extremely low protection (33.3–46.7%). Meanwhile, Kilany et al. (162) reported 80% protection against heterologous challenge. Regarding the tracheal and cloacal shedding, all aforementioned studies revealed variable quantitative (0.6–6.6 log base 10 and two higher challenge up to 8.5) positive shedding from the second day up to 10th day post-challenge with genotype VII–velogenic NDV (Supplementary Table 5), which establishes the field virus transmissibility particularly after heterologous vaccination and non-strict hygienic measures.
In an unprecedentedly swift response to develop and manufacture an anti-NDV–genotype VII vaccine, inactivated virus vaccine was prepared from currently circulating velogenic NDV genotype VII, national and regional companies and academic institutions are exploring the numerous strategies. Subsequently, the second vaccine scheme was anticipated to be based only on homologous vaccination tested experimentally since 2015. Regarding the use of one killed vaccine S/C in chickens containing GVII strains, the results revealed 100% protection against clinical signs and mortality post-challenge with a similar genotype (162, 164, 168), which was accompanied with reduced viral shedding to <2.2 till the 10th day post virus challenge. Other findings included the stoppage at the fifth day (165) or the complete absence of virus shedding (154). Nevertheless, Sedeik et al. (169) used an inactivated NDV genotype VII vaccine to immunize the birds against a challenged virus from the same genotype, where they showed protection of only 53.3% with presence of clinical signs and virus shedding. The low protection was explained by using an inactivated vaccine prepared from a Korean NDV strain (KBNP-C4152R2L strain, INC., Korea) that is different from the circulating NDV strains in Egypt, which was used as a challenge virus. Conversely, a mucosal inactivated vaccine containing genotype VII failed to protect when used once (0%), which increased to 60% upon booster/second vaccination and 100% when inactivated oil-based vaccine was applied with the mucosal one (163).
The Third Scheme was based on the conjunction of GII and GVII to get the benefit from a gift of naturally attenuated viruses of genotype II (Lentogenic vaccines) and broaden the scope of immunity by inclusion of killed genotype VII. The trials of Bastami et al. (170) of mixed use of different permutations from live vaccines, HB1 and LaSota with either native or foreign inactivated genotype VII to protect chickens against challenge with virulent GVII-NDV, had a range of 90–100%. The best trial gave 100% protection from deaths and the lowest shedding (22.2%) when primed at day 5 of age with simultaneous live HB1 and native inactivated and then two doses of live LaSota (10th and 21st days of age). Also, 100% protection with non-significance as low as less than one log10 was reported by Hassan et al. (167) when using Live LaSota (clone 79) (genotype II) in WOW (water in oil in water inactivated vaccine) (VIIj) against a virus challenge by velogenic ND (VIIj). In another study, even with more than five doses of both live (GII) and inactivated (GVII), the virus was detected in tracheal swabs of chickens through 3–5 days post-challenge with velogenic NDV genotype VII (159). This establishes that not only the diversity of vaccine and field viruses or number of doses but also efficient administration, dose, and other factors may limit the scheme's efficacy. In a different study, both live LaSota and inactivated genotype VII could not fully protect against challenge with velogenic genotype VII from teal (NDV/Teal/Egypt or egret, and NDV/Cattle egret/Egypt; 60 and 40%, respectively). However, the virus was shed from challenged chicken (4–6 dpi), which set the need for more epidemiological investigations of the impact of non-chicken strains of NDV genotype VII (59).
Concerning PPMV-1, limited investigations (Supplementary Table 6) were set to evaluate the vaccine strategies against the virus. Studies concluded that complete protection could be achieved using a homologous inactivated vaccine with good adjuvant to be administered either SC or IM. Amer (171) prepared inactivated cell culture PPMV-1, which induced HI antibodies (GMT: 64) on the third week post-vaccination. Amer et al. (172), Khedr et al. (173), and Soliman et al. (174) succeeded in protecting 100% of challenged pigeons using inactivated PPMV-1 vaccine. Combined live HB1 or LaSota vaccines with inactivated PPMV-1 gave high protection (100%). It was proved that both virulent NDV and PPMV-1 circulate among pigeons in the Sharkia province of Egypt (131), necessitating the review of vaccine strategy of both viruses at risk.
Despite the diversity of virulent genotypes reported along the recent history of the disease in Egypt, all NDV strains are clustered into one serotype. This explains that under laboratory conditions, a vaccine prepared from any strain or genotype can induce antibodies to protect birds (decrease/prevent the clinical signs and deaths) against challenge with the virulent viruses (149–152, 175). However, other components such as cellular immunity should be taken into consideration, as it is not defined by serotype and could not be achieved by using inactivated vaccines, which is an available option for genotype VII.
Successful protection against circulating diverse NDVs in Egypt is a multifactorial issue. Under experimental and some field vaccination programs containing NDV genotype II, the vaccine could provide good protection (96–100%) against infection with heterologous Newcastle viral strain (genotype VII) and reducing the amount of viral shedding (45, 155, 159, 160, 176). However, upon field conditions, multiple factors could contribute to a considerable reduction in the effectiveness of vaccination, including (i) the frequent incidence of NDV infection, even in vaccinated birds, as being vaccinated does not prevent infection or virus shedding; (ii) improper vaccination; (iii) immune suppression of infectious or non-infectious origin; (iv) faulty program that may cause loss of cell-mediated immunity; and (v) prejudging viral mutation/changes in the genomic sequence of the virus, which can lead to presence of many serological variants (3, 159). Also, other factors include insufficient biosecurity procedures and the probable threats for disease transmission from wild/migratory birds to domesticated birds and vice versa. Strict and improved biosecurity must be at the solution's core to minimize the environmental virus load and halt its mutation. Contingent to the presented data, it is essential to have a more detailed analysis of the biological and antigenic characters of currently circulating NDV strains and the efficacy of commonly used NDV vaccines for protection against the NDV isolates in a frame of national plan utilizing the accumulated knowledge and unifying the system of investigation for better controlling the NDV in Egypt.
ND is a highly prevalent viral disease, which is caused by various strains of NDV. The disease has potential intercontinental endemicity that further leads to economic crises due to high morbidity and mortality, long-lasting limitations on international trading operations, and increased costs of veterinary management. As a result of its extensive spread, countries like Egypt is trying to manage effective preventive and control measures, which preferably includes (i) culling and slaughter, (ii) active surveillance in domestic and wild birds, (iii) intensive biosafety measures, and (iv) strong vaccination programs. In Egypt, it is difficult to control the backyard/rural breeding of birds, which makes it relatively problematic to follow such regulations. Culling is not always an option due to the lost cost. The reliance on genetically dissimilar vaccines may not eliminate the persistent NDV viruses in commercial poultry sector, probably due to the presence of some antigenic variations. However, it is not the sole cause of vaccination unreliability in immunocompetent birds (177), particularly in several Egyptian studies in current commentary revealed protection with heterologous and mixed genotype with both live and inactivated vaccination (159). In accordance with field isolates, the usage of vaccine matched strains will probably help in extra control of NDV (178) with priming of live lentogenic strains to induce specific cell-mediated immunity, which could not be induced by a homologous one being inactivated virulent genotype VII. The NDV epidemiological gap in Egypt is still expanding with time, and accordingly, we advise the following recommendations for better outcomes:
(1) Following the OIE recommendations for identification of isolated NDV strains, which includes using at least two pathogenicity indices (ICPI and MDT at best), plus sequencing F protein cleavage site or monoclonal antibody HI matching using a reference panel (2). This shall avoid mischaracterization of the pathogenic nature (velogenic/mesogenic/lentogenic) of the isolated strains, especially with increased detection of lentogenic strains that possess a velogenic motif at their F protein cleavage site.
(2) For NDV isolates from birds rather than chicken, pathogenicity indices shall be performed in both actual bird of origin and chicken, as different indices may implicate the adaptation of the virus in both hosts, despite the close genetic relatedness (179).
(3) Full epidemiological data should be reported in future studies, including size of investigated flock, age of birds, the total number of raised birds, morbidity and mortality, the scientific name of bird involved, type of production, history of vaccination, and province (geographical site) in Egypt.
(4) We also advise to use the criteria proposed by Dimitrov et al. (17), particularly regarding full F protein sequencing and epidemiological parameters. Partial F protein sequence misleads accurate characterization, in either phylogenetics or detection of emergent and/or antigenic escape mutant NDVs. When possible, also the HN protein should be fully sequenced.
(5) We encourage broader observations of NDV viruses in free-ranging, aquatic, and migratory birds in Egypt and also not to ignore the possibility of viral or bacterial mixed (co)-infection in flocks under study.
(6) Application of biosecurity is a compulsory solution since vaccination alone cannot completely prevent field virus shedding.
(7) When applied, we advise to prime live lentogenic (genotype II) with a subsequent inactivated (homologous or heterologous) vaccine to achieve specific cell-mediated and humoral immunity and to broaden the scope of protection.
AAE, SM, and FM: conceptualization. SM, RE, FM, and AAE: data curation. SM, RE, FM, and AAE: formal analysis. SM, RE, FM, ARE, EH, MA, AAE, HA, and MI: investigation. AAE, ARE, and SM: supervision. SM, RE, and FM: writing—original draft. SM, RE, FM, EH, MA, MI, HA, AAE, and ARE: writing—review and editing. All authors contributed to the article and approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.647462/full#supplementary-material
Supplementary Table 1. Fusion protein analysis in Pigeons.
Supplementary Table 2. Fusion protein analysis in chickens and other birds.
Supplementary Table 3. Functional domains and epitopes in HN of Egyptian NDV strains.
Supplementary Table 4. Head-stalk linker region involved in fusion triggering.
Supplementary Table 5. Vaccination strategies in chickens (NDV).
Supplementary Table 6. Vaccination strategies in pigeons (PPMV-1).
1. Alexander DJ, Senne DA. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections. In: Saif YM. Fadly AM, Glisson JR, McDougald LR, Nolan LK, Swayne D, editors. Diseases of Poultry. Ames, IA: Blackwell Publishing (2008). p. 750–98.
2. World Health Organization. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (Chapter 2.3.14). (2012). Available online at: https://www.oie.int/doc/ged/d12009.pdf (accessed March 8, 2014).
3. Kattenbelt JA, Stevens MP, Gould AR. Sequence variation in the Newcastle disease virus genome. Virus Res. (2006) 116:168–84. doi: 10.1016/j.virusres.2005.10.001
4. Hassan KE, Shany S, Ali A, Dahshan A, El-Sawah AA, El-kady M. Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult Sci. (2016) 95:1271–80. doi: 10.3382/ps/pew068
5. Daubney R, Mansy W. The occurrence of Newcastle disease in Egypt. J. Comp. Pathol Therap. (1948) 58:189–200. doi: 10.1016/S0368-1742(48)80019-6
6. Orabi A, Hussein A, Saleh AA, Abu El-Magd M, Munir M. Evolutionary insights into the fusion protein of Newcastle disease virus isolated from vaccinated chickens in 2016 in Egypt. Arch Virol. (2017) 162:3069–79. doi: 10.1007/s00705-017-3483-1
7. Mansour SM, Mohamed FF, Eid AA, Mor SK, Goyal SM. Co-circulation of paramyxo-and influenza viruses in pigeons in Egypt. Avian Pathol. (2017) 46:367–75. doi: 10.1080/03079457.2017.1285391
8. Shehata AA, Sedeik ME, Elbestawy AR, Zain El-Abideen MA, Kilany WH, Ibrahim HH, et al. Co-infections, genetic, and antigenic relatedness of avian influenza H5N8 and H5N1 viruses in domestic and wild birds in Egypt. Poult Sci. (2019) 98:2371–9. doi: 10.3382/ps/pez011
9. Moharam I, Razik AAE, Sultan H, Ghezlan M, Meseko C, Franzke K, et al. Investigation of suspected Newcastle disease (ND) outbreaks in Egypt uncovers a high virus velogenic ND virus burden in small-scale holdings and the presence of multiple pathogens. Avian Pathol. (2019) 48:406–15. doi: 10.1080/03079457.2019.1612852
10. Abd El-Hamid HS, Shafi ME, Albaqami NM, Ellakany HF, Abdelaziz NM, Abdelaziz MN, et al. Sequence analysis and pathogenicity of Avian Orthoavulavirus 1 strains isolated from poultry flocks during 2015–2019. BMC Vet Res. (2020) 16:1–15. doi: 10.1186/s12917-020-02470-9
11. International Committee on Taxonomy of Viruses (ICTV). Virus Taxonomy. (2018). Available online at: https://talk.ictvonline.org/taxonomy (accessed December 20, 2020).
12. de Leeuw OS, Koch G, Hartog L, Ravenshorst N, Peeters BPH. Virulence of Newcastle disease virus is determined by the cleavage site of the fusion protein and by both the stem region and globular head of the haemagglutinin–neuraminidase protein. J Gen Virol. (2005) 86:1759–69. doi: 10.1099/vir.0.80822-0
13. Czeglédi A, Ujvári D, Somogyi E, Wehmann E, Werner O, Lomniczi B. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. (2006) 120:36–48. doi: 10.1016/j.virusres.2005.11.009
14. Liu X, Wang X, Wu S, Hu S, Peng Y, Xue F, et al. Surveillance for avirulent Newcastle disease viruses in domestic ducks (Anas platyrhynchos and Cairina moschata) at live bird markets in Eastern China and characterization of the viruses isolated. Avian Pathol. (2009) 38:377–91. doi: 10.1080/03079450903183637
15. Diel DG, da Silva LH, Liu H, Wang Z, Miller PJ, Afonso CL. Genetic diversity of avian paramyxovirus type 1: proposal for a unified nomenclature and classification system of Newcastle disease virus genotypes. Infect Genet Evol. (2012) 12:1770–9. doi: 10.1016/j.meegid.2012.07.012
16. Snoeck CJ, Owoade AA, Couacy-Hymann E, Alkali BR, Okwen MP, Adeyanju AT, et al. High genetic diversity of Newcastle disease virus in poultry in West and Central Africa: cocirculation of genotype XIV and newly defined genotypes XVII and XVIII. J Clin Microbiol. (2013) 51:2250–60. doi: 10.1128/JCM.00684-13
17. Dimitrov KM, Abolnik C, Afonso CL, Albina E, Bahl J, Berg M, et al. Updated unified phylogenetic classification system and revised nomenclature for Newcastle disease virus. Infect Genet Evol. (2019) 74:103917. doi: 10.1016/j.meegid.2019.103917
18. Alexander DJ. Newcastle disease in the European Union 2000 to 2009. Avian Pathol. (2011) 40:547–58. doi: 10.1080/03079457.2011.618823
19. Panda A, Huang Z, Elankumaran S, Rockemann DD, Samal SK. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb Pathog. (2004) 36:1–10. doi: 10.1016/j.micpath.2003.07.003
20. Fujii Y, Sakaguchi T, Kiyotani K, Yoshida T. Comparison of substrate specificities against the fusion glycoprotein of virulent Newcastle disease virus between a chick embryo fibroblast processing protease and mammalian subtilisin-like proteases. Microbiol Immunol. (1999) 43:133–40. doi: 10.1111/j.1348-0421.1999.tb02384.x
21. Marlier D, Vindevogel H. Viral infections in pigeons. Vet J. (2006) 172:40–51. doi: 10.1016/j.tvjl.2005.02.026
22. Guo H, Liu X, Han Z, Shao Y, Chen J, Zhao S, et al. Phylogenetic analysis and comparison of eight strains of pigeon paramyxovirus type 1 (PPMV-1) isolated in China between 2010 and 2012. Arch Virol. (2013) 158:1121–31. doi: 10.1007/s00705-012-1572-8
23. Ujvari D, Wehmann E, Kaleta EF, Werner O, Savic V, Nagy E, et al. Phylogenetic analysis reveals extensive evolution of avian paramyxovirus type 1 strains of pigeons (Columba livia) and suggests multiple species transmission. Virus Res. (2003) 96:63–73. doi: 10.1016/S0168-1702(03)00173-4
24. Miller P, Koch G. Newcastle disease, other avian paramyxoviruses, and avian metapneumovirus infections. In: Swayne DE, editor. Diseases of Poultry. Hoboken, NJ: John Wiley and Sons, Inc. (2013). p. 89–138.
25. Tantawi HH, Al Falluji MM, Al Sheikhly. F. Viral encephalomyelitis of pigeons: identification and characterization of the virus. Avian Dis. (1979) 23:785–93. doi: 10.2307/1589594
26. Kaleta E, Alexander D, Russell P. The first isolation of the avian PMV-1 virus responsible for the current panzootic in pigeons. Avian Pathol. (1985) 14:553–7. doi: 10.1080/03079458508436258
27. Aldous EW, Fuller CM, Ridgeon JH, Irvine RM, Alexander DJ, Brown IH. The evolution of pigeon paramyxovirus type 1 (PPMV-1) in Great Britain: a molecular epidemiological study. Transboundary Emerg Dis. (2014) 61:134–9. doi: 10.1111/tbed.12006
28. Alexander DJ, Manvell RJ, Frost KM, Pollitt WJ, Welchman D, Perry K. Newcastle disease outbreak in pheasants in Great Britain in May 1996. Vet Rec. (1997) 140:20–2. doi: 10.1136/vr.140.1.20
29. Aldous EW, Alexander DJ. Detection and differentiation of Newcastle disease virus (avian paramyxovirus type 1). Avian Pathol. (2001) 30:117–28. doi: 10.1080/03079450120044515
30. Aldous EW, Fuller CM, Mynn JK, Alexander DJ. A molecular epidemiological investigation of isolates of the variant avian paramyxovirus type 1 virus (PPMV-1) responsible for the 1978 to present panzootic in pigeons. Avian Pathol. (2004) 33:258–69. doi: 10.1080/0307945042000195768
31. Irvine RM, Aldous EW, Manvell RJ, Cox WJ, Fuller CM, Wood AM, et al. Outbreak of Newcastle disease due to pigeons paramyxovirus type 1 in grey partridgees (Perdix perdix) in Scotland in October 2006. Vet Rec. (2009) 165:531–5. doi: 10.1136/vr.165.18.531
32. KrapeŽ U, Steyer AF, Slavec B, Barlič-Maganja D, Dovč A, Račnik J, et al. Molecular characterization of avian paramyxovirus type 1 (Newcastle disease) viruses isolated from pigeons between 2000 and 2008 in Slovenia. Avian Dis. (2010) 54:1075–80. doi: 10.1637/9161-111709-ResNote.1
33. Alexander DJ. Newcastle disease, other avian paramyxo-viruses, pneumovirus infection. In: Barnes HJ, Saif YM, Glisson JR, Fadly AM, McDougald LR, Swayne DE editors. Disease of Poultry. Oxford: Blackwell (2003). p. 75–100.
34. Alexander DJ. Newcastle disease other avian para-myxoviruses. In: Swayne DE, Glisson JR, Jackwood MW, Pearson JE, Reed WM, editors. A Laboratory Manual for the Isolation, Identification Characterization of Avian Pathogens. Kenneth Square, IA: American Association of Avian Pathologists (1998). p. 156–63.
35. Terregino C, Capua I. Conventional diagnosis of Newcastle disease virus infection. In: Capua I, Alexander DJ, editors. Avian Influenza and Newcastle Disease. Milan: Springer Milan (2009). p. 123–5.
36. Pearson JE, Senne DA, Alexander DJ, Taylor WD, Peterson LA, Russell PH. Characterization of Newcastle disease virus (avian paramyxovirus-1) isolated from pigeons. Avian Dis. (1987) 31:105–11. doi: 10.2307/1590781
37. Alexander DJ, Parsons G. Protection of chickens against challenge with the variant virus responsible for Newcastle disease in 1984 by conventional vaccination. Vet Rec. (1986) 118:176–7. doi: 10.1136/vr.118.7.176
38. Eid AAM. The epidemiology of ND in Sharkia (Master thesis). Department of Poultry and Fish, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt (1988).
39. Saad AM, Arafa A, Moatez MS, Hassan MK, Sultan AH, Hussein AH. Phylogenetic analysis and pathogenicity study of field Newcastle disease virus. Anim. Health Res. Rev. (2016) 4:145–54.
40. Abdel-Moneim AS, El-Sawah AA, Kandil MA. Characterization of variant strain of Newcastle disease virus in Egypt. J Vet Med Res. (2006) 16:12–7. doi: 10.21608/jvmr.2006.77914
41. Mohamed MH, Kumar S, Paldurai A, Megahed MM, Ghanem IA, Lebdah MA, et al. Complete genome sequence of a virulent Newcastle disease virus isolated from an outbreak in chickens in Egypt. Virus Genes. (2009) 39:234–7. doi: 10.1007/s11262-009-0385-7
42. Mohamed MH, Kumar S, Paldurai A, Samal SK. Sequence analysis of fusion protein gene of Newcastle disease virus isolated from outbreaks in Egypt during 2006. Virol J. (2011) 8:237. doi: 10.1186/1743-422X-8-237
43. Osman N, Sultan S, Ahmed AI, Ibrahim RS, Sabra M. Isolation and pathotyping of Newcastle disease viruses from field outbreaks among chickens in the Southern part of Egypt 2011-2012. Glob. Vet. (2014). 12:237–43. doi: 10.5829/idosi.gv.2014.12.02.82104
44. Shalaby AG, Erfan AM, Reheem MF, Selim AA, Al Husseny HM, Nasef AS. Avian influenza virus and Newcastle virus surveillance and characterization in broiler and layer chicken flocks in Egypt. Assiut Vet Med J. (2014) 60:142. doi: 10.21608/avmj.2014.170980
45. Saad AM, Samy A, Soliman MA, Arafa A, Zanaty A, Hassan MK, et al. Genotypic and pathogenic characterization of genotype VII Newcastle disease viruses isolated from commercial farms in Egypt and evaluation of heterologous antibody responses. Arch Virol. (2017) 162:1985–94. doi: 10.1007/s00705-017-3336-y
46. El-Bagoury GF, El-Habbaa AS, El-Adaway SF, El-Mahdy SS. Isolation, identification and pathotyping of Newcastle disease virus from chickens in Egypt. BVMJ. (2015) 29:196–204. doi: 10.21608/bvmj.2015.31819
47. El-Habbaa AS, El-Bagoury GF, El-Adaway SF, ElMahdy SS. Isolation, pathotyping and genotyping of Newcastle disease virus from broiler chickens in Egypt. J Virol Sci. (2017) 1:114–22.
48. Mouhamed AA, Mohamed MA, Bakheet BM, Aziz El-Din KA, Song C. Molecular studies on newcastle disease virus isolates in relation to field vaccine strains in Egypt (2012-2015). Int J Poult Sci. (2020) 19:193–209. doi: 10.3923/ijps.2020.193.209
49. Abd El Aziz M, El-Hamid A, Ellkany H, Nasef S, Nasr S, El Bestawy. A. Biological and molecular characterization of Newcastle disease virus circulating in chicken flocks, Egypt, during 2014-2015. Zag Vet J. (2016) 44:9–20. doi: 10.21608/zvjz.2016.7827
50. Selim KM, Selim A, Arafa A, Hussein HA, Elsanousi AA. Molecular characterization of full fusion protein (F) of Newcastle disease virus genotype VIId isolated from Egypt during 2012-2016. Vet World. (2018) 11:930. doi: 10.14202/vetworld.2018.930-938
51. Nagy A, Ali A, Zain El-Abideen MA, Kilany W, Elsayed M. Characterization and genetic analysis of recent and emergent virulent Newcastle disease viruses in Egypt. Transbound Emerg Dis. (2020) 67:2000–12. doi: 10.1111/tbed.13543
52. Zanaty AM, Hagag NM, Rabie N, Saied M, Selim K, Mousa SA, et al. Epidemiological, phylogenetic analysis and pathogenicity of newcastle disease virus circulating in poultry farms, Egypt during 2015-2018. Hosts Viruses. (2019) 6:50. doi: 10.17582/journal.hv/2019/6.3.50.59
53. Naguib M, Höper D, El-Kady M, Afify M, Erfan A, Abozeid H, et al. Genomic and antigenic properties of Newcastle disease virus genotypes 2. XX1 and 2. VII from Egypt do not point to antigenic drift as a driving force of spread. Authorea Preprints. (2020). doi: 10.22541/au.160637962.28915681/v1. [Epub ahead of print].
54. Rohaim MA, El Naggar RF, Helal AM, Hussein HA, LeBlanc N. Genetic characterization of pigeon paramyxovirus type 1 in Egypt. Br J Virol. (2016) 3:27–32. doi: 10.17582/journal.bjv/2016.3.2.27.32
55. Sabra M, Dimitrov KM, Goraichuk IV, Wajid A, Sharma P, Williams-Coplin D, et al. Phylogenetic assessment reveals continuous evolution and circulation of pigeon-derived virulent avian avulaviruses 1 in Eastern Europe, Asia, and Africa. BMC Vet Res. (2017) 13:291. doi: 10.1186/s12917-017-1211-4
56. Samy A, Tolba HMN, Farag GK, Halim AAE. Genetic and pathogenic characterization of a Newcastle disease virus isolated from pigeons in Egypt. Hosts Viruses. (2018) 5:63–71. doi: 10.17582/journal.hv/2018/5.5.63.71
57. Ahmed AAS, Sabban MS, Ibrahim AMM, Amin A, Khafagi AR, Sheble A. Some properties of Newcastle disease virus isolates recovered from migratory birds to Egypt. Zentralblatt für Veterinärmedizin Reihe B. (1980) 27:313–9. doi: 10.1111/j.1439-0450.1980.tb01696.x
58. El-Tarabili MM, El-Shahiedy MS, Hammouda MS, Fetaih HA, Abdel-Wahab SA, Ramzy NM. Natural and experimental infections of quails (couturnix couturnix japonica) with Newcastle disease virus. SCVMJ. (2009) 16:67–80.
59. El Naggar RF, Rohaim MA, Bazid AH, Ahmed KA, Hussein HA, Munir M. Biological characterization of wild-bird-origin avian avulavirus 1 and efficacy of currently applied vaccines against potential infection in commercial poultry. Arch Virol. (2018) 163:2743–55. doi: 10.1007/s00705-018-3916-5
60. Higgins DA. Nine disease outbreaks associated with myxoviruses among ducks in Hong Kong. Trop Anim Health Prod. (1971) 3:232–40. doi: 10.1007/BF02359585
61. Zhang S, Wang X, Zhao C, Liu D, Hu Y, Zhao J, et al. Phylogenetic and pathotypical analysis of two virulent Newcastle disease viruses isolated from domestic ducks in China. PLoS ONE. (2011) 6:e25000. doi: 10.1371/journal.pone.0025000
62. Chen SY, Qi-lin H, Shi-long C, Xiao-xia C, Nan-yang WN, Feng-qiang L, et al. Isolation and identification of duck paramyxovirus. Chin J Prev Vet Med. (2004) 26:118–20.
63. Cheng LF, Fu GH, Huang Y, Shi SH, Peng CX. Isolation and identification and sequence analysis of F protein gene of paramyxovirus type 1 strain FM01 from semi-muscovy duck. Chin J Prev Vet Med. (2006) 28:499–502.
64. Dai Y, Cheng XU, Liu M, Shen X, Li J, Yu S, et al. Experimental infection of duck origin virulent Newcastle disease virus strain in ducks. BMC Vet Res. (2014) 10:164. doi: 10.1186/1746-6148-10-164
65. Xu Q, Sun J, Gao M, Zhao S, Liu H, Zhang T, et al. Genetic, antigenic, and pathogenic characteristics of Newcastle disease viruses isolated from geese in China. J Vet Diagn Invest. (2017) 29:489–98. doi: 10.1177/1040638717709855
66. Aziz-Ul-Rahman N, Munir M, Shabbir MZ. Comparative evolutionary and phylogenomic analysis of Avian avulaviruses 1-20. Mol Phylogenetics Evol. (2018) 127:931–51. doi: 10.1016/j.ympev.2018.06.040
67. Elbestawy AR, Ellakany HF, Abd El-Hamid HS, Zidan RE, Gado AR, Mahmoud E, et al. Muscovy ducks infected with velogenic Newcastle disease virus (genotype VIId) act as carriers to infect in-contact chickens. Poult Sci. (2019) 98:4441–8. doi: 10.3382/ps/pez276
68. Hassan KE, El-Kady MF, EL-Sawah AA, Luttermann C, Parvin R, Shany S, et al. Respiratory disease due to mixed viral infections in poultry flocks in Egypt between 2017 and 2018: upsurge of highly pathogenic avian influenza virus subtype H5N8 since 2018. Transbound Emerg Dis. (2021) 68:21–36. doi: 10.1111/tbed.13281
69. Meng C, Rehman ZU, Liu K, Qiu X, Tan L, Sun Y, et al. Potential of genotype VII Newcastle disease viruses to cause differential infections in chickens and ducks. Transbound Emerg Dis. (2018) 65:1851–62. doi: 10.1111/tbed.12965
70. DaPalma T, Doonan BP, Trager NM, Kasman LM. A systematic approach to virus–virus interactions. Virus Res. (2010) 149:1–9. doi: 10.1016/j.virusres.2010.01.002
71. El Zowalaty ME, Chander Y, Redig PT, El Latif HKA, El Sayed MA, Goyal SM. Selective isolation of avian influenza virus (AIV) from cloacal samples containing AIV and Newcastle disease virus. J Vet Diagn Invest. (2011) 23:330–2. doi: 10.1177/104063871102300222
72. Samy A, Naguib MM. Avian respiratory coinfection and impact on avian influenza pathogenicity in domestic poultry: field and experimental findings. Vet Sci. (2018) 5:23. doi: 10.3390/vetsci5010023
73. Hassan TIR. Viral agents associated with complicated chronic respiratory disease in broilers (Master thesis). Department of Avian and Rabbit Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt (2015).
74. Gado HAA, Eid AAM, Ghanem IA, Moursi MK. Occurrence of avian influenza virus (H9 subtype) in broiler and layer chickens at sharkia governorate. Zag Vet J. (2017) 45:79–90. doi: 10.21608/zvjz.2017.29243
75. El-Aried TA, Mansour SM, ElBakrey RM, Ismail ANE, Eid AA. Infectious bursal disease virus: molecular epidemiologic perspectives and impact on vaccine efficacy against avian influenza and Newcastle disease viruses. Avian Dis. (2019) 63:606–18. doi: 10.1637/aviandiseases-D-19-00086
76. Zaher KS, Girh ZMS. Concurrent infectious bronchitis and Newcastle disease infection in Egypt. Br Poult Sci. (2014) 3:1–5. doi: 10.5829/idosi.bjps.2014.3.1.83211
77. Eid AA. Studies on some avian viruses with special reference to a heat resistant Newcastle disease virus (PhD thesis). Department of Poultry and Fish, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt (1994).
78. El Nagar A, Ali I. Case study of the Egyptian poultry sector. In: Proceedings of the International Poultry Conference, Vol. 31 Cairo (2007).
79. Radwan MM, Darwish SF, El-Sabagh IM, El-Sanousi AA, Shalaby MA. Isolation and molecular characterization of Newcastle disease virus genotypes II and VIId in Egypt between 2011 and 2012. Virus Genes. (2013) 47:311–6. doi: 10.1007/s11262-013-0950-y
80. Tran GTH, Sultan S, Osman N, Hassan MI, Van Dong H, Dao TD, et al. Molecular characterization of full genome sequences of Newcastle disease viruses circulating among vaccinated chickens in Egypt during 2011–2013. J Vet Med Sci. (2020) 19:0623. doi: 10.1292/jvms.19-0623
81. Amer MM. Isolation and identification of H9N2 avian influenza and Newcastle disease viruses co-infections in chicken. Egypt J Vet Sci. (2018) 49:135–46. doi: 10.21608/ejvs.2018.4963.1043
82. Ferreira HL, Taylor TL, Dimitrov KM, Sabra M, Afonso CL, Suarez DL. Virulent Newcastle disease viruses from chicken origin are more pathogenic and transmissible to chickens than viruses normally maintained in wild birds. Vet Microbiol. (2019) 235:25–34. doi: 10.1016/j.vetmic.2019.06.004
83. Hicks JT, Dimitrov KM, Afonso CL, Ramey AM, Bahl J. Global phylodynamic analysis of avian paramyxovirus-1 provides evidence of inter-host transmission and intercontinental spatial diffusion. BMC Evol Biol. (2019) 19:1–15. doi: 10.1186/s12862-019-1431-2
84. Wang Y, Bi Y, Yu W, Wei N, Wang W, Wei Q, et al. Two mutations in the HR2 region of Newcastle disease virus fusion protein with a cleavage motif “RRQRRL” are critical for fusogenic activity. Virol J. (2017) 14:1–7. doi: 10.1186/s12985-017-0851-0
85. Liu H, de Almeida RS, Gil P, Albina E. Cleavage site of Newcastle disease virus determines viral fitness in persistent infection cells. Vet Microbiol. (2018) 216:123–31. doi: 10.1016/j.vetmic.2018.02.006
86. Samal S, Kumar S, Khattar SK, Samal SK. A single amino acid change, Q114R, in the cleavage-site sequence of Newcastle disease virus fusion protein attenuates viral replication and pathogenicity. J Gen Virol. (2011) 92:2333–8. doi: 10.1099/vir.0.033399-0
87. Sergel TA, McGinnes LW, Morrison TG. A single amino acid change in the Newcastle disease virus fusion protein alters the requirement for HN protein in fusion. J Virol. (2000) 74:5101–7. doi: 10.1128/JVI.74.11.5101-5107.2000
88. Sergel TA, McGinnes LW, Morrison TG. Mutations in the fusion peptide and adjacent heptad repeat inhibit folding or activity of the Newcastle disease virus fusion protein. J Virol. (2001) 75:7934–43. doi: 10.1128/JVI.75.17.7934-7943.2001
89. Sergel-Germano T, McQuain C, Morrison T. Mutations in the fusion peptide and heptad repeat regions of the Newcastle disease virus fusion protein block fusion. J Virol. (1994) 68:7654–8. doi: 10.1128/jvi.68.11.7654-7658.1994
90. Yusoff K, Nesbit M, McCartney H, Meulemans G, Alexander DJ, Collins MS, et al. Location of neutralizing epitopes on the fusion protein of Newcastle disease virus strain Beaudette C. J Gen Virol. (1989) 70:3105–9. doi: 10.1099/0022-1317-70-11-3105
91. Gravel KA, McGinnes LW, Reitter J, Morrison TG. The transmembrane domain sequence affects the structure and function of the Newcastle disease virus fusion protein. J Virol. (2011) 85:3486–97. doi: 10.1128/JVI.02308-10
92. Samal S, Khattar SK, Paldurai A, Palaniyandi S, Zhu X, Collins PL, et al. Mutations in the cytoplasmic domain of the Newcastle disease virus fusion protein confer hyperfusogenic phenotypes modulating viral replication and pathogenicity. J Virol. (2013) 87:10083–93. doi: 10.1128/JVI.01446-13
93. Ren S, Xie X, Wang Y, Tong L, Gao X, Jia Y, et al. Molecular characterization of a Class I Newcastle disease virus strain isolated from a pigeon in China. Avian Pathol. (2016) 45:408–17. doi: 10.1080/03079457.2016.1153036
94. Wang Y, Yu W, Huo N, Wang W, Guo Y, Wei Q, et al. Comprehensive analysis of amino acid sequence diversity at the F protein cleavage site of Newcastle disease virus in fusogenic activity. PLoS ONE. (2017) 12:e0183923. doi: 10.1371/journal.pone.0183923
95. Connaris H, Takimoto T, Russell R, Crennell S, Moustafa I, Portner A, et al. Probing the sialic acid binding site of the hemagglutinin-neuraminidase of Newcastle disease virus: identification of key amino acids involved in cell binding, catalysis, and fusion. J Virol. (2002) 76:1816–24. doi: 10.1128/JVI.76.4.1816-1824.2002
96. Connolly SA, Leser GP, Jardetzky TS, Lamb RA. Bimolecular complementation of paramyxovirus fusion and hemagglutinin-neuraminidase proteins enhances fusion: implications for the mechanism of fusion triggering. J Virol. (2009) 83:10857–68. doi: 10.1128/JVI.01191-09
97. Lamb RA, Kolakofsky D. The paramyxoviruses. In: Fields BN, Knipe DM, Howley PM, editors. Fields Virology. Philadelphia, PA: Lippincott-Raven Publishers (1996). p. 577–604.
98. Huang Z, Panda A, Elankumaran S, Govindarajan D, Rockemann DD, Samal SK. The hemagglutinin-neuraminidase protein of Newcastle disease virus determines tropism and virulence. J Virol. (2004) 78:4176–84. doi: 10.1128/JVI.78.8.4176-4184.2004
99. Stone-Hulslander J, Morrison TG. Mutational analysis of heptad repeats in the membrane-proximal region of Newcastle disease virus HN protein. J Virol. (1999) 73:3630–7. doi: 10.1128/JVI.73.5.3630-3637.1999
100. Jin J, Zhao J, Ren Y, Zhong Q, Zhang G. Contribution of HN protein length diversity to Newcastle disease virus virulence, replication and biological activities. Sci Rep. (2016) 6:36890. doi: 10.1038/srep36890
101. McGinnes L, Sergel T, Morrison T. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology. (1993) 196:101–10. doi: 10.1006/viro.1993.1458
102. Iorio RM, Syddall RJ, Sheehan JP, Bratt MA, Glickman RL, Riel AM. Neutralization map of the hemagglutinin-neuraminidase glycoprotein of Newcastle disease virus: domains recognized by monoclonal antibodies that prevent receptor recognition. J Virol. (1991) 65:4999–5006. doi: 10.1128/jvi.65.9.4999-5006.1991
103. Cho SH, Kim SJ, Kwon HJ. Genomic sequence of an antigenic variant Newcastle disease virus isolated in Korea. Virus Genes. (2007) 35:293–302. doi: 10.1007/s11262-007-0078-z
104. Hu S, Wang T, Liu Y, Meng C, Wang X, Wu Y, et al. Identification of a variable epitope on the Newcastle disease virus hemagglutinin–neuraminidase protein. Vet Microbiol. (2010) 140:92–7. doi: 10.1016/j.vetmic.2009.07.029
105. Liu Y, Chi M, Liu Y, Wen H, Zhao L, Song Y, et al. Roles of the highly conserved amino acids in the second receptor binding site of the Newcastle disease virus HN protein. Virol J. (2019) 16:1–13. doi: 10.1186/s12985-019-1273-y
106. Gravel KA, Morrison TG. Interacting domains of the HN and F proteins of Newcastle disease virus. J Virol. (2003) 77:11040–9. doi: 10.1128/JVI.77.20.11040-11049.2003
107. Chi M, Liu Y, Wen H, Zhao L, Song Y, Liu N, et al. Role of the head stalk linker region (aa115-122) of Newcastle disease virus haemagglutinin neuraminidase in regulating fusion triggering. J Gen Virol. (2020) 101:5–20. doi: 10.1099/jgv.0.001349
108. Gu M, Liu W, Xu L, Cao Y, Yao C, Hu S, et al. Positive selection in the hemagglutinin-neuraminidase gene of Newcastle disease virus and its effect on vaccine efficacy. Virol J. (2011) 8:150. doi: 10.1186/1743-422X-8-150
109. Ruan B, Zhang X, Zhang C, Du P, Meng C, Guo M, et al. Residues 315 and 369 in HN protein contribute to the thermostability of Newcastle disease virus. Front Microbiol. (2020) 11:2223. doi: 10.3389/fmicb.2020.560482
110. Abdel-Glil MY, Mor SK, Sharafeldin TA, Porter RE, Goyal SM. Detection and characterization of Newcastle disease virus in formalin-fixed, paraffin-embedded tissues from commercial broilers in Egypt. Avian Dis. (2014) 58:118–23. doi: 10.1637/10616-071813-Reg.1
111. Ewies SS, Ali A, Tamam SM, Madbouly HM. Molecular characterization of Newcastle disease virus (genotype VII) from broiler chickens in Egypt. BJBAS. (2017) 6:232–7. doi: 10.1016/j.bjbas.2017.04.004
112. Saleh A, Ali R, Fawzy M, Eltarabily M. Characterization of Recent Isolates of Newcastle disease virus. SCVMJ. (2016) 21:235–48. doi: 10.21608/scvmj.2016.62839
113. Magdy H, Amer MM, Elbayoumi KM, Kutkat MA. Isolation and identification of genotype VII of newcastle disease virus from chicken flocks in six egyptian governorates. Spec J Med Res Health Sci. (2017) 2:1–7.
114. Awad AM, Sedeik ME, Abdelkariem AA. Isolation, molecular characterization and pathotyping of Newcastle disease viruses from field outbreaks among broiler flocks in Egypt from 2014-2015. Int J Curr Res. (2015) 7:12925–34.
115. Megahed M, Eid A, Mohamed W, Hassanin O. Genetic characterization of Egyptian Newcastle disease virus strains isolated from flocks vaccinated against Newcastle disease virus, 2014-2015. Slov Vet Res. (2018) 55:17–29. doi: 10.26873/SVR-623-2018
116. Taher MT, Amer MM, Arafa A, Saad FE. Epidemiology of viral components causing respiratory problems in broilers in six Egyptian governorates. J Vet Med Res. (2017) 24:308–20. doi: 10.21608/jvmr.2017.43266
117. El-Shall NA, Sedeik ME, El-Nahas AF, Abdel-salam RA, Awad AM. Epidemiological surveillance of some avian respiratory viral diseases in broiler chickens. AJVS. (2019) 61:185–94. doi: 10.5455/ajvs.9184
118. Lilienfeld DE, Stolley PD. Foundations of Epidemiology. New York, NY: Oxford University Press (1994).
119. Kim LM, King DJ, Curry PE, Suarez DL, Swayne DE, Stallknecht DE, et al. Phylogenetic diversity among low-virulence newcastle disease viruses from waterfowl and shorebirds and comparison of genotype distributions to those of poultry-origin isolates. J Virol. (2007) 81:12641–53. doi: 10.1128/JVI.00843-07
120. Brown VR, Bevins SN. A review of virulent Newcastle disease viruses in the United States and the role of wild birds in viral persistence and spread. Vet Res. (2017) 48:77. doi: 10.1186/s13567-017-0485-7
121. Toro H, Hoerr FJ, Farmer K, Dykstra CC, Roberts SR, Perdue M. Pigeon paramyxovirus: association with common avian pathogens in chickens and serologic survey in wild birds. Avian Dis. (2005) 49:92–8. doi: 10.1637/7268-083104R1
122. Carrasco ADOT, Seki MC, de Freitas Raso T, Paulillo AC, Pinto AA. Experimental infection of Newcastle disease virus in pigeons (Columba livia): humoral antibody response, contact transmission and viral genome shedding. Vet Microbiol. (2008) 129:89–96. doi: 10.1016/j.vetmic.2007.11.012
123. Mansour SM, ElBakrey RM, Ali H, Knudsen DE, Eid AA. Natural infection with highly pathogenic avian influenza virus H5N1 in domestic pigeons (Columba livia) in Egypt. Avian Pathol. (2014) 43:319–24. doi: 10.1080/03079457.2014.926002
124. Elgendy EM, Watanabe Y, Daidoji T, Arai Y, Ikuta K, Ibrahim MS, et al. Genetic characterization of highly pathogenic avian influenza H5N1 viruses isolated from naturally infected pigeons in Egypt. Virus Genes. (2016) 52:867–71. doi: 10.1007/s11262-016-1369-z
125. Shakal MA. Studies on paramyxovirus infection in pigeons in Egypt (PhD thesis). Department of Poultry Diseases, Faculty of Veterinary Medicine, Cairo University, Cairo, Egypt (1989).
126. Moustafa SA, Tantawy AA, Ibrahim MF. Pathological studies on outbreak of paramyxovirus infection in pigeons at Kaluobia province. BVMJ. (2003) 14:103–16.
127. Abou Hashem NA. Studies on some Paramyxovirus infection in pigeons (Master thesis). Department of Poultry and Fish, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt (1993).
128. Ali SEE. Studies on avian paramyxovirus type 1 in pigeons (Master thesis). Department of Virology, Faculty of Veterinary Medicine, Mansoura University, Mansoura, Egypt (2004).
129. Amer MIS, El-Bagoury GF, Khodeir MH. Studies on experimental infection of pigeons with Newcastle disease virus. BVMJ. (2013) 25:143–7.
130. El-Shazly MM, Ahmed BM, El-Sanousi AA, Youssef YI. Isolation and molecular characterization of newcastle disease virus genotype VI from pigeons in Egypt. AJVS. (2016) 50:145–9. doi: 10.5455/ajvs.230284
131. Hamouda EE, Mohamed NA, Eid AA, Ismail AN, El-Sisi MA, Hassanin OA, et al. Utilization of RT-PCR and restriction enzyme analysis in detection and differentiation of pigeon paramyxovirus-1 and Newcastle disease virus in pigeons. Zag Vet J. (2017) 45:37–47. doi: 10.21608/zvjz.2017.29239
132. Teske L, Ryll M, Rautenschlein S. Epidemiological investigations on the role of clinically healthy racing pigeons as a reservoir for avian paramyxovirus-1 and avian influenza virus. Avian Pathol. (2013) 42:557–65. doi: 10.1080/03079457.2013.852157
133. Wang J, Liu H, Liu W, Zheng D, Zhao Y, Li Y, et al. Genomic characterizations of six pigeon paramyxovirus Type 1 viruses isolated from live bird markets in China during 2011 to 2013. PLoS ONE. (2015) 10:e0124261. doi: 10.1371/journal.pone.0124261
134. Ellakany HF, Elbestawy AR, Abd El-Hamid HS, Zidan RE, Gado AR, Taha AE, et al. Role of pigeons in the transmission of avian avulavirus (Newcastle disease-genotype VIId) to chickens. Animal. (2019) 9:338–53. doi: 10.3390/ani9060338
135. Hamouda EE. Pathogenicity and Pathogenesis of Pigeon Paramyxovirus-1 in Pigeons. Non-published work. Department of Avian and Rabbit Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt. (2020).
136. Sabban MS, Zied AA, Basyouni A, Nadiem S, Barhouma N, Habashi YZ. Susceptibility and possible role of doves in transmission of Newcastle disease in Egypt. Zentralblatt für Veterinärmedizin Reihe B. (1982) 29:193–8. doi: 10.1111/j.1439-0450.1982.tb01215.x
137. Lima FS, Santin E, Paulillo AC, Doretto L Jr, De Moraes VRM, Schocken RP. Japanese quail (Coturnix coturnix japonica) as Newcastle disease virus carrier. Int J Poult Sci. (2004) 3:483–4. doi: 10.3923/ijps.2004.483.484
138. Czirják GÁ, Köbölkuti LB, Cadar D, Ungvári A, Niculae M, Bolfă P. An outbreak of the Newcastle disease in Japanese quail (Coturnix coturnix japonica). Bull USAMV-CN. (2007) 64:1–2. ?
139. El-Zanaty K, Abd-El-Motelib TY. Viscerotropic velogenic Newcastle disease in quails (Coturnix coturnix). Assiut Vet Med J. (1993) 29:264–75.
140. Aly BAA. Studies on pigeon paramyxovirus in quails (Master thesis). Department of Virology, Faculty of Veterinary Medicine, Benha University, Benha, Egypt (2014).
141. Abd El Aziem A, Abd-Ellatieff H, Elbestawy A, Belih S, El-Hamid A, Abou-Rawash AR. Susceptibility of Japanese quail and chickens to infection with newcastle disease virus genotype VIId. DJVS. (2020) 3:27–31. doi: 10.5455/DJVS.2020.27706.1013
142. Wakamatsu N, King DJ, Kapczynski DR, Seal BS, Brown CC. Experimental pathogenesis for chickens, turkeys, and pigeons of exotic Newcastle disease virus from an outbreak in California during 2002-2003. Vet Pathol. (2006) 43:925–33. doi: 10.1354/vp.43-6-925
143. Heckert RA, Collins MS, Manvell RJ, Strong I, Pearson JE, Alexander DJ. Comparison of Newcastle disease viruses isolated from cormorants in Canada and the USA in 1975, 1990 and 1992. Can J Vet Res. (1996) 60:50–4.
144. Lee EK, Jeon WJ, Kwon JH, Yang CB, Choi KS. Molecular epidemiological investigation of Newcastle disease virus from domestic ducks in Korea. Vet Microbiol. (2009) 134:241–8. doi: 10.1016/j.vetmic.2008.08.020
145. Sharawi S, El-Habbaa AS, Heba MZ, Khodeir MH. Experimental infection of quail by NDV and its immune response to vaccination. BVMJ. (2015) 29:218–24. doi: 10.21608/bvmj.2015.31712
146. Mohamed MA, Hafez MSA. The susceptibility of Japanese quails to the infection with chicken originated Newcastle disease virus. J Adv Vet Res. (2016) 6:37–43.
147. Ibrahim HMM. Studies on the role of migratory birds in the epidemiology of Newcastle disease in Egypt (Master thesis). Faculty of Veterinary Medicine, Cairo University, Cairo, Egypt (1979).
148. Rohaim MA, El Naggar RF, Helal AM, Hussein HA, Munir M. Reverse spillover of avian viral vaccine strains from domesticated poultry to wild birds. Vaccine. (2017) 35:3523–7. doi: 10.1016/j.vaccine.2017.05.033
149. Liu XF, Wan HQ, Ni XX, Wu YT, Liu WB. Pathotypical and genotypical characterization of strains of Newcastle disease virus isolated from outbreaks in chicken and goose flocks in some regions of China during 1985-2001. Arch Virol. (2003) 148:1387–403. doi: 10.1007/s00705-003-0014-z
150. Miller PJ, King DJ, Afonso CL, Suarez DL. Antigenic differences among Newcastle disease virus strains of different genotypes used in vaccine formulation affect viral shedding after a virulent challenge. Vaccine. (2007) 25:7238–46. doi: 10.1016/j.vaccine.2007.07.017
151. Miller PJ, Estevez C, Yu Q, Suarez DL, King DJ. Comparison of viral shedding following vaccination with inactivated and live Newcastle disease vaccines formulated with wild-type and recombinant viruses. Avian Dis. (2009) 53:39–49. doi: 10.1637/8407-071208-Reg.1
152. Dimitrov KM, Afonso CL, Yu Q, Miller PJ. Newcastle disease vaccines—a solved problem or a continuous challenge?. Vet Microbiol. (2017) 206:126–36. doi: 10.1016/j.vetmic.2016.12.019
153. Ellakany H, El-Hamid A, Nasef S, Abdel Aziz M, Gado A, Zedan R. Evaluation of the protection of commercial live and inactivated NDV vaccines against Newcastle virus genotype VIId circulating in the field. DJVS. (2019) 1:17–20. doi: 10.5455/DJVS.25271
154. El-Dabae WH, Hussein HA, Rohaim MA, El-Safty MM, Ata NS, Reda IM. Saponin-adjuvanted vaccine protects chickens against velogenic Newcastle disease virus. Arch Virol. (2018) 163:2423–32. doi: 10.1007/s00705-018-3917-4
155. Mahmoud NK, El-Deeb AH, Emara MM, Abd El-Khaleck MA, Hussein HA. Genotypes II and VIId-based inactivated Newcastle disease vaccine reduces virus shedding. Virus Dis. (2019) 30:453–61. doi: 10.1007/s13337-019-00537-2
156. Mohamed MHA. Current status of Newcastle disease at Sharkia governorate with special reference to characterization and diagnosis using PCR (Master thesis). Department of Avian and Rabbit Medicine, Faculty of Veterinary Medicine, Zagazig University, Zagazig, Egypt (2006).
157. Lebdah MA, Megahed MM, Hassanin OA, Ali AM. The negative impact of chicken infectious anemia virus infection on immune responses to different Newcastle disease virus vaccination programs. Zag Vet J. (2016) 44:138–48. doi: 10.21608/zvjz.2016.7856
158. Mohamed MH, Abdelaziz AM, Kumar S, Al-Habib MA, Megahed MM. Effect of phylogenetic diversity of velogenic Newcastle disease virus challenge on virus shedding post homologous and heterologous DNA vaccination in chickens. Avian Pathol. (2016) 45:228–34. doi: 10.1080/03079457.2016.1144870
159. Ayoub MA, Elfeil WK, El Boraey D, Hammam H, Nossair MA. Evaluation of some vaccination programs in protection of experimentally challenged broiler chicken against Newcastle disease virus. Am J Anim Vet Sci. (2019) 14:197–206. doi: 10.3844/ajavsp.2019.197.206
160. Shahin R, Yousef Y, Ibrahim M. Protective efficacy of some Newcastle disease virus (NDV) vaccination programs against velogenic NDV genotype VII in broiler chickens. JCVR. (2019) 1:103–14. doi: 10.21608/jcvr.2019.36559
161. Said M, Soliman AM, Mousa S, Arafa A, Hussein AH, Amarin N, et al. Efficacy of bivalent inactivated vaccine containing insect cell–expressed avian influenza H5 and egg-based Newcastle disease virus (NDV) against dual infection with highly pathogenic H5N1 and velogenic NDV in chickens. Avian Dis. (2019) 63:474–80. doi: 10.1637/12017-122618-Reg.1
162. Kilany WH, Ali A, Bazid AHI, ZainEl-Abideen MA, Elsayed M. Evaluation of two inactivated Newcastle disease virus vaccines (genotype II and VII) against challenge of Newcastle disease genotype VII infection in chicken. J Anim Vet Adv. (2015) 14:211–8. doi: 10.3923/javaa.2015.211.218
163. Ismail NM, El-Deeb AH, Emara MM, Tawfik HI, Wanis NA, Hussein HA. Prime-boost vaccination strategy against avian influenza and Newcastle disease viruses reduces shedding of the challenge viruses. Virus Dis. (2018) 29:324–32. doi: 10.1007/s13337-018-0463-3
164. Fawzy M, Ali RR, Elfeil WK, Saleh AA, El-Tarabilli MMA. Efficacy of inactivated velogenic Newcastle disease virus genotype VII vaccine in broiler chickens. In Vet Res Forum. (2020) 11:113–20. doi: 10.30466/vrf.2019.95311.2295
165. Shawky A, Ahmed BM, El-Makaky HM, El-Sanousi AA. A matching multivalent vaccine candidate combining velogenic NDV genotype vii and variant ibv protects chicken from virulent challenge and eliminates virus shedding. Adv Anim Vet Sci. (2020) 8:1380–7. doi: 10.17582/journal.aavs/2020/8.12.1380.1387
166. Ahmed H, Khodier M, Kasem S, El-Gohary AEG. Protective efficacy of commercial Newcastle disease vaccines against virulent genotype Vii Newcastle disease virus. KVMJ. (2017) 15:89–103. doi: 10.21608/kvmj.2017.112753
167. Hassan MI, Abd El-Azeem MW, Selim A, Sultan S. Molecular and biological characterization of the immunological potency of Newcastle disease virus oil emulsion–inactivated vaccines prepared from field isolate obtained from vaccinated chickens outbreak. Braz J Microbiol. (2020) 51:815–26. doi: 10.1007/s42770-019-00203-1
168. Nemr MHM. Anti-Viral activity of nanoparticles based chemotherapeutic system (PhD thesis). Department of Virology, Faculty of Veterinary Medicine, Suez Canal University, Ismalia, Egypt (2018).
169. Sedeik ME, Elbestawy AR, El-Shall NA, Abd El-Hack ME, Saadeldin IM, Swelum AA. Comparative efficacy of commercial inactivated Newcastle disease virus vaccines against Newcastle disease virus genotype VII in broiler chickens. Poult Sci. (2019) 98:2000–7. doi: 10.3382/ps/pey559
170. Bastami MA, Afifi MA, El-Beheiry MA, ZouElfakar SA, Rafik HS, Ahmed KA, et al. Evaluation of Some Vaccination Programs against Field Strain of Genotype VIID of Newcastle Disease in Broilers. Biosci. Res. (2018) 15:2171–84.
171. Amer MIS. Trial for preparation of an inactivated pigeon paramyxo virus vaccine (Master thesis). Department of Virology, Faculty of Veterinary Medicine, Benha University, Benha, Egypt (2008).
172. Amer MIS, El-Bagoury GF, Khodeir MH. Evaluation of the immune response of pigeons to Newcastle disease and pigeon paramyxo virus vaccines. BVMJ. (2013) 24:148–56.
173. Khedr AA, Abdel Latef AA, Sayed ML, Abdrabo MA, Nagy N, Radwan AA, et al. Efficacy of an experimental combined inactivated salmonella typhimurium and paramyxovirus vaccine in pigeons. Zag Vet J. (2016) 44:149–55. doi: 10.21608/zvjz.2016.7857
174. Soliman YA, El-Nagar E, Abd-wanees NA, El-Safty M. Efficacy of prepared oil inactivated pigeon paramyxo vaccine. Alex J Vet Sci. (2019) 60:30–40. doi: 10.5455/ajvs.5378
175. Dimitrov KM, Sharma P, Volkening JD, Goraichuk IV, Wajid A, Rehmani SF, et al. A robust and cost-effective approach to sequence and analyze complete genomes of small RNA viruses. Virol J. (2017) 14:72. doi: 10.1186/s12985-017-0741-5
176. Ali A, Safwat M, Kilany WH, Nagy A, Shehata AA, El-Abideen MAZ, et al. Combined H5ND inactivated vaccine protects chickens against challenge by different clades of highly pathogenic avian influenza viruses subtype H5 and virulent Newcastle disease virus. Vet World. (2019) 12:97. doi: 10.14202/vetworld.2019.97-105
177. Liu H, de Almeida RS, Gil P, Majó N, Nofrarías M, Briand FX, et al. Can genotype mismatch really affect the level of protection conferred by Newcastle disease vaccines against heterologous virulent strains?. Vaccine. (2018) 36:3917–25. doi: 10.1016/j.vaccine.2018.05.074
178. Cardenas-Garcia S, Diel DG, Susta L, Lucio-Decanini E, Yu Q, Brown CC, et al. Development of an improved vaccine evaluation protocol to compare the efficacy of Newcastle disease vaccines. Biologicals. (2015) 43:136–45. doi: 10.1016/j.biologicals.2014.11.003
Keywords: APMV-1, Newcastle disease virus, phylogenetic analysis, deduced amino acid, cleavage site, fusion protein, genotypes
Citation: Mansour SMG, ElBakrey RM, Mohamed FF, Hamouda EE, Abdallah MS, Elbestawy AR, Ismail MM, Abdien HMF and Eid AAM (2021) Avian Paramyxovirus Type 1 in Egypt: Epidemiology, Evolutionary Perspective, and Vaccine Approach. Front. Vet. Sci. 8:647462. doi: 10.3389/fvets.2021.647462
Received: 29 December 2020; Accepted: 14 June 2021;
Published: 15 July 2021.
Edited by:
Vinay G. Joshi, Lala Lajpat Rai University of Veterinary and Animal Sciences, IndiaReviewed by:
Madhuri Subbiah, National Institute of Animal Biotechnology (NIAB), IndiaCopyright © 2021 Mansour, ElBakrey, Mohamed, Hamouda, Abdallah, Elbestawy, Ismail, Abdien and Eid. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amal A. M. Eid, YW1hbGFlaWR2ZXRAZ21haWwuY29t; Shimaa M. G. Mansour, c21nYWxhbEB6dS5lZHUuZWc=; c2hpbWFhZ2FsYWxAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.