- 1Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX, United States
- 2Department of Entomology, Texas A&M AgriLife Research, College Station, TX, United States
- 3Department of Ecology and Conservation Biology, Texas A&M University, College Station, TX, United States
- 4Department of Entomology, Cornell University, Ithaca, NY, United States
- 5Department of Ecology and Evolutionary Biology, University of Tennessee, Knoxville, Knoxville, TN, United States
- 659th Medical Wing, Joint Base San Antonio, Lackland, San Antonio, TX, United States
- 7Texas Parks and Wildlife Department, Government Canyon State Natural Area, San Antonio, TX, United States
Tick-host bloodmeal associations are important factors when characterizing risks of associated pathogen transmission and applying appropriate management strategies. Despite their biological importance, comparatively little is known about soft tick (Argasidae) host associations in the United States compared to hard ticks (Ixodidae). In this study, we evaluated a PCR and direct Sanger sequencing method for identifying the bloodmeal hosts of soft ticks. We collected 381 cave-associated Ornithodoros turicata near San Antonio, Texas, USA, and also utilized eight colony-reared specimens fed artificially on known host blood sources over 1.5 years ago. We correctly identified the vertebrate host bloodmeals of two colony-reared ticks (chicken and pig) up to 1,105 days post-feeding, and identified bloodmeal hosts from 19 out of 168 field-collected soft ticks, including raccoon (78.9%), black vulture (10.5%), Texas black rattlesnake (5.3%), and human (5.3%). Our results confirm the retention of vertebrate blood DNA in soft ticks and advance the knowledge of argasid host associations in cave-dwelling O. turicata.
Introduction
The identification of arthropod host-feeding patterns through bloodmeal analysis can provide key information for vertebrate host contact and pathogen transmission networks (1–4). Bloodmeal analysis methods based on the detection of vertebrate DNA left in the residual bloodmeal are widely used across diverse arthropod taxa. For example, reservoir hosts of Leishmania were identified by studying previous bloodmeals of sand flies (5), and host-feeding patterns in mosquitoes allowed for an enhanced understanding of the reservoirs of West Nile virus (6, 7). Molecular analysis of bloodmeals has also been used to identify a broad host community for Culicoides, vectors of avian Haemosporida infections (8), and of triatomines, vectors of Trypanosoma cruzi, agent of Chagas disease (9).
Bloodmeal analysis, applied to ticks, has repeatedly been associated with limited success (10, 11), likely owing to DNA degradation during the molt and many months since prior bloodmeal acquisition. Given their importance as vectors of human pathogens, several studies have conducted bloodmeal analysis of hard ticks (Ixodidae), identifying vertebrate hosts in 20–93% of analyzed ticks (12, 13). Given the challenges of PCR-Sanger sequencing-based bloodmeal analysis of hard ticks, alternative strategies have been evaluated to identify bloodmeal hosts, including analysis of the variation in stable isotopes in fed ticks (10, 14, 15), reverse line blot (16, 17), and proteomics (18). In comparison, relatively few studies have attempted to identify the bloodmeal hosts of argasid ticks (soft ticks).
Ornithodoros turicata, found in the southwestern United States and Florida (19), is a vector of human and animal pathogens. O. turicata is a known vector of tick-borne relapsing fever caused by Borrelia turicatae (20), and is also a putative vector for transmission of African swine fever virus, an emerging disease in Africa, Europe, and most recently Asia (21, 22). This DNA virus is transmitted by soft ticks of the Ornithodoros genus and is highly pathogenic to domestic swine (23). While African swine fever has yet to be detected in the U.S., recent studies have identified O. turicata as a most likely vector should the virus reach the US (24, 25).
Ornithodoros turicata is found in caves or burrows occupied or visited by diverse vertebrate hosts (19). Larvae, nymphs and adults attach, blood-feed, and drop from a host quickly (typically 15–20 min); thus, they are seldom collected from hosts during blood feeding events, complicating knowledge of tick-host associations (26). Further, they can survive for years between bloodmeals as nymphs and adults (27).
Identification of the host community that supports O. turicata populations could be useful in providing an ecological basis for vector control and disease management. We previously conducted a bloodmeal analysis study using quantitative PCR for the identification of vertebrates in experimentally fed O. turicata (28). The results demonstrated vertebrate DNA could be detected 330 days post-feeding and through multiple molts, suggesting longer retention of bloodmeal DNA in soft ticks compared to hard ticks. The current study builds on these prior results by conducting a PCR-Sanger sequencing bloodmeal analysis protocol on O. turicata fed experimentally on known hosts as well as field-collected specimens from cave environments in Texas near the location of recent outbreaks of TBRF in humans (29, 30).
Materials and Methods
Tick Collections
We obtained eight adult O. turicata specimens (five male, three female) from an established colony at Texas A&M University, previously described (31). The feeding history of colony O. turicata was known only to one author (PDT) while the rest of the authors remained blinded to the bloodmeal species identification. We collected soft ticks from three caves in Government Canyon State Natural Area, San Antonio, TX, USA, (Lat: 29.549316, Lon:−98.764715) in March 2019 (Figure 1). Seven dry-ice baited sticky traps were set out inside or near the openings of three caves over 2 days for 18 and 21.5 h, respectively (Figure 2). These caves are closed to park visitors and were selected based on a previous study which demonstrated robust O. turicata populations in these caves (31). Traps consisted of 1.9L coolers filled with dry ice (cooler spout open) bolted through the bottom to a 0.41 m2 untreated 3-ply pine plywood with edges cut at 45° angles to improve tick access to the surface of the board. Double sided carpet tape (Roberts, Boca Raton, FL) was applied to the surface of the board, and insect glue boards (Bell Laboratories, Madison, WI) were cut into strips and applied to the carpet tape and under the corners of the plywood board. The ticks were removed from the sticky tape on-site and placed into ethanol-filled 1.5 ml tubes; each vial contained 3–10 ticks depending on the number of ticks caught at each trap each day. Samples were transported to the laboratory and stored at 4°C until DNA extraction.
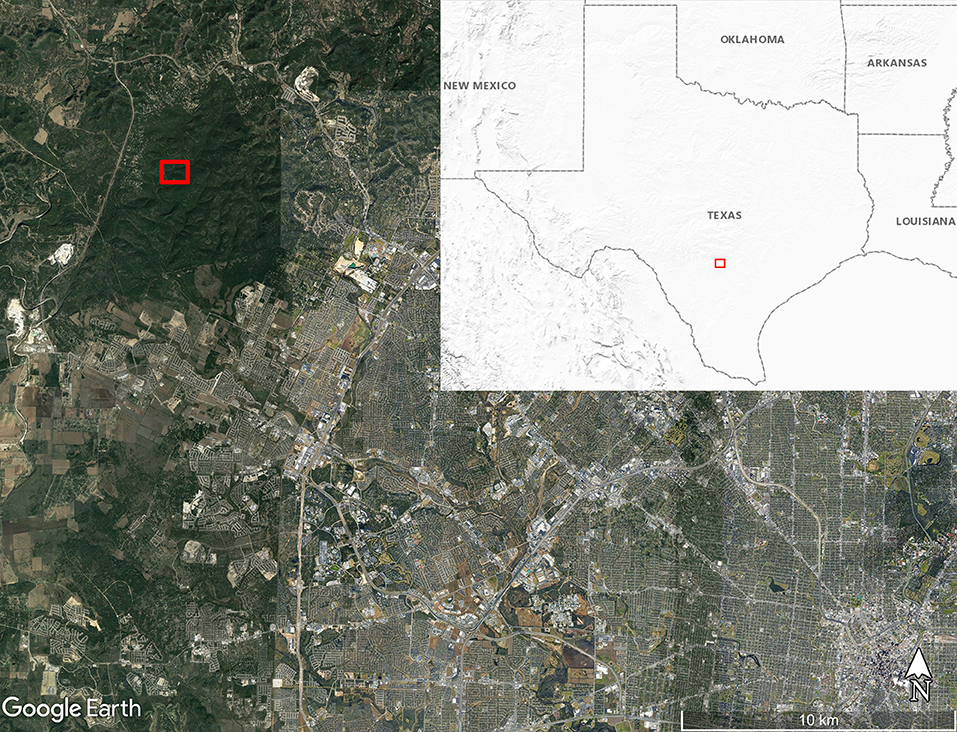
Figure 1. Map of soft tick collection location along Government Canyon Creek (red box) inside Government Canyon State Natural Area on the northwest side of San Antonio, Texas, USA. Map made using Google Earth Pro version 7.3.3.7786. Inset map shows the United States Geological Survey Shaded Relief map of the region around Texas with the map boundary near San Antonio outlined with the red box.

Figure 2. Pictures of dry ice-baited soft tick traps deployed inside and outside of caves in Government Canyon State Natural Area, Texas, USA. (A, left) Soft tick traps were placed with a cooler full of dry ice in the mouths of caves, and the cooler spout was left open for dry ice sublimation. (B, bottom middle) O. turicata soft ticks can be seen attached to the perimeter of the sticky tape. (C, right) Additional portions of sticky tape were added to parts of the perimeter of the trap to enhance collections.
Tick Processing and DNA Extraction
O. turicata specimens were measured, identified to species and life stage by morphological features, and sexed if adults (32). A subset of 124 ticks were photographed to serve as a reference for confirming life stage and sex (Supplemental Data Sheet 1). Photos were taken early in the identification process and throughout tick cataloging as needed when life stage or sex was difficult to determine. We attempted to record the size of the bloodmeal based on the shape of the abdomen and presence of blood; however, the storage in ethanol produced a dark red color in all specimens and visual bloodmeal scoring was not reliable.
The first 20 ticks collected at each site, followed by equal numbers of the largest ticks from each cave (based on length), were processed further using the following methods until a threshold of 40% of the total number of collected ticks had been processed. To minimize exogenous DNA on the exterior surface of ticks, ticks were washed in ethanol for 5 s, then a 10% bleach solution for 15 s, and finally two consecutive 15 s DNA-free water rinses immediately after (10, 33, 34). On a sterile microscope slide over ice, the legs were then removed, placed in ethanol, and stored at −40°C to decrease the amount of tick DNA processed and to preserve samples of each tick for future use. The tick bodies were placed in clean tubes, flash-frozen in liquid nitrogen, and crushed with a sterile pestle (Wards Sciences, Rochester, NY), which was discarded after each use. The crushed tissue was then lysed, and DNA was extracted using the MagMAX CORE Nucleic Acid Purification Kit (Applied Biosystems ThermoFisher Scientific, Waltham, Massachusetts) according to the manufacturer's protocol. Negative and positive controls, including blood from sheep, tiger, and crane (vertebrate species not expected to occur around the cave environment), were included during DNA extraction procedures. The eluted tick DNA from each tick was stored in two tubes at −40°C until PCR amplification. For a small subset of samples from two early extractions (n = 6), we quantified DNA using a spectrophotometer (Epoch, BioTek Instruments, Inc.) to confirm the presence of DNA in the extracted samples (average = 32.39 ng/μL, range = 9.07–68.07 ng/μL).
PCR Amplification and Sanger Sequencing
DNA extracts from ticks were subjected to multiple vertebrate barcoding primers targeting different genes in an iterative process to identify the most successful primers (Supplementary Table 1). The Failsafe PCR Enzyme Mix with PreMix E (Epicenter Biotechnologies, Madison, WI) and primer pairs from Integrated DNA Technologies (IDT) were used for PCR amplification. Primers were used at varying concentrations of 0.33 or 0.4 μM (see Supplementary Table 1) in 25 μL reactions, including 2 μL of tick DNA. PCR products were visualized on e-gels (ThermoFisher Scientific, Waltham, Massachusetts) and all products were purified by Exo-SAP-IT (Applied Biosystems, Foster City, CA) and sequenced in forward directions by Eton Biosciences (San Diego, California). Sequences were trimmed to at least 215 base pairs long, and chromatographs were manually scrutinized for quality. Sequences were blasted to the NCBI database using Geneious software (Newark, New Jersey) to identify the closest match of the unknown sequence to a known organism. Sequences with >90% similarity were interpreted as a match, in which case the bloodmeal host was identified. All samples that produced sequences with double peaks were re-run with the “mammal c” primer pair. Because of the chance for contamination of samples with human DNA, any sample that produced a sequence that matched to human was tested a second time with an independent PCR, and in these cases, two matches to the same species were needed to confirm a result.
Results
Colony Ticks
Bloodmeal analysis was conducted on eight O. turicata from a colonized population with known prior bloodmeals, with personnel conducting the molecular work blinded to the vertebrate species. We successfully detected chicken (Gallus gallus) in an adult tick 1,105 days (last fed 27 January 2016) post bloodmeal and pig (Sus scrofa) in an adult tick 622 days (last fed 24 May 2017) days post bloodmeal. We were unable to obtain a PCR amplicon or sequence from six ticks that had fed 622–1,109 days post-bloodmeal (last fed on dates through January 23, 2016 to May 24, 2017).
Field-Collected Ticks
A total of 381 soft ticks were collected in March 2019, and all were identified by morphology as O. turicata. Mad Crow Cave yielded the highest number of soft ticks trapped (n = 184), followed by Bone Pile Cave (n = 109) and Little Crevice Cave (n = 88). We identified 32 females, 55 males, 285 nymphs, four adults that were damaged and unable to be sexed, and five other specimens that were damaged and we were unable to determine either sex or life stage (Table 1). The average length of males was 3.53 mm (n = 54, SD: 0.98), 5.28 mm for females (n = 32, SD: 1.70), and 2.23 mm (n = 284; SD: 0.82) for nymphs. One male and one nymph were damaged and unable to be measured.
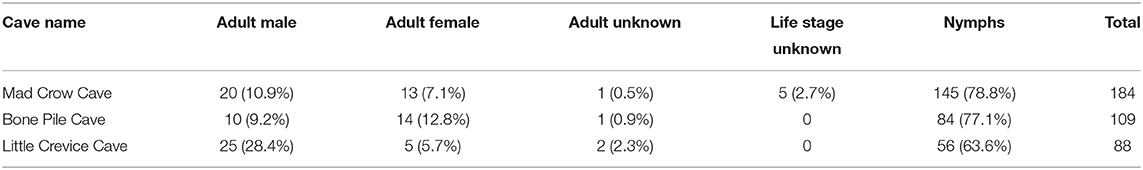
Table 1. Demographic data of field-collected Ornithodoros turicata collected from Government Canyon State Natural Area, San Antonio, TX, 2019.
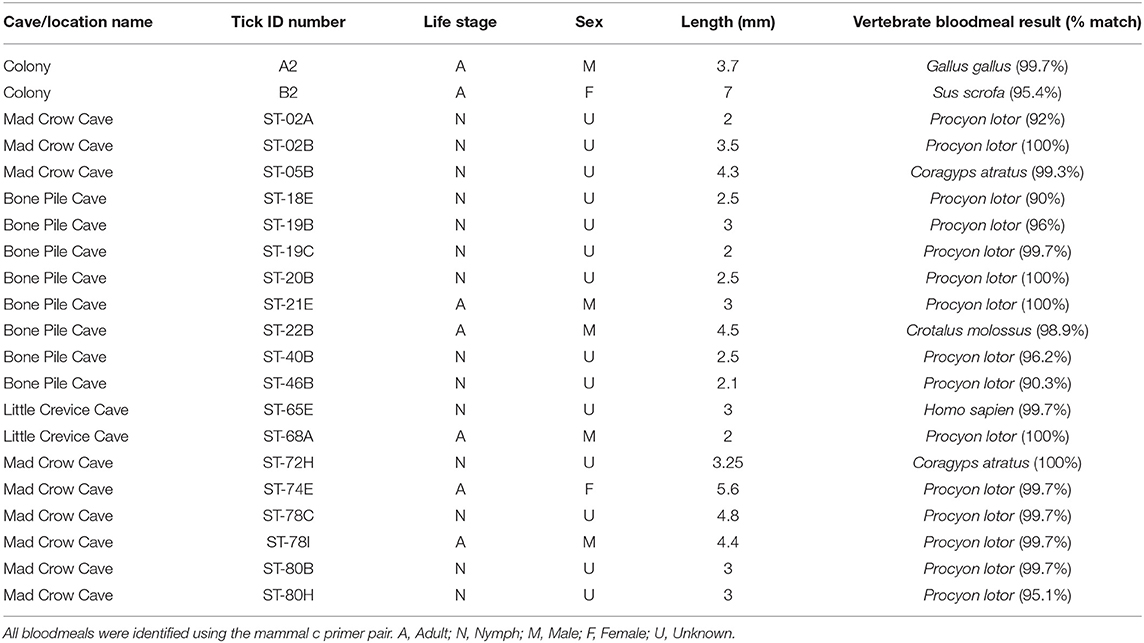
A subset of 168 field-collected ticks were processed for bloodmeal analysis. Most PCR primers amplified exclusively Ornithodoros sp. DNA, including the primer pairs mammal c, 0066/0067, 0035/0049, 0033/0049, and Herp/BM1. The primer pair which had the best success at minimizing tick DNA amplification and maximizing vertebrate DNA amplification was “mammal c” targeting a 395 base pair region of cytochrome b (4, 35). The bloodmeals from 19 ticks (11.3%) were identified to species using the “mammal c” primer pair (Table 2). Of this subset, 15 ticks (78.9%) contained raccoon (Procyon lotor) DNA, two ticks (10.5%) contained black vulture (Coragyps atratus) DNA, one tick (5.3%) contained black-tailed rattlesnake (Crotalus molossus) DNA, and one tick (5.3%) contained human (Homo sapiens) DNA. Of the 168 field-collected samples subjected to bloodmeal analysis, 69 were adults (39 male, mean size = 3.62 mm; 26 female, mean size = 5.67 mm), 97 were nymphs (mean size = 2.64 mm, n = 96 as one tick length was unreliable), and two were unable to be determined. The 19 ticks with identifiable bloodmeals included four males (mean size = 3.48 mm), one female (5.6 mm), and 14 nymphs (mean size = 2.96 mm).
Discussion
This study builds on prior results, which demonstrated that vertebrate DNA detected by quantitative PCR in prior bloodmeals of O. turicata persists for long periods post-feeding and through molts (27, 31). In the current study, we adopted PCR and direct Sanger sequencing and confirmed that, for experimentally fed ticks in the laboratory, we were able to detect bloodmeals that were up to 1,105 days old. However, a challenge encountered by the molecular approach used in this study was that Ornithodoros sp. DNA was amplified consistently using five different primer pairs (mammal c, 0066/0067, 0035/0049, 0033/0049, and Herp/BM1). Amplification of vector DNA has not been an issue during mosquito bloodmeal analysis studies, which served as a main source of bloodmeal primers used in this study (3, 34, 36). Many of the chromatographs from the sequences suggested double-nucleotide peaks in the amplicons that matched to Ornithodoros sp., and repeated PCRs with the same or different primers were unable to resolve the amplified sequences. We suspect this non-target amplification of tick DNA is attributed to the barcoding primer design that minimizes non-target amplification of Insecta but perhaps not Ixodida. The size of the argasid tick genome (1.2 Gbp) is also 2.2x larger than the genome of Culex pipiens (0.54 Gbp), a common mosquito in which bloodmeal primers are developed, which further increases the opportunity for non-target amplification (35, 37, 38).
Despite the challenges posed by the non-target amplification of soft tick DNA, we were still able to produce repeatable bloodmeal host identification results in 19 samples. We suspect the success in the vertebrate ID in these samples was possible when sufficient blood was present. The most common vertebrate ID for these cave-dwelling soft ticks was raccoon, followed by black vulture, black-tailed rattlesnake, and human. The presence of these species in these caves was confirmed during a prior camera trapping study (31). Although these caves are off-limits to the public, camera traps documented unauthorized human access to these caves, supporting the potential for a human bloodmeal in this study and also identifying a risk associated with exposure to B. turicatae, which has been documented for central Texas (29, 39, 40). A study in Iowa documented large infestations of the soft tick, Carios kelleyi, in human dwellings using capillary electrophoresis to conduct bloodmeal analysis and identified one nymphal soft tick that had fed on a female human (1). Additionally, Argas cooleyi invaded a hospital in Arizona from their resident bird nests outside and fed on humans, with 17% of the analyzed bloodmeals belonging to humans (41). These studies, along with our study of a wild O. turicata population, indicate the importance of studying soft ticks and their potential associations with humans as hosts.
This cross-sectional study also informs the population structure of O. turicata in the caves of the region. Of the 381 collected soft ticks, 74.8% were found to be in the nymphal stage and no larvae were collected. These skewed demographics of the soft tick community composition illustrate a large proportion of immature ticks in early March compared to adults. Some soft tick species molt to future nymphal instars based on ambient temperature, and in some species, larvae do not need to feed on a bloodmeal, and receive all the nutrients they need to molt into first instar nymphs from the egg (26). However, in colony, O. turicata have been recorded to feed between larval and nymphal stages, and nymphs reared in similar environmental conditions can molt to adults as either 4, 5, or 6th nymphal instars (42). Due to the limited literature available on soft tick ecology, this study may indicate that in spring months, nymphs are more abundant than adults. Alternately, larvae may have been present in the caves yet less likely to be trapped using the dry ice/sticky trap stations that we deployed. Population demographic data is key in understanding the natural cycles of soft tick populations.
Limited prior studies have documented vertebrate host feeding patterns of soft ticks collected in the field. A study in Portugal performed bloodmeal analysis on Ornithodoros erraticus collected in two pig pens and were able to identify vertebrate hosts including pigs, humans, bovines, sheep, rodents, and birds through bloodmeal analysis in 23% of the analyzed ticks (4). We used many of the same primer pairs from this previous study, and the lower success of the bloodmeal identification was likely also due to non-target amplification of tick DNA.
The challenges of non-target amplification of tick DNA could be resolved by multiple modifications in the future. All specimens collected in this study were host-seeking as they approached the CO2-baited traps. Accordingly, very few likely had fresh bloodmeals, which made it difficult to distinguish bloodmeal contents in the abdomen, especially when ethanol added a red coloration to all specimens. Future studies processing field-collected soft ticks should consider the use of morphological features such as the size and depth of inter-mammillary grooves to judge the state of repletion. A future sampling approach could use an aspirator (43) which would increase the chances of obtaining specimens with fresh bloodmeals. One modification would be to specifically design primers to avoid tick genomes and amplify exclusively vertebrate DNA (35, 44). Another modification could be to insert a clone of the amplicon into a bacterial vector, and then select several colonies per sample to sequence, in hopes of detecting the vertebrate host sequence even if tick DNA was preferentially amplified. This technique is used routinely in bloodmeal analysis studies (45), although this method is labor intensive and limited in resolution. A third option would be to perform amplicon deep sequencing, which would provide thousands of sequences of each amplicon and is an approach recently adopted for arthropod bloodmeal analysis studies (46–48). This method of metabarcoding and deep sequencing would be advantageous given soft tick biology, including multiple bloodmeals obtained during immature development and multiple gonotropic cycles of adults. The amplicon deep sequencing approach would allow the ID of not just the most recent bloodmeal but also the potential to detect prior vertebrate bloodmeals (49, 50).
Given the continued emergence of human and animal diseases vectored by soft ticks, further studies of the ecology of argasid ticks- including their vertebrate host associations- are critical for informing tick-host-pathogen transmission networks, vector management efforts, and disease risk assessment.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://hdl.handle.net/1969.1/191880, OAKTrust, Texas A&M University Libraries.
Author Contributions
PT, SH, and GH conceived study, supervised field, and lab work. RB, VM, ED, TE, TLC, TMC, and GH conducted field collections. RB, MO, VM, ED, CB, DE, BH, and BB conducted lab work. PT provided colony ticks. RB, VM, and ED wrote initial manuscript. All authors contributed to revisions.
Funding
Funding for this study was provided in part by the Montague Center for Teaching Excellence Scholar Program, Texas A&M University Department of Veterinary Integrative Biosciences Teaching Enhancement Funds, Texas A&M University Department of Entomology, and a Seed Fund from the Coalition for Epi Response, Engagement, and Science (CERES).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We appreciate permission from Texas Parks and Wildlife Division and Government Canyon State Natural Area for providing access to the campgrounds and caves to facilitate this study. We would also like to acknowledge the students of VIBS/ENTO 426/626 that participated in field work.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.639400/full#supplementary-material
References
1. Gill JS, Rowley WA, Bush PJ, Viner JP, Ginchrist MJR. Detection of human blood in the bat tick Carios (Ornithodoros) kelleyi (Acari: Argasidae) in Iowa. J Med Entomol. (2004) 41:1179. doi: 10.1603/0022-2585-41.6.1179
2. Pichon B, Rogers M, Egan D, Gray J. Blood-meal analysis for the identification of reservoir hosts of tick-borne pathogens in Ireland. Vector Borne Zoonotic Dis. (2005) 5:172–80. doi: 10.1089/vbz.2005.5.172
3. Hamer GL, Kitron U, Goldberg TL, Brawn JD, Loss SR, Ruiz MO, et al. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am J Trop Med Hyg. (2009) 80:268–78. doi: 10.4269/ajtmh.2009.80.268
4. Palma M, Lopes de Carvalho I, Osorio H, Ze-Ze L, Cutler SJ, Nuncio MS. Portuguese hosts for Ornithodoros erraticus ticks. Vector Borne Zoonotic Dis. (2013) 13:775–7. doi: 10.1089/vbz.2012.1070
5. Haouas N, Pesson B, Boudabous R, Dedet JP, Babba H, Ravel C. Development of a molecular tool for the identification of Leishmania reservoir hosts by blood meal analysis in the insect vectors. Am J Trop Med Hyg. (2007) 77:1054–9. doi: 10.4269/ajtmh.2007.77.1054
6. Apperson CS, Hassan HK, Harrison BA, Savage HM, Aspen SE, Farajollahi A, et al. Host feeding patterns of established and potential mosquito vectors of West Nile virus in the eastern United States. Vector Borne Zoonotic Dis. (2004) 4:71–82. doi: 10.1089/153036604773083013
7. Hamer GL, Kitron U, Brawn JD, Loss SR, Ruiz MO, Goldberg TL, et al. Culex pipiens (Diptera: Culicidae): a bridge vector of West Nile virus to humans. J Med Entomol. (2008) 45:125–8. doi: 10.1093/jmedent/45.1.125
8. Santiago-Alarcon D, Havelka P, Schaefer HM, Segelbacher G. Bloodmeal analysis reveals avian Plasmodium infections and broad host preferences of Culicoides (Diptera: Ceratopogonidae) vectors. PLoS ONE. (2012) 7:e31098. doi: 10.1371/journal.pone.0031098
9. Meyers AC, Meinders M, Hamer SA. Widespread Trypanosoma cruzi infection in government working dogs along the Texas-Mexico border: discordant serology, parasite genotyping and associated vectors. PLoS Negl Trop Dis. (2017) 11:e0005819. doi: 10.1371/journal.pntd.0005819
10. Hamer SA, Weghorst AC, Auckland LD, Roark EB, Strey OF, Teel PD. Comparison of DNA and carbon and nitrogen stable isotope-based techniques for identification of prior vertebrate hosts of ticks. J Med Entomol. (2015) 52:1043–9. doi: 10.1093/jme/tjv063
11. Leger E, Liu X, Masseglia S, Noel V, Vourc'h G, Bonnet S, et al. Reliability of molecular host-identification methods for ticks: an experimental in vitro study with Ixodes ricinus. Parasit Vectors. (2015) 8:433. doi: 10.1186/s13071-015-1043-7
12. Moran Cadenas F, Rais O, Humair PF, Douet V, Moret J, Gern L. Identification of host bloodmeal source and Borrelia burgdorferi sensu lato in field-collected Ixodes ricinus ticks in Chaumont (Switzerland). J Med Entomol. (2007) 44:1109–17. doi: 10.1603/0022-2585(2007)441109:iohbsa2.0.co;2
13. Kent RJ. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour. (2009) 9:4–18. doi: 10.1111/j.1755-0998.2008.02469.x
14. LoGiudice K, Kurchena K, Christopher K, Scott N. Exploration of stable isotope analysis for tick host identification. Ticks Tick Borne Dis. (2018) 9:151–4. doi: 10.1016/j.ttbdis.2017.08.010
15. Heylen D, Schmidt O, Dautel H, Gern L, Kampen H, Newton J, et al. Host identification in unfed ticks from stable isotope compositions (delta(13) C and delta(15) N). Med Vet Entomol. (2019) 33:360–6. doi: 10.1111/mve.12372
16. Allan BF, Goessling LS, Storch GA, Thach RE. Blood meal analysis to identify reservoir hosts for Amblyomma americanum ticks. Emerg Infect Dis. (2010) 16:433–40. doi: 10.3201/eid1603.090911
17. Scott MC, Harmon JR, Tsao JI, Jones CJ, Hickling GJ. Reverse line blot probe design and polymerase chain reaction optimization for bloodmeal analysis of ticks from the eastern United States. J Med Entomol. (2012) 49:697–709. doi: 10.1603/me11162
18. Onder O, Shao W, Kemps BD, Lam H, Brisson D. Identifying sources of tick blood meals using unidentified tandem mass spectral libraries. Nat Commun. (2013) 4:1746. doi: 10.1038/ncomms2730
19. Donaldson TG, Perez de Leon AA, Li AY, Castro-Arellano I, Wozniak E, Boyle WK, et al. Assessment of the geographic distribution of Ornithodoros turicata (Argasidae): climate variation and host diversity. PLoS Negl Trop Dis. (2016) 10:e0004383. doi: 10.1371/journal.pntd.0004383
20. Dworkin MS, Schwan TG, Anderson DE. Tick-borne relapsing fever in North America. Med Clin North Am. (2002) 86:417–33. doi: 10.1016/s0025-7125(03)00095-6
21. Sanchez-Cordon PJ, Montoya M, Reis AL, Dixon LK. African swine fever: a re-emerging viral disease threatening the global pig industry. Vet J. (2018) 233:41–8. doi: 10.1016/j.tvjl.2017.12.025
22. Wang T, Sun Y, Qiu HJ. African swine fever: an unprecedented disaster and challenge to China. Infect Dis Poverty. (2018) 7:111. doi: 10.1186/s40249-018-0495-3
23. Hess WR, Endris RG, Haslett TM, Monahan MJ, McCoy JP. Potential arthropod vectors of African swine fever virus in North America and the Caribbean Basin. Vet Parasitol. (1987) 26:145–55. doi: 10.1016/0304-4017(87)90084-7
24. Golnar AJ, Martin E, Wormington JD, Kading RC, Teel PD, Hamer SA, et al. Reviewing the potential vectors and hosts of African swine fever virus transmission in the United States. Vector Borne Zoonotic Dis. (2019) 19:512–24. doi: 10.1089/vbz.2018.2387
25. Wormington JD, Golnar A, Poh KC, Kading RC, Martin E, Hamer SA, et al. Risk of African swine fever virus sylvatic establishment and spillover to domestic swine in the United States. Vector Borne Zoonotic Dis. (2019) 19:506–11. doi: 10.1089/vbz.2018.2386
26. Apanaskevich DA, Oliver JH. Life cycles and natural history of ticks. In: DE Soneshine, RM Roe, editors. Biology of Ticks. New York, NY: Oxford University Press. (2014). p. 59–73.
27. Francis E. Longevity of the tick Ornithodoros turicata and of Spirochaeta recurrentis within this tick. Public Health Rep. (1938) 53:2220–41. doi: 10.2307/4582740
28. Kim HJ, Hamer GL, Hamer SA, Lopez JE, Teel PD. Identification of host bloodmeal source in Ornithodoros turicata Dugès (Ixodida: Argasidae) using DNA-based and stable isotope-based techniques. Front Vet Sci. (2021) 8:620441. doi: 10.3389/fvets.2021.620441
29. Bissett JD, Ledet S, Krishnavajhala A, Armstrong BA, Klioueva A, Sexton C, et al. Detection of tickborne relapsing fever spirochete, Austin, Texas, USA. Emerg Infect Dis. (2018) 24:2003–9. doi: 10.3201/eid2411.172033
30. Campbell SB, Klioueva A, Taylor J, Nelson C, Tomasi S, Replogle A, et al. Evaluating the risk of tick-borne relapsing fever among occupational cavers-Austin, TX, 2017. Zoonoses Public Health. (2019) 66:579–86. doi: 10.1111/zph.12588
31. Kim HJ. Habitat-host-vector interactions of Ornithodoros turicata Dugès (Ixodida: Argasidae) in Texas, USA. Doctoral dissertation. College Station (TX): Texas A&M University (2017).
32. Pratt H, Stojanovich C. Acarina: illustrated key to some common adult female mites and adult ticks. In: H Pratt, editor. Mites of Public Health Importance and Their Control. Washington, DC: US Government Printing Office (1963). p. 26–37.
33. Graham CB, Black WC, Boegler KA, Montenieri JA, Holmes JL, Gage KL, et al. Combining real-time polymerase chain reaction using SYBR Green I detection and sequencing to identify vertebrate bloodmeals in fleas. J Med Entomol. (2012) 49:1442–52. doi: 10.1603/me12071
34. Olson MF, Ndeffo-Mbah ML, Juarez JG, Garcia-Luna S, Martin E, Borucki MK, et al. High rate of non-human feeding by Aedes aegypti reduces Zika virus transmission in South Texas. Viruses. (2020) 12:40453. doi: 10.3390/v12040453
35. Molaei G, Andreadis TG, Armstrong PM, Anderson JF, Vossbrinck CR. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, Northeastern United States. Emerg Infect Dis. (2006) 12:468–74. doi: 10.3201/eid1203.051004
36. Medeiros MC, Ricklefs RE, Brawn JD, Hamer GL. Plasmodium prevalence across avian host species is positively associated with exposure to mosquito vectors. Parasitology. (2015) 142:1612–20. doi: 10.1017/S0031182015001183
37. Jost E, Mameli M. DNA content in nine species of Nematocera with special reference to the sibling species of the Anopheles maculipennis group and the Culex pipiens group. Chromosoma. (1972) 37:201–8. doi: 10.1007/BF00284939
38. Geraci NS, Spencer Johnston J, Paul Robinson J, Wikel SK, Hill CA. Variation in genome size of argasid and ixodid ticks. Insect Biochem Mol Biol. (2007) 37:399–408. doi: 10.1016/j.ibmb.2006.12.007
39. Christensen AM, Pietralczyk E, Lopez JE, Brooks C, Schriefer ME, Wozniak E, et al. Diagnosis and management of Borrelia turicatae infection in febrile soldier, Texas, USA. Emerg Infect Dis. (2017) 23:883–4. doi: 10.3201/eid2305.162069
40. Gettings JR, Lopez JE, Krishnavahjala A, Armstrong BA, Thompson AT, Yabsley MJ. Antibodies to Borrelia turicatae in experimentally infected dogs cross-react with Borrelia burgdorferi serologic assays. J Clin Microbiol. (2019) 57:19. doi: 10.1128/JCM.00628-19
41. Beatty NL, Klotz SA, Elliott SP. Hematophagous ectoparasites of cliff swallows invade a hospital and feed on humans. Clin Infect Dis. (2017) 65:2119–21. doi: 10.1093/cid/cix697
42. Beck AF, Holscher KH, Butler JF. Life cycle of Ornithodoros turicata americanus (Acari: Argasidae) in the laboratory. J Med Entomol. (1986) 23:313–9. doi: 10.1093/jmedent/23.3.313
43. Diatta G, Duplantier J-M, Granjon L, Bâ K, Chauvancy G, Ndiaye M, et al. Borrelia infection in small mammals in West Africa and its relationship with tick occurrence inside burrows. Acta Trop. (2015) 152:131–40. doi: 10.1016/j.actatropica.2015.08.016
44. Cupp EW, Zhang D, Yue X, Cupp MS, Guyer C, Sprenger TR, et al. Identification of reptilian and amphibian blood meals from mosquitoes in an eastern equine encephalomyelitis virus focus in Central Alabama. Am J Trop Med Hyg. (2004) 7:272–6. doi: 10.1056/NEJMp1914328
45. Stevens L, Dorn PL, Hobson J, de la Rua NM, Lucero DE, Klotz JH, et al. Vector blood meals and Chagas disease transmission potential, United States. Emerg Infect Dis. (2012) 18:646–9. doi: 10.3201/eid1804.111396
46. Logue K, Keven JB, Cannon MV, Reimer L, Siba P, Walker ED, et al. Unbiased characterization of Anopheles mosquito blood meals by targeted high-throughput sequencing. PLoS Negl Trop Dis. (2016) 10:e0004512. doi: 10.1371/journal.pntd.0004512
47. Dumonteil E, Ramirez-Sierra MJ, Perez-Carrillo S, Teh-Poot C, Herrera C, Gourbiere S, et al. Detailed ecological associations of triatomines revealed by metabarcoding and next-generation sequencing: implications for triatomine behavior and Trypanosoma cruzi transmission cycles. Sci Rep. (2018) 8:4140. doi: 10.1038/s41598-018-22455-x
48. Dumonteil E, Pronovost H, Bierman EF, Sanford A, Majeau A, Moore R, et al. Interactions among Triatoma sanguisuga blood feeding sources, gut microbiota and Trypanosoma cruzi diversity in southern Louisiana. Mol Ecol. (2020) 29:3747–61. doi: 10.1111/mec.15582
49. Arias-Giraldo LM, Munoz M, Hernandez C, Herrera G, Velasquez-Ortiz N, Cantillo-Barraza O, et al. Identification of blood-feeding sources in Panstrongylus, Psammolestes, Rhodnius and Triatoma using amplicon-based next-generation sequencing. Parasit Vectors. (2020) 13:434. doi: 10.1186/s13071-020-04310-z
Keywords: soft ticks, Argasidae, Ornithodoros turicata, blood meal, host identification
Citation: Busselman RE, Olson MF, Martinez V, Davila E, Briggs C, Eldridge DS, Higgins B, Bass B, Cropper TL, Casey TM, Edwards T, Teel PD, Hamer SA and Hamer GL (2021) Host Bloodmeal Identification in Cave-Dwelling Ornithodoros turicata Dugès (Ixodida: Argasidae), Texas, USA. Front. Vet. Sci. 8:639400. doi: 10.3389/fvets.2021.639400
Received: 08 December 2020; Accepted: 20 January 2021;
Published: 15 February 2021.
Edited by:
Sebastián Muñoz-Leal, University of Concepcion, ChileReviewed by:
Markéta Nováková, Masaryk University, CzechiaDeon Bakkes, Agricultural Research Council of South Africa, South Africa
Copyright © 2021 Busselman, Olson, Martinez, Davila, Briggs, Eldridge, Higgins, Bass, Cropper, Casey, Edwards, Teel, Hamer and Hamer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriel L. Hamer, Z2hhbWVyQHRhbXUuZWR1
 Rachel E. Busselman
Rachel E. Busselman Mark F. Olson2
Mark F. Olson2 Pete D. Teel
Pete D. Teel Sarah A. Hamer
Sarah A. Hamer Gabriel L. Hamer
Gabriel L. Hamer