- 1College of Animal Sciences and Technology and Farm Animal Genetic Resources Exploration and Innovation Key Laboratory of Sichuan Province, Sichuan Agricultural University, Chengdu, China
- 2China Conservation and Research Center for the Giant Panda, Chengdu, China
- 3Department of Theriogenology, Riphah College of Veterinary Sciences, Lahore, Pakistan
Cryopreservation induces sperm cryoinjuries, including physiological and functional changes. However, the molecular mechanisms of sperm cryoinjury and cryoresistance are still unknown. Cryoresistance or the freeze tolerance of sperm varies across species, and boar sperm is more susceptible to cold stress. Contrary to boar sperm, giant panda sperm appears to be strongly freeze-tolerant and is capable of surviving repeated cycles of freeze-thawing. In this study, differentially expressed (DE) PIWI-interacting RNAs (piRNAs) of fresh and frozen-thawed sperm with different freeze tolerance capacity from giant panda and boar were evaluated. The results showed that 1,160 (22 downregulated and 1,138 upregulated) and 384 (110 upregulated and 274 downregulated) DE piRNAs were identified in giant panda and boar sperm, respectively. Gene ontology (GO) enrichment analysis revealed that the target DE messenger RNAs (mRNAs) of DE piRNAs were mainly enriched in biological regulation, cellular, and metabolic processes in giant panda and boar sperm. Moreover, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis indicated that the target DE mRNAs of DE piRNAs were only distributed in DNA replication and the cyclic adenosine monophosphate (cAMP) signaling pathway in giant panda, but the cAMP, cyclic guanosine monophosphate (cGMP), and mitogen-activated protein kinase (MAPK) signaling pathways in boar sperm were considered as part of the olfactory transduction pathway. In conclusion, we speculated that the difference in the piRNA profiles and the DE piRNAs involved in the cAMP signaling pathway in boar and giant panda may have contributed to the different freeze tolerance capacities between giant panda and boar sperm, which helps to elucidate the molecular mechanism behind sperm cryoinjury and cryoresistance.
Introduction
Sperm cryopreservation is widely used to manage and preserve male fertility in human and domestic animals (1). Then, artificial insemination (AI) is extensively employed with frozen-thawed sperm to enhance the rate of genetic improvement, especially in cattle (2). However, sperm cryoresistance or freeze tolerance and the post-thawed sperm quality vary across species. Less than 1% AI with frozen-thawed boar sperm was carried out due to the low conception rate and litter size (3, 4). It is well-known that various factors during cryopreservation, including rapid temperature transitions, osmotic stress, and ice crystal formation, affect the post-thaw quality of semen (5). Furthermore, the transcriptomics, epigenetics, and proteomics of sperm were also modified during cryopreservation (6–8). Despite the extensive progress that has been achieved in optimizing the cryopreservation process through the selection of friendly cryoprotectants and the design of better freezing and thawing procedures to ameliorate cryodamage, the underlying mechanisms of freeze tolerance or freezability involved in cryopreservation have not been completely elucidated yet.
Compared with other mammals' sperm, the higher level of phospholipids and the lower level of cholesterol in the plasma membrane of boar sperm contribute to the susceptibility to cold shock or cold stress (9). Cold shock causes the rearrangement of phospholipids, destruction of acrosomal integrity, and functional damage to ion transporters and channels in sperm (9, 10). However, compared with boar sperm, giant panda sperm shows a higher freeze tolerance capacity and can sustain repeated freeze–thaw cycles (11). Cryopreservation has no significant impact on sperm viability and motility, and the acrosome integrity and functional capacitation of giant panda sperm were also not affected after repeated freeze–thaw cycles (12). Our previous studies have shown that the transcriptomic profiles were significantly different between boar and giant panda sperm during cryopreservation (13). Furthermore, comparative analysis of the transcriptomic modifications between boar and giant panda sperm during cryopreservation indicated that differentially expressed (DE) messenger RNAs (mRNAs) were mainly distributed in inflammatory-related pathways, the cytokine–cytokine receptor interaction pathway, and membrane signal transduction-related pathways (14). These previous studies demonstrated that cryopreservation induces different transcriptomic modifications and may explain why sperm with different freeze tolerance or cryoresistance capacities are susceptible to cold stress.
PIWI-interacting RNAs (piRNAs) are small non-coding RNAs which are germline-specific and are required to protect genomic integrity from deleterious effects and to preserve RNA homeostasis during male gametogenesis; they are also associated with sperm morphology, motility, and fertility (15). The expression of piRNAs in human sperm was correlated with the sperm concentration and fertilization rate (16). Moreover, a panel of piRNAs discovered in seminal plasma can serve as fertility or infertility markers in males (17). Recently, 79 putative piRNAs were found to be differentially expressed between low and high motile bovine sperm after cryopreservation (18). Therefore, we speculated that piRNAs may be involved in post-thawed sperm cryoinjury or cryoresistance, motility, and fertility during cryopreservation. Thus, in this study, we first evaluated the differences in the piRNA profiles of fresh and frozen-thawed boar and giant panda sperm, which will help to uncover the underlying molecular mechanisms of sperm cryoresistance and freeze tolerance and improve post-thawed sperm quality and fertility.
Materials and Methods
Ethical Statement, Semen Collection, and Treatment
Fresh ejaculates from five sexually mature giant pandas with normal physiological parameters were obtained by electrical stimulation from the Bifengxia Base of China Conservation and Research Center for the Giant Panda (Yaán, Sichuan, China) according to a previous protocol (11). Briefly, giant pandas were anesthetized by an intramuscular injection of 10 mg/kg ketamine HCl and maintained with 0–5% isofluorane gas. Electroejaculation was conducted by using an electroejaculator (Boring, OR, USA); the period of electrical stimuli (2–8 V, repeated three times) was 2 s following an intermittent break of 2 s. When penile erection occurs during stimulation, semen was collected into a sterile glass container. Fresh ejaculates from 11 boars were collected with the glove-handed technique. All procedures were carried out while strictly following the Regulations of the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004) and were accredited by the Institutional Animal Care and Use Committee in the College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China (under permit no. 2019202012).
All ejaculates from giant panda and boar were pooled separately and two equal groups were generated (fresh sperm and cryopreserved sperm). Direct RNA extraction was performed with fresh sperms, and the other group was cryopreserved according to a previously procedure (19). Briefly, TES–Tris (TEST) egg yolk buffer was used to dilute the giant panda sperm (Irvine Scientific, Santa Ana, CA) to obtain 5% concentration of glycerol. This diluted material was filled into 0.25-ml semen straws and gradually cooled to 4°C in 4 h, then kept at 7.5 cm for 1 min over liquid nitrogen (LN) to obtain the cooling rate of −40°C/min and then at 2.5 cm for 1 min above LN (approximate cooling rate was −100°C/min), before plunging in LN until further processing. Thawing was performed by immersing the semen straws for 30 s in a water bath with constant temperature of 37°C. Semen was diluted with an equal volume of Ham's F10 (HF10) containing 5% fetal calf serum and 25 mM HEPES. Boar sperm was cryopreserved according to the following procedure; firstly, the sperm was centrifuged (for 5 min at 1,800 rpm and 17°C) and then diluted with a lactose–egg yolk (LEY) extender containing 10 ml hen's egg yolk and 40 ml 11% β-lactose. Secondly, the sperm and the extender mixture were cooled to 4°C (at 0.2°C/min), and further dilution with LEY was performed to obtain a final 3% concentration of glycerol. Lastly, the 0.25-ml semen straws (FHK, Tokyo, Japan) were loaded with this mixture, sealed, and kept 3 cm above LN for 10 min before being submerged into it until future use.
RNA Extraction, Library Preparation, and Sequencing
Before RNA extraction, seminal plasma was removed from all the samples by washing with RNase-free water three times. Then, 0.5% Triton (X-100) was employed in accordance with a previous study (16) to minimize the somatic cell count as they hinder the spermatic RNA extraction process. Then, the TRIzol LS Reagent kit (Invitrogen, Carlsbad, CA, USA) was utilized to extract total RNA from all sperm samples according to the manufacturer's instructions. The RNA samples were pooled together equally in their respective groups before constructing RNA libraries. Furthermore, a Nanodrop (Thermo Fisher Scientific, Wilmington, DE, USA) equipment was used to determine the purity and concentration of the RNA and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) was employed to check its integrity. Then, a NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB, E7490, Ipswich, MA, USA) was utilized to isolate mRNA. The small RNA libraries were built by using the NEB Next Ultra RNA Library Prep Kit for Illumina (NEB, E7530, Ipswich, MA, USA) and the NEBNext Multiplex Oligos for Illumina (NEB, E7500, Ipswich, MA, USA) according to the manufacturer's guidelines. After confirming the quality using Qubit 2.0 and the Agilent Bioanalyzer 2100 system (Agilent Technologies), all libraries were sequenced with the Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA).
piRNA Identification and Expression Analysis
After removal of low-quality, poly-N, and adapter-containing reads and sequences with <18 or >34 nt, clean reads were acquired. The sequence alignments of giant panda and pig were carried out with their reference genomes (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/004/335/GCF_000004335.2_AilMel_1.0 and ftp://ftp.ensembl.org/pub/release-75/fasta/sus_scrofa/, respectively). Furthermore, to compare the clean reads with the Silva database, Rfam database, Repbase, and the GtRNAdb database and filtering out non-coding RNAs (ncRNAs) such as ribosomal RNA (rRNA), transport RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA), and repetitive sequences, Bowtie analysis was performed (20). Novel and known piRNAs were sorted out by comparing the obtained piRNA sequences with miRbase RNA sequences using proTRAC (21). Differential expression of piRNAs in the fresh and frozen-thawed groups was determined with the DESeq R package (v. 1.10.1) based on the reads per kilobase million (TPM) and fragments per kilobase million (FPKM) algorithms (22). The piRNAs between both sperm groups were analyzed by iDEG (23), and those with adjusted p < 0.01 and absolute value of log2 fold change (FC) >1 were classified as DE piRNAs. Then, hierarchical clustering analysis was performed by R heatmap.2 on the selected DE piRNAs; piRNAs with similar expressions were clustered based on the log10(TPM + 1) value.
piRNA Target Prediction, GO, and KEGG Enrichment Analyses
The prediction of potential piRNA targets was performed by BLAST with non-redundant (NR) (20), Gene Ontology (GO) (20), Kyoto Encyclopedia of Genes and Genomes (KEGG) (24), and EuKaryotic Orthologous Group (KOG) (25) databases to obtain annotation information of the target genes. KEGG pathways and GO enriched in predicted DE piRNA target genes were elucidated using KOBAS software (26) and the GOseqR package (27), respectively.
Comparison of DE piRNAs in Fresh and Post-thawed Boar and Giant Panda Sperm During Cryopreservation
piRBase (http://www.regulatoryrna.org/database/piRNA/) was used to browse the common piRNAs and annotations. The homology of piRNAs was predicted between various species by considering the similarity and conserved sequences of the piRNAs to determine the piRNAs. The software Python 2.7 was used for comparing the sequence similarities of the DE piRNAs in giant panda sperm and boar sperm. During sequence alignment, 1–18 bases were perfectly matched; one mismatch base was allowed after the 19th base to select the best pairing sequence (28).
Statistical Analysis
All data were shown as the means ± SEM. SPSS (v. 20.0) with independent samples t test was used to determine statistical differences, and p < 0.05 were considered as statistically significant.
Results
piRNA Profiles of Fresh and Cryopreserved Boar and Giant Panda Sperm
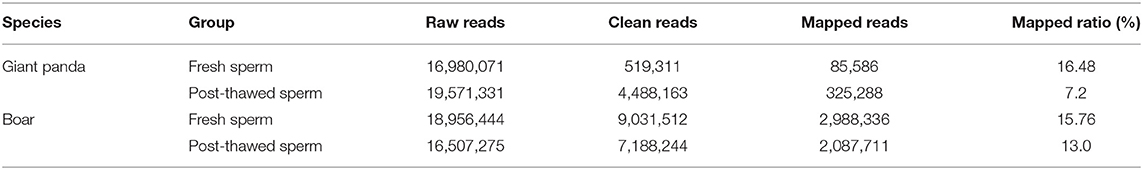
A total of 16,980,071 and 19,571,331 raw reads were obtained from fresh and cryopreserved sperm groups of giant pandas, respectively. Similarly, respective boar sperm groups generated 18,956,444 and 16,507,275 raw reads. After removal of low-quality reads, ploy-N, adapter, and sequences with <24 or >32 nt, 519,311 and 4,488,163 clean reads were generated in respective fresh and frozen-thawed giant panda sperm. Similarly, 9,031,512 and 7,188,244 clean reads were generated in fresh and frozen-thawed boar sperm, respectively (Table 1). The 24-nt (21.76%) and 31-nt (34.21%) piRNAs were the most abundant in fresh and frozen-thawed giant panda sperms, respectively. Similarly, the 30-nt (25%) and 32-nt (1.96%) piRNAs showed the highest and the lowest respective abundances, respectively, in boar sperm.
A total of 88 (containing 116,706 piRNAs) and 133 (containing 21,5835 piRNAs) piRNA clusters were identified after mapping to the designated reference genomes of giant panda and boar sperm, respectively. Compared to the 125,435 and 112,708 piRNAs expressed in fresh and frozen-thawed boar sperm, respectively, 49,393 and 87,670 piRNAs were expressed in fresh and frozen-thawed giant panda sperm, respectively. Differential analysis depicted the differential expression of 1,160 piRNAs (1,138 upregulated and 22 downregulated) between fresh and frozen-thawed giant panda sperm (Figure 1A). In contrast to the giant panda sperm, 384 DE piRNAs (110 upregulated and 274 downregulated) were identified in boar sperm (Figure 1A). Moreover, hierarchical clustering analysis was performed for the clustering of all DE piRNAs (Figure 1B).
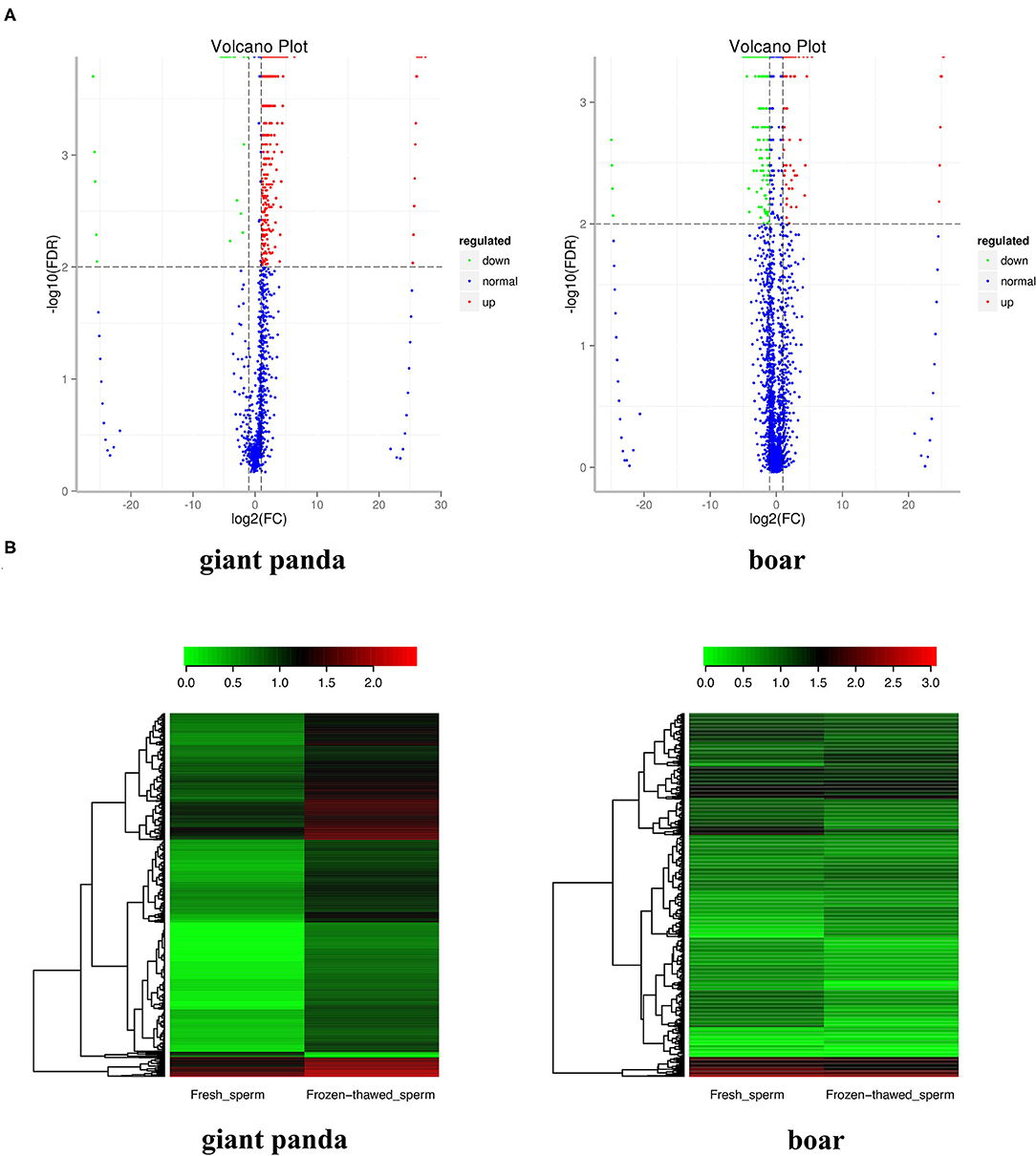
Figure 1. Volcano plot and clustering analysis of differentially expressed PIWI-interacting RNAs (DE piRNAs) in fresh and frozen-thawed giant panda and boar sperm. (A) Volcano plot of DE piRNAs in fresh and frozen-thawed giant panda sperm. Blue dots represent normal expressed, green dots represent the downregulated, and red dots represent the upregulated piRNAs. (B) Heat maps of the cluster analysis of piRNAs. Red indicates high expression while green means low expression of piRNAs.
Combined Analysis of piRNAs and Target mRNAs in Boar and Giant Panda Sperm
Two hundred fifty-three (seven upregulated and 246 downregulated) and 453 target DE mRNAs (366 upregulated and 87 downregulated) of the DE piRNAs were obtained between fresh and post-thawed sperm in giant panda and boar, respectively (Figure 2A). Twenty-eight DE piRNAs were identified to be the common piRNAs by joint analysis of the DE piRNAs of giant panda and boar sperm (Figure 2B). Therefore, 1,132 and 356 DE piRNAs were selected and regarded as the unique piRNAs in giant panda and boar sperm, respectively (Data Sheet 1, 2). Based on the similarity and conservation of the piRNA sequences, 28 DE piRNAs were considered as the homologous piRNAs between giant panda and boar sperm according to the piRBase database (Data Sheet 3). However, no target DE mRNAs were found for these common DE piRNAs.
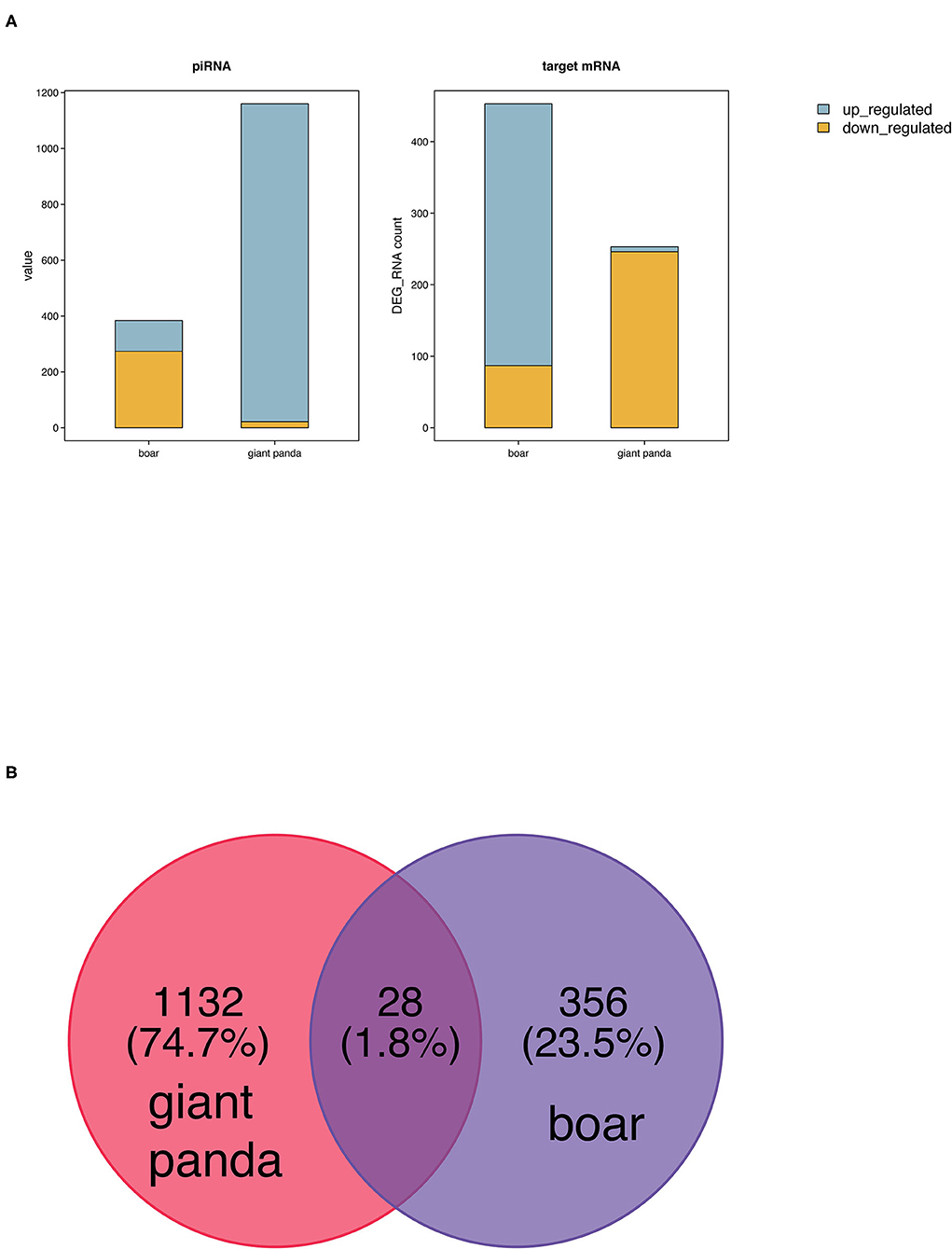
Figure 2. Comparative analysis of differentially expressed PIWI-interacting RNAs (DE piRNAs) in fresh and frozen-thawed giant panda and boar sperm. (A) Comparison of DE piRNAs and target DE mRNAs. (B) Unique and common DE piRNAs.
Comparative GO and KEGG Analysis of DE piRNAs in Giant Panda and Boar Sperm
GO enrichment analysis showed that 106 and 3,251 target DE mRNAs of the DE piRNAs were annotated with 41 and 59 GO terms in giant panda and boar sperm, respectively. Most of the target mRNAs of the DE piRNAs were seen to be distributed in cell, cell part, binding and biological regulation, and metabolic terms in giant panda and boar sperm, which are strictly associated with the structural and functional modifications of sperm. The GO term distributions of the target DE mRNAs of DE piRNAs were significantly different in fresh and frozen-thawed giant panda and boar sperm (Figure 3A).
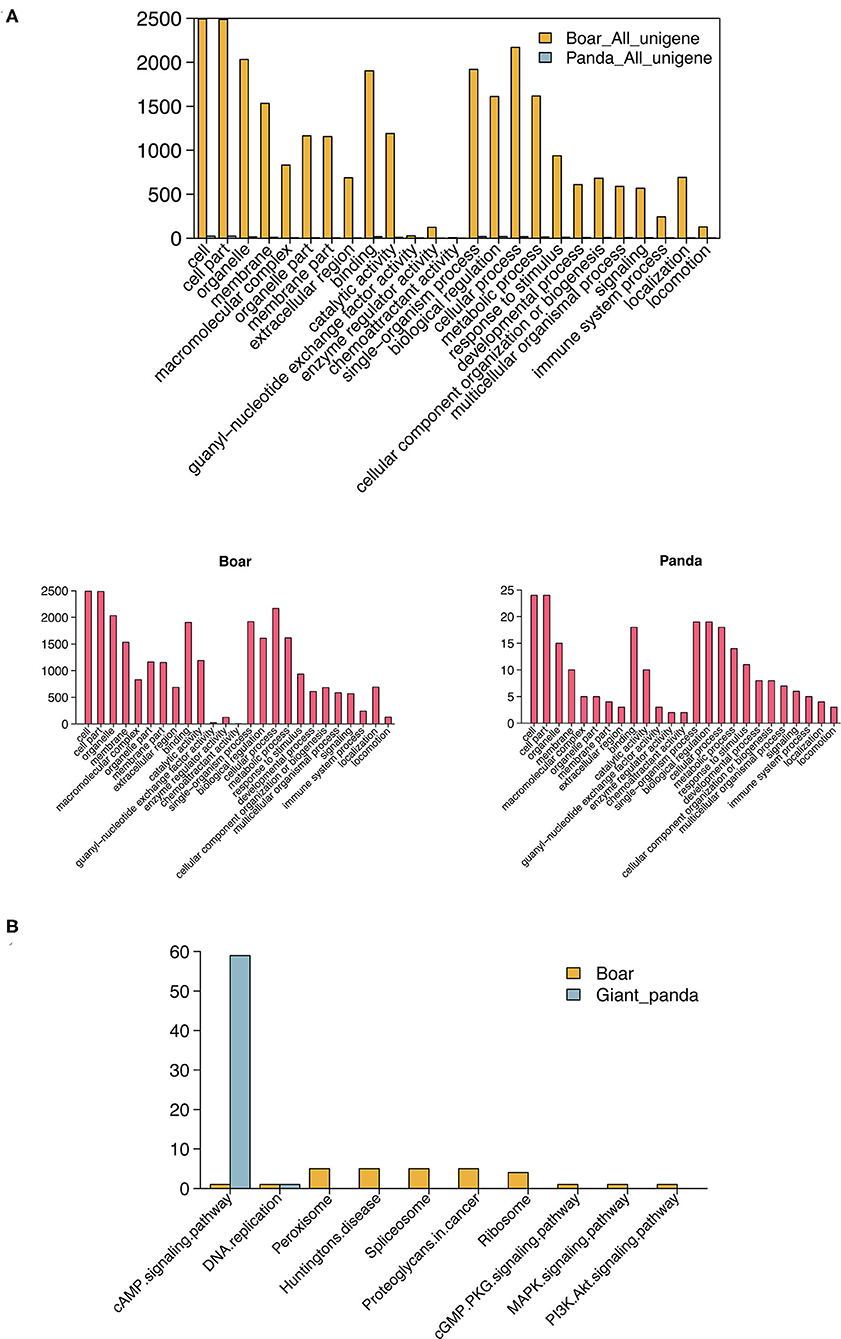
Figure 3. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of differentially expressed PIWI-interacting RNAs (DE piRNAs) in giant panda and boar sperm. (A) GO analysis of the target DE messenger RNAs (mRNAs) of DE piRNAs. (B) Top 10 KEGG pathways of the target DE mRNAs of DE piRNAs.
Notably, most of the target DE mRNAs of DE piRNAs were distributed in the cyclic adenosine monophosphate (cAMP) signaling pathway in giant panda sperm, except for DNA replication (Figure 3B). However, the target mRNAs of the DE piRNAs in boar sperm were mainly distributed in the peroxisome and spliceosome, followed by the membrane-related pathway, such as the cAMP, cyclic guanosine monophosphate (cGMP), mitogen-activated protein kinase (MAPK), and PI3K–Akt signaling pathways. Moreover, the cAMP pathway was found in both giant panda and boar sperm, but was extremely enriched in giant panda sperm. Further analysis indicated that DE piRNAs involved in the cAMP signaling pathway may regulate the post-thawed sperm function by targeting cyclic nucleotide-gated (CNG) ion channel-related genes.
Discussion
It is well-known that differences in the size, shape, and the lipid–protein composition of sperm across various species result in different sensitivities to freezing (29, 30). Esmaeili and colleagues have demonstrated that cryotolerance shows a relation to the ratio of polyunsaturated fatty acids (PUFAs) (omega-3/omega-6) (30). The plasma membrane of boar sperm contains a higher concentration of phospholipids and a lower concentration of cholesterol (9). In addition, the head size of boar sperm is larger than that of the giant panda and is more sensitive to freezing (31). Sperm with smaller heads are usually less cryopreservation-sensitive; thus, the freeze tolerance capacity of giant panda sperm is higher than that of boar sperm after cryopreservation (14).
Nowadays, electroejaculation is the preferred method to collect semen from giant panda. Previous studies have reported that electroejaculation may have an impact on semen parameters, increasing semen osmolarity, disrupting plasma membrane integrity, acrosomal damage, and acrosomal exocytosis (32, 33). However, the sperm morphology remained within acceptable standards (34). Compared with the quality parameters of fresh feline ejaculates collected using three different techniques—urethral catheterization after medetomidine administration, electroejaculation, and epididymal slicing after orchiectomy—the highest quality semen parameters were achieved using electroejaculation (35). Spindler et al. reported that most sperm of giant panda were morphologically normal using electroejaculation, and the sperm parameters (seminal volume, concentration, initial motility, acrosomal integrity, etc.) were consistent with previous reports (11). Therefore, the fertility of frozen-thawed giant panda sperm will be similar to that following the use of fresh sperm (11).
The process of freeze–thawing induces apoptotic-like changes in sperm, and these changes may affect the plasma membrane and acrosomal activity (36) and the mitochondrial activity (37) and also cause abnormal expressions of genes and proteins associated with cryoinjury (38). Moreover, sperm genomic epigenetic elements may be altered during cryopreservation. Previous studies have demonstrated that some genes play critical roles in freezing, such as PRM1, FSHB, ADD1, ARNT, and SNORD116/PWSAS (39, 40). Some proteins, such as TPI1, ACRBP, HSP90AA1, and PHGPx, were proven to be markers of sperm cryoresistance (38, 41, 42). Furthermore, certain mRNA transcripts encoding related proteins were affected during cryopreservation; for instance, PRM1 mRNA transcripts were reduced in boar, cattle, and human sperm (43–45). Beyond that, some sperm mRNA transcripts associated with early embryo development were downregulated in embryos fertilized with frozen horse sperm compared to those with fresh sperm (46). In fact, some microRNAs (miRNAs) associated with cryopreservation, or named CryomiRs, may affect the expressions of the mRNA transcripts during cryopreservation, which ultimately affects the expressions of genes and proteins associated with sperm metabolism and apoptosis (14, 47).
According to our previous study, the DE miRNAs and target mRNAs of giant panda sperm were mainly enriched in olfactory transduction pathways, including the cAMP and cGMP signaling pathways (14). In the present study, we found that the target DE mRNAs of DE piRNAs in giant panda sperm were mainly distributed in the cAMP signaling pathway and partially involved in DNA replication. Similarly, few targets of the DE piRNAs in boar sperm were also enriched in the cAMP signaling pathway, but the ratio was much lower than that of giant panda sperm. Therefore, we speculated that the 1,132 specific piRNAs involved in the cAMP signaling pathway in giant panda sperm may be closely related to the freeze tolerance of sperm. Therefore, we speculated that cryopreservation can affect the expression levels of olfactory transduction pathway-related genes and is probably involved in the regulation of capacitation, motility, fertility, and even the freeze tolerance of post-thawed sperm. However, the regulatory mechanism of the olfactory transduction signaling pathway on post-thawed sperm is still unknown.
It is well-known that olfactory receptors or odorant receptors are associated with sperm motility and chemotaxis. In the olfactory transduction pathway, after the attachment of odorant molecules with the G protein-coupled receptor (GPCR) in sperm, the concentration of cAMP increases, leading to the opening of CNG ion channels (48). Notably, CNG channels play an important role in the regulation of the intracellular Ca2+ level, which causes influx of Ca2+, and then induce sperm hyperactivity (49). In mature sperm, cAMP binding with a target protein is essential for those events during sperm capacitation, including sperm plasma membrane hyperactivation (50, 51), tyrosine phosphorylation (52), and increasing intracellular Ca2+ and pH (53–55). It was demonstrated that the intracellular concentrations of cAMP and Ca2+ play a primary role in sperm capacitation, motility, acrosomal reaction, lipid remodeling, and hyperpolarization of the plasma membrane (55–58). Furthermore, cAMP is known to be an important second messenger for steroid (hormones) biosynthesis, and the specific role of its downstream protein kinase A (PKA) pathway is regulating steroid biosynthesis (59). Steroid hormones induce sperm capacitation and acrosomal response (60). cAMP–PKA signaling pathways induce steroid biosynthesis in stromal cells by activating certain transcription factors, such as CREB, CREM, and GATA4, and regulating the expressions of downstream target proteins (58, 61). In addition, the synthesis of cAMP also activates a Ca2+ signal regulated by PKA or protein kinase C (PKC), which upregulates Nur77 expression, and causes StAR transcription, promoting steroid hormone biosynthesis (62). Previous studies indicated that the intracellular Ca2+, 1,2-diacylglycerol (DAG), and cAMP levels in buffalo sperm were increased significantly after cryopreservation as compared to fresh ejaculates, and the addition of taurine or trehalose reduced the extent of capacitation-like changes in buffalo sperm (56). Likewise, cryopreservation negatively affected the PKA and AMP-activated protein kinase (AMPK) activity in Atlantic salmon sperm (63), and when AMPK was inhibited, the sperm motility decreased accordingly. In this study, the target mRNAs of the DE piRNAs in giant panda sperm are mainly enriched in the cAMP pathway, which indicates that cAMP and calcium may be associated with frozen-thawed sperm quality of giant panda. Differences in the cAMP pathway-related piRNAs and mRNAs between the giant panda and boar sperm may have contributed to sperm cryotolerance. Therefore, our study first revealed that piRNAs might be regulating the cAMP signaling pathway to regulate post-thawed sperm quality, which provides new insights into the cryoinjury, cryoresistance, or the freeze tolerance mechanisms of sperm varying across species. Future exploration should focus on the biological roles of these DE piRNAs in sperm freeze tolerance or cryoresistance and their association with post-thawed sperm quality, which may provide some insights regarding the molecular mechanisms of cryoinjury.
Conclusion
In this study, we first conducted a comparative analysis of the piRNAs and target mRNAs between giant panda sperm and boar sperm during cryopreservation. The differentially expressed piRNAs and their target DE mRNAs are mainly involved in the cAMP signaling pathway and DNA replication, which indicated that these piRNAs play a critical role in sperm cryoresistance and cryoinjury during cryopreservation. Our study provides new insights into the cryoinjury, cryoresistance, or freeze tolerance mechanisms of sperm varying across species.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: Gene Expression Omnibus, GSE163128.
Ethics Statement
The animal study was reviewed and approved by the Regulations of the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, revised in June 2004) the Institutional Animal Care and Use Committee in the College of Animal Science and Technology, Sichuan Agricultural University, Sichuan, China, under permit No: 2019202012. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
YW, YZho, MA, JZ, WW, and ZQ collected samples, performed the experiments, analyzed the data, and drafted the manuscript. BL, YH, and HZ contributed to samples collection, data analysis, and revised the manuscript. MZ, GZ, and YZha revised the manuscript critically and given final approval to be published. CZ granted, concept, designed the experiment and revised, given final approval version of the manuscript to be published. All authors reviewed and approved the final manuscript.
Funding
This research was supported by National Natural Science Foundation of China (No: 31872356 and No: 31570533).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We offer special thanks to the Sichuan TIEQILISHI group for semen collection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.635013/full#supplementary-material
Data Sheet 1. All DE piRNAs in giant panda and boar sperm.
Data Sheet 2. DE piRNAs and their target DE mRNAs in giant panda and boar sperm.
Data Sheet 3. The common DE piRNAs in giant panda and boar sperm.
References
1. Yeste M, Rodríguez-Gil JE, Bonet S. Artificial insemination with frozen-thawed boar sperm. Mol Reprod Dev. (2017) 84:802–13. doi: 10.1002/mrd.22840
2. Flores E, Ramió-Lluch L, Bucci D, Fernández-Novell JM, Peña A, Rodríguez-Gil JE. Freezing-thawing induces alterations in histone H1-DNA binding and the breaking of protein-DNA disulfide bonds in boar sperm. Theriogenology. (2011) 76:1450–64. doi: 10.1016/j.theriogenology.2011.05.039
3. Yeste M. Recent advances in boar sperm cryopreservation: State of the art and current perspectives. Reprod Domest Anim. (2015) 50(Suppl 2):71–9. doi: 10.1111/rda.12569
4. Kim S, Lee YJ, Kim YJ. Changes in sperm membrane and ROS following cryopreservation of liquid boar semen stored at 15°c. Anim Reprod Sci. (2011) 124:118–24. doi: 10.1016/j.anireprosci.2011.01.014
5. Hezavehei M, Sharafi M, Kouchesfahani HM, Henkel R, Agarwal A, Esmaeili V, et al. Sperm cryopreservation: a review on current molecular cryobiology and advanced approaches. Reprod Biomed Online. (2018) 37:327–39. doi: 10.1016/j.rbmo.2018.05.012
6. Yeste M, Estrada E, Casas I, Bonet S, Rodríguez-Gil JE. Good and bad freezability boar ejaculates differ in the integrity of nucleoprotein structure after freeze-thawing but not in ROS levels. Theriogenology. (2013) 79:929–39. doi: 10.1016/j.theriogenology.2013.01.008
7. Flores E, Cifuentes D, Fernández-Novell JM, Medrano A, Bonet S, Briz MD, et al. Freeze-thawing induces alterations in the protamine-1/DNA overall structure in boar sperm. Theriogenology. (2008) 69:1083–94. doi: 10.1016/j.theriogenology.2008.01.022
8. Mostek A, Westfalewicz B, Słowińska M, Dietrich MA, Judycka S, Ciereszko A. Differences in sperm protein abundance and carbonylation level in bull ejaculates of low and high quality. PLoS ONE. (2018) 13:e0206150. doi: 10.1371/journal.pone.0206150
9. Cerolini S, Maldjian A, Surai P, Noble R. Viability, susceptibility to peroxidation and fatty acid composition of boar semen during liquid storage. Anim Reprod Sci. (2000) 58:99–111. doi: 10.1016/S0378-4320(99)00035-4
10. Bailey JL, Lessard C, Jacques J, Brèque C, Dobrinski I, Zeng WX, et al. Cryopreservation of boar semen and its future importance to the industry. Theriogenology. (2008) 70:1251–9. doi: 10.1016/j.theriogenology.2008.06.014
11. Spindler RE, Huang Y, Howard JG, Wang P, Zhang H, Zhang G, et al. Acrosomal integrity and capacitation are not influenced by sperm cryopreservation in the giant panda. Reproduction. (2004) 127:547–56. doi: 10.1530/rep.1.00034
12. Santiago-Moreno J, Esteso MC, Pradiee J, Castaño C, Toledano-Díaz A, O'brien E, et al. Giant panda (Ailuropoda melanoleuca) sperm morphometry and function after repeated freezing and thawing. Andrologia. (2016) 48:470–4. doi: 10.1111/and.12468
13. Ran MX, Li Y, Zhang Y, Liang K, Ren YN, Zhang M, et al. Transcriptome sequencing reveals the differentially expressed lncRNAs and mRNAs involved in cryoinjuries in frozen-thawed giant panda (Ailuropoda melanoleuca) sperm. Int J Mol Sci. (2018) 19:3066. 19. doi: 10.3390/ijms19103066
14. Ran MX, Zhou YM, Liang K, Wang WC, Zhang Y, Zhang M, et al. Comparative analysis of microRNA and mRNA profiles of sperm with different freeze tolerance capacities in boar (Sus scrofa) and giant panda (Ailuropoda melanoleuca). Biomolecules. (2019) 9:432. doi: 10.3390/biom9090432
15. Heejin C, Wang Z, Dean J. Sperm Acrosome Overgrowth and Infertility in Mice Lacking Chromosome 18 Pachytene piRNA. BioRxiv (2020).
16. Cui L, Fang L, Shi B, Qiu S, Ye Y. Spermatozoa expression of piR-31704, piR-39888, and piR-40349 and their correlation to sperm concentration and fertilization rate after ICSI. Reprod Sci. (2018) 25:733–9. doi: 10.1177/1933719117725822
17. Hong YT, Wang C, Fu Z, Liang HW, Zhang SY, Lu ML, et al. Systematic characterization of seminal plasma piRNAs as molecular biomarkers for male infertility. Sci Rep. (2016) 6:24229. doi: 10.1038/srep24229
18. Capra E, Turri F, Lazzari B, Cremonesi P, Gliozzi TM, Fojadelli I, et al. Small RNA sequencing of cryopreserved semen from single bull revealed altered miRNAs and piRNAs expression between High- and Low-motile sperm populations. BMC Genomics. (2017) 18:14. doi: 10.1186/s12864-016-3394-7
19. Huang Y, Li D, Zhou Y, Zhou Q, Li R, Wang C, et al. Factors affecting the outcome of artificial insemination using cryopreserved spermatozoa in the giant panda (Ailuropoda melanoleuca). Zoo Biol. (2012) 31:561–73. doi: 10.1002/zoo.20421
20. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene ontology consortium. Nat Genet. (2000) 25:25–9. doi: 10.1038/75556
21. Rosenkranz D, Zischler H. proTRAC–a software for probabilistic piRNA cluster detection, visualization and analysis. BMC Bioinformat. (2012) 13:5. doi: 10.1186/1471-2105-13-5
22. Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. (2007) 2:e219. doi: 10.1371/journal.pone.0000219
23. Romualdi C, Bortoluzzi S, D'alessi F, Danieli GA. IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. (2003) 12:159–62. doi: 10.1152/physiolgenomics.00096.2002
24. Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. (2004) 32:D277–80. doi: 10.1093/nar/gkh063
25. Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. (2004) 5:R7. doi: 10.1186/gb-2004-5-2-r7
26. Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. (2005) 21:3787–93. doi: 10.1093/bioinformatics/bti430
27. Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. (2009) 10:R25. doi: 10.1186/gb-2009-10-3-r25
28. Rosenkranz D. piRNA cluster database: a web resource for pirna producing loci. Nucleic Acids Res. (2016) 44:D223–30. doi: 10.1093/nar/gkv1265
29. Lv CR, Wu GQ, Hong QH, Quan GB. Spermatozoa cryopreservation: state of art and future in small ruminants. Biopreserv Biobank. (2019) 17:171–82. doi: 10.1089/bio.2018.0113
30. Esmaeili V, Shahverdi AH, Moghadasian MH, Alizadeh AR. Dietary fatty acids affect semen quality: a review. Andrology. (2015) 3:450–61. doi: 10.1111/andr.12024
31. Garde JJ, Soler AJ, Cassinello J, Crespo C, Malo AF, Espeso G, et al. Sperm cryopreservation in three species of endangered gazelles (Gazella cuvieri, G. dama mhorr, and G. dorcas neglecta). Biol Reprod. (2003) 69:602–11. doi: 10.1095/biolreprod.102.012914
32. Dooley MP, Pineda MH. Effect of method of collection on seminal characteristics of the domestic cat. Am J Vet Res. (1986) 47:286–92.
33. Aitken RJ, Wang YF, Liu J, Best F, Richardson DW. The influence of medium composition, osmolarity and albumin content on the acrosome reaction and fertilizing capacity of human spermatozoa: development of an improved zona-free hamster egg penetration test. Int J Androl. (1983) 6:180–93. doi: 10.1111/j.1365-2605.1983.tb00337.x
34. Benavides F, Sutovsky P, López V, Kennedy C, Echevarría L. Semen parameters of fertile guinea pigs (cavia porcellus) collected by transrectal electroejaculation. Animals. (2020) 10:767. doi: 10.3390/ani10050767
35. Jelinkova K, Vitasek R, Novotny R, Bartoskova A. A comparison of quality parameters of fresh feline ejaculates collected by three different collection techniques. Reprod Domest Anim. (2018) 53:1068–74. doi: 10.1111/rda.13205
36. Raad G, Lteif L, Lahoud R, Azoury J, Azoury J, Tanios J, et al. Cryopreservation media differentially affect sperm motility, morphology and DNA integrity. Andrology. (2018) 6:836–45. doi: 10.1111/andr.12531
37. Meyers S Bulkeley E and Foutouhi A. Sperm mitochondrial regulation in motility and fertility in horses. Reprod Domest Anim. (2019) 54(Suppl 3):22–8. doi: 10.1111/rda.13461
38. Chen XL, Zhu HB, Hu CH, Hao HS, Zhang JF, Li KP, et al. Identification of differentially expressed proteins in fresh and frozen-thawed boar spermatozoa by iTRAQ-coupled 2D LC-MS/MS. Reproduction. (2014) 147:321–30. doi: 10.1530/REP-13-0313
39. Valcarce DG, Cartón-García F, Riesco MF, Herráez MP, Robles V. Analysis of DNA damage after human sperm cryopreservation in genes crucial for fertilization and early embryo development. Andrology. (2013) 1:723–30. doi: 10.1111/j.2047-2927.2013.00116.x
40. Dai LS, Zhao ZH, Zhao RF, Xiao SQ, Jiang H, Yue XP, et al. Effects of novel single nucleotide polymorphisms of the FSH beta-subunit gene on semen quality and fertility in bulls. Anim Reprod Sci. (2009) 114:14–22. doi: 10.1016/j.anireprosci.2008.08.021
41. Casas I, Sancho S, Ballester J, Briz M, Pinart E, Bussalleu E, et al. The HSP90AA1 sperm content and the prediction of the boar ejaculate freezability. Theriogenology. (2010) 74:940–50. doi: 10.1016/j.theriogenology.2010.04.021
42. Vilagran I, Castillo J, Bonet S, Sancho S, Yeste M, Estanyol JM, et al. Acrosin-binding protein (ACRBP) and triosephosphate isomerase (TPI) are good markers to predict boar sperm freezing capacity. Theriogenology. (2013) 80:443–50. doi: 10.1016/j.theriogenology.2013.05.006
43. Seshagiri PB, Mariappa D, Aladakatti RH. Tyrosine phosphorylated proteins in mammalian spermatozoa: molecular and functional aspects. Soc Reprod Fertil Suppl. (2007) 63:313–25.
44. Ganguly I, Gaur GK, Kumar S, Mandal DK, Kumar M, Singh U, et al. Differential expression of protamine 1 and 2 genes in mature spermatozoa of normal and motility impaired semen producing crossbred frieswal (HF × Sahiwal) bulls. Res Vet Sci. (2013) 94:256–62. doi: 10.1016/j.rvsc.2012.09.001
45. Zeng CJ, Peng WP, Ding L, He L, Zhang Y, Fang DH, et al. A preliminary study on epigenetic changes during boar spermatozoa cryopreservation. Cryobiology. (2014) 69:119–27. doi: 10.1016/j.cryobiol.2014.06.003
46. Ortiz-Rodriguez JM, Ortega-Ferrusola C, Gil MC, Martín-Cano FE, Gaitskell-Phillips G, Rodríguez-Martínez H, et al. Transcriptome analysis reveals that fertilization with cryopreserved sperm downregulates genes relevant for early embryo development in the horse. PLoS ONE. (2019) 14:e0213420. doi: 10.1371/journal.pone.0213420
47. Zhang Y, Dai DH, Chang Y, Li Y, Zhang M, Zhou GB, et al. Cryopreservation of boar sperm induces differential microRNAs expression. Cryobiology. (2017) 76:24–33. doi: 10.1016/j.cryobiol.2017.04.013
48. Gibson AD, Garbers DL. Guanylyl cyclases as a family of putative odorant receptors. Annu Rev Neurosci. (2000) 23:417–39. doi: 10.1146/annurev.neuro.23.1.417
49. Luddi A, Governini L, Wilmskötter D, Gudermann T, Boekhoff I, Piomboni P. Taste receptors: new players in sperm biology. Int J Mol Sci. (2019) 20:967. doi: 10.3390/ijms20040967
50. Alonso CAI, Osycka-Salut CE, Castellano L, Cesari A, Di Siervi N, Mutto A, et al. Extracellular cAMP activates molecular signalling pathways associated with sperm capacitation in bovines. Mol Hum Reprod. (2017) 23:521–34. doi: 10.1093/molehr/gax030
51. Zapata-Carmona H, Barón L, Zuñiga LM, Díaz ES, Kong M, Drobnis EZ, et al. The activation of the chymotrypsin-like activity of the proteasome is regulated by soluble adenyl cyclase/cAMP/protein kinase A pathway and required for human sperm capacitation. Mol Hum Reprod. (2019) 25:587–600. doi: 10.1093/molehr/gaz037
52. Matamoros-Volante A, Moreno-Irusta A, Torres-Rodriguez P, Giojalas L, Gervasi MG, Visconti PE, et al. Semi-automatized segmentation method using image-based flow cytometry to study sperm physiology: the case of capacitation-induced tyrosine phosphorylation. Mol Hum Reprod. (2018) 24:64–73. doi: 10.1093/molehr/gax062
53. Brukman NG, Nuñez SY, Puga Molina LDC, Buffone MG, Darszon A, Cuasnicu PS, et al. Tyrosine phosphorylation signaling regulates Ca2+ entry by affecting intracellular pH during human sperm capacitation. J Cell Physiol. (2019) 234:5276–88. doi: 10.1002/jcp.27337
54. Stival C, Puga Molina Ldel C, Paudel B, Buffone MG, Visconti PE, Krapf D. Sperm capacitation and acrosome reaction in mammalian sperm. Adv Anat Embryol Cell Biol. (2016) 220:93–106. doi: 10.1007/978-3-319-30567-7_5
55. Buffone MG, Wertheimer EV, Visconti PE, Krapf D. Central role of soluble adenylyl cyclase and cAMP in sperm physiology. Biochim Biophys Acta. (2014) 1842:2610–20. doi: 10.1016/j.bbadis.2014.07.013
56. Singh VK, Atreja SK, Kumar R, Chhillar S, Singh AK. Assessment of intracellular Ca2+, cAMP and 1,2-diacylglycerol in cryopreserved buffalo (Bubalus bubalis) spermatozoa on supplementation of taurine and trehalose in the extender. Reprod Domest Anim. (2012) 47:584–90. doi: 10.1111/j.1439-0531.2011.01922.x
57. Orta G, De La Vega-Beltran JL, Martín-Hidalgo D, Santi CM, Visconti PE, Darszon A. Catsper channels are regulated by protein kinase A. J Biol Chem. (2018) 293:16830–41. doi: 10.1074/jbc.RA117.001566
58. Balbach M, Beckert V, Hansen JN, Wachten D. Shedding light on the role of cAMP in mammalian sperm physiology. Mol Cell Endocrinol. (2018) 468:111–20. doi: 10.1016/j.mce.2017.11.008
59. Selvaraj V, Stocco DM, Clark BJ. Current knowledge on the acute regulation of steroidogenesis. Biol Reprod. (2018) 99:13–26. doi: 10.1093/biolre/ioy102
60. Gimeno-Martos S, González-Arto M, Casao A, Gallego M, Cebrián-Pérez JA, Muiño-Blanco T, et al. Steroid hormone receptors and direct effects of steroid hormones on ram spermatozoa. Reproduction. (2017) 154:469–81. doi: 10.1530/REP-17-0177
61. Ravnskjaer K, Madiraju A, Montminy M. Role of the cAMP pathway in glucose and lipid metabolism. Handb Exp Pharmacol. (2016) 233:29–49. doi: 10.1007/164_2015_32
62. Costa RR, Reis RI, Aguiar JF, Varanda WA. Luteinizing hormone (LH) acts through PKA and PKC to modulate T-type calcium currents and intracellular calcium transients in mice leydig cells. Cell Calcium. (2011) 49:191–9. doi: 10.1016/j.ceca.2011.02.003
63. Lee-Estevez M, Herrera L, Díaz R, Beltrán J, Figueroa E, Dumorné K, et al. Effects of cryopreservation on cAMP-dependent protein kinase and AMP-activated protein kinase in Atlantic salmon (salmo salar) spermatozoa: relation with post-thaw motility. Anim Reprod Sci. (2019) 209:106133. doi: 10.1016/j.anireprosci.2019.106133
Keywords: piRNAs, sperm cryopreservation, freezability, boar, giant panda
Citation: Wang Y, Zhou Y, Ali MA, Zhang J, Wang W, Huang Y, Luo B, Zhang H, Qin Z, Zhang Y, Zhang M, Zhou G and Zeng C (2021) Comparative Analysis of piRNA Profiles Helps to Elucidate Cryoinjury Between Giant Panda and Boar Sperm During Cryopreservation. Front. Vet. Sci. 8:635013. doi: 10.3389/fvets.2021.635013
Received: 29 November 2020; Accepted: 17 February 2021;
Published: 22 April 2021.
Edited by:
Manuel Alvarez Rodriguez, Linköping University, SwedenReviewed by:
Gregorio Miguel Ferreira De Camargo, Federal University of Bahia, BrazilJulián Santiago-Moreno, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), Spain
Copyright © 2021 Wang, Zhou, Ali, Zhang, Wang, Huang, Luo, Zhang, Qin, Zhang, Zhang, Zhou and Zeng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changjun Zeng, emVuZ2NoakBzaWNhdS5lZHUuY24=
†These authors have contributed equally to this work
 Yihan Wang1†
Yihan Wang1† Malik Ahsan Ali
Malik Ahsan Ali Guangbin Zhou
Guangbin Zhou Changjun Zeng
Changjun Zeng