
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 16 December 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.614513
 Vanessa C. Johanns1
Vanessa C. Johanns1 Lennard Epping2
Lennard Epping2 Torsten Semmler2
Torsten Semmler2 Fereshteh Ghazisaeedi3
Fereshteh Ghazisaeedi3 Antina Lübke-Becker3
Antina Lübke-Becker3 Yvonne Pfeifer4
Yvonne Pfeifer4 Inga Eichhorn3
Inga Eichhorn3 Roswitha Merle5
Roswitha Merle5 Astrid Bethe3
Astrid Bethe3 Birgit Walther1*†
Birgit Walther1*† Lothar H. Wieler6†
Lothar H. Wieler6†To prevent economic losses due to post-weaning diarrhea (PWD) in industrial pig production, zinc (Zn) feed additives have been widely used, especially since awareness has risen that the regular application of antibiotics promotes buildup of antimicrobial resistance in both commensal and pathogenic bacteria. In a previous study on 179 Escherichia coli collected from piglets sacrificed at the end of a Zn feeding trial, including isolates obtained from animals of a high-zinc fed group (HZG) and a corresponding control group (CG), we found that the isolate collection exhibited three different levels of tolerance toward zinc, i.e., the minimal inhibitory concentration (MIC) detected was 128, followed by 256 and 512 μg/ml ZnCl2. We further provided evidence that enhanced zinc tolerance in porcine intestinal E. coli populations is clearly linked to excessive zinc feeding. Here we provide insights about the genomic make-up and phylogenetic background of these 179 E. coli genomes. Bayesian analysis of the population structure (BAPS) revealed a lack of association between the actual zinc tolerance level and a particular phylogenetic E. coli cluster or even branch for both, isolates belonging to the HZG and CG. In addition, detection rates for genes and operons associated with virulence (VAG) and bacteriocins (BAG) were lower in isolates originating from the HZG (41 vs. 65% and 22 vs. 35%, p < 0.001 and p = 0.002, resp.). Strikingly, E. coli harboring genes defining distinct pathotypes associated with intestinal disease, i.e., enterotoxigenic, enteropathogenic, and Shiga toxin-producing E. coli (ETEC, EPEC, and STEC) constituted 1% of the isolates belonging to the HZG but 14% of those from the CG. Notably, these pathotypes were positively associated with enhanced zinc tolerance (512 μg/ml ZnCl2 MIC, p < 0.001). Taken together, zinc excess seems to influence carriage rates of VAGs and BAGs in porcine intestinal E. coli populations, and high-zinc feeding is negatively correlated with enteral pathotype occurrences, which might explain earlier observations concerning the relative increase of Enterobacterales considering the overall intestinal microbiota of piglets during zinc feeding trials while PWD rates have decreased.
The gastrointestinal microbiota of pigs is a large, multifaceted and complex microbial community which has been estimated to comprise of 1010-1011 bacteria per gram of gut content (1). Different stress factors are capable of altering the healthy gut microbiome of a piglet, e.g., weaning from the sow, which is among the most stressful events in a piglet's life (2): The prompt change in diet, social and environmental life conditions affects the gut microbiota in composition and structure and prevalently enhances vulnerability for onset of enteral post-weaning infections (3). Considering feed additives, different products are marketed to assist in boosting the pigs' immune system, regulate gut microbiota, and reduce negative impacts of weaning and other environmental challenges, including pharmacological levels of zinc and copper (4). However, a recent review concluded that it is not possible to recommend a specific additive that will have positive effects in all diets, since the effects strongly depend on the health status of the animals (4). Therefore, maintaining a physiological/healthy gut microbiota composition and equilibrium during and after weaning is important to prevent attachment, proliferation and spread of pathogenic microorganisms, especially enterotoxigenic Escherichia coli, which is a frequent cause of post-weaning diarrhea (PWD) in piglets (5).
E. coli causing intestinal diseases usually express virulence factors which induce and/or support an inflammation of the gut often leading to diarrhea, while the number of intestinal commensals decreases (6). Complex mechanisms of interaction between commensal and pathogenic bacteria have evolved within the gut, including competition for nutrients, shielding from the activated enteral immune response and induction of competitor-eliminating bacteriocin production (7). These bacteriocins are polypeptide toxins comprising colicins and microcins which exhibit a broad range of different cytotoxic mechanisms (8–10). Both colicins and microcins are capable of killing a narrow spectrum of competing Enterobacterales, including other E. coli lineages (9). Porcine pathogenic E. coli have been shown to produce predominantly colicins, especially colicin BM and Ib (11).
To prevent or mend PWD which is most commonly caused by enterotoxigenic E. coli (12), high-level dietary zinc oxide supplementation is used in the pig production sector in different parts of the world (5, 13, 14). Although the particular effects of zinc on the enteral microbiota are not fully understood yet, piglets fed with high-zinc supplemented diets clearly showed changes with respect to the overall composition and abundance of distinct gut-associated bacteria (15–17), which are probably associated with bacteriocin-producing bacteria as well. We therefore hypothesized that the occurrence of genes encoding bacteriocins might have an impact on the composition of the E. coli population in zinc-fed piglets.
The manifestation of clinical symptoms and pathology of E. coli-induced enteral diseases is closely associated with the occurrence of certain virulence associated genes (VAGs) (18, 19). Mainly based on the presence of distinct VAGs but also additional, especially phenotypical characteristics, diarrheagenic E. coli of importance for the pig production sector are often classified as enterotoxigenic E. coli (ETEC). Furthermore, enteropathogenic E. coli (EPEC), Shiga toxin-producing E. coli (STEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) have been reported as a probable cause of diarrhea in pigs as well (5, 19–21).
While we have recently shown that high-zinc supplemented diets foster accumulation of E. coli associated with increased zinc-tolerance in weaned piglets (22), the effects of that particular feed additive on the occurrence of VAGs and bacteriocin associated genes (BAGs) among the intestinal E. coli population of these piglets were not investigated so far. In the current study, we analyzed the phylogenetic relationship and make up of a broad collection of E. coli obtained from a former piglet zinc-feeding trial together with strains isolated from pigs suffering from clinical disease in order to investigate the presence of a putative correlation between feeding group and the zinc tolerance level of the isolates with the occurrence of VAGs and/or BAGs.
The representative set of E. coli isolates characterized here was selected based on a previous feeding trial (23) carried out in accordance with the principles of the Basel Declaration following the institutional and national guidelines for the care and use of animals. The protocol was approved by the local state office of occupational health and technical safety “Landesamt für Gesundheit und Soziales, Berlin” (LaGeSo Reg. Nr. 0296/13) as described before (23).
Briefly, 32 landrace piglets of regular commercial origin weaned at day 25 ± 1 were separated into two groups for 4 weeks: the first group of piglets, designated here as the high-zinc group (HZG) was fed with a diet supplemented with a comparatively high amount of zinc oxide (2,103 mg zinc/kg diet), while the second group served as the control group. This control group (CG) received a common piglet diet containing a concentration of zinc oxide (72 mg zinc/kg diet) sufficient to meet the nutritional requirements to avoid trace metal malnutrition (23). The trial started with 32 piglets, which were sacrificed mid-trial (38 ± 2 days of age, n = 16, 8 per group) and at the trial's end (52 ± 2 days of age, n = 16, 8 per group). No symptoms of disease were observed within the two feeding groups during the entire trial. Here we focus on E. coli obtained from three different sampling sites (feces, digesta and mucosa obtained from the colon ascendens) of those piglets sacrificed at the end (52 ± 2 days of age; n = 16) of the feeding trial. In total, 179 E. coli were collected as a stratified random sample (HZG = 99, CG = 80), with an average number of 11 E. coli isolates investigated per piglet (all three sampling sites). These isolates were previously studied with regard to their respective zinc tolerance levels and susceptibility patterns for antibiotics and biocides (22).
To compare, we chose six additional porcine strains isolated from clinically ill piglets [diarrhea (n = 4), edema disease (n = 2)] available in the strain collection of the Institute of Microbiology and Epizootics (IMT) as representatives for non-commensal isolates and a high-zinc tolerant isolate from a clinical human case (RKI6122) (24).
E. coli were sequenced using Illumina MiSeq® 300 bp paired-end whole genome sequencing (WGS) with an obtained coverage of >90x. The Illumina short reads were hybrid assembled using unicycler v4.4 (25). Adapter-trimmed reads were used for de novo assembly into contiguous sequences (contigs) and subsequently into scaffolds using SPAdes v3.11. All draft genomes were annotated using Prokka v1.14.5 (26). Illumina raw read data sequenced for this study is available at NCBI under Bioproject ID PRJNA552271. The determination of the maximum common genome (MCG) (27) alignment was done comprising those genes present in all 179 genomes. To obtain this, we clustered the coding sequences based on the parameters sequence similarity (min. 90%) and coverage (min. 90%) and defined the genes that were present in each genome, fulfilling the threshold parameters as MCG. This resulted in 2,804 orthologous genes that we used for the comparisons. We extracted the allelic variants of these genes from all genomes by a BLAST-based approach, aligned them individually for each gene and concatenated them, which result in an alignment of 2.762 Mbp for these 179 strains. This alignment was used to generate a phylogenetic tree with RAxML v 8.2.10 (28) using a General Time Reversible model and gamma correction for among site rate variation. Bayesian analysis of population structure (BAPS) (29) was applied to identify genetically distinct linages based on the constructed phylogeny.
An in-house BLAST-pipeline with the general gene identity threshold of 95 and 90% minimum coverage was used to identify 25 VAGs selected because of their (putative) importance in intestinal pathogenicity (Table 1). Further genotype characterization included the determination of multilocus sequence type (ST) and sequence type complex (STC) using MLST 2.0 (35), and serotype prediction using SerotypeFinder 2.0 (36).
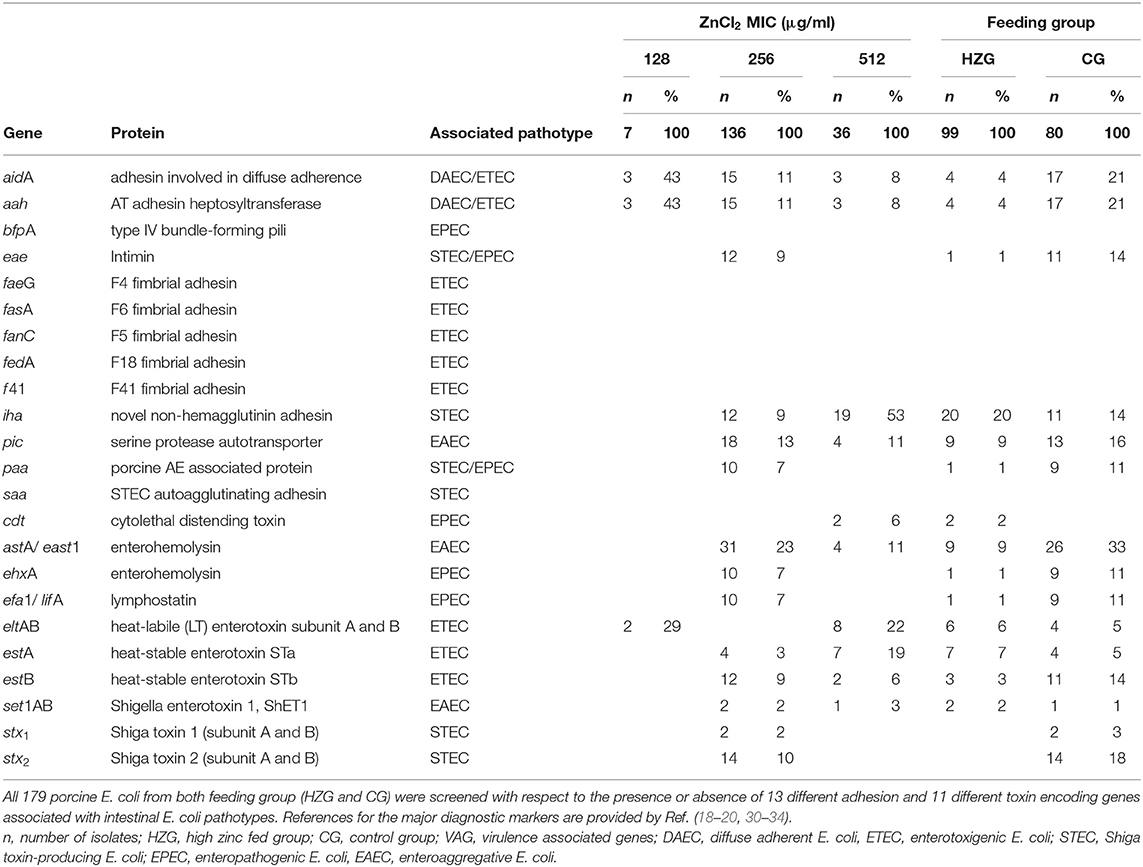
Table 1. Occurrence and distribution of VAGs associated with different types of intestinal pathogenic E. coli of relevance for pigs.
The screening procedure for bacteriocin associated genes included colicin types depending on either a Tol-dependent translocation system (group A) such as colicin A, E1–E4, E6–E9, K, N, S4, U, Y or a Ton system (group B) including colicin B, Ia, Ib, E5, E7, 5, 10, G, H, Js, D, and M (37–41). An in-house BLAST-pipeline was set-up for each of the corresponding genes with the general identity threshold of 95 and 90% minimum gene coverage (Supplementary Figure 1) (9, 41).
Each gene encoding a bacteriocin (colicin, microcin) was further investigated with respect to its predicted amino acid (aa) sequence coverage and identity using corresponding reference/prototype aa sequences of the particular E. coli protein from NCBI (Supplementary Figure 1). An in silico comparative analysis was performed on these aa sequences using Geneious Prime (version 2019.0.04).
A detailed overview of the characteristics of all animal trial isolates and the strains included for comparative purposes are provided in Supplementary Tables 1, 2.
Data were analyzed using SPSS software version 25.0 (IBM, New York, NY, USA). P-values < 0.05 were considered statistically significant.
A mixed-model regression approach was used to test whether the feeding group and E. coli ZnCl2 MICs had an effect on the total number of different VAGs, with the individual pig as random factor. Since the dependent variable showed a Poisson distribution, Poisson regression and logarithmic link function were applied, with feeding group and ZnCl2 MICs (< vs. > = 512) included as factors into the model. Interactions were tested and removed when the p-value was > 0.05. Variance components analysis was used to determine the proportion of variance that accounted for differences between individual animals. In a second mixed logistic regression model, the individual effects of the above mentioned influence factors on the presence of major diagnostic markers was tested. Interactions were also tested as described above.
Considering the overall phylogenetic diversity, we identified 2,804 orthologous genes representing the “maximum common genome” (MCG) of 186 genomic sequences representing isolates from both feeding groups (HZG = 99, CG = 80), six isolates previously collected from diagnostic samples of severely ill piglets (diarrhea, edema disease) and one further isolate of human origin [RKI6122 (24)] which exhibited a MIC value of 1,024 μg/ml ZnCl2.
The phylogeny was constructed from the MCG alignments of 186 genomes (Figure 1) and further investigated using Bayesian Analysis of Population Structure (BAPS). Seven distinct phylogenetic groups were identified, namely the BAPS cluster I-VII (Figure 1). BAPS cluster I comprised 54 isolates of both feeding groups (HZG: n = 35; CG: n = 19) distributed among eight STs, half of which containing strains with a ZnCl2 MIC of 256 μg/ml [ST21 (n = 10), ST56 (n = 4), ST58 (n = 8), and ST1308 (n = 2)]. ST101 (n = 4) and ST4577 (n = 6) isolates showed a ZnCl2 MIC of 512 μg/ml, including the only two CG-isolates with this MIC. ST154 (n = 14) isolates were associated with ZnCl2 MICs of either 256 (n = 12) or 512 μg/ml (n = 2), and ST40 (n = 6) isolates showed ZnCl2 MICs of 128 μg/ml (n = 1) and 256 μg/ml (n = 5).
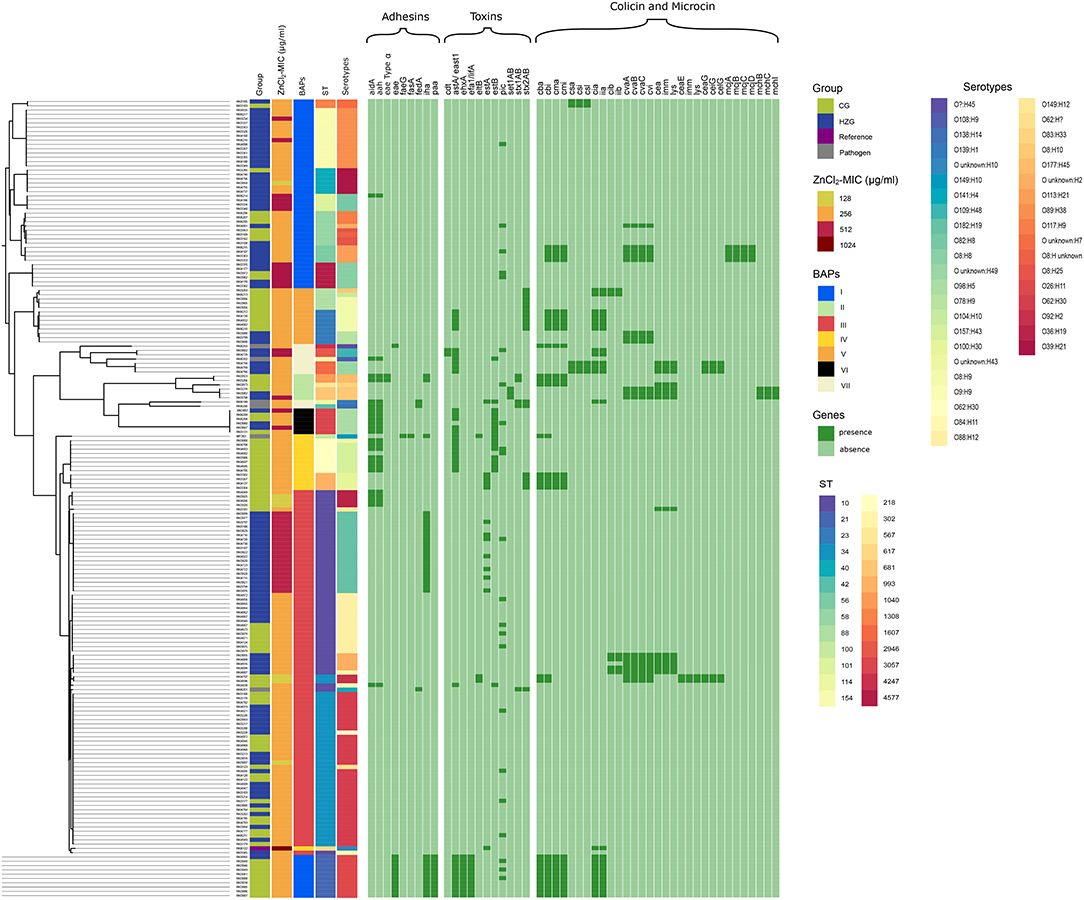
Figure 1. Maximum Common Genome Alignment of 179 commensal E. coli isolates, six porcine pathogenic E. coli and a high-zinc tolerant reference. Information about feeding group, ZnCl2 MIC values, BAPS (Bayesian analysis of the population structure (BAPS) clusters, sequence types, serotypes, presence (dark green) and absence (light green) of genes encoding adhesins, toxins, colicins, and microcins are presented from left to right.
Cluster II (n = 6; HZG: n = 2; CG: n = 4) contained five isolates associated with the 256 μg/ml ZnCl2 MIC including ST567 (n = 1), ST681 (n = 2), and ST1040 (n = 2). A third ST681 isolate of cluster II showed a 512 μg/ml ZnCl2 MIC.
With 84 isolates, BAPS cluster III formed the largest phylogenic group and consisted exclusively of isolates belonging to STC10 (HZG: n = 51; CG: n = 32; pathogenic isolate: n = 1). In more detail, ST10 (n = 45) isolates with distinct ZnCl2 MICs [128 μg/ml (n = 3), 256 mg/ml (n = 23), and 512 μg/ml (n = 19) ZnCl2], and ST34 (n = 38) isolates with either 128 μg/ml (n = 3) or 256 μg/ml (n = 35) ZnCl2 MICs, respectively, belonged to cluster III. In addition, one STEC isolate (IMT4632) isolated from a pig suffering from diarrhea (ST10; 256 μg/ml ZnCl2 MIC) clustered here, too.
Because of “long-branch attraction” artifacts (42), 12 CG isolates belonging to two different phylogenetic lineages (ST218, n = 8 and ST993, n = 4) with a ZnCl2 MIC value of 256 μg/ml were assigned to BAPS cluster IV. Also, two of the strains included for comparative reasons, i.e., one porcine ETEC-isolate (IMT203; ST100; 256 μg/ml ZnCl2 MIC) and the high-zinc tolerant human isolate (RKI6122; ST617; 1,024 μg/ml ZnCl2 MIC), clustered in BAPS IV, too.
Cluster V included 13 STC23-isolates (ST23, n = 8 [HZG: n = 3; CG: n = 5] and ST88, n = 5 [all CG]) associated with the 256 μg/ml ZnCl2 MIC. Six isolates of ST3057 (HZG: n = 3, CG: n = 3) were assigned to BAPS cluster VI (256 μg/ml ZnCl2 MIC, n = 4; 512 ZnCl2 MIC, n = 2).
Last, BAPS cluster VII consisted of ST1607 (n = 3) isolates associated with 256 μg/ml ZnCl2 MIC and ST2946 (n = 2) isolates associated with 512 μg/ml ZnCl2 MIC, all originating from animals of the HZG. Four of the pathogenic E. coli isolates included in our analysis (IMT19–ST42, STEC; IMT20–ST114, STEC; IMT6655–ST4247, ETEC; and IMT8071–ST302, EPEC; all associated with 256 μg/ml ZnCl2 MIC) clustered here, too (Supplementary Table 3).
In other words, the BAPS cluster analysis mirrored the predicted serotypes and STs, irrespective of the individual isolate's zinc tolerance level, clearly rejecting the idea of a directional association between a certain E. coli phylogenetic lineage and a particular zinc tolerance level or feeding group.
To assess the virulence potential of the isolate collection, we screened all genomes for the presence of genes that have frequently been reported as being associated with porcine enteral diseases (Supplementary Table 1). As a result, 18 out of 24 targeted genes were identified in at least one of the screened genomes (Table 1), with the number of VAGs per isolate ranging from 0 VAGs (n = 86 isolates) to a maximum of 7 VAGs (n = 3). Interestingly, 93% of the isolates originating from the HZG but 72% of the CG isolates harbored none, one, or two VAG(s) only. Isolates harboring more than two VAGs constituted 7% of the HZG and 28% of the CG isolates, indicating an influence of high-zinc diets on the intestinal microbiota with respect to VAG-carrying E. coli. Regression analysis revealed that the feeding group (p < 0.001) as well as the respective ZnCl2 MIC (p < 0.001) were associated with the number of VAGs present in an isolate. For instance, isolates belonging to the CG had 4.4 times higher odds for harboring higher numbers of VAGs (95% confidence interval: 2.0–9.8) compared to E. coli from the HZG. However, the odds ratio to carry higher numbers of VAGs was 4.2 (95% confidence interval 2.4–7.4) for E. coli associated with the 512 μg/ml ZnCl2 MIC when compared to the lower MICs.
Since most of the VAGs frequently associated with enteral diseases in piglets are carried by highly mobile genetic elements (43–46), divergent distribution of VAGs even beyond ST/serotype boundaries might occur. While there was an obvious association between ST/serotypes and VAG pattern, we also noted variation, even between isolates of the same serotype obtained from one piglet (e.g., pig 1, ST21 O26:H11 or pig 4, ST10 O182:H19) (Supplementary Table 1, Figure 1).
Since all isolates were originally obtained from healthy piglets kept under the defined conditions of an animal experiment, the overall low detection rates for enterotoxin and adhesion encoding genes, especially for ETEC-associated fimbriae (0%), was not surprising. However, all genes encoding the 11 toxins of interest here were identified among the 179 E. coli (Table 1). Again, isolates belonging to the CG harbored more toxin encoding genes than those obtained from the HZG (56 vs. 18%).
Eight of the 179 isolates were positive for the genes encoding the heat-labile enterotoxin eltAB (HZG, n = 6; CG, n = 2). Twenty-five isolates carried either the heat-stable enterotoxin gene estA (HZG, n = 7; CG, n = 4) or estB (HZG, n = 3; CG, n = 11) (Table 1).
Shiga toxin encoding genes were completely absent in E. coli representing the HZG, while 16 isolates of the CG were positive (20%, n = 16: stx1 n = 2 and stx2e n = 14). Genes encoding other toxins including astA encoding the enteroaggregative E. coli heat-stable enterotoxin EAST-1 (HZG, n = 9; CG, n = 26), enterohemolysin-encoding ehxA (HZG, n = 1; CG n = 9) or efa-1/lifA (HZG, n = 1; CG, n = 9) were detected in genomes from isolates of both feeding groups (Table 1, Supplementary Table 1).
Considering additional factors involved in the onset of PWD in piglets (47), 21 isolates (HZG, n = 4; CG, n = 17) harbored genes encoding an adhesion-involved-in-diffuse-adherence (AIDA-I) autotransporter and the AIDA-associated heptosyltransferase (aah) required to fully activate AIDA-I (Table 1). While 14% (n = 11) of the CG-isolates were found positive for the eae- encoded intimin associated with attaching and effacing, only one (1%) HZG isolate (RKI4865, ST21) was eae-positive, too.
According to the VAG profile, the six clinical porcine isolates included comprised two enterotoxigenic E. coli (ETEC—eltAB- and/or estB-positive), two Shiga toxin-producing E. coli (STEC—stx1− and stx2-positive), one ETEC/STEC-hybrid (estA, estB, stx1, stx2), and one enteropathogenic E. coli (aEPEC—eae-positive), respectively (Supplementary Table 2).
The E. coli isolates obtained from piglets fed with a high-zinc supplemented diet showed generally a lower VAG frequency. Based on this finding, we further investigated whether a specific pathotype is associated with a particular zinc tolerance level considering the previously determined ZnCl2 MIC values of 128, 256, or 512 μg/ml (22, 24) or not.
Our results showed that major diagnostic markers defining an isolate as ETEC were associated with all three different zinc tolerance levels and both feeding groups (Table 2). Moreover, ETEC showed a broad heterogeneity with respect to their phylogenetic backgrounds. However, genes encoding for ETEC-associated fimbriae F4, F5, F6, F18, and F41 were not identified. VAG combinations (eae, paa) that define porcine atypical EPEC (aEPEC) expressed the 256 μg/ml ZnCl2 MIC only, and were predominately detected among CG isolates (one exception) (Table 2). Isolates showing STEC characteristics were associated with the 256 μg/ml ZnCl2 MIC and the CG only.
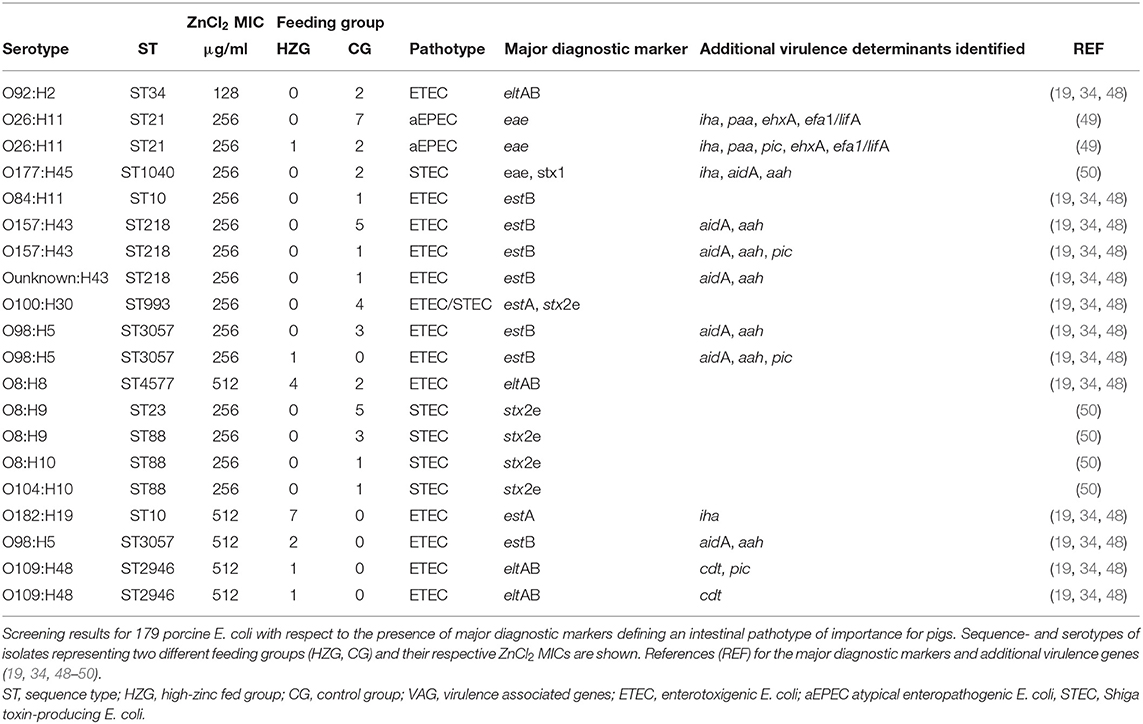
Table 2. Distribution of VAG profiles including major diagnostic markers and additional determinants associated with intestinal E. coli pathotypes.
The mixed logistic regression model showed that both, the ZnCl2 MIC and the feeding group were associated with the occurrence of major diagnostic markers defining an enteral pathotype in the present E. coli collection. The probability for harboring major diagnostic markers defining an enteral pathotype was lower in the HZG than in the CG [p = 0.002, odds ratio 47.6 (95%-confidence interval: 4.3–500)]. However, the ZnCl2 MIC 512 resulted in 58.6-fold higher odds for the presence of a major diagnostic marker (p < 0.001; 95% confidence interval 7.4–467.7). Nevertheless, 77% of the total variation was associated with variation between individual animals, indicating a greater similarity of E. coli obtained from one animal than between those of different pigs.
Amongst others, to kill or at least suppress the growth of non-host specific bacteria is one of the most important functions of the host-microbiota, a challenge which is fostered, beyond others, by production of plasmid-borne colicins and/or microcins (37, 51). To answer the question about whether genes required for colicin or microcin production were diversely distributed in E. coli obtained from different feeding groups, we screened the genomes under investigation accordingly. A detailed overview of information on aa sequence with the respective reference sequences used here for BAG identification is provided in Supplementary Figure 1.
Overall, 28 of 80 (35%) E. coli obtained from piglets belonging to the CG and 22 of 99 (22%) from the HZG harbored at least one BAG (Table 3). The most frequently detected BAGs were operons encoding for colicin B (cba+cbi: HZG n = 1; CG n = 18; cbi only, HZG n = 4; CG n = 5) and colicin M (HZG n = 5, CG n = 21) which are commonly co-located on a Col plasmid [(52), 35]. In addition, the plasmid carrying the colicin Ia encoding operon had a lower occurrence among the HZG isolates (HZG n = 6 vs. CG n = 16). Contrarily, the plasmid carrying the operon encoding for microcin V, one of the few actively secreted microcins (53), was more frequently associated with HZG isolates (HZG n = 15 vs. CG n = 3).
Other types were only rarely detected, including colicin S4 (HZG n = 4; CG n = 1), colicin Ib (HZG, n = 4; CG, n = 2), colicin E5 (HZG, n = 0; CG, n = 2), colicin E7 (HZG, n = 3; CG, n = 2), microcin H47 (HZG, n = 2; CG, n = 1), and microcin J25 (HZG, n = 4; CG, n = 0).
Considering the pathogenic isolates investigated in this study, IMT203 (ETEC) and IMT8071 (EPEC) harbor genes encoding colicin B und M (Supplementary Table 2), while IMT19 (STEC) and the high-zinc tolerant reference strain RKI6122 possessed genes for colicin Ia.
As mentioned before, the focus of our analysis was on the three different ZnCl2 MIC values determined for the E. coli collection in a previous study (22).
Of the seven 128 μg/ml ZnCl2 MIC isolates, two harbored BAGs. Both belonged to ST34, serotype O92:H2 and were positive for genes encoding colicin B, V, E5, and E7, while the remaining E. coli with 128 μg/ml ZnCl2 MIC did not carry any of the BAGs investigated.
One third of the isolates with MICs of 256 μg/ml ZnCl2 (n = 136, 17 STs) carried at least one bacteriocin gene (n = 46, 34%). In addition, colicin M, colicin S4, colicin Ib and the microcin J25 were found to be exclusively associated with isolates showing the MIC value of 256 μg/ml for ZnCl2 in a wide range of genomic backgrounds (Figure 1, Table 3). Isolates with a higher level of zinc tolerance (512 μg/ml MIC; n = 36, 7 STs) showed, by comparison, only rarely colicin encoding genes (n = 3, 8.33%) (Figure 1, Table 3).
Here we showed an analysis of E. coli genomes representing intestinal isolates collected from piglets fed with either a zinc-rich or common piglet diet (23) which exhibited three distinct zinc tolerance levels (22). A combination of different in silico analyses of the WGS data provided insights into the phylogenetic structure of these 179 porcine E. coli and their virulence and bacteriocin associated gene profiles. At present, there is only limited information available on the genetic diversity and relatedness of intestinal E. coli from pigs receiving high-zinc supplemented feed (13, 16, 23, 54).
Our results confirm that a certain phylogenetic background does not contribute to a particular zinc tolerance level in E. coli, since the tolerance levels obviously differed even within the STs of a single BAPS cluster (Figure 1). Despite their clearly phylogenetic heterogeneity (Figure 1), all six pathogenic porcine E. coli included for comparative purposes were assigned to the same “middle class” zinc tolerance level (256 μg/ml ZnCl2 MIC).
Serogroups frequently reported as being associated with diarrhea in pigs are O8, O138, O139, O141, O147, O149, and O157 (5, 20, 44, 55, 56). Our E. coli collection included a total of n = 23 isolates belonging to O8 (different H-types), one O149:H12 and seven O157:H43 isolates (Supplementary Table 1). Besides its association with porcine diarrhea, O157 isolates are of clinical importance for human patients, since Shiga toxin-producing E. coli of this particular serotype have been reported as a cause of severe foodborne illnesses (57, 58). Considering the collection investigated here, strains of this serotype were exclusively found in the CG, associated with the 256 μg/ml ZnCl2 MIC, harboring two to five VAGs, but all were negative for genes encoding Shiga toxins.
Isolates belonging to the O8 serogroup were detected in samples from both feeding groups, but more frequently in those representing the CG (HZG, n = 8; CG, n = 15). Interestingly, these CG isolates harbored none to two VAGs, including n=9 that are stx2e-positive, while Shiga toxin-encoding genes were completely absent from isolates of HZG samples.
Despite the high standards of the animal trial performed and the good health status of the animals during the whole trial period (23), our strain collection apparently included E. coli belonging to serotypes frequently associated with disease. In addition, isolates harboring key pathogenic markers defining pathotypes (Table 2) were identified as well, but they were less frequent among isolates representing the HZG, indicating a possible prophylactic effect of the feed additive. It also underlines the importance of the husbandry conditions to ensure a healthy gut microbiome (59), and, in line with this -to minimize the necessity of antibiotic consumption affecting the natural composition of the intestinal microbiota (60).
Furthermore, we identified serotype O26 : H11 belonging to ST21 (Supplementary Table 1), a strain background mainly reported for isolates of human and bovine origin and presumptively associated with EPEC and STEC/EHEC pathotypes (61–65). Previously, aEPEC strains of ST21-O26:H11 showed a close phylogenetic relationship to STEC/EHEC and the results of lysogenic conversion experiments using stx-depleted bacteriophage identified aEPEC (ST21-O26:H11) as progenitors of typical EHEC (66). Atypical EPEC (ST21-O26:H11) were also identified in our isolate collection, harboring a combination of VAGs (eae-positive, stx-negative, bfp-negative) meeting criteria for possible progenitor of human-pathogenic STEC/EHEC (67, 68).
While there is generally a strong association between genetic background and accessory gene content including VAGs and BAGs in our isolate collection, their occurrence and distribution were not solely lineage-specific: Some of the VAGs investigated here, for instance iha, estA and estB were present in different (Figure 1, Table 1). The EAEC associated enterotoxin EAST-1 (astA) was detected in E. coli from both feeding groups. While it has been assumed that there is an association between this toxin and diarrhea in humans and pigs (30, 31, 69), a further study found higher astA rates in E. coli from healthy pigs compared to isolates obtained from clinically ill pigs, questioning the particular role of astA as a virulence factor in porcine E. coli (32). In addition, the occurrence of one or a few VAGs alone does not necessarily reflect the pathogenic potential of E. coli, unless the strain acquires a compatible combination of VAGs able to cause disease in a specific host species (70).
The logistic regression applied in the present study revealed a positive association between the presence of E. coli major diagnostic markers and the 512 μg/ml ZnCl2 MIC. However, whether these E. coli are able to cause enteral disorders in piglets needs to be further studied. The heat stable enterotoxin I encoded by estA, which is the most common pathogenic marker associated with the 512 μg/ml ZnCl2 MIC, has been reported as a cause of neonatal diarrhea in different animals. This enterotoxin seems to be associated with ETEC causing PWD as well, but rarely as the sole enterotoxin (5). However, none of the six porcine pathogenic E. coli variants (aEPEC, ETEC, STEC; ETEC/STEC) included in our study for comparative purposes exhibited the 512 μg/ml ZnCl2 MIC (Table 2), possibly suggesting that pathogenic field strains commonly lack enhanced zinc tolerance.
Following this line of thought, it seems likely that zinc does not only affect the overall composition of the porcine E. coli intestinal population favoring phenotypes with enhanced zinc tolerance (22, 71), but also reduces the abundance of potentially harmful (field) strains. These effects possibly provide an explanation for former observations indicating an increase in relative abundance of intestinal Enterobacterales among piglets fed with high amounts of zinc (72, 73), while the occurrence of PWD and detection of pathogenic bacteria seemed to decrease coincidentally (74).
Our findings might also indicate the presence of a zinc-tolerant subpopulation of E. coli carrying genetic marker for the intestinal pathotypes. In the environment of an animal trial with high-standard hygienic conditions, this subpopulation might not be able to proliferate well enough to cause disease, but the presence of this subpopulation indicates the importance of housing conditions, feed and water hygiene and prophylactic measures like vaccinations to maintain a healthy animal population and produce a save food product.
Since fimbriae are key features of pathogenic E. coli in pigs (20, 30, 75–77), we were not surprised that our collection of isolates obtained from healthy piglets lacked these particular genes (Table 1). Notably, a former study investigating 844 E. coli isolates from PWD-affected pig farms in Europe showed that the prevalence of genes encoding fimbriae was low even among PWD-associated ETEC (9% F4 fimbriae; 9% F18 fimbriae) from Germany (19), possibly indicating that further factors might play a role in the pathogenesis of E. coli in PWD.
Moreover, previous research has also shown that the bacteriophage-encoded genes stx1 and stx2 are upregulated by DNA-damage induced activation of the SOS-response (78, 79), while zinc excess has been shown to mute the SOS-response via inhibition of recA (80). In addition, zinc is also known to inhibit expression of further VAGs (81), including those promoting adherence to the gut epithelium (i.e., esp genes) (82). In consequence, pathogenic strains might lose their ability to harm the host, which has exemplarily been demonstrated for STEC as well as EPEC before (82).
Since bacteriocins promote the producing bacterial cells while competing for resources in a distinct ecological niche with other E. coli strains and closely related bacteria, they probably play an important role in structuring microbial communities residing in the gut (83). In addition, activity of particular bacteriocin-producing E. coli toward competing bacteria, e.g., STEC O157:H7, has been reported (84, 85). This led us to investigate if HZG-E. coli and/or those isolates associated with the 512 μg/ml ZnCl2 MIC harbor either more BAGs or a certain BAG pattern (Figure 1, Table 3). Yet, similar to the results for VAGs, BAGs seem to be less common among isolates representing the HZG and especially less common among the group of E. coli expressing the 512 μg/ml ZnCl2 MIC.
The general structural organization of the colicin encoding operons includes at least two, usually three genes (i.e., activity gene, gene encoding the immunity protein and gene encoding the lysis protein), which are commonly co-located on a large conjugative plasmid in E. coli (52, 86, 87). Colicins B and M are among the most common colicins in porcine E. coli (12, 19, 23, 88). While most of the colicins are released by cell death of the producing cell only, these colicins are among the few actively secreted bacteriocins (53) and their production might therefore be less “costly” (89). In our dataset, colicin B and M were frequently associated with isolates belonging to the CG, while rarely detected among isolates of the HZG (Table 3). For most colicins, induction is assumed to be associated with the DNA-damage induced SOS response (90). However, recent research indicated the production of biologically significant amounts of several colicins in the absence of such stress (91). Furthermore, colicin M was found to be secreted despite the absence of the SOS box believed to regulate its production (88). Consequently, higher detection rates of genes encoding colicin M might be a result from its less severe “production consequences” for the individual cell, since cell death is not a prerequisite for its release. Interestingly, genes encoding colicin BM were found in BAPS cluster I, II, IV and V (Figure 1), but all these isolates lack zinc tolerance above the mid-range 256 μg/ml ZnCl2 MIC (Supplementary Table 1). The complete absence of colicin BM genes in isolates associated with enhanced zinc tolerance (isolates distributed within BAPS cluster I, II, III, VI and VII) suggests that co-occurrence of enhanced zinc tolerance with colicin BM production lacks a beneficial outcome for intestinal E. coli in piglets fed with high amounts of zinc. However, more investigation on this subject is clearly needed.
Contrarily, the operon encoding microcin V was more associated with isolates from the HZG than the CG, however it was also rarely associated with isolates with a 512 μg/ml ZnCl2 MIC. The expression of the immunity protein (Cvi) of this operon depends on the presence of iron [reviewed in (53)], since the cvi promoter region is associated with a previously identified binding site for the ferric uptake regulation protein (Fur) (53, 92). Interestingly, zinc excess was reported to increase the bacterial demand for iron (93), and microcin V activation might increase the chances of bacteria to benefit from iron released by dead competing populations. However, more research on functional interactions of bacteria under zinc-induced stress is needed, which left speculation only considering the interlinkages of microcin V expression and zinc excess.
Our analysis comprising 179 E. coli obtained from piglets of two different feeding groups using Bayesian analysis of the genomes showed a population structure (BAPS) which lacked an association between a particular zinc tolerance level and a phylogenetic cluster or even branch for both, isolates belonging to the HZG and CG, suggesting that zinc tolerance is not a characteristic of a particular phylogenetic background. In addition, detection rates for genes and operons associated with virulence (VAG) and bacteriocins (BAG) were lower in isolates originating from the HZG (41 vs. 65% and 22 vs. 35%, p < 0.001 and p = 0.002, resp.), indicating an effect of high-zinc supplementation of the piglets' diet on the occurrences of these genes among intestinal E. coli populations.
This effect seems to be more even more important considering E. coli harboring genes defining distinct pathotypes associated with intestinal disease, i.e., enterotoxigenic, enteropathogenic and Shiga toxin-producing E. coli (ETEC, EPEC and STEC), which constituted only 1% of the isolates belonging to the HZG but 14% of those from the CG, supporting previous observations that high zinc supplemented diets for piglets were associated with a decrease of PWD.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. Illumina raw read data for 186 E. coli are available at NCBI Bioproject ID PRJNA552271.
The representative set of E. coli isolates investigated here was selected based on a previous feeding trial (23) carried out in accordance with the principles of the Basel Declaration following the institutional and national guidelines for the care and use of animals. The animal study was reviewed and approved by the local state office of occupational health and technical safety Landesamt für Gesundheit und Soziales, Berlin (LaGeSo Reg. Nr. 0296/13) as described before (23).
LW, AL-B, and AB designed the project. VJ and BW conceived and designed the experiments. IE sequenced the isolates. VJ performed laboratory analysis. VJ, BW, LE, AB, TS, FG, and RM analyzed the data. VJ, AB, BW, and LW wrote the article. All authors have read and approved the final draft of the manuscript. All authors contributed to the article and approved the submitted version.
This work was funded by the Deutsche Forschungsgemeinschaft (DFG no. WI 1436/12-1). AL-B, AB, and BW were supported by the project #1Health-PREVENT (Grant Nos. 01KI1727F and 01KI1727D) from the German Federal Ministry of Education and Research (BMBF). LE was supported by the project PAC-CAMPY (Grant No. 01KI1725F) within the German Research Network of Zoonotic Diseases. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thanked Julia Assmann, Charlotte Huber, Anne Kauter, and the colleagues from the Advanced Light and Electron Microscopy (ZBS-4) department of the Robert Koch Institute for their individual contribution and support.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.614513/full#supplementary-material
1. Guevarra RB, Lee JH, Lee SH, Seok M-J, Kim DW, Kang BN, et al. Piglet gut microbial shifts early in life: causes and effects. J Anim Sci Biotechnol. (2019) 10:1. doi: 10.1186/s40104-018-0308-3
2. Campbell JM, Crenshaw JD, Polo J. The biological stress of early weaned piglets. J Anim Sci Biotechnol. (2013) 4:19–19. doi: 10.1186/2049-1891-4-19
3. Gresse R, Chaucheyras-Durand F, Fleury MA, Van De Wiele T, Forano E, Blanquet-Diot S. Gut microbiota dysbiosis in postweaning piglets: understanding the keys to health. Trends Microbiol. (2017) 25:851–73. doi: 10.1016/j.tim.2017.05.004
4. Liu Y, Espinosa CD, Abelilla JJ, Casas GA, Lagos LV, Lee SA, et al. Non-antibiotic feed additives in diets for pigs: a review. Anim Nutr. (2018) 4:113–25. doi: 10.1016/j.aninu.2018.01.007
5. Fairbrother JM, Nadeau É, Gyles CL. Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim Health Res Rev. (2005) 6:17–39. doi: 10.1079/AHR2005105
6. Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. (2007) 2:119–29. doi: 10.1016/j.chom.2007.06.010
7. Pickard JM, Zeng MY, Caruso R, Núñez G. Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol Rev. (2017) 279:70–89. doi: 10.1111/imr.12567
8. Destoumieux-Garzón D, Peduzzi J, Rebuffat S. Focus on modified microcins: structural features and mechanisms of action. Biochimie. (2002) 84:511–9. doi: 10.1016/S0300-9084(02)01411-6
9. Cascales E, Buchanan SK, Duché D, Kleanthous C, Lloubès R, Postle K, et al. Colicin biology. Microbiol. Mol. Biol. Rev. (2007) 71:158–229. doi: 10.1128/MMBR.00036-06
10. Flaherty RA, Freed SD, Lee SW. The wide world of ribosomally encoded bacterial peptides. PLoS Pathog. (2014) 10:e1004221. doi: 10.1371/journal.ppat.1004221
11. Bosák J, Hrala M, Pirková V, Micenková L, CíŽek A, Smola J, et al. Porcine pathogenic Escherichia coli strains differ from human fecal strains in occurrence of bacteriocin types. Vet Microbiol. (2019) 232:121–7. doi: 10.1016/j.vetmic.2019.04.003
12. Rhouma M, Fairbrother JM, Beaudry F, Letellier A. Post weaning diarrhea in pigs: risk factors and non-colistin-based control strategies. Acta Vet Scand. (2017) 59:31–31. doi: 10.1186/s13028-017-0299-7
13. Bednorz C, Oelgeschläger K, Kinnemann B, Hartmann S, Neumann K, Pieper R, et al. The broader context of antibiotic resistance: zinc feed supplementation of piglets increases the proportion of multi-resistant Escherichia coli in vivo. Int J Med Microbiol. (2013) 303:396–403. doi: 10.1016/j.ijmm.2013.06.004
14. Kloubert V, Blaabjerg K, Dalgaard TS, Poulsen HD, Rink L, Wessels I. Influence of zinc supplementation on immune parameters in weaned pigs. J Trace Elem Med Biol. (2018) 49:231–40. doi: 10.1016/j.jtemb.2018.01.006
15. Broom LJ, Miller HM, Kerr KG, Knapp JS. Effects of zinc oxide and Enterococcus faecium SF68 dietary supplementation on the performance, intestinal microbiota and immune status of weaned piglets. Res Vet Sci. (2006) 80:45–54. doi: 10.1016/j.rvsc.2005.04.004
16. Starke IC, Pieper R, Neumann K, Zentek J, Vahjen W. The impact of high dietary zinc oxide on the development of the intestinal microbiota in weaned piglets. FEMS Microbiol Ecol. (2014) 87:416–27. doi: 10.1111/1574-6941.12233
17. Wang W, Van Noten N, Degroote J, Romeo A, Vermeir P, Michiels J. Effect of zinc oxide sources and dosages on gut microbiota and integrity of weaned piglets. J Anim Physiol Anim Nutr. (2019) 103:231–41. doi: 10.1111/jpn.12999
18. Chapman TA, Wu X-Y, Barchia I, Bettelheim KA, Driesen S, Trott D, et al. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol. (2006) 72:4782–95. doi: 10.1128/AEM.02885-05
19. Luppi A, Gibellini M, Gin T, Vangroenweghe F, Vandenbroucke V, Bauerfeind R, et al. Prevalence of virulence factors in enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea in Europe. Porcine Health Manage. (2016) 2:20. doi: 10.1186/s40813-016-0039-9
20. Frydendahl K. Prevalence of serogroups and virulence genes in Escherichia coli associated with postweaning diarrhoea and edema disease in pigs and a comparison of diagnostic approaches. Vet Microbiol. (2002) 85:169–82. doi: 10.1016/S0378-1135(01)00504-1
21. Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Micro. (2004) 2:123–40. doi: 10.1038/nrmicro818
22. Johanns VC, Ghazisaeedi F, Epping L, Semmler T, Lübke-Becker A, Pfeifer Y, et al. Effects of a four-week high-dosage zinc oxide supplemented diet on commensal Escherichia coli of weaned pigs. Front Microbiol. (2019) 10:2734. doi: 10.3389/fmicb.2019.02734
23. Ciesinski L, Guenther S, Pieper R, Kalisch M, Bednorz C, Wieler LH. High dietary zinc feeding promotes persistence of multi-resistant E. coli in the swine gut. PLoS ONE. (2018) 13:e0191660. doi: 10.1371/journal.pone.0191660
24. Deus D, Krischek C, Pfeifer Y, Sharifi AR, Fiegen U, Reich F, et al. Comparative analysis of the susceptibility to biocides and heavy metals of extended-spectrum β-lactamase-producing Escherichia coli isolates of human and avian origin, Germany. Diagn Microbiol Infect Dis. (2017) 88:88–92. doi: 10.1016/j.diagmicrobio.2017.01.023
25. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. (2017) 13:e1005595. doi: 10.1371/journal.pcbi.1005595
26. Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
27. Von Mentzer A, Connor TR, Wieler LH, Semmler T, Iguchi A, Thomson NR, et al. Identification of enterotoxigenic Escherichia coli (ETEC) clades with long-term global distribution. Nat Genet. (2014) 46:1321. doi: 10.1038/ng.3145
28. Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
29. Tang J, Hanage WP, Fraser C, Corander J. Identifying currents in the gene pool for bacterial populations using an integrative approach. PLoS Comput Biol. (2009) 5:e1000455. doi: 10.1371/journal.pcbi.1000455
30. Vu-Khac H, Holoda E, Pilipcinec E, Blanco M, Blanco JE, Dahbi G, et al. Serotypes, virulence genes, intimin types and PFGE profiles of Escherichia coli isolated from piglets with diarrhoea in Slovakia. Vet J. (2007) 174:176–87. doi: 10.1016/j.tvjl.2006.05.019
31. Liu W, Yuan C, Meng X, Du Y, Gao R, Tang J, et al. Frequency of virulence factors in Escherichia coli isolated from suckling pigs with diarrhoea in China. Vet J. (2014) 199:286–9. doi: 10.1016/j.tvjl.2013.11.019
32. Zajacova ZS, Konstantinova L, Alexa P. Detection of virulence factors of Escherichia coli focused on prevalence of EAST1 toxin in stool of diarrheic and non-diarrheic piglets and presence of adhesion involving virulence factors in astA positive strains. Vet Microbiol. (2012) 154:369–75. doi: 10.1016/j.vetmic.2011.07.029
33. Schierack P, Steinruck H, Kleta S, Vahjen W. Virulence factor gene profiles of Escherichia coli isolates from clinically healthy pigs. Appl Environ Microbiol. (2006) 72:6680–6. doi: 10.1128/AEM.02952-05
34. Dubreuil JD, Isaacson RE, Schifferli DM. Animal enterotoxigenic Escherichia coli. EcoSal Plus. (2016) 7:1–47. doi: 10.1128/ecosalplus.ESP-0006-2016
35. Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, et al. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol. (2012) 50:1355–61. doi: 10.1128/JCM.06094-11
36. Joensen KG, Tetzschner AMM, Iguchi A, Aarestrup FM, Scheutz F. Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J Clin Microbiol. (2015) 53:2410–26. doi: 10.1128/JCM.00008-15
37. Hardy KG, Meynell GG, Dowman JE, Spratt BG. Two major groups of colicin factors: their evolutionary significance. Mol Gen Genet. (1973) 125:217–30. doi: 10.1007/BF00270744
38. Pilsl H, Smajs D, Braun V. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J Bacteriol. (1999) 181:3578–81. doi: 10.1128/JB.181.11.3578-3581.1999
39. Riley MA, Cadavid L, Collett MS, Neely MN, Adams MD, Phillips CM, et al. The newly characterized colicin Y provides evidence of positive selection in pore-former colicin diversification. Microbiology. (2000) 146:1671–7. doi: 10.1099/00221287-146-7-1671
40. Gordon DM, Oliver E, Littlefield-Wyer J. The diversity of bacteriocins in gram-negative bacteria. In: MA Riley, MA Chavan, editors. Bacteriocins: Ecology and Evolution. Berlin, Heidelberg: Springer Berlin Heidelberg. p. 5–18. doi: 10.1007/978-3-540-36604-1_2
41. Micenková L, Bosák J, Kucera J, Hrala M, Dolejšová T, Šedo O, et al. Colicin Z, a structurally and functionally novel colicin type that selectively kills enteroinvasive Escherichia coli and Shigella strains. Sci Rep. (2019) 9:11127. doi: 10.1038/s41598-019-47488-8
42. Bergsten J. A review of long-branch attraction. Cladistics. (2005) 21:163–93. doi: 10.1111/j.1096-0031.2005.00059.x
43. Mainil JG, Daube G, Jacquemin E, Pohl P, Kaeckenbeeck A. Virulence plasmids of enterotoxigenic Escherichia coli isolates from piglets. Vet Microbiol. (1998) 62:291–301. doi: 10.1016/S0378-1135(98)00225-9
44. Nagy B, Fekete PZ. Enterotoxigenic Escherichia coli (ETEC) in farm animals. Vet Res. (1999) 30:259–84.
45. Muniesa M, Recktenwald J, Bielaszewska M, Karch H, Schmidt H. Characterization of a shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect Immun. (2000) 68:4850–5. doi: 10.1128/IAI.68.9.4850-4855.2000
46. Fekete PZ, Gerardin J, Jacquemin E, Mainil JG, Nagy B. Replicon typing of F18 fimbriae encoding plasmids of enterotoxigenic and verotoxigenic Escherichia coli strains from porcine postweaning diarrhoea and oedema disease. Vet Microbiol. (2002) 85:275–84. doi: 10.1016/S0378-1135(01)00515-6
47. Niewerth U, Frey A, Voss T, Le Bouguénec C, Baljer G, Franke S, et al. The AIDA autotransporter system is associated with F18 and stx2e in Escherichia coli isolates from pigs diagnosed with edema disease and postweaning diarrhea. Clin Diagn Lab Immunol. (2001) 8:143–9. doi: 10.1128/CDLI.8.1.143-149.2001
48. Moredo FA, Piñeyro PE, Márquez GC, Sanz M, Colello R, et al. Enterotoxigenic Escherichia coli subclinical infection in pigs: bacteriological and genotypic characterization and antimicrobial resistance profiles. Foodborne Pathog Dis. (2015) 12:704–11. doi: 10.1089/fpd.2015.1959
49. García-Meniño I, García V, Mora A, Díaz-Jiménez D, Flament-Simon SC, Alonso MP, et al. Swine Enteric Colibacillosis in Spain: Pathogenic Potential of mcr-1 ST10 and ST131 E. coli isolates. Front Microbiol. (2018) 9:2659. doi: 10.3389/fmicb.2018.02659
50. Tseng M, Fratamico PM, Bagi L, Delannoy S, Fach P, Manning SD, et al. Diverse virulence gene content of Shiga toxin-producing Escherichia coli from finishing swine. Appl Environ Microbiol. (2014) 80:6395–402. doi: 10.1128/AEM.01761-14
51. Pultz NJ, Stiefel U, Subramanyan S, Helfand MS, Donskey CJ. Mechanisms by which anaerobic microbiota inhibit the establishment in mice of intestinal colonization by vancomycin-resistant Enterococcus. J Infect Dis. (2005) 191:949–56. doi: 10.1086/428090
52. Ölschläger T, Schramm E, Braun V. Cloning and expression of the activity and immunity genes of colicins B and M on ColBM plasmids. Mol Gen Genet. (1984) 196:482–7. doi: 10.1007/BF00436196
53. Baquero F, Lanza VF, Baquero M-R, Del Campo R, Bravo-Vázquez DA. Microcins in Enterobacteriaceae: peptide antimicrobials in the eco-active intestinal chemosphere. Front Microbiol. (2019) 10:2261. doi: 10.3389/fmicb.2019.02261
54. Ahmed S, Olsen JE, Herrero-Fresno A. The genetic diversity of commensal Escherichia coli strains isolated from non-antimicrobial treated pigs varies according to age group. PLoS ONE. (2017) 12:e0178623. doi: 10.1371/journal.pone.0178623
55. Harel J, Lapointe H, Fallara A, Lortie LA, Bigras-Poulin M, Larivière S, et al. Detection of genes for fimbrial antigens and enterotoxins associated with Escherichia coli serogroups isolated from pigs with diarrhea. J Clin Microbiol. (1991) 29:745–52. doi: 10.1128/JCM.29.4.745-752.1991
56. Salajka E, Salajkova Z, Alexa P, Hornich M. Colonization factor different from K88, K99, F41 and 987P in enterotoxigenic Escherichia coli strains isolated from postweaning diarrhoea in pigs. Vet Microbiol. (1992) 32:163–75. doi: 10.1016/0378-1135(92)90103-Z
57. Park S, Worobo RW, Durst RA. Escherichia coli O157:H7 as an emerging foodborne pathogen: a literature review. Crit Rev Biotechnol. (2001) 21:27–48. doi: 10.1080/20013891081674
58. Devleesschauwer B, Pires SM, Young I, Gill A, Majowicz SE, Study T. Associating sporadic, foodborne illness caused by Shiga toxin-producing Escherichia coli with specific foods: a systematic review and meta-analysis of case-control studies. Epidemiol Infect. (2019) 147:e235. doi: 10.1017/S0950268819001183
59. Pluske JR, Turpin DL, Kim J-C. Gastrointestinal tract (gut) health in the young pig. Anim Nutr. (2018) 4:187–96. doi: 10.1016/j.aninu.2017.12.004
60. Fouhse JM, Zijlstra RT, Willing BP. The role of gut microbiota in the health and disease of pigs. Anim Front. (2016) 6:30–6. doi: 10.2527/af.2016-0031
61. Eichhorn I, Heidemanns K, Semmler T, Kinnemann B, Mellmann A, Harmsen D, et al. Highly virulent non-O157 Enterohemorrhagic Escherichia coli (EHEC) serotypes reflect similar phylogenetic lineages, providing new insights into the evolution of EHEC. Appl Environ Microbiol. (2015) 81:7041–7. doi: 10.1128/AEM.01921-15
62. Januszkiewicz A, Wołkowicz T, Chróst A, Szych J. Characterization of the Shiga toxin-producing Escherichia coli O26 isolated from human in Poland between 1996 and 2014. Lett Appl Microbiol. (2015) 60:605–8. doi: 10.1111/lam.12413
63. Ferdous M, Friedrich AW, Grundmann H, De Boer RF, Croughs PD, Islam MA, et al. Molecular characterization and phylogeny of Shiga toxin-producing Escherichia coli isolates obtained from two Dutch regions using whole genome sequencing. Clin Microbiol Infect. (2016) 22:642.e641-649. doi: 10.1016/j.cmi.2016.03.028
64. Gonzalez-Escalona N, Toro M, Rump LV, Cao G, Nagaraja TG, Meng J. Virulence gene profiles and clonal relationships of Escherichia coli O26:H11 isolates from feedlot cattle as determined by whole-genome sequencing. Appl Environ Microbiol. (2016) 82:3900–12. doi: 10.1128/AEM.00498-16
65. Ishijima N, Lee K-I, Kuwahara T, Nakayama-Imaohji H, Yoneda S, Iguchi A, et al. Identification of a new virulent clade in enterohemorrhagic Escherichia coli O26:H11/H- sequence type 29. Sci Rep. (2017) 7:43136. doi: 10.1038/srep43136
66. Eichhorn I, Heidemanns K, Ulrich RG, Schmidt H, Semmler T, Fruth A, et al. Lysogenic conversion of atypical enteropathogenic Escherichia coli (aEPEC) from human, murine, and bovine origin with bacteriophage Φ3538 Δstx(2)::cat proves their enterohemorrhagic E. coli (EHEC) progeny. Int J Med Microbiol. (2018) 308:890–8. doi: 10.1016/j.ijmm.2018.06.005
67. Bielaszewska M, Köck R, Friedrich A, Eiff C, Zimmerhackl L, Karch H, et al. Shiga toxin-mediated hemolytic uremic syndrome: time to change the diagnostic paradigm? PLoS ONE. (2007) 2:e1024. doi: 10.1371/journal.pone.0001024
68. Ferdous M, Zhou K, Mellmann A, Morabito S, Pd C, Boer DR, et al. Are Shiga toxin negative Escherichia coli O157:H7 enteropathogenic or 1 enterohaemorrhagic Escherichia coli? A comprehensive molecular analysis using whole genome sequencing. J Clin Microbiol. (2015) 53:3530–8. doi: 10.1128/JCM.01899-15
69. Beutin L, Krüger U, Krause G, Miko A, Martin A, Strauch E. Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl Environ Microbiol. (2008) 74:4806–16. doi: 10.1128/AEM.00623-08
70. Leimbach A, Hacker J, Dobrindt U. E. coli as an all-rounder: the thin line between commensalism and pathogenicity. In: U Dobrindt, JH Hacker, C Svanborg, editors. Between Pathogenicity and Commensalism (Berlin, Heidelberg: Springer Berlin Heidelberg). p. 3–32. doi: 10.1007/82_2012_303
71. Ghazisaeedi F, Ciesinski L, Bednorz C, Johanns V, Pieper L, Tedin K, et al. Phenotypic zinc resistance does not correlate with antimicrobial multi-resistance in fecal E. coli isolates of piglets. Gut Pathogens. (2020) 12:4. doi: 10.1186/s13099-019-0342-5
72. Pieper R, Vahjen W, Neumann K, Van Kessel AG, Zentek J. Dose-dependent effects of dietary zinc oxide on bacterial communities and metabolic profiles in the ileum of weaned pigs. J Anim Physiol Anim Nutr. (2012) 96:825–33. doi: 10.1111/j.1439-0396.2011.01231.x
73. Yu T, Zhu C, Chen S, Gao L, Lv H, Feng R, et al. Dietary high zinc oxide modulates the microbiome of ileum and colon in weaned piglets. Front Microbiol. (2017) 8:825. doi: 10.3389/fmicb.2017.00825
74. Sales J. Effects of pharmacological concentrations of dietary zinc oxide on growth of post-weaning pigs: a meta-analysis. Biol Trace Elem Res. (2013) 152:343–9. doi: 10.1007/s12011-013-9638-3
75. Wu X-Y, Chapman T, Trott DJ, Bettelheim K, Do TN, Driesen S, et al. Comparative analysis of virulence genes, genetic diversity, and phylogeny of commensal and enterotoxigenic Escherichia coli isolates from weaned pigs. Appl Environ Microbiol. (2007) 73:83–91. doi: 10.1128/AEM.00990-06
76. Smith MG, Jordan D, Chapman TA, Chin JJC, Barton MD, Do TN, et al. Antimicrobial resistance and virulence gene profiles in multi-drug resistant enterotoxigenic Escherichia coli isolated from pigs with post-weaning diarrhoea. Vet Microbiol. (2010) 145:299–307. doi: 10.1016/j.vetmic.2010.04.004
77. Luise D, Lauridsen C, Bosi P, Trevisi P. Methodology and application of Escherichia coli F4 and F18 encoding infection models in post-weaning pigs. J Anim Sci Biotechnol. (2019) 10:53–53. doi: 10.1186/s40104-019-0352-7
78. Pacheco AR, Sperandio V. Shiga toxin in enterohemorrhagic E. coli: regulation and novel anti-virulence strategies. Front Cell Infect Microbiol. (2012) 2:81. doi: 10.3389/fcimb.2012.00081
79. Berger M, Aijaz I, Berger P, Dobrindt U, Koudelka G. Transcriptional and translational inhibitors block SOS response and shiga toxin expression in Enterohemorrhagic Escherichia coli. Sci Rep. (2019) 9:18777. doi: 10.1038/s41598-019-55332-2
80. Bunnell BE, Escobar JF, Bair KL, Sutton MD, Crane JK. Zinc blocks SOS-induced antibiotic resistance via inhibition of RecA in Escherichia coli. PLoS ONE. (2017) 12:e0178303. doi: 10.1371/journal.pone.0178303
81. Xue Y, Osborn J, Panchal A, Mellies JL. The RpoE stress response pathway mediates reduction of the virulence of enteropathogenic Escherichia coli by zinc. Appl Environ Microbiol. (2015) 81:3766–74. doi: 10.1128/AEM.00507-15
82. Crane JK, Broome JE, Reddinger RM, Werth BB. Zinc protects against Shiga-toxigenic Escherichia coli by acting on host tissues as well as on bacteria. BMC Microbiol. (2014) 14:145. doi: 10.1186/1471-2180-14-145
83. Zhao Z, Orfe LH, Liu J, Lu S-Y, Besser TE, Call DR. Microcin PDI regulation and proteolytic cleavage are unique among known microcins. Sci Rep. (2017) 7:42529. doi: 10.1038/srep42529
84. Sawant AA, Casavant NC, Call DR, Besser TE. Proximity-dependent inhibition in Escherichia coli isolates from cattle. Appl Environ Microbiol. (2011) 77:2345. doi: 10.1128/AEM.03150-09
85. Cameron A, Zaheer R, Adator EH, Barbieri R, Reuter T, Mcallister TA. Bacteriocin occurrence and activity in Escherichia coli isolated from bovines and wastewater. Toxins. (2019) 11:475. doi: 10.3390/toxins11080475
86. Săsărman A, Massie B, Zollinger M, Gagne-Tellier H, Shareck F, Garzon S, et al. Naturally occurring R.ColBM plasmids belonging to the IncFIII incompatibility group. J. Gen. Microbiol. (1980) 119:475–83. doi: 10.1099/00221287-119-2-475
87. Gordon DM, O'Brien CL. Bacteriocin diversity and the frequency of multiple bacteriocin production in Escherichia coli. Microbiology. (2006) 152:3239–44. doi: 10.1099/mic.0.28690-0
88. Christenson JK, Gordon DM. Evolution of colicin BM plasmids: the loss of the colicin B activity gene. Microbiology. (2009) 155:1645–55. doi: 10.1099/mic.0.026666-0
89. Harkness RE, Olschläger T. The biology of colicin M. FEMS Microbiol Rev. (1991) 8:27–41. doi: 10.1016/0168-6445(91)90004-2
90. Lu FM, Chak KF. Two overlapping SOS-boxes in ColE operons are responsible for the viability of cells harboring the Col plasmid. Mol Gen Genet. (1996) 251:407–11. doi: 10.1007/BF02172368
91. Mavridou DaI, Gonzalez D, Kim W, West SA, Foster KR. Bacteria use collective behavior to generate diverse combat strategies. Curr Biol. (2018) 28:345–355. doi: 10.1016/j.cub.2017.12.030
92. Boyer AE, Tai PC. Characterization of the cvaA and cvi Promoters of the Colicin V export system: iron-dependent transcription of cvaA is modulated by downstream sequences. J Bacteriol. (1998) 180:1662. doi: 10.1128/JB.180.7.1662-1672.1998
Keywords: E. coli, zinc, pig, virulence associated genes, bacteriocins, gut
Citation: Johanns VC, Epping L, Semmler T, Ghazisaeedi F, Lübke-Becker A, Pfeifer Y, Eichhorn I, Merle R, Bethe A, Walther B and Wieler LH (2020) High-Zinc Supplementation of Weaned Piglets Affects Frequencies of Virulence and Bacteriocin Associated Genes Among Intestinal Escherichia coli Populations. Front. Vet. Sci. 7:614513. doi: 10.3389/fvets.2020.614513
Received: 10 October 2020; Accepted: 20 November 2020;
Published: 16 December 2020.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Qiangde Duan, Yangzhou University, ChinaCopyright © 2020 Johanns, Epping, Semmler, Ghazisaeedi, Lübke-Becker, Pfeifer, Eichhorn, Merle, Bethe, Walther and Wieler. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Birgit Walther, d2FsdGhlcmJAcmtpLmRl
†These authors share last authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.