- 1Heilongjiang Key Laboratory for Animal Disease Control and Pharmaceutical Development, Northeast Agricultural University, Harbin, China
- 2Department of Basic Veterinary Science, College of Veterinary Medicine, Northeast Agricultural University, Harbin, China
Objective: Antibiotics play an essential role in the treatment and prevention of diseases in pig farms. However, the irrational use of antibiotics leads to the emergence of multi-drug resistance of bacteria, which poses a critical threat to the efficacy of antibiotic treatments. Therefore, the study is designed to analyze the drug resistance of pathogenic Escherichia coli isolated from large-scale pig farms in East China, which provides a theoretical basis for precisely targeted clinical drugs in swine farms.
Method: The pathogenic E. coli were isolated and identified from clinical samples of swine farms, and the drug resistance of pathogenic E. coli was detected by antimicrobial susceptibility test (AST) and minimum inhibitory concentration test (MIC). Moreover, the prevalence of plasmid-mediated β-lactam resistance genes was analyzed by PCR.
Results: A total of 67 pathogenic E. coli were isolated from 152 samples collected from 20 large-scale pig farms in East China. All isolated pathogenic E. coli are associated with severe drug resistance. Moreover, 70% of isolated pathogenic E. coli is resistant to more than four antibiotics. Besides, there were 19 serotypes including O2, O4, O5, O6, O14, O26, O38, O42, O49, O57, O92, O93, O95, O101, O121, O131, O143, O158, and O161, of which the O4 and O92 serotype were the main serotypes in swine farms. The main extended-spectrum beta-lactamases (ESBLs)-encoding genes in East China were blaCTX−M, blaTEM, and blaOXA by the detection of the ESBLs encoding genes of porcine pathogenic E. coli. The conjugation assays showed that a total of 30 transconjugants were obtained by conjugation, which indicated that drug resistance genes could be transmitted horizontally through conjugative plasmids.
Conclusion: The isolated pathogenic E. coli were all multi-drug resistant, and especially O4 and O92 were the main serotypes. The β-lactam resistance genes were prevalent in large-scale pig farms in East China, which provided a theoretical basis for the prevention and control of pig-derived pathogenic E. coli in the future.
Introduction
Escherichia coli, gram-negative bacteria, is one representative number of the genus E. coli. It is the leading microbial community in the intestinal tract of common animals, and its survival mode is mainly heterotrophic (1). Escherichia coli is an opportunistic pathogen. Livestock and poultry infected by pathogens often have local or systemic inflammatory symptoms, severe diarrhea, and even sepsis (2). Colibacillosis has become a stubborn disease in the modern breeding industry because of its high morbidity and mortality (3–5). Antibacterial drugs can effectively treat colibacillosis. However, overuse and misuse of broad-spectrum antibiotics accelerate the growing resistance and even emergence of a superbug (6–8).
Since the 1940s, antibiotics have transformed medicine, and most infectious diseases can be effectively controlled by the appropriate use of the correct drugs (9, 10). After that, a large number of natural β-lactam antibiotics were discovered. Also, a series of semi-artificial and synthetic β-lactam antibiotics have been obtained through the structural transformation of antibiotics. Penicillin and cephalosporins are the most common β-lactam antibiotics. In addition, there are some new types, such as cephalosporins, oxycephalosporins, monocyclic β-lactams, carbapenems and β-lactamase inhibitors, etc. (11–13). The mechanism of action of β-lactam drugs is relatively similar by inhibiting the production of mucopeptide synthase in the cell wall of the bacteria to prevent the production of the mucopeptides, and then achieve the effect of destroying the cell wall of the bacteria, and ultimately cause the death of the bacteria (14, 15). In addition, the autolysin enzyme activity of bacteria can also be stimulated by β-lactam drugs to achieve the purpose of inhibiting bacterial growth (16). However, bacterial resistance has gradually emerged with the large-scale use of these drugs (17–19).
These bacteria containing extended-spectrum beta-lactamases (ESBLs) are resistant to a variety of β-lactam drugs (20). ESBLs are mostly produced by Enterobacteriaceae bacteria, which are relatively sensitive to some β-lactamase inhibitors such as clavulanic acid, and are mainly derived from β-lactamase genes (21, 22). It is called extended-spectrum β-lactamase because of its complete hydrolysis substrate and strong drug resistance (23). In K. pneumoniae, the SHV-type ESBLs through the 1990s were successful and widely distributed (24). Currently, many types of plasmid-mediated ESBLs have been found and divided into five categories: TEM type, SHV type, CTX-M type, OXA type, and other types (25). ESBLs-producing E. coli strains have multi-drug resistance, mainly because the resistance genes encoding ESBLs are often associated with other resistance genes, such as aminoglycosides, chloramphenicol, fluoroquinolones, sulfonamides, and tetracyclines resistance genes, on the same plasmid (26). According to research, the transfer frequency of drug-resistant plasmids is about 10−8–10−4 (27). However, due to the widespread and common existence of E. coli and drug-resistant plasmids, the transfer and spread of drug resistance in E. coli are convenient and easy.
In this study, 20 large-scale pig farms were monitored in East China. The multi-drug resistance, drug resistance rate and drug resistance spectrum were analyzed, respectively. The research can provide scientific and sufficient theoretical basis for accurate and precise monitoring of E. coli drug resistance and the spread of drug resistance in the East China. The drug resistance analysis of pig-derived pathogenic E. coli can provide a scientific theoretical basis for rational drug use and prevention of E. coli diseases.
Materials and Methods
Sample Collection
Anal and nasal swabs were collected from a total of 152 sick pigs and stored in Eswabs (Copan, Brescia, Italy) from 20 large-scale pig farms in East China (Supplementary Data 1). Nasal swab and anal swab which were collected from the same pig were counted as one sample. Escherichia coli was isolated and identified using eosin methylene blue agar and MacConkey agar plates and biochemical identification methods, respectively. The protocols used during this study were approved by the Northeast Agricultural University Institutional Animal Care and Use Committee, and all the animal care and treatment methods complied with the standards described in the Laboratory Animal Management Regulations (revised 2016) of Heilongjiang Province, China.
Detection the Pathogenicity of E. coli
The E. coli solution was diluted to 5 × 108 CFU/mL with normal saline and was inoculated into abdominal cavity of Kunming mice (Vital River, Beijing, China) with 0.01 mL/g body weight. A total of 760 mice were randomly assigned to 152 groups and each group was inoculated with one E. coli isolates. Some mice died due to the infection caused by pathogenic E. coli. The heart blood and liver of dead mice were collected under aseptic conditions. Escherichia coli in the samples were separated and purified. In addition, they were identified by eosin-methylene blue medium. The isolates that meet the growth characteristics of E. coli were isolated. Finally, the isolates were subjected to gram staining, microscopic examination, and biochemical identification.
Antimicrobial Susceptibility Test and Minimum Inhibitory Concentration Test
Multidrug resistance was defined as the resistant to three or more antimicrobial subclasses. The AST of pig-derived pathogenic E. coli isolates was carried out according to the Kirby Bauer method recommended by the World Health Organization (WHO) (28). The following antimicrobial agents were purchased from the Chinese Institute of Veterinary Drug Control (Beijing, China): Streptomycin (S), gentamicin (GM), amikacin (AN), enrofloxacin (ENR), ciprofloxacin (CIP), doxycycline (DOX), ceftriaxone (CRO), ceftiofur (CTF), florfenicol (FLR), chloramphenicol (C), amoxicillin (AMX), sulfamethoxazole (SXT), fosfomycin (FOS), and polymyxin B (PB). Escherichia coli ATCC 25922 was used as a reference strain. Inhibitory zone diameter of 14 antimicrobial agents is shown in Supplementary Data 2. Besides, detected the MIC of these isolates, and the results were judged based on National Committee for Clinical Laboratory Standards (NCCLS) (Supplementary Data 3) (29). The reference strain E. coli ATCC 25922 was used as quality control for MICs.
Determination the Serotype of Pig-Derived Pathogenic E. coli
The O-serotypes of E. coli of the isolates were determined via a slide agglutination test following the manufacturer's 2016 instructions (Ningbo Tianrun Biotechnology Company, Ningbo, Republic of China).
Detection of ESBLs-Encoding Genes of Pig-Derived Pathogenic E. coli
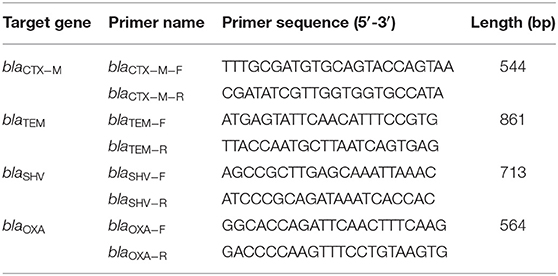
The DNA extraction kit (TaKaRa, Dalian, China) was used to extract the DNA of pig-derived pathogenic E. coli, and the primers of ESBLs-encoding genes were designed according to the ESBLs-encoding gene sequence published by GenBank and reference reports. Designed and synthesized primers of TEM type (GenBank: KM434126.1), SHV type (GenBank: EU376967.1), CTX-M type (GenBank: KM211691.1), and OXA type (GenBank: KX894452.1), respectively. The sequence of the primers is shown in Table 1. DNA sequencing using purified PCR products was provided by ABI PRISM 3730XL Analyzer (Applied Biosystems, Foster City, CA, United States) in Shanghai Sangon Biotech, Co., Ltd., China. The database similarity searches for nucleotide sequences performed using the BLAST tool at the National Center for Biotechnology Information (NCBI) website 1.
Conjugation Assay
The transferability of the ESBLs-carrying plasmid among different E. coli strains was examined using E. coli strain J53 as the recipient (30). Briefly, the donor and recipient strains were mixed at a ratio of 1:3 on a microporous membrane and incubated for 16 h. Then, the mixed bacterial solution was inoculated on tryptone soy agar plate containing 4 μg/ml ceftriaxone and 200 μg/ml sodium azide to select transconjugants. Transconjugants were confirmed by PCR and sequencing (31).
Results
Isolation and Identification of Pig-derived E. coli
As shown in (Supplementary Data 4), all isolates can ferment glucose, and most isolates can ferment lactose and xylose, but only some isolates can ferment sucrose, oxidase, urease, and VP. In addition, all isolates were negative in the inositol test but positive in MR and indole tests. The results of biochemical identification showed that 78 isolates were consistent with the biochemical characteristics of E. coli. These 78 isolates were, respectively, inoculated into mice, and then 67 isolates were found to cause diarrhea 6 h, and death of mice 36 h after injection. These isolates conform to the growth characteristics of E. coli, so they can be judged as pathogenic E. coli.
AST and MIC of Pig-derived Pathogenic E. coli
Through the AST, it was found that all isolates showed multi-drug resistance (Table 2). The results of the drug resistance spectrum showed that 67 pig-derived pathogenic E. coli isolates showed multi-drug resistance. These isolates mainly showed resistance ranged from 4 to 11 drugs, of which 8 drug-resistant isolates accounted for 28.36%. The second most is resistant to 7 drugs, reaching 16.42%. Furthermore, 71.64% (48/67) showed resistance to 7 or more drugs.
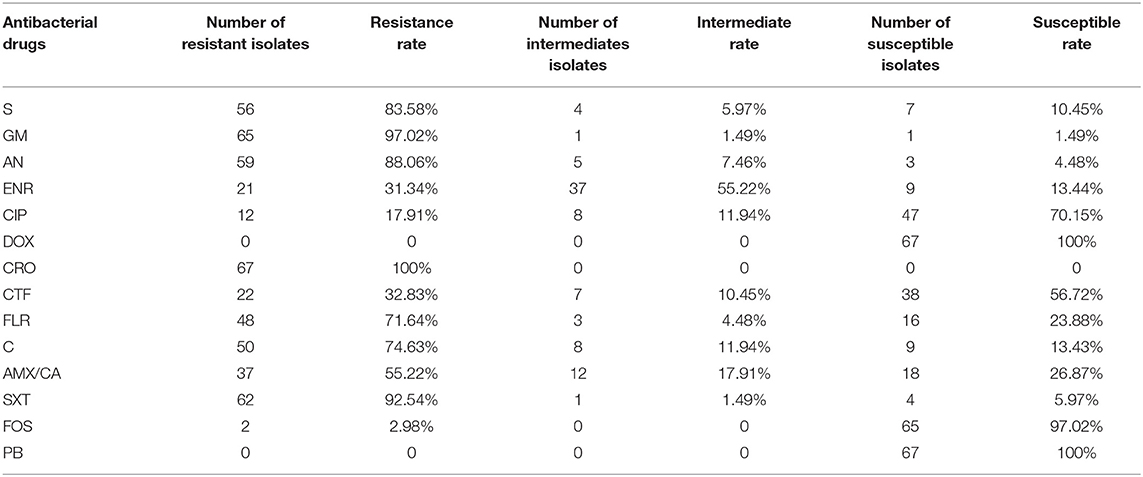
As showed in the Table 3, the resistance rate test results showed that 67 E. coli isolates are completely resistant to CRO, and all isolates were susceptible to DOX and PB. The results of MIC are consistent with the AST, which further confirms the drug resistance phenotype and drug resistance rate of each strain. The above results indicate that the drug resistance of pathogenic E. coli from pigs in East China is relatively serious.
The Serotype of Pig-Derived Pathogenic E. coli
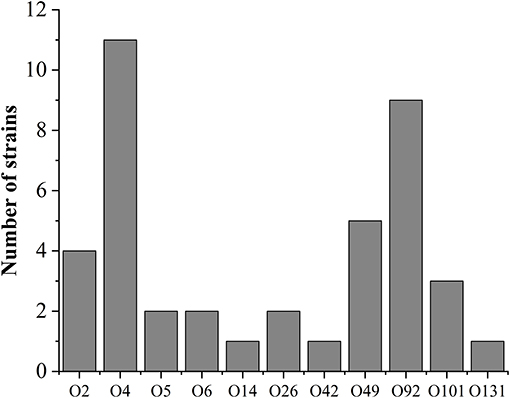
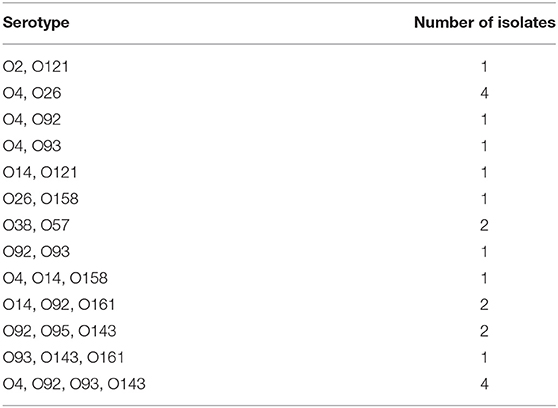
The serotypes of pig-derived pathogenic E. coli were identified. The results showed that the others were successfully serotyped except for 4 isolates, accounting for 94.03% (63/67) of all pathogenic isolates. The serotype distribution is shown in Figure 1. There are 19 serotypes in 63 isolates, of which O4 and O92 are the main serotypes. Furthermore, some isolates were determined as mixed serotypes. All isolates of mixed serotypes were re-purified, and then the slide agglutination test was repeated. According to Table 4, the 34.92% (22/63) pathogenic isolates were mixed serotype.
Detection of ESBLs-Encoding Genes of Pig-Derived Pathogenic E. coli
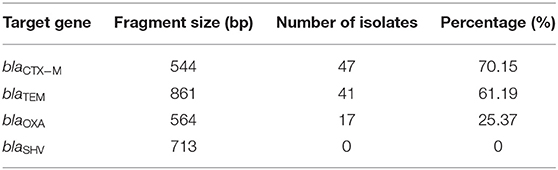
The detection rates of different ESBLs-encoding genes of 67 pathogenic E. coli isolates are shown in Table 5. The ESBLs-encoding genes with the highest detection rate were blaCTX−M (70.15%), followed by blaTEM (61.19%) and blaOXA (25.37%), and blaSHV were not detected.
Transferability of ESBLs Genes
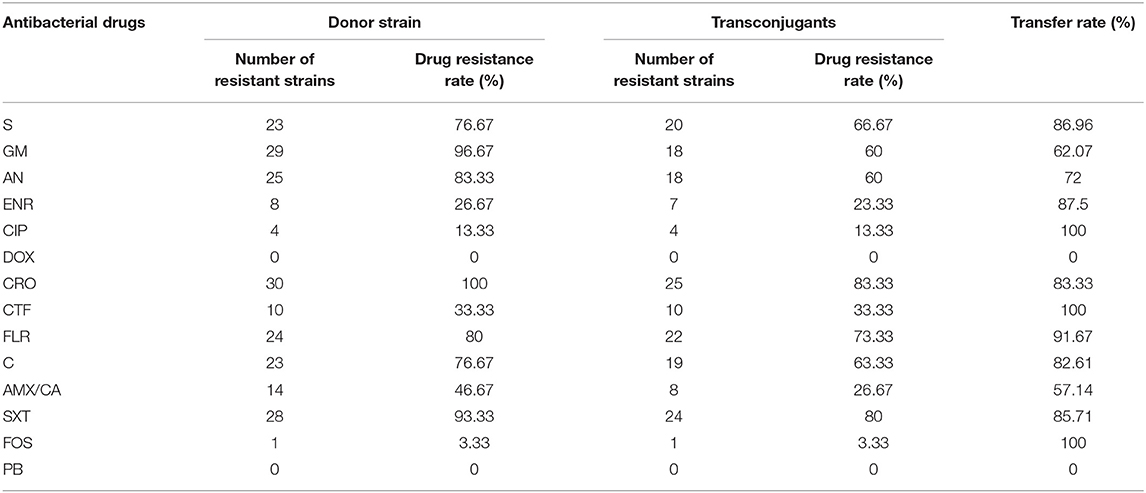
As shown in Table 6, the conjugation assay results showed that 30 isolates successfully conjugated, and the success rate of conjugation was as high as 44.78%, all 30 transconjugants showed 4 or more kinds of drug resistance. The resistance phenotypes of DOX and PB were not detected in the transconjugants, but the resistance phenotypes of the other 12 antibiotics were detected. In addition, the drug-resistant phenotypes of 7 transconjugants were completely consistent with those of the donor isolates. According to Table 7, the transfer rate of resistant phenotype to CIP, CTF and FOS reached 100%. One transconjugant showed a new drug-resistant phenotype, and the probability of mutation of the drug-resistant phenotype was 3.3%.
According to Tables 8, 9, the detection rate of blaCTX−M was the highest among the 30 donor strains, and the detection rates of blaTEM and blaOXA were 43.33 and 26.67%, respectively. Among the 30 transconjugants, the detection rates of blaCTX−M, blaTEM, and blaOXA were 23.33, 23.33, and 6.67%, respectively. Comparing the donor and the transconjugants, the transfer rates of the three-drug resistance genes blaCTX−M, blaTEM, and blaOXA were 43.75, 53.85, and 25%, respectively.
Discussion
Antibiotics are the foundation of modern medicine, and their use reduces mortality and prolongs life expectancy (32). However, Antibiotic-resistant bacteria that are difficult or impossible to treat are becoming increasingly common and are causing a global health crisis (33, 34). In this study, identified the pathogenic E. coli isolated from large-scale pig farms in East China, and analyzed its drug resistance to 14 commonly used antibiotics. The proportion of multi-drug resistant isolates accounted for 100% of the resistant isolates, and these isolates mainly showed resistance to varied from 4 to 11 drugs. These data all indicate that the drug resistance of E. coli in East China is mainly manifested as multidrug resistance. Accordingly, the phenomenon of drug resistance has been very serious.
CRO, GM, SXT, AN, S, C, FLR, and AMX/CA resistance rates have reached more than 50%, which may be due to the relatively common clinical use of these antibiotics (35–37). We found that 2.98% of the isolates were resistant to FOS, and all isolates were susceptible to DOX and PB. This is also due to the late availability of these drugs and the relatively small application range (38–40). These drugs might play a vital role in the prevention and treatment of multi-drug resistant E. coli in the future.
Escherichia coli is an opportunistic pathogen with many serotypes and large variations. At present, in pathogenic E. coli, the antigen structure of pathogenic bacteria is mainly divided into four types: O, K, H, and F antigens type, of which O antigen is the most common type (41). The serotypes of animal-derived E. coli prevalent in various regions are quite different, and the advantages of different pathogenic E. coli O antigen serotypes are different (42). There are currently 196 types of known E. coli O antigens, and the serotypes are divided into O1 to O187 types. Most of the genes related to the synthesis of O antigen in E. coli are arranged tightly on the chromosome, thus forming an O antigen gene cluster (43).
The serotype identification results showed that a total of 19 serotypes were included in these isolates, 34.92% (22/67) isolates had two or more serotypes, indicating the existence of O antigen cross-reaction. Serotypes are related to the pathogenicity of E. coli. Moreover, the predominant serotypes of pathogenic E. coli were different in different areas. Due to the weak antigen cross-protection between different serotypes, it is impossible to prepare an extended-spectrum vaccine that can cover all serotypes. This phenomenon will increase the difficulty of prevention of E. coli disease to a certain extent. In addition, the results showed that there was no direct correlation between serotype and genotype of drug resistance gene. Therefore, the diversity, variability and complexity of serotypes have become the factors that need to be carefully considered in the prevention of E. coli disease.
The transconjugants test results showed that 30 of the 67 pathogenic E. coli isolates were successfully conjugated, with a success rate of 44.78%. In addition, the transconjugants also showed multi-drug resistance, and the drug-resistant phenotype was reduced compared with the donor strains. However, compared with the donor isolates, the transconjugants showed a tendency that its resistance rate to 14 kinds of antibacterial drugs was exactly the same as that of the donor isolates. The above phenomenon shows that the resistance genes of E. coli located in the plasmid were transferred to the recipient isolates along with the plasmid, and therefore it showed resistance to antibiotics. Moreover, according to the statistical results of the drug-resistant phenotype transfer rate of donor isolates, drug-resistant genes can be horizontally transmitted and expressed with plasmids. In the experiment, one transconjugant showed a new drug-resistant phenotype. Compared with the donor isolates, it had a chloramphenicol-resistant phenotype. The reason for this phenomenon may be the result of gene mutation.
The production of ESBLs in Enterobacteriaceae is a well-known mechanism for resistance to antibiotics. In China, increasing studies reported the increasing detection rates of ESBLs-producing E. coli collected from infected patients in the clinic, and CTX-M types were identified to be the most common ESBLs in E. coli (44, 45). We found that transconjugants have transferred resistance genes, and 9 of them have the same resistance genotype as the donor isolates. In addition, among the 30 transconjugants, 7 recipient strains had TEM genes,7 recipient strains had CTX-M genes, and 2 recipient strains amplified OXA genes. This phenomenon should be attributed to the fact that the resistance genes of the donor strain were carried on the conjugative plasmid, and the resistance genes were transferred to the transconjugants with the conjugation of donor and recipient bacteria.
All findings suggested that these drug resistance genes of E. coli were located on the conjugate plasmid and can be transmitted and expressed horizontally in the plasmid. Among the various routes for the spread of drug-resistant genes, horizontal gene transfer by plasmid-mediated conjugation is regarded as one of the primary mechanisms. Through conjugation, DNA can be transferred by mobile plasmids between genera, phyla, and even significant domains. For example, E. coli shows resistance after obtaining exogenous drug-resistant genes. Conjugative plasmids of E. coli can be transmitted and diffused among the same species or different species, which leads to the spread of antibiotic resistance. The horizontal transmission of genes is directly affected by the success rate of conjugation between strains. In this study, the high conjugation rate and the high transfer rate of drug-resistant genes in this study also confirmed the fact that the drug-resistant transmission of pig-derived pathogenic E. coli in East China is relatively severe.
Conclusion
The result of this study showed that 67 pathogenic E. coli isolates showed multidrug resistance, which indicated that the drug resistance of E. coli in East China was severe. Besides, the results showed that there was no direct correlation between serotype and genotype of drug resistance gene. Therefore, the diversity, variability and complexity of serotypes have become factors that need to be carefully considered in the prevention of E. coli disease. Finally, by analyzing the transfer of ESBLs-encoding genes between donor and recipient isolates, it was found that antibiotic resistance of porcine pathogenic E. coli could be transmitted to recipient isolates through conjugation.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by Northeast Agricultural University Institutional Animal Care and Use Committee. Laboratory Animal Management Regulations (Revised 2016) of Heilongjiang Province, China.
Author Contributions
XL conceived and designed the experiments and wrote the manuscript. HL, SC, PC, FL, and JS provided assistance of the experiments and collected samples. MI revised the manuscript. XZ supervised the experiments and revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National 13th Five-Year Key R&D Program Special, Project under Grant: 2018YFD0500306.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.614651/full#supplementary-material
References
1. Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. (2000) 403:339–42. doi: 10.1038/35002131
2. Stoll BJ, Hansen NI, Sánchez PJ, Faix RG, Poindexter BB, Van Meurs KP, et al. Early onset neonatal sepsis: the burden of group b streptococcal and E. coli disease continues. Pediatrics. (2011) 127:817–26. doi: 10.1542/peds.2010-2217
3. Guabiraba R, Schouler C. Avian colibacillosis: still many black holes. Fems Microbiol Lett. (2015) 362:118. doi: 10.1093/femsle/fnv118
4. Kim NH, Cho TJ, Rhee MS. Current interventions for controlling pathogenic Escherichia coli. Adv Appl Microbiol. (2017) 100:1–47. doi: 10.1016/bs.aambs.2017.02.001
5. Gao J, Duan X, Li X, Cao H, Wang Y, Zheng SJ. Emerging of a highly pathogenic and multi-drug resistant strain of Escherichia coli causing an outbreak of colibacillosis in chickens. Infect Genet Evol. (2018) 65:392–8. doi: 10.1016/j.meegid.2018.08.026
6. Begum J, Mir NA, Dev K, Khan IA. Dynamics of antibiotic resistance with special reference to Shiga toxin-producing Escherichia coli infections. J Appl Microbiol. (2018) 125:1228–37. doi: 10.1111/jam.14034
7. Merlin C. Reducing the consumption of antibiotics: would that be enough to slow down the dissemination of resistances in the downstream environment? Front Microbiol. (2020) 11:33. doi: 10.3389/fmicb.2020.00033
8. Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Dis Clin North Am. (2016) 30:313–22. doi: 10.1016/j.idc.2016.02.001
9. Sahin U, Unlü M, Demirci M, Akkaya A, Turgut E. Penicillin resistance in Streptococcus pneumoniae in Isparta. Respirology. (2001) 6:23–6. doi: 10.1046/j.1440-1843.2001.00292.x
10. Spratt BG. Properties of the penicillin. Eur J Biochem. (1977) 72:341–52. doi: 10.1111/j.1432-1033.1977.tb11258.x
11. Carlier M, Stove V, Wallis SC, De Waele JJ, Verstraete AG, Lipman J, et al. Assays for therapeutic drug monitoring of β-lactam antibiotics: a structured review. Int J Antimicrob Agents. (2015) 46:367–75. doi: 10.1016/j.ijantimicag.2015.06.016
12. Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present, and future. Antimicrob Agents Chemothe. (2011) 55:4943–60. doi: 10.1128/AAC.00296-11
13. Mohr KI. History of antibiotics research. Curr Top Microbiol Immunol. (2016) 398:237–72. doi: 10.1007/82_2016_499
14. Kitano K, Tomasz A. Triggering of autolytic cell wall degradation in Escherichia coli by beta-lactam antibiotics. Antimicrob Agents Chemother. (1979) 16:838–48. doi: 10.1128/aac.16.6.838
15. Cho H, Uehara T, Bernhardt TG. Beta-lactam antibiotics induce a lethal malfunctioning of the bacterial cell wall synthesis machinery. Cell. (2014) 159:1300–11. doi: 10.1016/j.cell.2014.11.017
16. Jensen C, Bæk KT, Gallay C, Thalsø-Madsen I, Xu L, Jousselin A, et al. The ClpX chaperone controls autolytic splitting of Staphylococcus aureus daughter cells, but is bypassed by β-lactam antibiotics or inhibitors of WTA biosynthesis. PLoS Pathog. (2019) 15:e1008044. doi: 10.1371/journal.ppat.1008044
17. Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum beta-lactamases (ESBLs) in the community. J Antimicrob Chemother. (2005) 56:52–9. doi: 10.1093/jac/dki166
18. Hu FP, Zhu DM, Wang F, Jiang XF, Wu L. CHINET 2013 surveillance of bacterial resistance in China. Chin J Infect Chemother. (2014) 12:321–9. doi: 10.16718/j.1009-7708.2014.05.004
19. Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem. (2014) 6:25–64. doi: 10.4137/PMC.S14459
20. Ghafourian S, Sadeghifard N, Soheili S, Sekawi Z. Extended spectrum beta-lactamases: definition, classification and epidemiology. Curr Issues Mol Biol. (2015) 17:11–22. doi: 10.21775/cimb.017.011
21. Zhanel GG, Chung P, Adam H, Zelenitsky S, Denisuik A, Schweizer F, et al. Ceftolozane/tazobactam: a novel cephalosporin/β-lactamase inhibitor combination with activity against multidrug-resistant gram-negative bacilli. Drugs. (2014) 74:31–51. doi: 10.1007/s40265-013-0168-2
22. Zhanel GG, Lawrence CK, Adam H, Schweizer F, Zelenitsky S, Zhanel M, et al. Imipenem-relebactam and meropenem-vaborbactam: two novel carbapenem-β-lactamase inhibitor combinations. Drugs. (2018) 78:65–98. doi: 10.1007/s40265-017-0851-9
23. Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. (2008) 8:159–66. doi: 10.1016/S1473-3099(08)70041-0
24. Paterson DL, Bonomo RA. Extended-spectrum beta-lactamases: a clinical update. Clin Microbiol Rev. (2005) 18:657–86. doi: 10.1128/CMR.18.4.657-686.2005
25. Silva J, Aguilar C, Ayala G, Estrada MA, Garza-Ramos U, Lara-Lemus R, et al. TLA-1: a new plasmid-mediated extended-spectrum beta-lactamase from Escherichia coli. Antimicrob Agents Chemother. (2000) 44:997–1003. doi: 10.1128/aac.44.4.997-1003.2000
26. Zhai R, Fu B, Shi X, Sun C, Liu Z, Wang S, et al. Contaminated in-house environment contributes to the persistence and transmission of NDM-producing bacteria in a Chinese poultry farm. Environ Int. (2020) 139:105715. doi: 10.1016/j.envint.2020.105715
27. Banderas A, Carcano A, Sia E, Li S, Lindner AB. Ratiometric quorum sensing governs the trade-off between bacterial vertical and horizontal antibiotic resistance propagation. PLoS Biol. (2020) 18:e3000814. doi: 10.1371/journal.pbio.3000814
28. Wouter VDB, Schijffelen MJ, Bosboom RW, Cohen Stuart J, Diederen B, Kampinga G, et al. Susceptibility of ESBL Escherichia coli and Klebsiella pneumoniae to fosfomycin in the Netherlands and comparison of several testing methods including Etest, MIC test strip, Vitek2, Phoenix and disc diffusion. J Antimicrob Chemother. (2018) 73:2380–7. doi: 10.1093/jac/dky214
29. Brook I, Wexler HM, Goldstein EJ. Antianaerobic antimicrobials: spectrum and susceptibility testing. Clin Microbiol Rev. (2013) 26:526–46. doi: 10.1128/CMR.00086-12
30. Liu Z, Wang Y, Walsh TR, Liu D, Shen Z, Zhang R, et al. Plasmid-mediated novel bla(NDM−17) gene encoding a carbapenemase with enhanced activity in a sequence type 48 Escherichia coli strain. Antimicrob Agents Chemother. (2017) 61:e02233–16. doi: 10.1128/AAC.02233-16
31. Nordmann P, Poirel L, Carrër A, Toleman MA, Walsh TR. How to detect NDM-1 producers. J Clin Microbiol. (2011) 49:718–21. doi: 10.1128/JCM.01773-10
32. Arias CA, Murray BE. Antibiotic-resistant bugs in the 21st century–a clinical super-challenge. N Engl J Med. (2009) 360:439–43. doi: 10.1056/NEJMp0804651
33. Paitan Y. Current trends in antimicrobial resistance of Escherichia coli. Curr Top Microbiol Immunol. (2018) 416:181–211. doi: 10.1007/82_2018_110
34. Blair JMA, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJV. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. (2015) 13:42–51. doi: 10.1038/nrmicro3380
35. Aarestrup FM, Oliver Duran C, Burch DGS. Antimicrobial resistance in swine production. Anim Health Res Rev. (2008) 9:135–48. doi: 10.1017/S1466252308001503
36. Pan X, Qiang Z, Ben W, Chen M. Residual veterinary antibiotics in swine manure from concentrated animal feeding operations in Shandong Province, China. Chemosphere. (2011) 84:695–700. doi: 10.1016/j.chemosphere.2011.03.022
37. Eltayb A, Barakat S, Marrone G, Shaddad S, Stålsby Lundborg C. Antibiotic use and resistance in animal farming: a quantitative and qualitative study on knowledge and practices among farmers in Khartoum, Sudan. Zoonoses Public Health. (2012) 59:330–8. doi: 10.1111/j.1863-2378.2012.01458.x
38. Ito R, Mustapha MM, Tomich AD, Callaghan JD, McElheny CL, Mettus RT, et al. Widespread fosfomycin resistance in gram-negative bacteria attributable to the chromosomal fosa gene. mBio. (2017) 8:e00749–17. doi: 10.1128/mBio.00749-17
39. Patyra E, Kwiatek K, Nebot C, Gavilán RE. Quantification of veterinary antibiotics in pig and poultry feces and liquid manure as a non-invasive method to monitor antibiotic usage in livestock by liquid chromatography mass-spectrometry. Molecules. (2020) 25:3265–79. doi: 10.3390/molecules25143265
40. Kaufmann A, Widmer M. Quantitative analysis of polypeptide antibiotic residues in a variety of food matrices by liquid chromatography coupled to tandem mass spectrometry. Anal Chim Acta. (2013) 797:81–8. doi: 10.1016/j.aca.2013.08.032
41. DebRoy C, Roberts E, Fratamico PM. Detection of O antigens in Escherichia coli. Anim Health Res Rev. (2011) 12:169–85. doi: 10.1017/S1466252311000193
42. Szmolka A, Anjum MF, La Ragione RM, Kaszanyitzky EJ, Nagy B. Microarray based comparative genotyping of gentamicin resistant Escherichia coli strains from food animals and humans. Vet Microbiol. (2012) 156:110–8. doi: 10.1016/j.vetmic.2011.09.030
43. Mellor GE, Fegan N, Duffy LL, McMillan KE, Jordan D, Barlow RS. National survey of Shiga toxin-producing Escherichia coli serotypes O26, O45, O103, O111, O121, O145, and O157 in Australian beef cattle feces. J Food Prot. (2016) 79:1868–74. doi: 10.4315/0362-028X.JFP-15-507
44. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. (2010) 34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x
Keywords: ESBLs, E. coli, multi-drug resistance, resistance gene, resistance phenotype
Citation: Li X, Liu H, Cao S, Cheng P, Li F, Ishfaq M, Sun J and Zhang X (2021) Resistance Detection and Transmission Risk Analysis of Pig-Derived Pathogenic Escherichia coli in East China. Front. Vet. Sci. 8:614651. doi: 10.3389/fvets.2021.614651
Received: 06 October 2020; Accepted: 07 April 2021;
Published: 30 April 2021.
Edited by:
Ioannis Magouras, City University of Hong Kong, Hong KongReviewed by:
Lucy Brunton, Royal Veterinary College (RVC), United KingdomZhang Wan Jiang, Chinese Academy of Agricultural Sciences (CAAS), China
Copyright © 2021 Li, Liu, Cao, Cheng, Li, Ishfaq, Sun and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiuying Zhang, emhhbmd4aXV5aW5nQG5lYXUuZWR1LmNu
 Xiaoting Li
Xiaoting Li Haibin Liu1,2
Haibin Liu1,2 Ping Cheng
Ping Cheng Jichao Sun
Jichao Sun Xiuying Zhang
Xiuying Zhang