
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 03 March 2021
Sec. Veterinary Infectious Diseases
Volume 8 - 2021 | https://doi.org/10.3389/fvets.2021.583759
This article is part of the Research TopicAntibiotic Resistance and its Continuity in the Environmental NicheView all 19 articles
Marine mammals are frequently considered good sentinels for human, animal and environmental health due to their long lifespan, coastal habitat, and characteristics as top chain predators. Using a One Health approach, marine mammals can provide information that helps to enhance the understanding of the health of the marine and coastal environment. Antimicrobial resistance (AMR) is the quintessential One Health problem that poses a well-recognised threat to human, animal, and ecosystem health worldwide. Treated and untreated sewage, hospital waste and agricultural run-off are often responsible for the spread of AMR in marine and freshwater ecosystems. Rescued seals (n = 25) were used as sentinels to investigate the levels of AMR in the Irish coastal ecosystem. Faecal swabs were collected from these animals and bacterial isolates (E. coli and cefotaxime-resistant non-E. coli) from each swab were selected for further investigation. E. coli isolates were characterised in terms of phylogenetic group typing, AMR, and virulence factors. All E. coli isolates investigated in this study (n = 39) were ampicillin resistant while 26 (66.6%) were multi-drug resistant (MDR). Resistance genes blaOXA−1 and blaTEM−1 were detected in 16/39 and 6/39 isolates, respectively. Additionally, virulence factors associated with adhesion (sfa, papA, and papC) and siderophores (fyuA and iutA) were identified. An additional 19 faecal cefotaxime-resistant non-E. coli isolates were investigated for the presence of β-lactamase encoding genes. These isolates were identified as presumptive Leclercia, Pantoea and Enterobacter, however, none were positive for the presence of the genes investigated. To the authors knowledge this is the first study reporting the detection of blaOXA−1 and blaTEM−1 in phocid faecal E. coli in Europe. These results highlight the importance of marine mammals as sentinels for the presence and spread of AMR in the marine and coastal environment.
Located in the North Atlantic region, Ireland offers an important habitat for marine mammals including harbour seals (Phoca vitulina) and grey seals (Halichoerus grypus) in search of haul-out sites during breeding and moulting (1). Grey seals are known to migrate between countries, but harbour seals tend to travel less widely. Nonetheless, both species usually return to their breeding areas (2).
Marine mammals are frequently considered good sentinels for human and environmental health because their position at the top of the food chain, their long-life span and their coastal habitat can provide an early warning system for public health issues (3, 4). Using a One Health approach, marine mammals can be seen as an important source of information that helps to enhance our understanding of the health of the marine and coastal environment (3).
Antimicrobial resistance (AMR) is the quintessential One Health problem (5) that poses a well-recognised threat to human, animal and ecosystem health worldwide (6). Much of this problem has been associated with the misuse of antimicrobials in human, veterinary and agricultural settings (7) leading to the increased emergence of antimicrobial-resistant bacteria (ARB) in marine and fresh water ecosystems (8). Treated and untreated sewage, hospital waste and agricultural run-off are often responsible for the spread of AMR in these ecosystems (9–11). Studies have shown that natural environments, such as soils, sediments, and surface waters have complex microbiomes which include clinically important ARBs and antimicrobial-resistant genes (ARGs) (12). ARGs can be transferred into soils and leached to groundwater or carried by runoff and erosion to surface water (13). In addition, dense bacterial populations in treatment plants facilitate frequent genetic exchange through mobile genetic elements (MGE), such as plasmids, integrons, and transposons (8, 14). For example, the spread of β-lactamases, enzymes responsible for decreasing the efficacy of critically important β-lactam antimicrobials against Gram negative bacteria, is frequently due to MGEs (15). These enzymes are currently the most important mechanism of resistance in Gram negative pathogens with more than 2,600 enzymes described to date. The most frequently described enzymes in E. coli include CTX-M, TEM and SHV, Ambler class A enzymes, and OXA, Ambler class D enzymes (16). Also disseminated by MGEs, virulence factors including adherence factors, invasion factors, iron acquisition systems, capsules and toxins facilitate bacterial colonisation of the host (17).
An Irish technical report identified high levels of resistant E. coli in urban wastewater (18). More recently, Mahon et al. were the first in Europe to report the isolation of New Delhi metallo-beta-lactamase (NDM)-producing Enterobacteriaceae from both fresh water and seawater sampled on 2 Irish beaches located near an untreated human sewage ocean discharge (19). This finding raises concerns regarding the potential of sewage discharges to contribute to the spread of ARBs and ARGs in the environment, especially when recent studies have shown that resistant bacteria can be selected at extremely low antibiotic concentrations, similar to concentrations found in some aquatic and soil environments (20, 21). Although it is recognised that proximity to human activities shapes the AMR profile of the gastrointestinal microbiome of wild mammals, the presence of ARBs and ARGs in the intestinal microbiota of wild animals has not been thoroughly investigated (8, 22, 23). Additionally, the role played by wildlife colonised with ARB in the dissemination of ARGs worldwide needs to be addressed (24, 25).
In light of the recent findings of ARB in Irish coastal waters and the complexity of the factors governing dissemination of AMR in the environment, a pilot study was conducted to characterise the faecal E. coli populations of pinnipeds living in coastal waters surrounding Ireland, and investigate the presence of β-lactamase encoding genes and virulence factors. Furthermore, the presence of β-lactamase encoding genes was examined in cefotaxime-resistant non-E. coli isolates.
In the summer breeding season of 2017, collection of faecal swabs from 23 harbour seals (P. vitulina) and two grey seals (H. grypus) was attempted at the premises of Seal Rescue Ireland (SRI), the only marine rehabilitation centre in the Republic of Ireland; however, three animals did not defecate during the visit. Convenience sampling was conducted on two occasions in July 2017, 22 days apart, to sample as many individual animals as possible (Table 1). Fourteen and eleven animals were sampled on the first and second sampling-days, respectively, which made up the total number of animals housed at SRI at the time (Table 1).
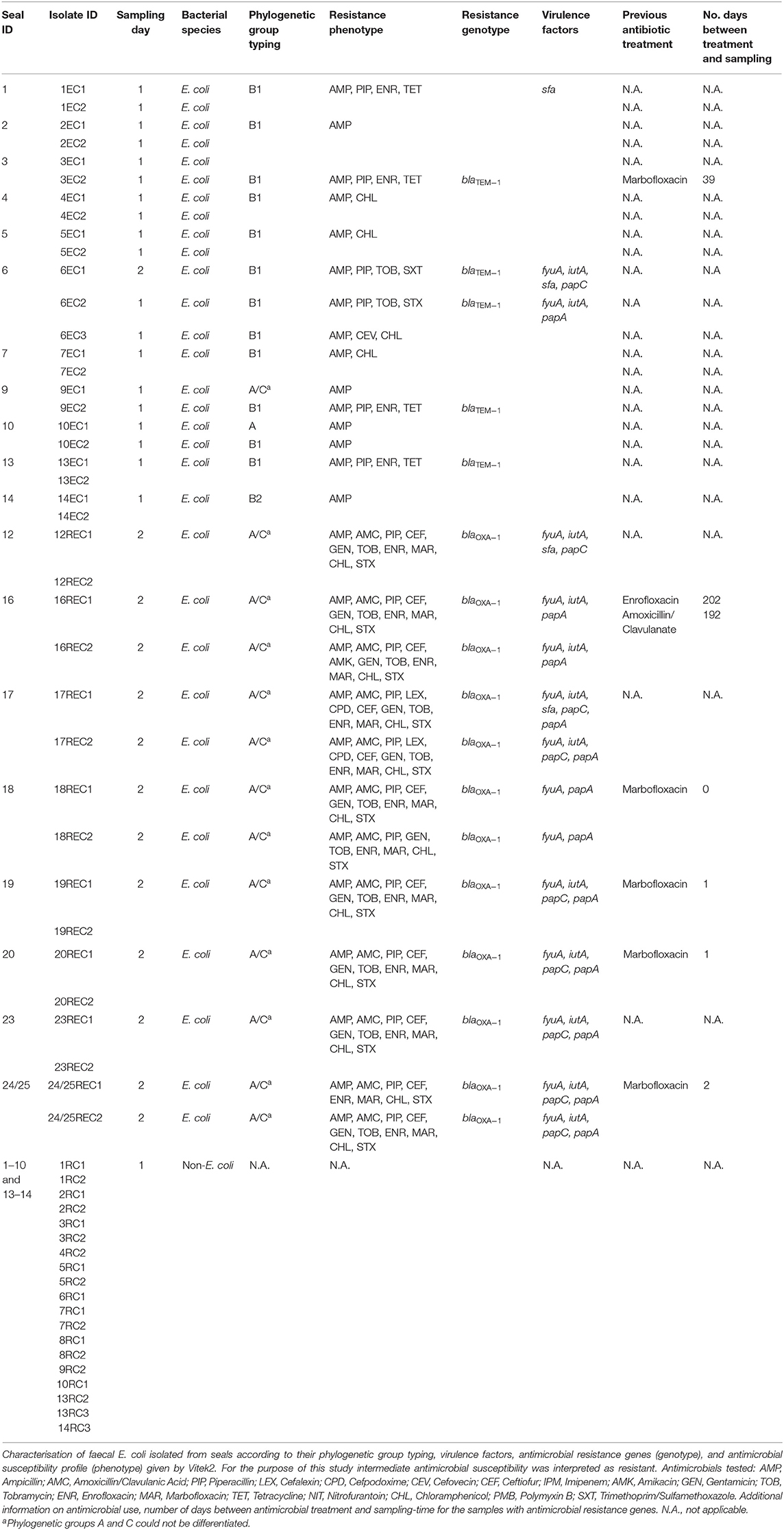
Table 1. Sampling details including animal identification, sampling day and bacteria isolated from faeces; E. coli isolated from TBX supplemented with cefotaxime (REC), E. coli isolated from TBX (EC) and non- E. coli isolated from TBX supplemented with cefotaxime (RC) according to sampling-day (3rd of July and 25th of July).
Sterile cotton swabs were used to collect freshly voided faeces from each animal's enclosure (individual pens with covered roof) without contacting the floor. Enclosures at SRI are cleaned daily; thorough washing and disinfection with bleach and Virkon© are performed before any new animal is moved into an enclosure.
Faecal swabs were kept refrigerated for a period no longer than 24 h before being processed in the laboratory.
At time of sampling, the age of animals ranged from 9 days to 10 months approximately. Samples were collected from animals between 24 h and 8 months after their arrival at the SRI facilities.
In the microbiology laboratory, the faecal swabs were placed into 20 mL sterile plastic tubes filled with 5 mL of buffered peptone water (BPW, Lab M) and vortexed for 10 s.
Aliquots of 0.1 mL of each initial suspension were plated onto the chromogenic selective medium Tryptone Bile X-glucuronide (TBX, Fisher Scientific) and TBX supplemented with cefotaxime (sc-202989 Cefotaxime Sodium Salt; 0.250 mg/L according to EUCAST epidemiological cut-off value (ECOFF) at the time of the study). TBX and TBX supplemented with cefotaxime were used to detect cefotaxime-susceptible and cefotaxime-resistant E. coli colonies (blue/green colonies), respectively. Sterile spreaders were used to evenly distribute the faecal suspension across the plates and then all plates were incubated at 37°C for a period of 20–24 h. Two to three colonies were isolated from each plate/sample if colonies differed phenotypically. E. coli ATCC 25922 and extended spectrum β-lactamase (ESBL)-producing isolate R5S (26) were used as negative and positive controls, respectively.
Thirty-nine E. coli isolates (selected from TBX and TBX supplemented with cefotaxime media) were grown on blood agar plates for 18 h at 37°C before testing for antimicrobial susceptibility by the VITEK 2 automated system (Biomerieux©), as recommended by the manufacturer (Table 1). Vitek 2 AST-GN65 cards (Biomerieux©) were used to investigate the susceptibility of the isolates to amoxicillin, ampicillin, amoxicillin and clavulanic acid, piperacillin, cefalotin, cefalexin, cefpodoxime, cefovecin, ceftiofur, imipenem, amikacin, gentamicin, tobramycin, enrofloxacin, marbofloxacin, tetracycline, nitrofurantoin, chloramphenicol, and trimethoprim/sulfamethoxazole. Results were interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. E. coli ATCC 25922 was used as control strain (27–31).
Rapid DNA extraction was performed on all isolates by the boiling method (32).
The phylogenetic group of 33 E. coli was investigated using an adapted version of the Clermont method (Table 2) (33). Each isolate was assigned to a group (A, B1, B2, C, D, E, F) according to the presence or absence of genes arpA, chuA, yjaA, and the DNA fragment TSPE4.C2 (33). Positive controls were provided by the Galway University Hospital National Microbiology Reference Laboratory.
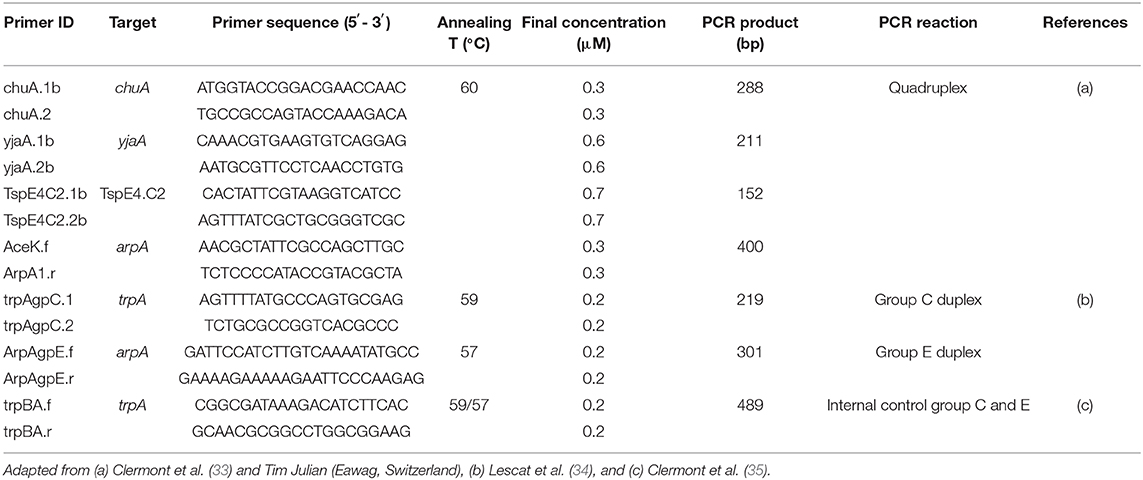
Table 2. List of primers, target genes, primer sequences, annealing temperatures, and primer concentrations used for E. coli phylogenetic group typing in this study.
Briefly, all PCR reactions were performed in a final volume of 25 μl containing 1× master mix [2× Qiagen Multiplex PCR Master Mix, final primer concentrations of 0.2–0.7 μM as appropriate (Table 2), PCR grade water] and 1.5 μl of bacterial lysate. PCR reactions were performed as follows: denaturation 15 min at 95°C, followed by 30 cycles of 20 s at 94°C, 20 s at 60°C, and 30 s at 72°C with a final extension step of 5 min at 72°C. PCR products were loaded on 2% agarose gels with SYBR® Safe DNA gel stain and run for 60 min at 100 V. DNA bands were visualised using a UV-transilluminator. For groups C and E, two further PCRs were performed using the previous protocol with 5× Q-Solution (Qiagen©) included in the master mix. PCR reactions were performed as follows: denaturation 15 min at 95°C, 30 cycles of 20 s at 94°C, 20 s at 59°C (group C) or 57°C (group E), respectively and 30 s at 72°C, with a final extension step of 5 min at 72°C.
Selected genes encoding virulence factors associated with adhesion (afaE8, papA, papC, and sfa), capsular antigen (kpsMFII), toxins (CNF1), and siderophores (fyuA and iutA) were also investigated as previously published (36–40). Positive PCR products were Sanger sequenced for identification of gene variants. The nucleotide sequence queries were loaded into the Virulence Factor Database (VFDB) (41).
Faecal samples collected on the first-sampling day did not yield any E. coli colonies that grew on medium supplemented with cefotaxime. However, 19 other colonies that were not E. coli were selected from this medium for further analysis (Table 1). Identification of these colonies was carried out by performing 16s rRNA PCR according to Marchesi et al. (42). PCR products (1,300 bp) were Sanger sequenced and the nucleotide sequence queries were loaded into the National Centre for Biotechnology Information (NCBI) Basic Local Alignment Search Tool (BLAST©). The highest query cover, identity and max score were used to determine the best fit for sequence alignment.
Thirty-three E. coli isolates yielding a phenotype of resistance to ampicillin (Table 1) were tested to see if they contained β-lactamase-coding genes SHV, TEM, and OXA (Multiplex I) while 16 isolates with susceptibility reported as intermediate or resistant to 3rd generation cephalosporins (Table 1) were further investigated for the presence of ESBL (CTX-M) (Multiplex II) and plasmid-mediated AmpC (ACC, FOX, MOX, CMY, DHA, LAT, ACT, BIL, MIR) (Multiplex III) encoding genes (32). Additionally, 19 cefotaxime-resistant non-E. coli isolates were investigated for the presence of β-lactamases. Positive controls were provided by the Galway University Hospital National Microbiology Reference Laboratory.
Briefly, for multiplex PCRs I, II and III, reactions were carried out in 25 μl of reaction mix containing master mix (2× Qiagen Multiplex PCR Master Mix, 5× Q-Solution (Qiagen), primers at concentration of 0.2–0.5 μM as appropriate, PCR grade water) and 1.0 μl of bacterial lysate. PCR reactions were performed as follows: denaturation 15 min at 95°C, 30 cycles of 30 s at 94°C, 90 s at 60°C, and 90 s at 72°C with a final extension step of 10 min at 72°C.
Agarose gels ranging between 1.2 and 2% (according to the size of PCR product) were run at 100 V for 60 min. A UV-transilluminator was used to visualise the PCR products.
Positive PCR products were Sanger sequenced. The nucleotide sequence queries were loaded into the Comprehensive Antibiotic Resistance Database (CARD) (43).
Faecal samples from 22 seals were collected over two sampling days. Samples were plated onto TBX and TBX supplemented with cefotaxime. After overnight incubation, samples were examined for growth and the results are shown in Table 1. E. coli were not retrieved from faeces sampled from 2 animals, while there was no E. coli growth on TBX supplemented with cefotaxime on samples collected on day 1. E. coli was recovered from all faecal samples from day 2 plated on TBX supplemented with cefotaxime.
In total, 39 E. coli isolates were investigated in the present study; 23 from the faeces collected on sampling-day 1 and 16 from sampling-day 2. All isolates were ampicillin-resistant, 16 of them were also resistant to amoxicillin and clavulanic acid (Table 1), while 22 of the isolates were intermediately susceptible or resistant to fluoroquinolones. From day 1 E. coli isolates, 10 (43.5%) were multidrug resistant (MDR) showing resistance to 3–4 different antimicrobial classes (penicillins, cephalosporins, tetracyclines, and potentiated sulphonamides) (44). In contrast, all E. coli isolates from sampling-day 2 were MDR, displaying resistance to 4–6 different antimicrobial classes including penicillins, fluoroquinolones, amphenicols, and potentiated sulphonamides (Table 1).
From a total of 39 E. coli isolates, 33 were selected for analysis by PCR and sequencing. Twenty-two carried β-lactamase encoding genes; blaTEM−1 was detected in six E. coli isolated from four seals on Day 1 while blaOXA−1 was detected in 16 E. coli isolated from nine animals on Day 2. Of four seals shedding TEM-1 E. coli, two originated from county Galway and one had been treated at SRI with marbofloxacin, 39 days before sampling. Of nine seals shedding E. coli carrying blaOXA−1, five had been medicated with marbofloxacin (Table 1). Reasons for medication included wounds, otitis, and umbilical abscess.
Additionally, the presence of CTX-M and AmpC encoding genes was investigated in the 16 isolates from day 2 (cefotaxime-resistant E. coli) using multiplex II and multiplex III PCRs; however, none of these genes was detected.
E. coli isolates belonged to phylogenetic groups A (n = 1), B1 (n = 13), B2 (n = 2), and A/C (n = 17) with the all 16 isolates from Day 2 belonging to group A/C. For isolates characterised as A/C it was not possible to further determine their phylogenetic group.
To further characterise the E. coli isolates investigation of selected virulence factors was performed. From day 1 samples, 3 of 17 E. coli isolates carried at least one virulence factor associated with adhesion (sfa, papA, and papC) and/or siderophores (fyuA and iutA) while all E. coli (16) isolated on the second sampling day carried at least two virulence factors. Some isolates carried multiple virulence factors including isolate 17REC1 that carried five of the eight virulence factors investigated.
Cefotaxime-resistant non-E. coli isolates were grown from samples collected on both sampling-days and 19 colonies recovered on the first sampling-day were selected for further investigation based on colony morphology.
16s rRNA PCR was used to amplify a specific region of the genome of each isolate. Despite the limitations of this method, sequence homology suggests that most of the isolates belong to the genera Leclercia, Enterobacter, Pantoea, and/or Psychrobacter (Supplementary File). Definitive species identification was not established. None of these isolates carried any of the β-lactamase encoding genes investigated (Table 1).
To the authors' knowledge this is the first study reporting the detection of blaOXA−1 and blaTEM−1 in phocid faecal E. coli in Europe. The presence of β-lactamase producing E. coli in the microbiota of wild seals, some of which had not been previously medicated with antimicrobials is a cause for concern and highlights their potential to serve as One Health sentinels when investigating AMR. There is also scope to explore zoonotic diseases including avian influenza and environmental contamination by heavy metals and domoic acid, among other things, using these species as sentinels.
All 39 E. coli isolated from seal faeces at the SRI marine rehabilitation centre were ampicillin resistant and 26 of 39 (66.6%) were MDR, which highlights the presence of MDR bacteria in the microbiome of marine mammals. Despite rigorous cleaning and disinfection protocols at the SRI centre, it cannot be proven that faecal isolates were not simply representative of the in-house flora of the centre however, the isolation of MDR E. coli from animals only recently arrived in the rescue centre strongly suggests that the organisms may have been present in the gastrointestinal tract of at least some of the seals before arrival at the centre. It is noteworthy that all E. coli (16) isolated on the second sampling-day were MDR which contrasts with 10 MDR E. coli of 23 E. coli recovered on the first sampling-day. Interestingly, the number of MDR E. coli isolates sampled on these two sampling-days differs considerably. The only major difference recorded between the 2 days was an outbreak of disease due to phocid herpes virus diagnosed soon after the second sampling-day. Whether this bears any relationship with the findings of this study is unclear. Research has shown that neurohormones released in the gut as a result of stressful events can increase the rate of horizontal gene transfer of genes encoding for AMR which can lead to an increase in shedding of resistant bacteria (45–47). Although little is known about the impact of acute viral infections on the composition and kinetics of the microbiome, these types of infections could be classified as systemic stressful events and therefore one could hypothesise that an increase in horizontal gene transfer may occur (48, 49). Fifty-six per cent of E. coli investigated showed resistance to fluoroquinolones, which may be associated with the use of marbofloxacin at the SRI, although not all isolates with resistance to fluoroquinolones came from animals with a history of marbofloxacin treatment.
In Ireland, a wide range of β-lactamases has been reported in bacteria isolated from humans, companion animals, production animals and wastewaters (26, 50–54). Karczmarczyk et al. (52) identified blaTEM in 89.2% of the E. coli examined in their study while blaOXA were detected in 1.35%. Additionally, Carroll et al. (55) identified blaTEM in faecal E. coli sampled from one Irish herring gull and one Irish black-headed gull while another study identified blaCTX−M group 1 in 4.5% of the samples collected from Irish gulls (55, 56). In line with this, isolates with either blaOXA or blaTEM were identified in the faecal samples collected, suggesting these genes are circulating in the marine environment also. ESBL-producing E. coli have been identified in more than 30 wild animal species (11) but none were identified in the E. coli or non-E. coli isolates investigated in this study. While this does not rule out their presence, it may suggest that these genes are not as prevalent in the environment of these seals as the ones that were identified.
This study further characterised E. coli isolated from seals in terms of phylogenetic groups and virulence factors. Due to the lack of information in the literature, a subset of virulence factors was selected for investigation, based on data available for E. coli isolated from domestic animals. In this study, virulence factors associated with adhesion and siderophores were detected in many isolates. Despite the constraints of the small number of animals investigated and further bias by the selection criteria of E. coli (1, 2, or 3 colonies per sample according to colony phenotype), a difference between the number of E. coli carrying virulence genes on each sampling day is clear. Horizontal gene transfer is an essential mechanim for the spread of virulence determinants between different bacterial strains and species (57). Moreover, studies have shown that stress can induce the release of norepinephrine in the gut and this catecholamine can promote horizontal gene transfer by conjugation and influence the production of virulence factors including toxins and adhesins in E. coli (46, 58–61). It is possible that the differences reported between sampling day 1 and day 2 were triggered by the herpes virus infection that was subsequently diagnosed (48, 62).
Studies have shown that ecological niches and life events impact the phylogenetic group dynamics and diversity of E. coli (63, 64). In the present study, phylogenetic groups A, B1, B2, and A/C were detected on the first sampling day while on the second sampling day only A/C E. coli were detected. Further characterisation of A/C isolates was not possible due to non-specific DNA amplification. Differences in the antimicrobial susceptibility profile and virulence factors exclude the possibility of clonal spread of A/C E. coli on the second sampling day. These suprising findings, including the number of MDR isolates, number of E. coli carrying β-lactamases and virulence factors and phylogenetic diversity detected on two different sampling days suggest differences in the population sampled on these two occasions again pointing to the impact a natural herpesvirus infection could have had on the profile of the samples. These data highlight the importance of examining the resistome of sentinal species throughout time.
Phocid faecal cefotaxime-resistant non-E. coli isolates were homologous to members of the Enterobacterales: Leclercia, Pantoea, and Enterobacter. Leclercia adecarboxylata is an opportunistic pathogen associated with water affecting both immunocompromised and immunocompetent patients (65). Studies have reported Leclercia adecarboxylata susceptibility to cephalosporins and blaSHV−12 has been identified in Leclercia adecarboxylata clinical samples (66, 67). Pantoea agglomerans may cause infections in humans and is variably susceptible to antimicrobials while Enterobacter ludwigii, previously included in the Enterobacter cloacae complex, is a MDR bacterium that can carry β-lactamase encoding genes (68–71). Because all the above bacteria belong to the Enterobacterales order, distinction between species is complex. For more accurate identification, PCR protocols investigating genetic characteristics other than 16S rRNA would be required.
Despite the fact that animals sampled in this study represented all live stranded seals in Ireland housed at SRI during the period of the trial, the relatively small sample size is a limitation of this study. A larger population would have given a better idea of the magnitude of the problem, but it is clear that even with a small sample size, this study has pinpointed issues and has provided justification and a roadmap for future studies in this area. Stringent cleaning and disinfection and other infection control protocols in place at SRI and a rigorous sampling technique greatly reduced the possibility of cross-contamination between different enclosures/pens.
It is difficult to determine the exact origin of β-lactamase encoding genes circulating in the population of young Irish seals as they share their costal habitat with different species, but the fact that blaTEM−1 and blaOXA−1 were present in their faeces and MDR E. coli were frequently detected, is concerning. The presence of β-lactamases jeopardises the use of critically important antimicrobials including penicillins and cephalosporins and the findings of this study indicate the spread of AMR mechanisms to bacteria in coastal areas.
Marine mammals can act as reservoirs, vectors, and bioindicators of resistant bacteria and AMR genes in the environment (72, 73). Treated and untreated sewage, hospital waste, aquaculture discharges and agricultural runoff provide means to deliver antibiotics, pollutants and resistant bacteria to the aquatic environment, thus playing a major role in driving ARG transfer, ecology, and evolution (14). Additionally, seals can interact with other marine wildlife and birds and engage in the transfer of ARB between populations across large parts of the world. Future investigations should acknowledge the presence of these issues and seek to understand the movement of ARGs between populations and the extent to which global spread of ARGs in human populations is reflected in wild animal populations.
This study shows that some isolates of E. coli carried β-lactamase encoding genes (blaOXA−1 or blaTEM−1) as well as virulence factors associated with adhesion (sfa, papA, and papC) and/or siderophores (fyuA and iutA). While harbour seals have the potential to migrate to different locations, they tend to return to the same breeding grounds (2) and young pups similar to the ones sampled in this study do not tend to migrate long distances. The presence of MDR bacteria in the seal pups indicates that were probably acquired locally, however, it is also possible that the adult seals or other migratory wildlife in the area may have acquired these resistant bacteria elsewhere, brought them to Ireland and passed them to the pups. At this point, although it is difficult to identify the geographic source exactly, the data presented in this study clearly establishes the presence of MDR E. coli circulating in the Irish marine environment at the time of sampling.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethical review and approval was not required for the animal study because the seals were not manipulated for the purpose of collecting faecal swabs as these were sampled from the floor. This study was part of a bigger project that got exemption from full ethical review according to the University College Dublin Animal Research Ethics Committee (AREC-E-17-24-Barry).
AV designed the study, conducted the experiments, and wrote the manuscript. LS and JC designed the study and conducted the experiments. FL and GB designed the study and critically reviewed the manuscript. All authors contributed to the manuscript and approved the submitted version.
This study was supported by the National Institutes of Medicine grant number 5T35OD010956 and by the University College Dublin School of Veterinary Medicine student scholarship scheme.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank the Seal Rescue Ireland staff for all their assistance during this study.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.583759/full#supplementary-material
1. Cronin MA. The conservation of seals in Irish waters: how research informs policy. Mar Policy. (2011) 35:748–55. doi: 10.1016/j.marpol.2011.01.006
2. Bonner W. The grey and common seal in European waters. Oceangr Mar Biol Ann Rev. (1972) 10:461–507.
3. Bossart GD. Marine mammals as sentinel species for oceans and human health. Vet Pathol. (2011) 48:676–90. doi: 10.1177/0300985810388525
4. Reif JS. Animal sentinels for environmental and public health. Public Health Rep. (2011) 126:50–7. doi: 10.1177/00333549111260S108
5. Robinson TP, Bu DP, Carrique-Mas J, Fèvre EM, Gilbert M, Grace D, et al. Antibiotic resistance is the quintessential One Health issue. Trans R Soc Trop Med Hyg. (2016) 110:377–80. doi: 10.1093/trstmh/trw048
6. WHO, FAO, OIE. The FAO-OIE-WHO Collaboration Sharing Responsibilities and Coordinating Global Activities to Address Health Risks at the Animal-Human-Ecosystems Interfaces. WHO, FAO, OIE (2010).
8. Allen HK, Donato J, Wang HH, Cloud-Hansen KA, Davies J, Handelsman J. Call of the wild: antibiotic resistance genes in natural environments. Nat Rev Microbiol. (2010) 8:251–9. doi: 10.1038/nrmicro2312
9. Baquero F, Martínez JL, Cantón R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol. (2008) 19:260–5. doi: 10.1016/j.copbio.2008.05.006
10. Taylor NGH, Verner-Jeffreys DW, Baker-Austin C. Aquatic systems: maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol Evol. (2011) 26:278–84. doi: 10.1016/j.tree.2011.03.004
11. Wellington EMH, Boxall ABA, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. (2013) 13:155–65. doi: 10.1016/S1473-3099(12)70317-1
12. Leonard AFC, Zhang L, Balfour AJ, Garside R, Gaze WH. Human recreational exposure to antibiotic resistant bacteria in coastal bathing waters. Environ Int. (2015) 82:92–100. doi: 10.1016/j.envint.2015.02.013
13. Zhang XX, Zhang T, Fang HHP. Antibiotic resistance genes in water environment. Appl Microbiol Biotechnol. (2009) 82:397–414. doi: 10.1007/s00253-008-1829-z
14. Marti E, Variatza E, Balcazar JL. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. (2014) 22:36–41. doi: 10.1016/j.tim.2013.11.001
15. Li XZ, Mehrotra M, Ghimire S, Adewoye L. β-Lactam resistance and β-lactamases in bacteria of animal origin. Vet Microbiol. (2007) 121:197–214. doi: 10.1016/j.vetmic.2007.01.015
16. De Oliveira D, Forde B, Kidd T, Harris P, Schembri M, Beatson S, et al. Antimicrobial resistance in ESKAPE pathogens. Clin Microbiol Rev. (2020) 33:e00181-19. doi: 10.1128/CMR.00181-19
17. Daga A, Koga V, Soncini J, Matos C, Marcia P, Pelisson M, et al. Escherichia coli bloodstream infections in patients at a University hospital: virulence factors and clinical characteristics. Front Cell Infect Microbiol. (2019) 9:191. doi: 10.3389/fcimb.2019.00191
18. Morris D, Harris S, Morris C, Commins E, Cormican M. Hospital Effluent: Impact on the Microbial Environment and Risk to Human Health. Dublin: EPA (2015).
19. Mahon BM, Brehony C, McGrath E, Killeen J, Cormican M, Hickey P, et al. Indistinguishable NDM-producing Escherichia coli isolated from recreational waters, sewage, and a clinical specimen in Ireland, 2016 to 2017. Eurosurveillance. (2017) 22:1–5. doi: 10.2807/1560-7917.ES.2017.22.15.30513
20. Kümmerer K. Antibiotics in the aquatic environment–a review–part, I. Chemosphere. (2009) 75:417–34. doi: 10.1016/j.chemosphere.2008.11.086
21. Gullberg E, Cao S, Berg OG, Ilbäck C, Sandegren L, Hughes D, et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog. (2011) 7:e1002158. doi: 10.1371/journal.ppat.1002158
22. Bruno C, Albino B, François-Xavier M. Wildlife, exotic pets, and emerging zoonoses. Emerg Infect Dis. (2009) 13:1–7. doi: 10.3201/eid1301.060480
23. Greig J, Rajić A, Young I, Mascarenhas M, Waddell L, LeJeune J. A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health. (2015) 62:269–84. doi: 10.1111/zph.12147
24. Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Herman L, et al. Broad-spectrum β-lactamases among Enterobacteriaceae of animal origin: molecular aspects, mobility and impact on public health. FEMS Microbiol Rev. (2009) 34:295–316. doi: 10.1111/j.1574-6976.2009.00198.x
25. Guenther S, Ewers C, Wieler LH. Extended-spectrum beta-lactamases producing E. coli in wildlife, yet another form of environmental pollution? Front Microbiol. (2011) 2:246. doi: 10.3389/fmicb.2011.00246
26. Wang J, Gibbons JF, Mcgrath K, Bai L, Li F, Leonard F, et al. Molecular characterization of bla ESBL-producing Escherichia coli cultured from pig farms in Ireland. J Antimicrob Chemother. (2016) 71:3062–5. doi: 10.1093/jac/dkw278
27. CLSI. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. CLSI Document M31-A3. Wayne: Clinical and Laboratory Standards Institute (2008).
28. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Nineteenth Informational Supplement; CLSI Document M100-S19. Wayne: Clinical and Laboratory Standards Institute (2009).
29. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Second Informational Supplement; CLSI Document M100-S22. Wayne: Clinical and Laboratory Standards Institute (2011).
30. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth Informational Supplement; CLSI Document M100-S25. Wayne: Clinical and Laboratory Standards Institute (2015).
31. Stegemann MR, Passmore CA, Sherington J, Lindeman CJ, Papp G, Weigel DJ, et al. Antimicrobial activity and spectrum of cefovecin, a new extended- spectrum cephalosporin, against pathogens collected from dogs and cats in Europe and North America. Antimicrob Agents Chemother. (2006) 50:2286–92. doi: 10.1128/AAC.00077-06
32. Dallenne C, Da Costa A, Decré D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J Antimicrob Chemother. (2010) 65:490–5. doi: 10.1093/jac/dkp498
33. Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. (2013) 5:58–65. doi: 10.1111/1758-2229.12019
34. Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, et al. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ Microbiol Rep. (2013) 5:49–57.
35. Clermont O, Lescat M, O'Brien CL, Gordon DM, Tenaillon O, Denamur E. Evidence for a human-specific Escherichia coli clone. Environ Microbiol. (2008) 10:1000–6. doi: 10.1111/j.1462-2920.2007.01520.x
36. Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol. (1995) 12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x
37. Gérardin J, Lalioui L, Jacquemin E, Le Bouguénec C, Mainil JG. The afa-related gene cluster in necrotoxigenic and other Escherichia coli from animals belongs to the afa-8 variant. Vet Microbiol. (2000) 76:175–84. doi: 10.1016/S0378-1135(00)00234-0
38. Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. (2000) 181:261–72. doi: 10.1086/315217
39. Tramuta C, Robino P, Nucera D, Salvarani S, Banche G, Malabaila A, et al. Molecular characterization and antimicrobial resistance of faecal and urinary Escherichia coli isolated from dogs and humans in Italy. Vet Ital. (2014) 50:23–30. doi: 10.12834/VetIt.1304.09
40. Liu X, Liu H, Li Y, Hao C. Association between virulence profile and fluoroquinolone resistance in Escherichia coli isolated from dogs and cats in China. J Infect Dev Ctries. (2017) 11:306–13. doi: 10.3855/jidc.8583
41. Chen L, Zheng D, Liu B, Yang J, Jin Q. VFDB 2016: Hierarchical and refined dataset for big data analysis−10 years on. Nucleic Acids Res. (2016) 44:D694–7. doi: 10.1093/nar/gkv1239
42. Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. (1998) 64:795–9. doi: 10.1128/AEM.64.2.795-799.1998
43. Jia B, Raphenya AR, Alcock B, Alcock B, Waglechner N, Guo P, et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2017) 45:D566–73. doi: 10.1093/nar/gkw1004
44. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2011) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
45. Morro M, Beran G, Hoffman L, Griffith R. Effects of cold stress on the antimicrobial drug resistance of Escherichia coli of the intestinal flora of swine. Lett Appl Microbiol. (1998) 27:251–4. doi: 10.1046/j.1472-765X.1998.t01-13-00449.x
46. Peterson G, Kumar A, Gart E, Narayanan S. Catecholamines increase conjugative gene transfer between enteric bacteria. Microb Pathog. (2011) 51:1–8. doi: 10.1016/j.micpath.2011.03.002
47. Verbrugghe E, Boyen F, Gaastra W, Bekhuis L, Leyman B, Van Parys A, et al. The complex interplay between stress and bacterial infections in animals. Vet Microbiol. (2012) 155:115–27. doi: 10.1016/j.vetmic.2011.09.012
48. Yildiz S, Mazel-Sanchez B, Kandasamy M, Manicassamy B, Schmolke M. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. (2018) 6:1–17. doi: 10.1186/s40168-017-0386-z
49. Yuan L, Hensley C, Mahsoub H, Ramesh A, Zhou P. Microbiota in viral infection and disease in humans and farm animals. Prog Mol Biol Transl Sci. (2020) 171:15–60. doi: 10.1016/bs.pmbts.2020.04.005
50. Morris D, O'Hare C, Glennon M, Maher M, Corbett-Feeney G, Cormican M. Extended-spectrum beta-lactamases in Ireland, including a novel enzyme, TEM-102. Antimicrob Agents Chemother. (2003) 47:2572–8. doi: 10.1128/AAC.47.8.2572-2578.2003
51. Galvin S, Boyle F, Hickey P, Vellinga A, Morris D, Cormican M. Enumeration and characterization of antimicrobial-resistant escherichia coli bacteria in effluent from municipal, hospital, and secondary treatment facility sources. Appl Environ Microbiol. (2010) 76:4772–9. doi: 10.1128/AEM.02898-09
52. Karczmarczyk M, Abbott Y, Walsh C, Leonard N, Fanning S. Characterization of multidrug-resistant Escherichia coli isolates from animals presenting at a University Veterinary Hospital. Appl Environ Microbiol. (2011) 77:7104–12. doi: 10.1128/AEM.00599-11
53. Burke L, Humphreys H, Fitzgerald-Hughes D. The revolving door between hospital and community: extended-spectrum beta-lactamase-producing Escherichia coli in Dublin. J Hosp Infect. (2012) 81:192–8. doi: 10.1016/j.jhin.2012.04.021
54. Burke L, Humphreys H, Fitzgerald-Hughes D. The molecular epidemiology of resistance in cefotaximase-producing Escherichia coli clinical isolates from Dublin, Ireland. Microb Drug Resist. (2016) 22:552–8. doi: 10.1089/mdr.2015.0154
55. Carroll D, Wang J, Fanning S, McMahon BJ. Antimicrobial resistance in wildlife: implications for public health. Zoonoses Public Health. (2015) 62:534–42. doi: 10.1111/zph.12182
56. Stedt J, Bonnedahl J, Hernandez J, Waldenström J, McMahon BJ, Tolf C, et al. Carriage of CTX-M type extended spectrum β-lactamases (ESBLs) in gulls across Europe. Acta Vet Scand. (2015) 57:74. doi: 10.1186/s13028-015-0166-3
57. Leimbach A, Hacker J, Dobrindt U. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol. (2013) 358:3–32. doi: 10.1007/82_2012_303
58. O'Donnell PM, Aviles H, Lyte M, Sonnenfeld G. Enhancement of in vitro growth of pathogenic bacteria by norepinephrine : importance of inoculum density and role of transferrin. Appl Environ Microbiol. (2006) 72:5097–9. doi: 10.1128/AEM.00075-06
59. Freestone PPE, Sandrini SM, Haigh RD, Lyte M. Microbial endocrinology: how stress influences susceptibility to infection. Trends Microbiol. (2007) 16:55–64. doi: 10.1016/j.tim.2007.11.005
60. Lyte M, Freestone PPE. Microbial Endocrinology: Interkingdom Signaling in Infectious Disease and Health. New York, NY: Springer-Verlag (2010).
61. Lyte M. Microbial endocrinology in the pathogenesis of infectious disease. Microbiol Spectr. (2016) 4:1–24. doi: 10.1128/microbiolspec.VMBF-0021-2015
62. Zhao N, Wang S, Li H, Liu S, Meng L, Luo M, et al. Influence of novel highly pathogenic avian influenza A (H5N1) virus infection on migrating whooper swans fecal microbiota. Front Cell Infect Microbiol. (2018) 8:46. doi: 10.3389/fcimb.2018.00046
63. Gordon DM, Cowling A. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology. (2003) 149:3575–86. doi: 10.1099/mic.0.26486-0
64. Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. (2010) 8:207–17. doi: 10.1038/nrmicro2298
65. Keren Y, Keshet D, Eidelman M, Geffen M, Raz-Pasteur A, Hussein K. Is leclercia adecarboxylata a new and unfamiliar marine pathogen? J Clin Microbiol. (2014) 52:1775–6. doi: 10.1128/JCM.03239-13
66. Mazzariol A, Zuliani J, Fontana R, Cornaglia G. Isolation from blood culture of a leclercia adecarboxylata strain producing an SHV-12 extended-spectrum beta-lactamase. J Clin Microbiol. (2003) 41:1738–9. doi: 10.1128/JCM.41.4.1738-1739.2003
67. Stock I, Burak S, Wiedemann B. Natural antimicrobial susceptibility patterns and biochemical profiles of Leclercia decarboxylata strains. Clin Microbiol Infect. (2004) 10:724–33. doi: 10.1111/j.1469-0691.2004.00892.x
68. Cruz AT, Cazacu AC, Allen CH. Pantoea agglomerans, a plant pathogen causing human disease. J Clin Microbiol. (2007) 45:1989–92. doi: 10.1128/JCM.00632-07
69. Deletoile A, Decre D, Courant S, Passet V, Audo J, Grimont P, et al. Phylogeny and identification of pantoea species and typing of pantoea agglomerans strains by multilocus gene sequencing. J Clin Microbiol. (2009) 47:300–10. doi: 10.1128/JCM.01916-08
70. Khajuria A, Praharaj AK, Grover N, Kumar M. First report of an enterobacter ludwigii isolate coharboring NDM-1. Antimicrob Agents Chemother. (2013) 57:5189–90. doi: 10.1128/AAC.00789-13
71. Flores-Carrero A, Labrador I, Paniz-Mondolfi A, Peaper DR, Towle D, Araque M. Nosocomial outbreak of extended-spectrum β-lactamase-producing Enterobacter ludwigii co-harbouring CTX-M-8, SHV-12 and TEM-15 in a neonatal intensive care unit in Venezuela. J Glob Antimicrob Resist. (2016) 7:114–8. doi: 10.1016/j.jgar.2016.08.006
72. Literak I, Dolejska M, Radimersky T, Klimes J, Friedman M, Aarestrup FM, et al. Antimicrobial-resistant faecal Escherichia coli in wild mammals in central Europe: multiresistant Escherichia coli producing extended-spectrum beta-lactamases in wild boars. J Appl Microbiol. (2009) 108:1702–11. doi: 10.1111/j.1365-2672.2009.04572.x
Keywords: antimicrobial resistance, β-lactamases, One Health, seals, virulence factors, E. coli
Citation: Vale AP, Shubin L, Cummins J, Leonard FC and Barry G (2021) Detection of blaOXA-1, blaTEM-1, and Virulence Factors in E. coli Isolated From Seals. Front. Vet. Sci. 8:583759. doi: 10.3389/fvets.2021.583759
Received: 15 July 2020; Accepted: 26 January 2021;
Published: 03 March 2021.
Edited by:
Marina Spinu, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Geetanjali Singh, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaCopyright © 2021 Vale, Shubin, Cummins, Leonard and Barry. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ana P. Vale, YW5hLnZhbGVAdWNkLmll
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.