- Center for Food Animal Health Research, College of Food, Agricultural and Environmental Sciences, The Ohio State University, Wooster, OH, United States
Coronaviruses (CoVs) possess the largest and most complex RNA genome (up to 32 kb) that encodes for 16 non-structural proteins regulating RNA synthesis and modification. Coronaviruses are known to infect a wide range of mammalian and avian species causing remarkably diverse disease syndromes. Variable tissue tropism and the ability to easily cross interspecies barriers are the well-known characteristics of certain CoVs. The 21st century epidemics of severe acute respiratory CoV (SARS-CoV), Middle East respiratory CoV and the ongoing SARS-CoV-2 pandemic further highlight these characteristics and emphasize the relevance of CoVs to the global public health. Bovine CoVs (BCoVs) are betacoronaviruses associated with neonatal calf diarrhea, and with winter dysentery and shipping fever in older cattle. Of interest, no distinct genetic or antigenic markers have been identified in BCoVs associated with these distinct clinical syndromes. In contrast, like other CoVs, BCoVs exist as quasispecies. Besides cattle, BCoVs and bovine-like CoVs were identified in various domestic and wild ruminant species (water buffalo, sheep, goat, dromedary camel, llama, alpaca, deer, wild cattle, antelopes, giraffes, and wild goats), dogs and humans. Surprisingly, bovine-like CoVs also cannot be reliably distinguished from BCoVs using comparative genomics. Additionally, there are historical examples of zoonotic transmission of BCoVs. This article will discuss BCoV pathogenesis, epidemiology, interspecies transmission, immune responses, vaccines, and diagnostics.
Introduction
Coronaviruses (CoVs) are enveloped viruses with the largest RNA genome (26.4–31.7 kb) that belong to the subfamily of Coronavirinae within the family Coronaviridae, order Nidovirales (1). Currently, CoVs are classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus, with alpha- and betacoronaviruses represented by mammalian CoVs, while all avian CoVs are members of the other two genera (1).
Coronaviruses infect a wide diversity of mammalian and avian species causing respiratory, enteric, neurologic and hepatic disorders (1). The 21st century has already provided abundant and well-documented evidence of the ability of CoVs to quickly adapt to new hosts and ecological niches. Examples of expansion of host and geographical ranges include the severe acute respiratory coronavirus (SARS-CoV) pandemic, as well as the ongoing Middle East respiratory coronavirus (MERS-CoV) epidemic and the SARS-CoV-2 pandemic; emergence of porcine epidemic diarrhea virus in both Americas, and porcine deltacoronavirus emergence and spread in Asia followed by its spread to the US (2–6). This ability to quickly adapt to novel hosts and ecological niches are attributed to the high mutation rate (number of mutations/genome/round of replication) caused by relatively low fidelity of the viral RNA polymerase (7), the large genomes of CoVs (8), and the high frequency of homologous recombination events during RNA replication well-documented for several porcine, feline and canine coronaviruses (9–13). CoVs are the only known RNA viruses that evolved a mechanism for proofreading their genomes (via activities of non-structural proteins 10–14) allowing them to escape lethal error catastrophe events (14) and to generate and maintain highly diverse and viable quasispecies pools.
Several human CoVs (HCoVs) associated with common colds and acute gastroenteritis, HCoV-229E, HCoV-HKU1, HCoV-NL63, HCoV-OC43, and human enteric CoV 44 (HECoV-44) were recognized (15) even before modern tools for molecular characterization and diagnostics were available and the epidemic/pandemic nature of CoVs was widely appreciated. Some of them were of suspect zoonotic origin (15, 16) including spillover from cattle (HCoV-OC43 and HECoV-44) (15, 17, 18).
Bovine coronavirus (BCoV) is a pneumoenteric virus that belongs to the species Betacoronavirus 1 (subgenus Embecovirus) of the Betacoronavirus genus along with wild ruminant CoVs, porcine hemagglutinating encephalomyelitis virus, equine coronavirus, HCoV-OC43, HECoV-44, and canine respiratory coronavirus (1). Due to their close antigenic and genetic relatedness, Betacoronavirus 1 species members appear to be host-range variants originating from the same parental virus as a result of multiple genetic recombination and interspecies transmission events (17, 19–21).
Bovine CoV particles are enveloped and pleomorphic, 65–210 nm in diameter (22). Bovine CoV particles possess 5 major structural proteins: the nucleocapsid protein (N, 50 kDa), the integral membrane (M, 25 kDa), the small membrane/envelope protein (E, 8 kDa), the haemagglutinin-esterase (HE, 120–140 kDa) and the spike (S, 190 kDa) (22). The latter consists of an S1 subunit that contains the dominant neutralizing epitopes and an S2 subunit that mediates viral membrane fusion. The HE acts as a receptor-destroying enzyme (esterase) to reverse hemagglutination. The N protein lies internal to the virus envelope and is associated with the viral RNA, the M spans the viral envelope while the S and HE project from the envelope. Additionally, 16 non-structural proteins (nsp1-16) have been identified in betacoronaviruses (23, 24). Like other enveloped viruses, BCoVs are sensitive to detergents and lipid solvents (including ether, chloroform) and are easily inactivated by most conventional disinfectants, formalin, and heat.
Bovine coronaviruses cause respiratory and enteric diseases in cattle and other ruminants but can be identified in the respiratory and intestinal tracts of healthy cattle (22, 25–27). BCoV is shed in feces and nasal secretions and is associated with 3 distinct clinical syndromes in cattle (27): (neonatal) calf diarrhea [(N)CD)] (22, 28), winter dysentery (WD) characterized by hemorrhagic diarrhea in adults (25, 28–30) and respiratory infections in cattle of different ages commonly as a part of the bovine respiratory disease complex (BRDC) or shipping fever of feedlot cattle (22, 26, 31, 32) (Figure 1). BRDC causes major economic losses to the beef and dairy cattle industries worldwide due to substantial morbidity and mortality. In North America, this complex represents the leading cause of morbidity and mortality in 6–10-month-old beef cattle after entry into feedlots (33).
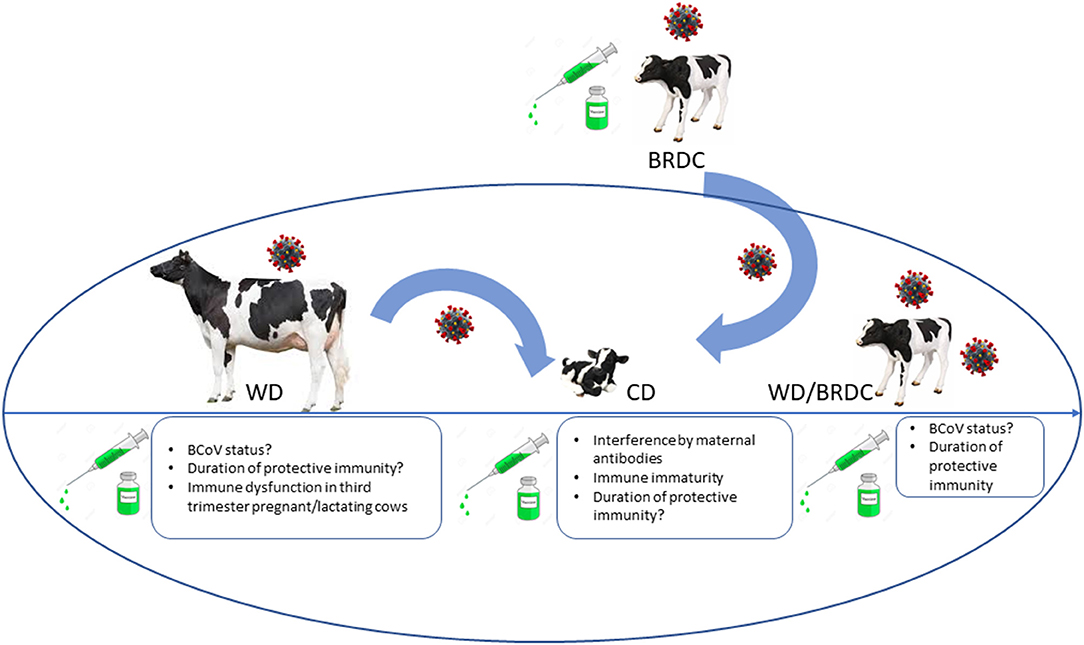
Figure 1. Summary diagram of different clinical syndromes associated with BCoV and challenges for successful vaccination associated with different ages/production status of cattle. CD, calf diarrhea; WD, winter dysentery; BRDC, bovine respiratory disease complex. Coronavirus image placed above animal represents potential carrier status. Rectangle boxes list unknown host and vaccine-associated factors that can result in suboptimal vaccine performance or lack of protection of cattle of different ages.
All BCoV isolates identified so far are shed in feces and nasal secretions and belong to a single serotype/genotype based on virus cross-neutralization and genotyping analyses regardless of clinical origin (27, 28, 34, 35). However, genotyping identified distinct sublineages and clusters based on the year and place of isolation but not on the disease type (19, 35–38) (Figure 2). Similarly, 2 to 3 subtypes of BCoV are recognized based on their biologic and antigenic characteristics identified in virus neutralization and ELISA [with monoclonal antibodies (MAbs)] tests without association with the different disease types (22, 26–28). While some studies identified mutations potentially associated with respiratory and enteric phenotypes (40–42); these findings have not been consistently confirmed by other groups in observational or experimental studies (36). Further, despite numerous genetic differences (point mutations and deletions) detected in the spike (S) gene between enteric and respiratory isolates or between BCoV and bovine-like CoVs from wild ruminants and humans, in vivo and in vitro experiments demonstrated high levels of cross-protection and cross-neutralization between such isolates (36, 43–48). Thus, no genetic or antigenic markers associated with the different disease manifestations have been identified, suggesting that the latter may result from the complex interplay between pathogens (CoV and other viruses or bacteria), host and environmental factors.
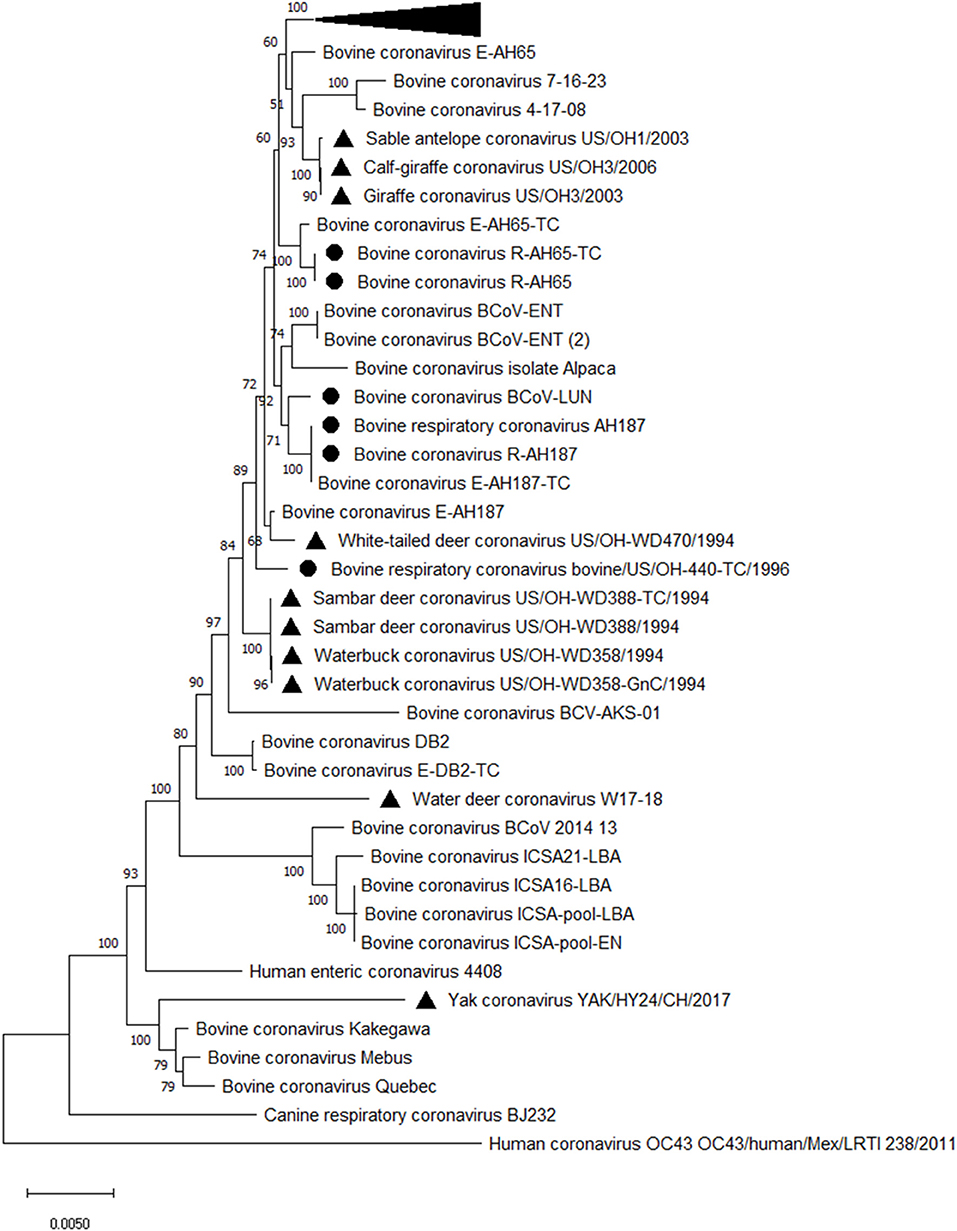
Figure 2. Phylogenetic analysis of complete genomes of enteric and respiratory BCoVs and bovine-like CoVs from wild ruminants. The evolutionary history was inferred by using the Maximum Likelihood method and General Time Reversible model. The tree with the highest log likelihood (−81,750.81) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis involved 53 nucleotide sequences. Evolutionary analyses were conducted in MEGA X (39). Black triangle markers are used for bovine-like CoVs isolated from wild ruminants, and black round markers are used to mark respiratory BCoVs. The collapsed branch includes a cluster of recent BCoV strains from Japan (35).
Receptors and Attachment Factors
BCoV attaches to N-acetyl-9-O-acetylneuraminic acid (Neu5,9Ac2) through HE and S proteins to initiate infection (49–51). BCoV attachment is blocked by acetylesterase or neuraminidase treatment and can be restored by resialylation. It is further hypothesized that after the initial binding to sialic acid (SA)-containing receptors, the BCoV S protein may interact with a specific cellular receptor that leads to a conformational change and the viral-cell membrane fusion (52). A study demonstrated that similar to HCoV-OC43, BCoV preferentially used α-2,6-(SA) and not α-2,3-SA; however, this effect was less pronounced for BCoV compared with HCoV-OC43 (53). The same study has shown that BCoV (similar to canine respiratory CoV) employs human leukocyte antigen class I (HLA-I) as an entry receptor. HLA class I molecules that belong to the immunoglobulin superfamily consist of three α chains linked with a β2-microglobulin molecule that adopt a standard immunoglobulin-like fold (54). The receptor-binding (lectin) and receptor-destroying (esterase) domains of the HE protein also play important roles for viral entry (55); although, these interactions may be weaker than for the S protein because HE is a less efficient hemagglutinin. Additionally, heparan sulfate was identified as an alternative attachment factor, but this may be related to the cell culture adaptation, not affecting the in vivo infection and pathogenesis (53).
Histo-blood group antigens (HBGAs) are known to play a role in cell attachment and pathogenesis of other betacoronaviruses, including SARS-CoV and SARS-CoV-2 (56, 57). Specifically, studies indicate that blood group A individuals are at higher risk of infection suggesting that HBGAs may serve as additional cellular receptors. It is currently unknown if BCoV can utilize HBGAs as cell entry receptors, which necessitates further research in this direction to understand the mechanisms of emergence of BCoV-like CoVs (such as HCoV-OC43 and HECoV-4408) into human population.
Overall, the complex interactions between BCoVs, their cellular receptors and mucosal microbiota need to be comprehensively evaluated to improve our understanding of BCoV epidemiology, pathogenesis and interspecies transmission.
Epidemiology and Pathogenesis of Respiratory and Enteric Bovine Coronaviruses
Bovine coronavirus is widespread in cattle of all ages, resulting in economic losses to the beef and dairy industry throughout the world. The virus presence has been confirmed on all continents, and seroprevalence studies demonstrate that over 90% of cattle have been exposed to BCoV during their lifetime. Moreover, BCoVs are commonly identified in the respiratory and intestinal tracts of healthy and diseased cattle (58). Recent evidence suggests that BCoV can persist in colostrum-deprived calves or colostrum-fed calves with repeated nasal shedding (59) and detectable BCoV antibodies for up to 3 years suggesting that active immune response does not always result in viral clearance (60).
Enteric BCoV Infections
After its accidental discovery in 1972 by Mebus et al. at the University of Nebraska (61), the virus was quickly isolated and characterized (62), and soon after recognized as a common cause of calf diarrhea (CD) (63). The virus plays a major role in the development of CD during the first 3 weeks of life in both dairy and beef cattle herds (27, 64). The disease results from the virus infecting both the small and large intestines, destroying villi and leading to severe, sometimes bloody, diarrhea and high mortality (65–67). Virus replication initially occurs in different sections of the small intestine subsequently spreading throughout the large intestine and causing a malabsorptive diarrhea. BCoV-infected intestinal epithelial cells (IECs) die, slough off, and are replaced by immature IECs. Stunted and fused small intestinal villi as well as atrophied colonic ridges are observed during pathological examination (65, 66, 68). The compensatory crypt hyperplasia and increased fluid secretion further exacerbates diarrhea (69). The loss of the mature IECs capable of absorption and digestive enzyme secretion greatly diminishes the absorptive, metabolic and secretory capacity of the intestinal tract (22, 69).
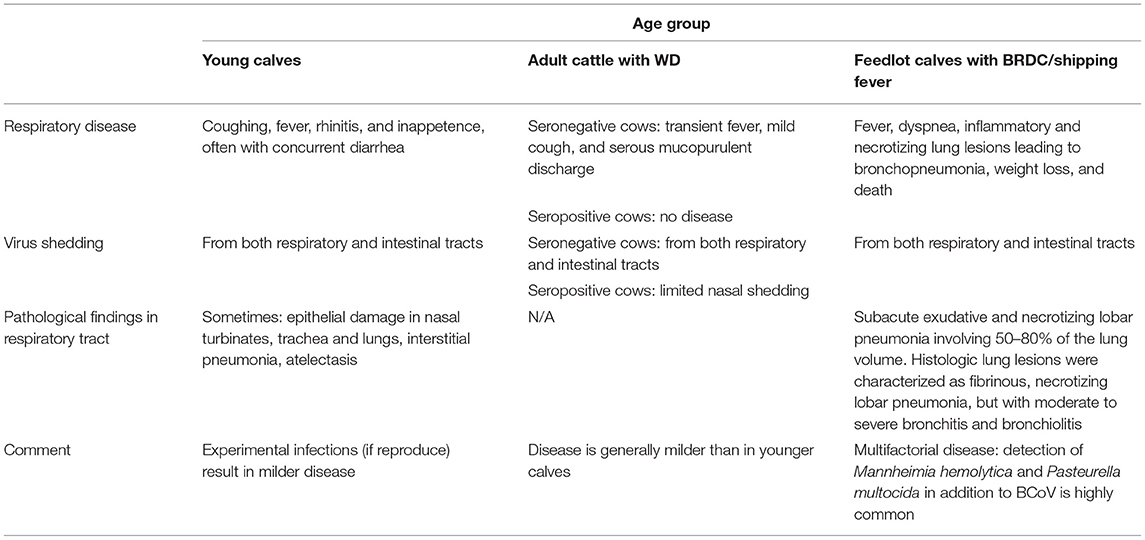
Continual feeding often provides more nutrients than the damaged small intestinal epithelium can absorb (70). This in turn leads to undigested nutrient fermentation in the large intestine, increased fluid accumulation, bacterial overgrowth and overproduction of organic acids, aggravating diarrhea (69, 71). Altogether, it leads to dehydration, metabolic acidosis and electrolyte imbalance (due to sodium, chloride, potassium, and bicarbonate loss) (72). While these pathological changes and the ensuing disease are most severe in younger animals, BCoV infection also contributes to winter dysentery (WD) development in adult dairy cattle that causes a dramatic decrease in milk production and significant economic losses (25, 27, 73).
BCoV causes enteritis in both dairy and beef herds, with most cases occurring within the first 30 days of life (22, 61, 64, 74). BCoV incidence in naturally occurring outbreaks of diarrheal disease reportedly may vary from 15 to 70% (68, 75, 76). CD may occur as early as 24 h of age in colostrum-deprived calves and as late as 5 months of age (59, 68, 77). Soon after becoming infected, calves begin shedding high amounts of BCoV (≤109 viral particles/ml of feces) which can continue for up to 2 weeks; while during the convalescent phase recovered calves generally shed lower amounts for several weeks (78). While BCoV is detected in the feces of both diarrheic and healthy calves, diarrheic calves tested positive more often (incidence: 8–69%) than healthy ones (incidence: 0–24%) (68, 69, 75). Low-level intermittent BCoV shedding can be observed in >70% of healthy cows despite the presence of serum and intestinal BCoV-specific antibodies (76, 79). Because BCoV is more stable at lower ambient temperature and reduced ultraviolet light levels, BCoV shedding rates increase by 50–60% during the winter months (22, 80) likely contributing to WD development in adult cattle (29). BCoV shedding also increased by 65% at parturition, and by 71% 2 weeks postpartum due to immunological and hormonal perturbations in cows (80). Thus, calves born to BCoV-positive cows have a significantly higher risk of developing diarrhea due to periparturient exposure to the contaminated perineum, teats, and the calving area (81).
Besides being an important enteric pathogen of cattle, BCoVs are capable of interspecies transmission and causing disease in adoptive/spillover hosts. For example, HECoV-4408 from a child with acute diarrhea is closely related to BCoV genetically and antigenically and is suggested to be a BCoV variant (17). It was confirmed that it can infect and cause diarrhea in gnotobiotic calves as well as induce complete cross-protective immunity against the virulent BCoV-DB2 enteric CD strain (47). Based on complete genome analysis, it is hypothesized that porcine hemagglutinating encephalomyelitis virus and HCoV-OC43 have evolved from ancestral BCoV strains at some point in the past (42). This analysis provides evidence that CoVs of bovine/ruminant origin can become endemic in adoptive/spillover hosts. Enteric BCoVs (bovine-like CoVs) can also infect dogs (experimentally) and various domestic and wild ruminant species (naturally and experimentally), causing clinical (82) or subclinical infections and seroconversion (83, 84). These data raise questions of whether dogs or wild ruminants could also be a reservoir for bovine-like CoVs transmissible to cattle, or conversely, if cattle can transmit CoVs to dogs, other ruminant species and humans. Overall, the existing evidence indicates that BCoVs are associated with serious and economically significant disease.
Respiratory BCoV Infections
In 1982 Thomas et al. was first to identify BCoV as one of contributing agents in calf pneumonia (85). Subsequently, numerous investigators have confirmed that enteric and respiratory BCoVs are members of the same quasispecies (86), despite genotypic and antigenic differences between individual isolates (26). Since 1995, the role of respiratory BCoV in BRDC development and reduced growth performance in feedlot cattle has been increasingly recognized (26, 27, 31, 87–90). Currently, respiratory tract infections in growing and feedlot calves are frequently attributed to BCoV (26, 27, 91, 92) and can manifest as mild respiratory disease (coughing, rhinitis) or pneumonia in 2–6-month-old calves (26, 27). A study demonstrated high respiratory BCoV nasal and fecal shedding rates of 84 and 96%, respectively (90). In another study, testing cattle 3-day pre-arrival demonstrated that nasal shedding consistently preceded fecal shedding (91). It was also demonstrated that many animals (61–74%) shed respiratory BCoV prior to shipping to feedlots (93). Fifty-eight to ninety-five percent of feedlot cattle seroconverted to BCoV by 3 weeks after arrival (87, 90, 94, 95). The widespread distribution of BCoV could be explained by two main factors: (1) shedding of high titers of the virus from respiratory and intestinal tracts (59, 92), and (2) existence of asymptomatic (carrier) animals within most herds. These carrier animals shed the virus in nasal secretions and feces and serve as a source of infection to neonates and other susceptible animals on the farm (59, 96).
BCoV seroprevalence studies that surveyed 135 Norwegian dairy herds have demonstrated that calves in BCoV-seropositive herds had an increased risk of respiratory disease development compared with BCoV-seronegative herds (97). Dairy calves in Ohio shed respiratory BCoV fecally and nasally with some of them developing diarrhea and/or respiratory disease (rhinitis) (59, 98). In a large feedlot study (n = 1,074 cattle), it was demonstrated that feedlot calves shedding respiratory BCoV nasally were more likely to have respiratory disease (1.6 times) and pulmonary lesions at slaughter (2.2 times) than animals that did not shed respiratory BCoV (31). In two subsequent studies, calves shedding respiratory BCoV nasally were 2.7 and 1.5 times more likely to have respiratory disease than those that did not shed (90, 91). In another report, nasal BCoV shedding or an antibody titer <20 correlated with increased risk of treatment for BRDC, while intranasal vaccination with BCoV vaccine mitigated this risk (99). Further, in 3 of 4 outbreaks in Italy, the classic signs of BRDC (dyspnea, fever, respiratory distress) were evident in 2–3-month-old calves positive for BCoV and negative for other respiratory viral pathogens, while presence of common bacterial pathogens of cattle (Staphylococcus spp. and Proteus mirabilis or Mannheimia haemolytica) was only confirmed in 2 of 4 outbreaks (92).
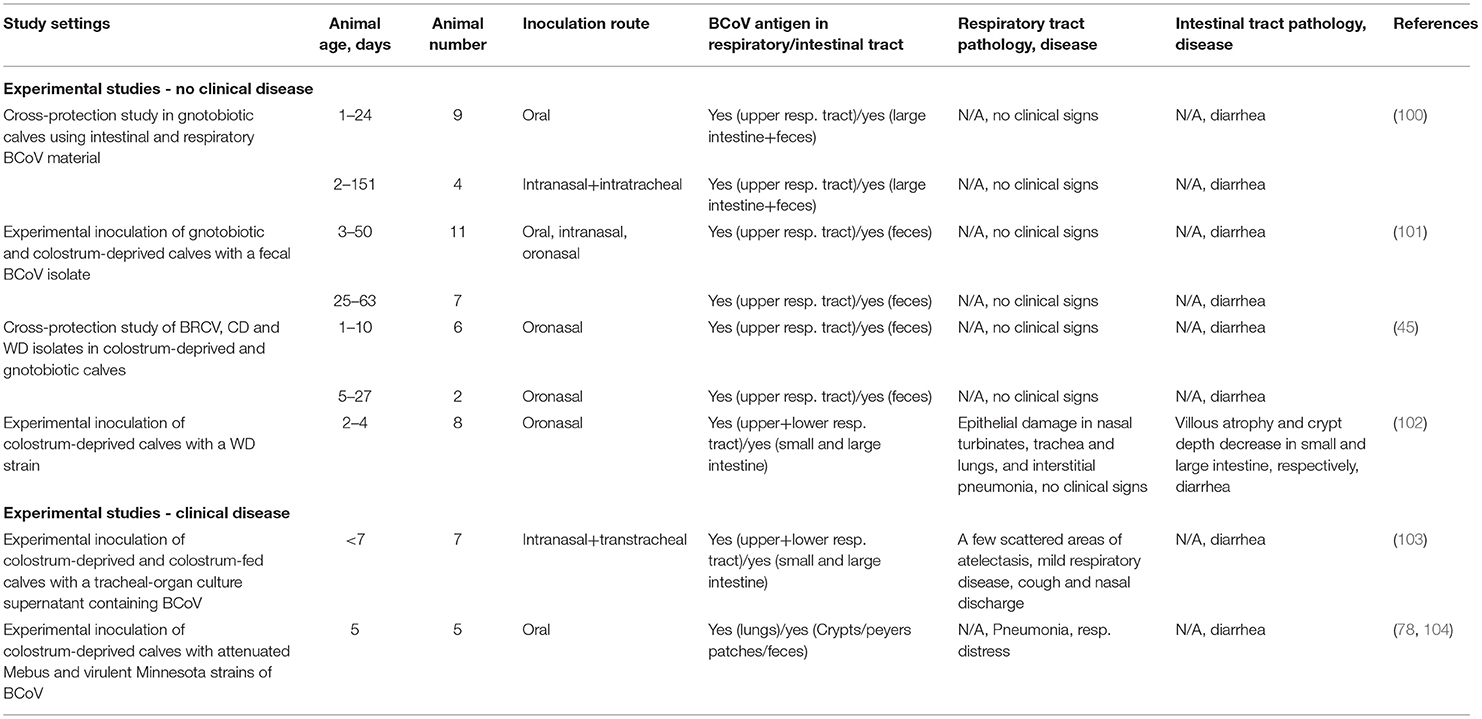
Despite this abundant circumstantial evidence derived from epidemiological studies and routine diagnostic testing for respiratory pathogens, the role of BCoV in the bovine respiratory disease complex (BRDC) has been debated for several decades (32). This is because attempts to experimentally reproduce clinical respiratory disease are often unsuccessful (45, 100–102) (Table 1). Of interest one out of the 4 studies that failed to reproduce clinical disease (Table 1) reported somewhat extensive pathological changes in the respiratory tract of the colostrum-deprived calves inoculated with KWD3 strain (102) (Table 1).
In two studies, investigators were able to reproduce some features of respiratory disease of variable severity (Table 1) (78, 103, 104). In the first one, 7 colostrum-deprived or colostrum-fed calves received tracheal-organ culture supernatant containing BCoV (from a field case) and developed cough, nasal discharge, and diarrhea with a few scattered areas of atelectasis observed in lungs of 3 calves at post-mortem (Table 1). In another study, 3 colostrum-deprived calves were inoculated with a field virulent pneumoenteric BCoV (Minnesota); all of them developed diarrhea, 2 had pneumonia/respiratory distress, and 1 died (Table 1).
Thus, the BCoV-induced respiratory clinical disease and the associated pathological changes remain less defined than those for enteric BCoV (Tables 1, 2) (26, 27). The consensus opinion is that the respiratory disease and pathology may vary with BCoV strain, animal age, co-infections with other pathogens (from BRDC complex), weather and additional environmental stress factors (Table 2) (26, 27). The list of other viral and bacterial pathogens identified in association with BRDC is rather extensive and includes: parainfluenza virus, type 3, bovine respiratory syncytial virus, adenovirus, enterovirus, reovirus, influenza D virus, bovine viral diarrhea virus, bovine herpesvirus 1 and 4, Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, Mycoplasma bovis, and Trueperella pyogenes (105). Multiagent experiments evaluated interactions between the respiratory pathogens and stress and emphasized their synergistic effects in the BRDC, but no such experiments have been reported for BCoV (32, 105, 106). A recent study has concurrently identified BCoV, H. somni, M. haemolytica, and P. multocida (but not other common BRDC pathogens) during a respiratory disease outbreak in pre-weaned beef calves emphasizing that these pathogens were likely the major contributors to the disease development (107). The multifactorial nature of BRDC makes treating it extremely challenging. For example, antibiotic treatment of animals with BRDC may lead to massive release of bacterial lipopolysaccharides (LPS) inducing proinflammatory cytokine response and further enhancing lung damage (27, 108) while treatment with corticosteroids can reduce the numbers of CD4 and CD8 T cells and certain cytokine levels compromising protective immunity and exacerbating disease (109, 110). Thus, current evidence suggests that BCoV alone may not be routinely associated with substantial infection/pathology of the lower respiratory tract, but likely is significant as a respiratory pathogen in the case of mixed infections and plays a major role in inciting the disease. However, experimental data on infection of SPF BCoV seronegative cattle with BCoV respiratory isolates is needed to better confirm the role of BCoV in respiratory disease.
BCOV Interspecies Transmission
BCoV-Like Infections of Wild and Domestic Ruminants
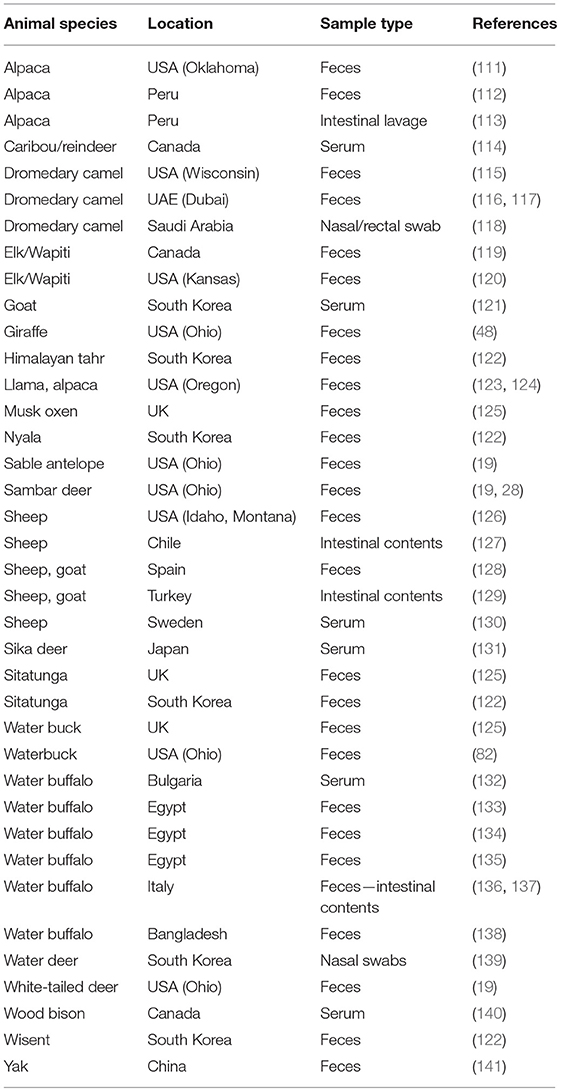
In the last several decades, numerous CoV strains sharing extensive biologic, antigenic and genetic similarities with BCoV (named therefore bovine-like CoVs) have been identified in the feces, intestinal contents or respiratory secretions of a diverse group of domestic and wild (captive or free-range) ruminant species (Table 3). Experimental inoculation of gnotobiotic (Gn) or colostrum-deprived calves demonstrated that many of the bovine-like CoVs were capable of efficient replication in these calves, produced enteric disease and generated cross-protective immune responses (48, 82). Additionally, 6.6, 8.7, and 13.3% of sera from white-tailed deer in Ohio, mule deer in Wyoming and caribous in Quebec, respectively, were seropositive for BCoV antibodies (82, 114) suggesting that natural infections are common.
Complete genome sequencing revealed that some of the bovine-like CoVs isolated from wild and captive ruminants (giraffe, sambar deer, white-tailed deer, waterbuck, water deer, sable antelope, camelids and water buffalo) share high (>98–99.6%) sequence identity with enteric and respiratory BCoV strains, supporting their close genetic relatedness (19, 48, 84). Similar to the lack of genetic markers for discriminating between enteric and respiratory BCoVs, no reliable genetic markers were identified to distinguish between BCoVs and ruminant bovine-like CoVs (19). Instead, phylogenetic analysis demonstrates clustering of BCoVs/bovine-like CoVs according to the year of detection/isolation (Figure 2), which suggests that there is likely a co-evolution with continuous exchange by the respective virus pools. Finally, no consistent changes were observed in the genomes of bovine-like CoVs after passage in gnotobiotic calves. These data suggest that wild ruminants may represent a natural reservoir for bovine-like CoVs and transmit them to cattle or vice versa; these BCoVs represent a single host-range CoV species (27). Such interspecies infections may result in generation of more genetically divergent, potentially recombinant strains that escape immune responses and could potentially spread to other species including humans based on an historical precedent (42). Further, similar to BCoVs, a number of bovine-like CoVs from wild/captive ruminants can readily replicate in human rectal tumor-18 (HRT-18) cells (19, 48, 82), which further suggests a possibility for zoonotic transmission events.
Of interest, while bovine-like CoVs are frequently detected in diarrheic or healthy wild ruminants, there are no reports of occurrence of CoV-related respiratory disease outbreaks in the wild ruminants. This once again emphasizes the multifactorial nature of BRDC in cattle and suggests that its development is likely related to existing herd management practices including crowded housing, transportation, constant influx of new animals, production-associated stresses in cows, etc.
BCoV-Like Infections of Non-ruminant Species
In addition to a broad range of domestic and wild ruminants, CoVs genetically/antigenically similar to BCoV have been detected in dogs with respiratory disease (142) and in humans (17). An enteropathogenic BCoV strain caused a subclinical infection and seroconversion in experimentally infected dogs (83). A recent report demonstrated that the HECoV-4408 strain infects upper respiratory and intestinal tracts causing diarrhea and intestinal lesions and conferring complete cross-protection against the virulent BCoV DB2 strain in gnotobiotic calves (47). Finally, while the most common ancestor has not been identified, it is assumed that HCoV-OC43 has emerged in the human population at the end of the 19th century likely originating form a BCoV ancestral strain via recombination events (42). Thus, detailed investigation of the endemic and emerging BCoV strains in the context of the host glycobiology is needed to identify and control potential pre-pandemic variants.
Diagnosis
Pneumoenteric BCoVs replicate in the upper respiratory and the intestinal tracts and are detected in nasal secretions and feces, with nasal often preceding fecal shedding (27, 90, 95, 100, 101, 143). BCoV has also been detected or isolated from lung in animals with respiratory disease (88). The complete list of post-mortem diagnostic samples from animals with suspected respiratory (or enteric) BCoV disease includes tissues from the respiratory tract (e.g., nasal, pharyngeal, tracheal and lung tissues) (34, 89, 93) and from the distal small intestine and colon (101). For live animals, nasal secretions collected with sterile polyester swabs and feces collected in sterile fecal cups should be chilled and transported to the diagnostic lab (95, 144). From live calves with acute respiratory disease, tracheobronchial lavage fluids that were previously shown to be positive for BCoV antigen by ELISA can be aspirated (145).
The acute transient nature of enteric BCoV infections in younger calves and seronegative cattle necessitates sample collection at disease onset or shortly thereafter. However, persistent or long-term intermittent shedding has been reported in recovered or healthy cattle, respectively (59, 60, 146). Due to the stress of shipping and comingling of animals from different sources, the peak of BCoV nasal (or fecal) shedding associated with BRDC infections occurs within 1 week after arrival to the feedlot (89–91, 95).
BCoV infection can be diagnosed by detection of virus, viral antigen, or viral RNA in tissues, or various animal secretions/excretions. Comprehensive diagnosis includes virus detection in nasal secretions, lung homogenates or feces, and virus isolation in HRT-18 cells during the acute phase and/or detection of BCoV-binding or virus neutralizing (VN) antibodies in the convalescent phase (27, 89, 93, 144, 147).
Immunofluorescent/immunohistochemical staining with hyperimmune antiserum or MAbs to BCoV is performed for viral antigen detection in respiratory (trachea, lung) or intestinal (ileum, colon) tissues (frozen or paraffin-embedded). Detection of BCoV in nasal secretions or feces can be done using immune electron microscopy, which has the advantage of detecting other viruses, but its sensitivity is relatively low (59, 82). BCoV antigens are commonly detected by ELISA using BCoV MAbs which can be used for fast and reliable analysis of large sample numbers. RT-PCR, nested RT-PCR, real-time qRT-PCR (targeting most conserved genomic regions—-ORF1ab or N gene) are the most sensitive assays currently available for BCoV detection (45, 146, 148). For fecal samples, internal controls or additional sample dilution may be needed to detect interference by PCR inhibitors (149).
Antibodies to BCoV in serum, nasal secretions, or feces can be quantitated in ELISA [overall or isotype-specific antibodies (IgM, IgA, IgG1, and IgG2)] or using VN or HI tests that measure functional neutralizing or hemagglutinating antibodies, respectively (34, 150, 151). Because BCoV antibodies are widespread in cattle, paired acute and convalescent serum samples are needed for serologic diagnosis of BCoV infections (26).
Immunity, Vaccines, and Other Prevention Strategies
The correlates of immune protection against BCoV infections remain poorly defined. In multiple studies the BCoV-binding, neutralizing and HI antibody levels in serum of naturally infected calves or cattle on arrival in feedlots were correlated with protection against enteric or respiratory disease (including pneumonia), or BCoV shedding (27, 91, 94, 95, 98, 151–153). Inoculation of gnotobiotic or colostrum-deprived calves with CD, WD, or respiratory BCoV strains induced complete protection against diarrhea following challenge with a CD strain (43, 45). However, whether the serum antibodies themselves confer protection against BCoV or they only reflect previous BCoV exposure is uncertain and requires further investigation. Limited data from some epidemiologic studies of BRDC infections in cattle suggest that BCoV serum antibody levels may be a marker for respiratory protection (26). So, Lin et al. demonstrated that cattle shedding respiratory BCoV at the start of the epizootic of BRDC/pneumonia had low levels of VN and HI antibodies, whereas cattle with high levels of antibodies against the HE and S viral glycoproteins remained negative for respiratory BCoV (151, 152). Furthermore, the levels of VN, HI and IgM, IgG1, and IgG2-BCoV antibody levels in serum were highly correlated with protection against respiratory BCoV infection. Finally, in cattle with fatal respiratory BCoV infections, only IgM antibody responses were detected. However, a recent longitudinal study demonstrated that BCoV serum antibody levels did not correlate with the incidence of BRDC or BCoV shedding (107).
Optimal vaccines against pneumoenteric mucosal pathogens should be delivered to and be effective at both sites of virus replication (respiratory and intestinal tracts) to provide optimal protection. Also, most vaccines against mucosal pathogens fail to induce sterilizing immunity or to prevent subsequent reinfections, as observed for natural or experimental BCoV infections (45, 59). Thus, the goal of vaccination is to confer broad protection against the severe enteric/respiratory disease that may lead to mortality and requires treatments. These objectives may be best accomplished by vaccinating calves on farms prior to shipping to auction barns or feedlots because BCoV infections occurring at feedlots necessitate rapid onset of protective immune responses. In support of this hypothesis, a recent study demonstrated that IN vaccination of feedlot calves with a modified live-BCoV calf vaccine on entry to a feedlot reduced the risk of treatment for BRDC in calves (99). These data confirm the results of experimental challenge studies of calves confirming cross-protection among BCoV strains of distinct clinical origin. Another recent study from Uruguay demonstrated that maternal vaccination against BCoV reduced BCoV shedding in their calves (154). Further studies are needed to determine whether inclusion of a mixture of CD/WD/BRDC strains could be an optimal strategy to develop a single broad-spectrum BCoV vaccine effective against BCoV infections associated with distinct clinical syndromes (26).
While several licensed modified live vaccines against BCoV are currently available (Bovilis, Merck/PBS Animal Health; Guardian, Merck; Calf-Guard, ScourGuard 4KC and BoviShield Gold 5, Zoetis; CattleWin BC, KyotoBiken Laboratories, Kyoto, Japan), observational data or field studies evaluating their protective efficacy against CD/WD remain scarce, while some of studies suggest they induce poor protection against BRDC viruses (155). Additionally, most studies evaluating candidate BCoV vaccines report on their safety and immunogenicity but not protective efficacy (156–159).
The ubiquitous nature of BCoV infections, lack of comprehensive data on vaccine efficacy, short duration of post-vaccine mucosal immunity, immunological immaturity of calves, and potential interference by maternal antibodies represent some of the existing challenges that likely contributed to the lack of optimal protective effects against BRDC and BCoV diarrheal disease. Moreover, the complexity of vaccine management (the need to target variable age groups of cattle, production status, newly arriving animals, etc.) and suboptimal cost-benefit ratio of vaccine use suggest that alternative control and prevention strategies need to be evaluated and employed. So far, Norway is the only country that has been implementing a joint biosecurity control program for BCoV and BRSV since 2016 (160). This program has the goal of reducing the occurrence of BCoV on a herd level by implementing strict biosecurity measures and financial reward system for compliant farmers/industries, while vaccination is not part of this program. The efficacy of such approach remains to be determined.
Concluding Remarks
An overview of the existing data suggests that additional research is needed to understand the basic mechanisms of BCoV respiratory and enteric disease pathogenesis and the variables and interactions between viral, host and environmental factors that exacerbate disease or can lead to enhanced shedding and transmission. Further additional extensive research is needed to identify correlates of immune protection and attributes required to generate effective vaccines/regimens that can prevent severe disease and limit virus spread. The limited knowledge of the mechanisms that regulate BCoV interspecies transmission and determine broad host ranges warrants further research. Finally, continuous investigation is needed to better understand the ecology of bovine-like CoVs present in wildlife reservoirs (including wild ruminants or other susceptible species) or the threats they represent for public or animal health.
Author Contributions
AV generated the initial draft. AV and LS have revised the final content critically. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. De Groot RJ, Baker SC, Baric R, Enjuanes L, Gorbalenya AE, Holmes KV, et al. Family coronaviridae. In: King AMQ, Adams M, Carstens E, Lefkowitz E, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. San Diego, CA: Elsevier Science Publishing Co., Inc. (2012).
2. Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. (2003) 348:1953–66. doi: 10.1056/NEJMoa030781
3. Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, et al. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science. (2003) 300:1394–9. doi: 10.1126/science.1085952
4. Drosten C, Muth D, Corman VM, Hussain R, Al Masri M, Hajomar W, et al. An observational, laboratory-based study of outbreaks of middle East respiratory syndrome coronavirus in Jeddah and Riyadh, kingdom of Saudi Arabia, 2014. Clin Infect Dis. (2015) 60:369–77. doi: 10.1093/cid/ciu812
5. Wang Q, Vlasova AN, Kenney SP, Saif LJ. Emerging and re-emerging coronaviruses in pigs. Curr Opin Virol. (2019) 34:39–49. doi: 10.1016/j.coviro.2018.12.001
6. Ralph R, Lew J, Zeng T, Francis M, Xue B, Roux M, et al. 2019-nCoV (Wuhan virus), a novel Coronavirus: human-to-human transmission, travel-related cases, vaccine readiness. J Infect Dev Ctries. (2020) 14:3–17. doi: 10.3855/jidc.12425
7. Drake JW, Holland JJ. Mutation rates among RNA viruses. Proc Natl Acad Sci USA. (1999) 96:13910–3. doi: 10.1073/pnas.96.24.13910
8. Woo PC, Lau SK, Huang Y, Yuen KY. Coronavirus diversity, phylogeny and interspecies jumping. Exp Biol Med. (2009) 234:1117–27. doi: 10.3181/0903-MR-94
9. Decaro N, Mari V, Elia G, Addie DD, Camero M, Lucente MS, et al. Recombinant canine coronaviruses in dogs, Europe. Emerg Infect Dis. (2010) 16:41–7. doi: 10.3201/eid1601.090726
10. Terada Y, Matsui N, Noguchi K, Kuwata R, Shimoda H, Soma T, et al. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE. (2014) 9:e106534. doi: 10.1371/journal.pone.0106534
11. Akimkin V, Beer M, Blome S, Hanke D, Hoper D, Jenckel M, et al. New chimeric porcine coronavirus in Swine Feces, Germany, 2012. Emerg Infect Dis. (2016) 22:1314–5. doi: 10.3201/eid2207.160179
12. Boniotti MB, Papetti A, Lavazza A, Alborali G, Sozzi E, Chiapponi C, et al. Porcine epidemic diarrhea virus and discovery of a recombinant swine enteric coronavirus, Italy. Emerg Infect Dis. (2016) 22:83–7. doi: 10.3201/eid2201.150544
13. Mandelik R, Sarvas M, Jackova A, Salamunova S, Novotny J, Vilcek S. First outbreak with chimeric swine enteric coronavirus (SeCoV) on pig farms in Slovakia - lessons to learn. Acta Vet Hung. (2018) 66:488–92. doi: 10.1556/004.2018.043
14. Lauber C, Goeman JJ, Parquet Mdel C, Nga PT, Snijder EJ, Morita K, et al. The footprint of genome architecture in the largest genome expansion in RNA viruses. PLoS Pathog. (2013) 9:e1003500. doi: 10.1371/journal.ppat.1003500
15. Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, et al. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. (2016) 24:490–502. doi: 10.1016/j.tim.2016.03.003
16. Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, et al. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol. (2012) 86:12816–25. doi: 10.1128/JVI.00906-12
17. Zhang XM, Herbst W, Kousoulas KG, Storz J. Biological and genetic characterization of a hemagglutinating coronavirus isolated from a diarrhoeic child. J Med Virol. (1994) 44:152–61. doi: 10.1002/jmv.1890440207
18. Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, et al. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol. (2005) 79:1595–604. doi: 10.1128/JVI.79.3.1595-1604.2005
19. Alekseev KP, Vlasova AN, Jung K, Hasoksuz M, Zhang X, Halpin R, et al. Bovine-like coronaviruses isolated from four species of captive wild ruminants are homologous to bovine coronaviruses, based on complete genomic sequences. J Virol. (2008) 82:12422–31. doi: 10.1128/JVI.01586-08
20. Lau SK, Lee P, Tsang AK, Yip CC, Tse H, Lee RA, et al. Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination. J Virol. (2011) 85:11325–37. doi: 10.1128/JVI.05512-11
21. Lau SK, Woo PC, Yip CC, Fan RY, Huang Y, Wang M, et al. Isolation and characterization of a novel betacoronavirus subgroup A coronavirus, rabbit coronavirus HKU14, from domestic rabbits. J Virol. (2012) 86:5481–96. doi: 10.1128/JVI.06927-11
23. Cox GJ, Parker MD, Babiuk LA. Bovine coronavirus nonstructural protein ns2 is a phosphoprotein. Virology. (1991) 185:509–12. doi: 10.1016/0042-6822(91)90810-X
24. Gustin KM, Guan BJ, Dziduszko A, Brian DA. Bovine coronavirus nonstructural protein 1 (p28) is an RNA binding protein that binds terminal genomic cis-replication elements. J Virol. (2009) 83:6087–97. doi: 10.1128/JVI.00160-09
25. Saif LJ. A review of evidence implicating bovine coronavirus in the etiology of winter dysentery in cows: an enigma resolved? Cornell Vet. (1990) 80:303–11.
26. Saif LJ. Bovine respiratory coronavirus. Vet Clin North Am Food Anim Pract. (2010) 26:349–64. doi: 10.1016/j.cvfa.2010.04.005
27. Saif LJ, Jung K. Comparative pathogenesis of bovine and porcine respiratory coronaviruses in the animal host species and SARS-CoV-2 in humans. J Clin Microbiol. (2020) 58:3291–8. doi: 10.1128/JCM.01355-20
28. Tsunemitsu H, Saif LJ. Antigenic and biological comparisons of bovine coronaviruses derived from neonatal calf diarrhea and winter dysentery of adult cattle. Arch Virol. (1995) 140:1303–11. doi: 10.1007/BF01322757
29. Cho KO, Halbur PG, Bruna JD, Sorden SD, Yoon KJ, Janke BH, et al. Detection and isolation of coronavirus from feces of three herds of feedlot cattle during outbreaks of winter dysentery-like disease. J Am Vet Med Assoc. (2000) 217:1191–4. doi: 10.2460/javma.2000.217.1191
30. Traven M, Naslund K, Linde N, Linde B, Silvan A, Fossum C, et al. Experimental reproduction of winter dysentery in lactating cows using BCV – comparison with BCV infection in milk-fed calves. Vet Microbiol. (2001) 81:127–51. doi: 10.1016/S0378-1135(01)00337-6
31. Lathrop SL, Wittum TE, Brock KV, Loerch SC, Perino LJ, Bingham HR, et al. Association between infection of the respiratory tract attributable to bovine coronavirus and health and growth performance of cattle in feedlots. Am J Vet Res. (2000) 61:1062–6. doi: 10.2460/ajvr.2000.61.1062
32. Ellis J. What is the evidence that bovine coronavirus is a biologically significant respiratory pathogen in cattle? Can Vet J. (2019) 60:147–52.
33. USDA. Feedlot 2011 Part IV: Health and Health Management on U.S. Feedlots with a Capacity of 1,000 or More Head (2011).
34. Hasoksuz M, Lathrop S, Al-Dubaib MA, Lewis P, Saif LJ. Antigenic variation among bovine enteric coronaviruses (BECV) and bovine respiratory coronaviruses (BRCV) detected using monoclonal antibodies. Arch Virol. (1999) 144:2441–7. doi: 10.1007/s007050050656
35. Suzuki T, Otake Y, Uchimoto S, Hasebe A, Goto Y. Genomic characterization and phylogenetic classification of bovine coronaviruses through whole genome sequence analysis. Viruses. (2020) 12:183. doi: 10.3390/v12020183
36. Zhang X, Hasoksuz M, Spiro D, Halpin R, Wang S, Vlasova A, et al. Quasispecies of bovine enteric and respiratory coronaviruses based on complete genome sequences and genetic changes after tissue culture adaptation. Virology. (2007) 363:1–10. doi: 10.1016/j.virol.2007.03.018
37. Bok M, Mino S, Rodriguez D, Badaracco A, Nunes I, Souza SP, et al. Molecular and antigenic characterization of bovine coronavirus circulating in Argentinean cattle during 1994-2010. Vet Microbiol. (2015) 181:221–9. doi: 10.1016/j.vetmic.2015.10.017
38. Gunn L, Collins PJ, O'connell MJ, O'shea H. Phylogenetic investigation of enteric bovine coronavirus in Ireland reveals partitioning between European and global strains. Ir Vet J. (2015) 68:31. doi: 10.1186/s13620-015-0060-3
39. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547–9. doi: 10.1093/molbev/msy096
40. Gelinas AM, Boutin M, Sasseville AM, Dea S. Bovine coronaviruses associated with enteric and respiratory diseases in Canadian dairy cattle display different reactivities to anti-HE monoclonal antibodies and distinct amino acid changes in their HE. S and ns4.9 protein. Virus Res. (2001) 76:43–57. doi: 10.1016/S0168-1702(01)00243-X
41. Gelinas AM, Sasseville AM, Dea S. Identification of specific variations within the HE. S1, and ORF4 genes of bovine coronaviruses associated with enteric and respiratory diseases in dairy cattle. Adv Exp Med Biol. (2001) 494:63–7. doi: 10.1007/978-1-4615-1325-4_9
42. Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, et al. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol. (2006) 80:7270–4. doi: 10.1128/JVI.02675-05
43. El-Kanawati ZR, Tsunemitsu H, Smith DR, Saif LJ. Infection and cross-protection studies of winter dysentery and calf diarrhea bovine coronavirus strains in colostrum-deprived and gnotobiotic calves. Am J Vet Res. (1996) 57:48–53.
44. Chouljenko VN, Kousoulas KG, Lin X, Storz J. Nucleotide and predicted amino acid sequences of all genes encoded by the 3' genomic portion (9.5 kb) of respiratory bovine coronaviruses and comparisons among respiratory and enteric coronaviruses. Virus Genes. (1998) 17:33–42. doi: 10.1023/A:1008048916808
45. Cho KO, Hasoksuz M, Nielsen PR, Chang KO, Lathrop S, Saif LJ. Cross-protection studies between respiratory and calf diarrhea and winter dysentery coronavirus strains in calves and RT-PCR and nested PCR for their detection. Arch Virol. (2001) 146:2401–19. doi: 10.1007/s007050170011
46. Hasoksuz M, Sreevatsan S, Cho KO, Hoet AE, Saif LJ. Molecular analysis of the S1 subunit of the spike glycoprotein of respiratory and enteric bovine coronavirus isolates. Virus Res. (2002) 84:101–9. doi: 10.1016/S0168-1702(02)00004-7
47. Han MG, Cheon DS, Zhang X, Saif LJ. Cross-protection against a human enteric coronavirus and a virulent bovine enteric coronavirus in gnotobiotic calves. J Virol. (2006) 80:12350–6. doi: 10.1128/JVI.00402-06
48. Hasoksuz M, Alekseev K, Vlasova A, Zhang X, Spiro D, Halpin R, et al. Biologic, antigenic, and full-length genomic characterization of a bovine-like coronavirus isolated from a giraffe. J Virol. (2007) 81:4981–90. doi: 10.1128/JVI.02361-06
49. Vlasak R, Luytjes W, Spaan W, Palese P. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci USA. (1988) 85:4526–9. doi: 10.1073/pnas.85.12.4526
50. Schultze B, Gross HJ, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. (1991) 65:6232–7. doi: 10.1128/JVI.65.11.6232-6237.1991
51. Schultze B, Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J Gen Virol. (1992) 73:901–6. doi: 10.1099/0022-1317-73-4-901
52. Schultze B, Herrler G. Recognition of cellular receptors by bovine coronavirus. Arch Virol Suppl. (1994) 9:451–9. doi: 10.1007/978-3-7091-9326-6_44
53. Szczepanski A, Owczarek K, Bzowska M, Gula K, Drebot I, Ochman M, et al. Canine respiratory coronavirus, bovine coronavirus, and human coronavirus OC43: receptors and attachment factors. Viruses. (2019) 11:328. doi: 10.3390/v11040328
54. Hassan I, Ahmad F. Structural diversity of class I MHC-like molecules and its implications in binding specificities. Adv Protein Chem Struct Biol. (2011) 83:223–70. doi: 10.1016/B978-0-12-381262-9.00006-9
55. Kienzle TE, Abraham S, Hogue BG, Brian DA. Structure and orientation of expressed bovine coronavirus hemagglutinin-esterase protein. J Virol. (1990) 64:1834–8. doi: 10.1128/JVI.64.4.1834-1838.1990
56. Guillon P, Ruvoen-Clouet N, Le Moullac-Vaidye B, Marchandeau S, Le Pendu J. Association between expression of the H histo-blood group antigen, alpha1,2fucosyltransferases polymorphism of wild rabbits, and sensitivity to rabbit hemorrhagic disease virus. Glycobiology. (2009) 19:21–8. doi: 10.1093/glycob/cwn098
57. Fan Q, Zhang W, Li B, Li DJ, Zhang J, Zhao F. Association between ABO blood group system and COVID-19 susceptibility in Wuhan. Front Cell Infect Microbiol. (2020) 10:404. doi: 10.3389/fcimb.2020.00404
58. Fulton RW, Herd HR, Sorensen NJ, Confer AW, Ritchey JW, Ridpath JF, et al. Enteric disease in postweaned beef calves associated with Bovine coronavirus clade 2. J Vet Diagn Invest. (2015) 27:97–101. doi: 10.1177/1040638714559026
59. Heckert RA, Saif LJ, Hoblet KH, Agnes AG. A longitudinal study of bovine coronavirus enteric and respiratory infections in dairy calves in two herds in Ohio. Vet Microbiol. (1990) 22:187–201. doi: 10.1016/0378-1135(90)90106-6
60. Kanno T, Ishihara R, Hatama S, Uchida I. A long-term animal experiment indicating persistent infection of bovine coronavirus in cattle. J Vet Med Sci. (2018) 80:1134–7. doi: 10.1292/jvms.18-0050
61. Mebus CA, White RG, Stair EL, Rhodes MB, Twiehaus MJ. Neonatal calf diarrhea: results of a field trial using a reo-like virus vaccine. Vet Med Small Anim Clin. (1972) 67:173–4.
62. Mebus CA, Stair EL, Rhodes MB, Twiehaus MJ. Neonatal calf diarrhea: propagation, attenuation, and characteristics of a coronavirus-like agent. Am J Vet Res. (1973) 34:145–50.
63. Woode GN, Bridger JC, Meyling A. Significance of bovine coronavirus infection. Vet Rec. (1978) 102:15–16. doi: 10.1136/vr.102.1.15
64. Boileau MJ, Kapil S. Bovine coronavirus associated syndromes. Vet Clin North Am Food Anim Pract. (2010) 26:123–46. doi: 10.1016/j.cvfa.2009.10.003
65. Mebus CA, Stair EL, Rhodes MB, Twiehaus MJ. Pathology of neonatal calf diarrhea induced by a coronavirus-like agent. Vet Pathol. (1973) 10:45–64. doi: 10.1177/030098587301000105
66. Mebus CA, Newman LE, Stair EL Jr. Scanning electron, light, and immunofluorescent microscopy of intestine of gnotobiotic calf infected with calf diarrheal coronavirus. Am J Vet Res. (1975) 36:1719–25.
67. Torres-Medina A, Schlafer DH, Mebus CA. Rotaviral and coronaviral diarrhea. Vet Clin North Am Food Anim Pract. (1985) 1:471–93. doi: 10.1016/S0749-0720(15)31297-4
68. Langpap TJ, Bergeland ME, Reed DE. Coronaviral enteritis of young calves: virologic and pathologic findings in naturally occurring infections. Am J Vet Res. (1979) 40:1476–8.
69. Moon HW. Mechanisms in the pathogenesis of diarrhea: a review. J Am Vet Med Assoc. (1978) 172:443–8.
70. Nappert G, Hamilton D, Petrie L, Naylor JM. Determination of lactose and xylose malabsorption in preruminant diarrheic calves. Can J Vet Res. (1993) 57:152–8.
71. Ewaschuk JB, Naylor JM, Palmer R, Whiting SJ, Zello GA. D-lactate production and excretion in diarrheic calves. J Vet Intern Med. (2004) 18:744–7. doi: 10.1111/j.1939-1676.2004.tb02615.x
72. Lewis LD, Phillips RW. Pathophysiologic changes due to coronavirus-induced diarrhea in the calf. J Am Vet Med Assoc. (1978) 173:636–42.
73. Natsuaki S, Goto K, Nakamura K, Yamada M, Ueo H, Komori T, et al. Fatal winter dysentery with severe anemia in an adult cow. J Vet Med Sci. (2007) 69:957–60. doi: 10.1292/jvms.69.957
74. Hoet AE, Smiley J, Thomas C, Nielsen PR, Wittum TE, Saif LJ. Association of enteric shedding of bovine torovirus (Breda virus) and other enteropathogens with diarrhea in veal calves. Am J Vet Res. (2003) 64:485–90. doi: 10.2460/ajvr.2003.64.485
75. Marsolais G, Assaf R, Montpetit C, Marois P. Diagnosis of viral agents associated with neonatal calf diarrhea. Can J Comp Med. (1978) 42:168–71.
76. Crouch CF, Acres SD. Prevalence of rotavirus and coronavirus antigens in the feces of normal cows. Can J Comp Med. (1984) 48:340–2.
77. Reynolds DJ, Morgan JH, Chanter N, Jones PW, Bridger JC, Debney TG, et al. Microbiology of calf diarrhoea in southern Britain. Vet Rec. (1986) 119:34–9. doi: 10.1136/vr.119.2.34
78. Kapil S, Trent AM, Goyal SM. Excretion and persistence of bovine coronavirus in neonatal calves. Arch Virol. (1990) 115:127–32. doi: 10.1007/BF01310629
79. Crouch CF, Bielefeldt Ohmann H, Watts TC, Babiuk LA. Chronic shedding of bovine enteric coronavirus antigen-antibody complexes by clinically normal cows. J Gen Virol. (1985) 66:1489–500. doi: 10.1099/0022-1317-66-7-1489
80. Collins JK, Riegel CA, Olson JD, Fountain A. Shedding of enteric coronavirus in adult cattle. Am J Vet Res. (1987) 48:361–5.
81. Bulgin MS, Ward AC, Barrett DP, Lane VM. Detection of rotavirus and coronavirus shedding in two beef cow herds in Idaho. Can Vet J. (1989) 30:235–9.
82. Tsunemitsu H, El-Kanawati ZR, Smith DR, Reed HH, Saif LJ. Isolation of coronaviruses antigenically indistinguishable from bovine coronavirus from wild ruminants with diarrhea. J Clin Microbiol. (1995) 33:3264–9. doi: 10.1128/JCM.33.12.3264-3269.1995
83. Kaneshima T, Hohdatsu T, Hagino R, Hosoya S, Nojiri Y, Murata M, et al. The infectivity and pathogenicity of a group 2 bovine coronavirus in pups. J Vet Med Sci. (2007) 69:301–3. doi: 10.1292/jvms.69.301
84. Amer HM. Bovine-like coronaviruses in domestic and wild ruminants. Anim Health Res Rev. (2018) 19:113–24. doi: 10.1017/S1466252318000117
85. Thomas LH, Gourlay RN, Stott EJ, Howard CJ, Bridger JC. A search for new microorganisms in calf pneumonia by the inoculation of gnotobiotic calves. Res Vet Sci. (1982) 33:170–82. doi: 10.1016/S0034-5288(18)32331-2
86. Domingo E, Martinez-Salas E, Sobrino F, De La Torre JC, Portela A, Ortin J, et al. The quasispecies (extremely heterogeneous) nature of viral RNA genome populations: biological relevance–a review. Gene. (1985) 40:1–8. doi: 10.1016/0378-1119(85)90017-4
87. Lathrop SL, Wittum TE, Loerch SC, Perino LJ, Saif LJ. Antibody titers against bovine coronavirus and shedding of the virus via the respiratory tract in feedlot cattle. Am J Vet Res. (2000) 61:1057–61. doi: 10.2460/ajvr.2000.61.1057
88. Storz J, Lin X, Purdy CW, Chouljenko VN, Kousoulas KG, Enright FM, et al. Coronavirus and Pasteurella infections in bovine shipping fever pneumonia and Evans' criteria for causation. J Clin Microbiol. (2000) 38:3291–8. doi: 10.1128/JCM.38.9.3291-3298.2000
89. Storz J, Purdy CW, Lin X, Burrell M, Truax RE, Briggs RE, et al. Isolation of respiratory bovine coronavirus, other cytocidal viruses, and Pasteurella spp from cattle involved in two natural outbreaks of shipping fever. J Am Vet Med Assoc. (2000) 216:1599–604. doi: 10.2460/javma.2000.216.1599
90. Hasoksuz M, Hoet AE, Loerch SC, Wittum TE, Nielsen PR, Saif LJ. Detection of respiratory and enteric shedding of bovine coronaviruses in cattle in an Ohio feedlot. J Vet Diagn Invest. (2002) 14:308–13. doi: 10.1177/104063870201400406
91. Thomas CJ, Hoet AE, Sreevatsan S, Wittum TE, Briggs RE, Duff GC, et al. Transmission of bovine coronavirus and serologic responses in feedlot calves under field conditions. Am J Vet Res. (2006) 67:1412–20. doi: 10.2460/ajvr.67.8.1412
92. Decaro N, Campolo M, Desario C, Cirone F, D'abramo M, Lorusso E, et al. Respiratory disease associated with bovine coronavirus infection in cattle herds in Southern Italy. J Vet Diagn Invest. (2008) 20:28–32. doi: 10.1177/104063870802000105
93. Storz J, Stine L, Liem A, Anderson GA. Coronavirus isolation from nasal swab samples in cattle with signs of respiratory tract disease after shipping. J Am Vet Med Assoc. (1996) 208:1452–5.
94. Martin SW, Nagy E, Shewen PE, Harland RJ. The association of titers to bovine coronavirus with treatment for bovine respiratory disease and weight gain in feedlot calves. Can J Vet Res. (1998) 62:257–61.
95. Cho KO, Hoet AE, Loerch SC, Wittum TE, Saif LJ. Evaluation of concurrent shedding of bovine coronavirus via the respiratory tract and enteric route in feedlot cattle. Am J Vet Res. (2001) 62:1436–41. doi: 10.2460/ajvr.2001.62.1436
96. Carman PS, Hazlett MJ. Bovine coronavirus infection in Ontario 1990-1991. Can Vet J. (1992) 33:812–4.
97. Gulliksen SM, Jor E, Lie KI, Loken T, Akerstedt J, Osteras O. Respiratory infections in Norwegian dairy calves. J Dairy Sci. (2009) 92:5139–46. doi: 10.3168/jds.2009-2224
98. Heckert RA, Saif LJ, Myers GW, Agnes AG. Epidemiologic factors and isotype-specific antibody responses in serum and mucosal secretions of dairy calves with bovine coronavirus respiratory tract and enteric tract infections. Am J Vet Res. (1991) 52:845–51.
99. Plummer PJ, Rohrbach BW, Daugherty RA, Daugherty RA, Thomas KV, Wilkes RP, et al. Effect of intranasal vaccination against bovine enteric coronavirus on the occurrence of respiratory tract disease in a commercial backgrounding feedlot. J Am Vet Med Assoc. (2004) 225:726–31. doi: 10.2460/javma.2004.225.726
100. Reynolds DJ, Debney TG, Hall GA, Thomas LH, Parsons KR. Studies on the relationship between coronaviruses from the intestinal and respiratory tracts of calves. Arch Virol. (1985) 85:71–83. doi: 10.1007/BF01317007
101. Saif LJ, Redman DR, Moorhead PD, Theil KW. Experimentally induced coronavirus infections in calves: viral replication in the respiratory and intestinal tracts. Am J Vet Res. (1986) 47:1426–32.
102. Park SJ, Kim GY, Choy HE, Hong YJ, Saif LJ, Jeong JH, et al. Dual enteric and respiratory tropisms of winter dysentery bovine coronavirus in calves. Arch Virol. (2007) 152:1885–900. doi: 10.1007/s00705-007-1005-2
103. Mcnulty MS, Bryson DG, Allan GM, Logan EF. Coronavirus infection of the bovine respiratory tract. Vet Microbiol. (1984) 9:425–34. doi: 10.1016/0378-1135(84)90063-4
104. Kapil S, Pomeroy KA, Goyal SM, Trent AM. Experimental infection with a virulent pneumoenteric isolate of bovine coronavirus. J Vet Diagn Invest. (1991) 3:88–9. doi: 10.1177/104063879100300123
105. Mcgill JL, Sacco RE. The immunology of bovine respiratory disease: recent advancements. Vet Clin North Am Food Anim Pract. (2020) 36:333–48. doi: 10.1016/j.cvfa.2020.03.002
106. Jakab GJ. Viral-bacterial interactions in pulmonary infection. Adv Vet Sci Comp Med. (1982) 26:155–71.
107. Workman AM, Kuehn LA, Mcdaneld TG, Clawson ML, Loy JD. Longitudinal study of humoral immunity to bovine coronavirus, virus shedding, and treatment for bovine respiratory disease in pre-weaned beef calves. BMC Vet Res. (2019) 15:161. doi: 10.1186/s12917-019-1887-8
108. Van Reeth K, Nauwynck H. Proinflammatory cytokines and viral respiratory disease in pigs. Vet Res. (2000) 31:187–213. doi: 10.1051/vetres:2000113
109. Jung K, Alekseev KP, Zhang X, Cheon DS, Vlasova AN, Saif LJ. Altered pathogenesis of porcine respiratory coronavirus in pigs due to immunosuppressive effects of dexamethasone: implications for corticosteroid use in treatment of severe acute respiratory syndrome coronavirus. J Virol. (2007) 81:13681–93. doi: 10.1128/JVI.01702-07
110. Zhang X, Alekseev K, Jung K, Vlasova A, Hadya N, Saif LJ. Cytokine responses in porcine respiratory coronavirus-infected pigs treated with corticosteroids as a model for severe acute respiratory syndrome. J Virol. (2008) 82:4420–8. doi: 10.1128/JVI.02190-07
111. Genova SG, Streeter RN, Simpson KM, Kapil S. Detection of an antigenic group 2 coronavirus in an adult alpaca with enteritis. Clin Vaccine Immunol. (2008) 15:1629–32. doi: 10.1128/CVI.00232-08
112. López WP, Chamorro ML, Garmendia AEB. Rapid detection of rotavirus and coronavirus in alpaca crias (Vicugna pacos) with diarrhea in the Cusco region, Peru. Rev Investig Vet Perú. (2011) 22:407–11.
113. Rojas M, Manchego A, Rocha CB, Fornells LA, Silva RC, Mendes GS, et al. Outbreak of diarrhea among preweaning alpacas (Vicugna pacos) in the southern Peruvian highland. J Infect Dev Ctries. (2016) 10:269–74. doi: 10.3855/jidc.7398
114. Elazhary MA, Frechette JL, Silim A, Roy RS. Serological evidence of some bovine viruses in the caribou (rangifer tarandus caribou) in Quebec. J Wildl Dis. (1981) 17:609–12. doi: 10.7589/0090-3558-17.4.609
115. Wunschmann A, Frank R, Pomeroy K, Kapil S. Enteric coronavirus infection in a juvenile dromedary (camelus dromedarius). J Vet Diagn Invest. (2002) 14:441–4. doi: 10.1177/104063870201400518
116. Woo PC, Lau SK, Wernery U, Wong EY, Tsang AK, Johnson B, et al. Novel betacoronavirus in dromedaries of the Middle East, 2013. Emerg Infect Dis. (2014) 20:560–72. doi: 10.3201/eid2004.131769
117. Woo PC, Lau SK, Fan RY, Lau CC, Wong EY, Joseph S, et al. Isolation and characterization of dromedary camel coronavirus UAE-HKU23 from dromedaries of the middle east: minimal serological cross-reactivity between MERS coronavirus and dromedary camel coronavirus UAE-HKU23. Int J Mol Sci. (2016) 17:691. doi: 10.3390/ijms17050691
118. Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. (2016) 351:81–4. doi: 10.1126/science.aac8608
119. Smits JE. A brief review of infectious and parasitic diseases of wapiti, with emphasis on western Canada and the northwestern United States. Can Vet J. (1991) 32:471–9.
120. Majhdi F, Minocha HC, Kapil S. Isolation and characterization of a coronavirus from elk calves with diarrhea. J Clin Microbiol. (1997) 35:2937–42. doi: 10.1128/JCM.35.11.2937-2942.1997
121. Yang DK, Hwang IJ, Kim BH, Kweon CH, Lee KW, Kang MI, et al. Serosurveillance of viral diseases in Korean native goats (Capra hircus). J Vet Med Sci. (2008) 70:977–9. doi: 10.1292/jvms.70.977
122. Chung JY, Kim HR, Bae YC, Lee OS, Oem JK. Detection and characterization of bovine-like coronaviruses from four species of zoo ruminants. Vet Microbiol. (2011) 148:396–401. doi: 10.1016/j.vetmic.2010.08.035
123. Cebra CK, Mattson DE, Baker RJ, Sonn RJ, Dearing PL. Potential pathogens in feces from unweaned llamas and alpacas with diarrhea. J Am Vet Med Assoc. (2003) 223:1806–8. doi: 10.2460/javma.2003.223.1806
124. Jin L, Cebra CK, Baker RJ, Mattson DE, Cohen SA, Alvarado DE, et al. Analysis of the genome sequence of an alpaca coronavirus. Virology. (2007) 365:198–203. doi: 10.1016/j.virol.2007.03.035
125. Chasey D, Reynolds DJ, Bridger JC, Debney TG, Scott AC. Identification of coronaviruses in exotic species of Bovidae. Vet Rec. (1984) 115:602–3. doi: 10.1136/vr.115.23.602
126. Harp JA, Myers LL, Rich JE, Gates NL. Role of Salmonella arizonae and other infective agents in enteric disease of lambs. Am J Vet Res. (1981) 42:596–9.
127. Reinhardt G, Zamora J, Tadich N, Polette M, Aguilar M, Riedemann S, et al. Diagnosis of coronavirus in sheep in Valdivia province, Xth Region, Chile. Arch Med Vet. (1995) 27:129–32.
128. Munoz M, Alvarez M, Lanza I, Carmenes P. Role of enteric pathogens in the aetiology of neonatal diarrhoea in lambs and goat kids in Spain. Epidemiol Infect. (1996) 117:203–11. doi: 10.1017/S0950268800001321
129. Ozmen O, Yukari BA, Haligur M, Sahinduran S. Observations and immunohistochemical detection of coronavirus, cryptosporidium parvum and giardia intestinalis in neonatal diarrhoea in lambs and kids. Schweiz Arch Tierheilkd. (2006) 148:357–64. doi: 10.1024/0036-7281.148.7.357
130. Traven M, Carlsson U, Lunden A, Larsson B. Serum antibodies to bovine coronavirus in Swedish sheep. Acta Vet Scand. (1999) 40:69–74. doi: 10.1186/BF03547042
131. Yokoi K, Okazaki H, Inahara K, Hatama S. Prevalence of eight bovine viruses in sika deer (cervus nippon yesoensis) in Japan. Vet Rec. (2009) 165:754–5.
132. Muniiappa L, Mitov BK, Kharalambiev Kh. E. [Demonstration of coronavirus infection in buffaloes]. Vet Med Nauki. (1985) 22:27–32.
133. Abd El-Karim IA, El-Sanousi AA, Aly NM, Reda IM. Detection of coronavirus antigen in fecal samples obtained from newborn buffalo calves. Vet Med J Giza. (1990) 38:77–85.
134. Byomi A, Herbst W, Baljer G. Some viral agents associated with neonatal calf diarrhea. Assuit Vet Med J. (1996) 35:1–9.
135. Abd El-Rahim IHA. Rota and/or coronavirus infections in newborn buffalo calves in Upper Egypt. In: Proceedings of the 4th Scientific Congress, Egyptian Society for Cattle Diseases. Asyut (1997). p. 210–20.
136. Decaro N, Martella V, Elia G, Campolo M, Mari V, Desario C, et al. Biological and genetic analysis of a bovine-like coronavirus isolated from water buffalo (Bubalus bubalis) calves. Virology. (2008) 370:213–22. doi: 10.1016/j.virol.2007.08.031
137. Decaro N, Cirone F, Mari V, Nava D, Tinelli A, Elia G, et al. Characterisation of bubaline coronavirus strains associated with gastroenteritis in water buffalo (Bubalus bubalis) calves. Vet Microbiol. (2010) 145:245–51. doi: 10.1016/j.vetmic.2010.04.010
138. Lau SK, Tsang AK, Shakeel Ahmed S, Mahbub Alam M, Ahmed Z, Wong PC, et al. First genome sequences of buffalo coronavirus from water buffaloes in Bangladesh. New Microbes New Infect. (2016) 11:54–6. doi: 10.1016/j.nmni.2016.02.011
139. Kim JH, Jang JH, Yoon SW, Noh JY, Ahn MJ, Kim Y, et al. Detection of bovine coronavirus in nasal swab of non-captive wild water deer, Korea. Transbound Emerg Dis. (2018) 65:627–31. doi: 10.1111/tbed.12847
140. Harms NJ, Jung TS, Andrew CL, Surujballi OP, Vanderkop M, Savic M, et al. Health status of reintroduced wood bison (bison bison athabascae): assessing the conservation value of an isolated population in northwestern Canada. J Wildl Dis. (2019) 55:44–53. doi: 10.7589/2017-09-235
141. He Q, Guo Z, Zhang B, Yue H, Tang C. First detection of bovine coronavirus in Yak (Bos grunniens) and a bovine coronavirus genome with a recombinant HE gene. J Gen Virol. (2019) 100:793–803. doi: 10.1099/jgv.0.001254
142. Erles K, Toomey C, Brooks HW, Brownlie J. Detection of a group 2 coronavirus in dogs with canine infectious respiratory disease. Virology. (2003) 310:216–23. doi: 10.1016/S0042-6822(03)00160-0
143. Oma VS, Traven M, Alenius S, Myrmel M, Stokstad M. Bovine coronavirus in naturally and experimentally exposed calves; viral shedding and the potential for transmission. Virol J. (2016) 13:100. doi: 10.1186/s12985-016-0555-x
144. Hasoksuz M, Lathrop SL, Gadfield KL, Saif LJ. Isolation of bovine respiratory coronaviruses from feedlot cattle and comparison of their biological and antigenic properties with bovine enteric coronaviruses. Am J Vet Res. (1999) 60:1227–33.
145. Uttenthal A, Jensen NP, Blom JY. Viral aetiology of enzootic pneumonia in Danish dairy herds: diagnostic tools and epidemiology. Vet Rec. (1996) 139:114–7. doi: 10.1136/vr.139.5.114
146. Gomez DE, Arroyo LG, Poljak Z, Viel L, Weese JS. Detection of bovine coronavirus in healthy and diarrheic dairy calves. J Vet Intern Med. (2017) 31:1884–91. doi: 10.1111/jvim.14811
147. Hasoksuz M, Vlasova A, Saif LJ. Detection of group 2a coronaviruses with emphasis on bovine and wild ruminant strains. Virus isolation and detection of antibody, antigen, nucleic acid. Methods Mol Biol. (2008) 454:43–59. doi: 10.1007/978-1-59745-181-9_5
148. Decaro N, Elia G, Campolo M, Desario C, Mari V, Radogna A, et al. Detection of bovine coronavirus using a TaqMan-based real-time RT-PCR assay. J Virol Methods. (2008) 151:167–71. doi: 10.1016/j.jviromet.2008.05.016
149. Acharya KR, Dhand NK, Whittington RJ, Plain KM. PCR inhibition of a quantitative PCR for Detection of Mycobacterium avium subspecies paratuberculosis DNA in feces: diagnostic implications and potential solutions. Front Microbiol. (2017) 8:115. doi: 10.3389/fmicb.2017.00115
150. Heckert RA, Saif LJ, Mengel JP, Myers GW. Mucosal and systemic antibody responses to bovine coronavirus structural proteins in experimentally challenge-exposed calves fed low or high amounts of colostral antibodies. Am J Vet Res. (1991) 52:700–8.
151. Lin X, O'reilly KL, Burrell ML, Storz J. Infectivity-neutralizing and hemagglutinin-inhibiting antibody responses to respiratory coronavirus infections of cattle in pathogenesis of shipping fever pneumonia. Clin Diagn Lab Immunol. (2001) 8:357–62. doi: 10.1128/CDLI.8.2.357-362.2001
152. Lin XQ, O'reilly KL, Storz J. Antibody responses of cattle with respiratory coronavirus infections during pathogenesis of shipping fever pneumonia are lower with antigens of enteric strains than with those of a respiratory strain. Clin Diagn Lab Immunol. (2002) 9:1010–3. doi: 10.1128/CDLI.9.5.1010-1013.2002
153. Bok M, Alassia M, Frank F, Vega CG, Wigdorovitz A, Parreno V. Passive immunity to control bovine coronavirus diarrhea in a dairy herd in Argentina. Rev Argent Microbiol. (2018) 50:23–30. doi: 10.1016/j.ram.2017.03.007
154. Castells M, Giannitti F, Caffarena RD, Casaux ML, Schild C, Castells D, et al. Bovine coronavirus in Uruguay: genetic diversity, risk factors and transboundary introductions from neighboring countries. Arch Virol. (2019) 164:2715–24. doi: 10.1007/s00705-019-04384-w
155. Fulton RW, D'offay JM, Landis C, Miles DG, Smith RA, Saliki JT, et al. Detection and characterization of viruses as field and vaccine strains in feedlot cattle with bovine respiratory disease. Vaccine. (2016) 34:3478–92. doi: 10.1016/j.vaccine.2016.04.020
156. Welter MW. Adaptation and serial passage of bovine coronavirus in an established diploid swine testicular cell line and subsequent development of a modified live vaccine. Adv Exp Med Biol. (1998) 440:707–11. doi: 10.1007/978-1-4615-5331-1_91
157. Takamura K, Matsumoto Y, Shimizu Y. Field study of bovine coronavirus vaccine enriched with hemagglutinating antigen for winter dysentery in dairy cows. Can J Vet Res. (2002) 66:278–81.
158. Decaro N, Campolo M, Mari V, Desario C, Colaianni ML, Di Trani L, et al. A candidate modified-live bovine coronavirus vaccine: safety and immunogenicity evaluation. New Microbiol. (2009) 32:109–13.
159. Jee J, Hoet AE, Azevedo MP, Vlasova AN, Loerch SC, Pickworth CL, et al. Effects of dietary vitamin A content on antibody responses of feedlot calves inoculated intramuscularly with an inactivated bovine coronavirus vaccine. Am J Vet Res. (2013) 74:1353–62. doi: 10.2460/ajvr.74.10.1353
Keywords: cattle, bovine coronavirus, respiratory, bovine respiratory disease complex, diarrhea, enteric, wild ruminants
Citation: Vlasova AN and Saif LJ (2021) Bovine Coronavirus and the Associated Diseases. Front. Vet. Sci. 8:643220. doi: 10.3389/fvets.2021.643220
Received: 17 December 2020; Accepted: 08 March 2021;
Published: 31 March 2021.
Edited by:
Suresh Varma Kuchipudi, Pennsylvania State University (PSU), United StatesReviewed by:
Diego G. Diel, Cornell University, United StatesLakshmi Kavitha Kommalapati, Sri Venkateswara Veterinary University, India
Copyright © 2021 Vlasova and Saif. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anastasia N. Vlasova, dmxhc292YS4xQG9zdS5lZHU=; Linda J. Saif, c2FpZi4yQG9zdS5lZHU=
 Anastasia N. Vlasova
Anastasia N. Vlasova Linda J. Saif
Linda J. Saif