
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci. , 10 November 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.582899
This article is part of the Research Topic Antibiotic Resistance and its Continuity in the Environmental Niche View all 19 articles
 Nwai Oo Khine1,2
Nwai Oo Khine1,2 Kittitat Lugsomya3
Kittitat Lugsomya3 Benjarong Kaewgun1
Benjarong Kaewgun1 Lertrob Honhanrob1
Lertrob Honhanrob1 Panupong Pairojrit1
Panupong Pairojrit1 Suthipat Jermprasert1
Suthipat Jermprasert1 Nuvee Prapasarakul1,4*
Nuvee Prapasarakul1,4*The presence of the plasmid-mediated colistin resistance encoding mcr gene family in the Enterobacteriaceae is one of the crucial global concerns. The use of colistin in livestock rearing is believed to be the cause of mcr gene spreading and is of impact to public health. The objective of this research was to detect the frequency and virulent genes of mcr-positive Escherichia coli (MCRPE) in fecal samples from healthy pigs in a contract farming system across Thailand. A total of 696 pooled samples were derived from 80 farms, located in 49 provinces across six regions of Thailand. The colistin-resistant E. coli were identified by MALDI-TOF mass spectrometry and antimicrobial susceptibility testing by broth microdilution. The antibiogram was determined using an automated susceptibility machine, and the genetic characteristics were investigated for mcr-1–5 genes, phylogenetic group, replicon types, and virulent genes. In total, 31 of 696 samples were positive, with E. coli containing mcr-1 or combination of mcr-1 and mcr-3 with incidence of 4.45 and 0.43%. Phylogenetic groups A and B1 and the IncF and IncFIB replicon types were predominantly found in the MCRPE located in the central area, with multidrug-resistant traits against 3–14 types of antimicrobials. Additionally, 19 of 31 isolates identified as enterotoxigenic E. coli were with the stap and stb (enterotoxin-encoding genes). In conclusion, a low carriage rate of mcr-positive E. coli was detected in the large-scale farming of healthy pigs. The association between multidrug-resistant MCRPE and their pathogenic potential should be of concern.
Antimicrobial resistance (AMR) is an emerging concern for both human and animal sectors of the world. The inappropriate use of antimicrobials in clinical settings and, most importantly, in livestock farming imposes social and economic burdens on society (1). The diminishing number of active (effective) antimicrobial agents to treat sick farm animals is accompanied by the downfall in food production and the likelihood of exposure of farmers to resistant bacteria. Escherichia coli, a commensal microbe, can accumulate resistance genes. It is widely used as a representative example for monitoring resistance genes, especially for horizontal gene transfer (2). Therefore, the assessment of mobile genetic elements from commensal E. coli could highlight the AMR transmission between hosts (3).
Colistin is a cationic antibiotic that has long been regarded as a last resort antibiotic for Enterobacteriaceae infections. However, the widespread use of colistin in animal production acts as a selective pressure for the spread of plasmid-mediated colistin resistance genes, which are in the mcr family. The first discovery of plasmid-mediated colistin resistance (mcr-1 gene) in E. coli from China raised an enormous attention globally and was followed by the subsequent discovery of other mcr resistance genes, including mcr-2, mcr-3, mcr-4, and mcr-5, in different geographical areas (4). Recently, another four colistin resistance genes (mcr-6, mcr-7, mcr-8, and mcr-9) were identified mainly from members in the Enterobacteriaceae family (5–8). Among them, mcr-1 is the most frequently detected in farmed animals and from Enterobacteriaceae infections in humans (9). These reports raised awareness upon colistin usage, especially in livestock animals.
In Thailand, over 80% of pig farming systems are contract farming between the primary producers and the agribusiness companies, for the latter to procure a certain pre-agreed quality and quantity of products at an economical price and is lesser from the primary producers. Antimicrobials including colistin are feed additives or prophylactic agents, including colistin, against bacterial infections in pig farms under veterinary prescription (10). Although there have been a few reports regarding a high prevalence (60–90%) of multidrug-resistant (MDR) E. coli in pigs in Thailand, the antimicrobials used on the farms have not always been clearly defined (11). Since colistin resistance is the crucial epidemiological data of public health concern, monitoring the prevalence of colistin-resistant E. coli and their characteristics is of high priority. The objective of this study was to characterize the antibiogram and virulent traits of mcr-positive E. coli (MCRPE) from the fecal samples of healthy pigs derived from the contract farming system across Thailand.
Samples were collected from 80 farms, in 49 provinces across six regions of Thailand, comprised of 15, 5, 12, 7, 4, and 6 provinces from central, northern, northeastern, eastern, western, and southern Thailand, respectively. Farms were selected based on the available management data, including the antimicrobial usage, housing, vaccination, feed type, and production cycle. However, all historical data was allowed as inclusion criteria for farm selection only but not allowed to be included in the analysis. A total of 696 pooled fecal samples (5–10 samples per farm) were collected from individual 18- to 20-weeks-old fattening pigs with a normal clinical appearance and no recent history of enteric disease or therapeutic antimicrobial treatment.
At least 5 g of feces per pig was collected into a sterile container and kept at 4°C until processed. Then, the fecal samples were homogenized and mixed to get pooled fecal samples with a total mass of 25 g. Then, 5 g of well-mixed feces was collected and diluted 10-fold using sterile 0.85% (w/v) NaCl. Dilutions of 10−7-10−8 were spread on eosin methylene blue agar (Oxoid, UK) plates containing 2 μg/ml colistin sulfate (Sigma-Aldrich, USA) to select for the presumptive colistin-resistant E. coli. The biohazard execution control was approved by the Institutional Biosafety Committee of the Faculty of Veterinary Science, Chulalongkorn University (IBC 1731021). One representative colony with typical E. coli morphology was picked and subcultured to get pure culture. The E. coli species was confirmed using matrix-assisted laser desorption ionization combined with time-of-flight analysis (MALDI Biotyper, Bruker, USA). The principle behind MALDI-TOF is based on mass spectrometry and “soft” ionization technique. Depending on the time of flight of each pathogen, the characteristic spectrum will be analyzed and displayed via the inbuilt software. Briefly, the bacterial colony sample was smeared as a thin film directly on a target plate and then coated with 1 μl polymeric matrix (a saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile and 2.5% trifluoroacetic acid) and air-dried at room temperature. This matrix could penetrate the cell wall of microorganisms and able to extract proteins. The target plate was placed into the mass spectrometer and irradiated by a laser. Afterwards, the molecules vaporized and ionized at the same time into the vacuum and transported to the detection device. Lastly, the computerized database results compared with the reference library database were generated with interpretations (12).
For colistin, the broth microdilution procedure was performed according to the Clinical and Laboratory Standards Institute (CLSI) recommendation (13). The plasmid-mediated colistin resistance genes (mcr-1–5) were detected by multiplex (m)PCR using GoTaq® Green Master Mix (Promega, USA) and the previously reported primers and PCR conditions (14). The E. coli strain CUP13 (15), which is positive for mcr-1 and mcr-3 (confirmed by Sanger sequencing), and ATCC25922 were used as positive and negative controls, respectively. Briefly, the thermocycling conditions were performed at 94°C for 15 min, followed by 25 cycles of 94°C for 30 s, 58°C for 90 s, and 72°C for 1 min, and then followed by 72°C for 10 min.
The minimal inhibitory concentration (MIC) of antimicrobial agents against the E. coli isolates was determined using the AST-GN 38 test kit in a Vitek2 compact automated susceptibility level detection apparatus (BioMérieux, France). The antimicrobial groups selected were synchronized with veterinary guidelines (16). Justification of the antibiotics chosen is for AMR monitoring and for the purpose of public health awareness such as the second generation of cephalosporin, aminoglycoside, fluoroquinolone, and carbapenem. E. coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, and Staphylococcus aureus ATCC 25913 were used as the control strains. The antimicrobials selected were amikacin (AK), amoxicillin (AMX), amoxicillin/clavulanic acid (AMC), ampicillin (AMP), cefalexin (CEX), cefpodoxime (CPD), cefovecin (INN), ceftiofur (XNL), chloramphenicol (C), enrofloxacin (ENR), gentamicin (GEN), imipenem (IMP), marbofloxacin (MBR), nitrofurantoin (NIT), piperacillin (PIP), tetracycline (TET), tobramycin (TOB), and trimethoprim/sulfamethoxazole (SXT). The MIC interpretations will be reported according to Food and Drug Administration (FDA) (17), CLSI (13), and EUCAST values (18). The isolates that presented an extended-spectrum beta-lactamase (ESBL) phenotype were confirmed with a double disc synergy test and phenotypic disc confirmatory test as previously reported (19).
The MCRPE isolates were determined using an approved mPCR identification of their phylogenetic groups and subgroups (A, B1, B2, C, D, E, and F) as reported (20). Each reaction was performed in a 25-μl mixture containing 12.5 μl of GoTaq® Green Master Mix (supplied with Taq polymerase), 20 pmol of each primer, and 200 ng of genomic DNA. The E. coli ATCC 25922 and E. fergusonii CUVET427 (21) strains were used as the controls.
The Enterobacteriaceae plasmid replicons IncF (IncFIA, IncFIB, IncFIC, and IncFrep), IncI1-Ig, IncN, IncP, IncW, IncHI1, IncHI2, IncL/M, IncT, IncA/C, IncK, IncB/O, IncX, and IncY were detected using five mPCR and three simplex PCR tests. The primers, PCR conditions, and thermal cycles were applied as previously reported (22). Briefly, PCR amplifications, except the F-simplex, were thermal cycled at 94°C for 5 min, followed by 30 cycles at 94°C for 1 min, 60°C for 30 s, and 72°C for 1 min, and then followed by 72°C for 5 min. The F-simplex PCR was performed with the same amplification program except at an annealing temperature of 52°C. Positive control samples were provided and used as reported (21).
The sets of mPCR and simplex PCRs were performed as previously reported (23), with the positive control strains taken from the previously sequenced enterotoxigenic E. coli (ETEC) and enterohemorrhagic E. coli (EHEC) strains (24). Primers specific for the StaP (heat-stable toxin a subdivide p), Stb (heat-stable toxin b), Stx2e (Shiga toxin), K88 (Fimbriae), F4 (Fimbriae), and Ltb (heat-labile enterotoxin b subunit) genes were used. The PCR assays were prepared with GoTaq® Green Master Mix (Promega, USA) and thermocycled at 94°C for 10 min, followed by 30 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1.5 min increasing by 3 s each cycle, and then followed by 72°C for 10 min.
The colistin resistance rates are presented as percentages divided by region and province in comparison of the rate with and without the mcr genes, and the antimicrobial resistance profiles are reported as the antibiogram patterns of mcr-positive E. coli. The patterns of virulence gene profiles among MCRPE isolates are presented in percentages. To define MDR and pathogenic traits among the colistin-resistant E. coli, the relation between AMR phenotypes and pathotype characteristics was analyzed using Fischer's exact test (p ≤ 0.05).
A total of 105 colistin-resistant E. coli from the 696 samples were isolated using the eosin methylene blue (EMB) media. From the broth microdilution method, the MCRPE isolates had MIC values of 4 (n = 17) or 8 (n = 14) μg/ml. From the PCR detection, the mcr-1 gene was found in 31 of these 105 colistin-resistant E. coli isolates, and among them, three isolates were found to also express mcr-3. The distributions of colistin-resistant E. coli were from central (5.4%) (Phetchabun, Nakhon Pathom, Ang-Thong, and Lopburi), western (0.4%) (Ratchaburi), and eastern (1.4%) (Chonburi) Thailand. The geographical distributions of E. coli with or without mcr genes are shown in Figure 1.
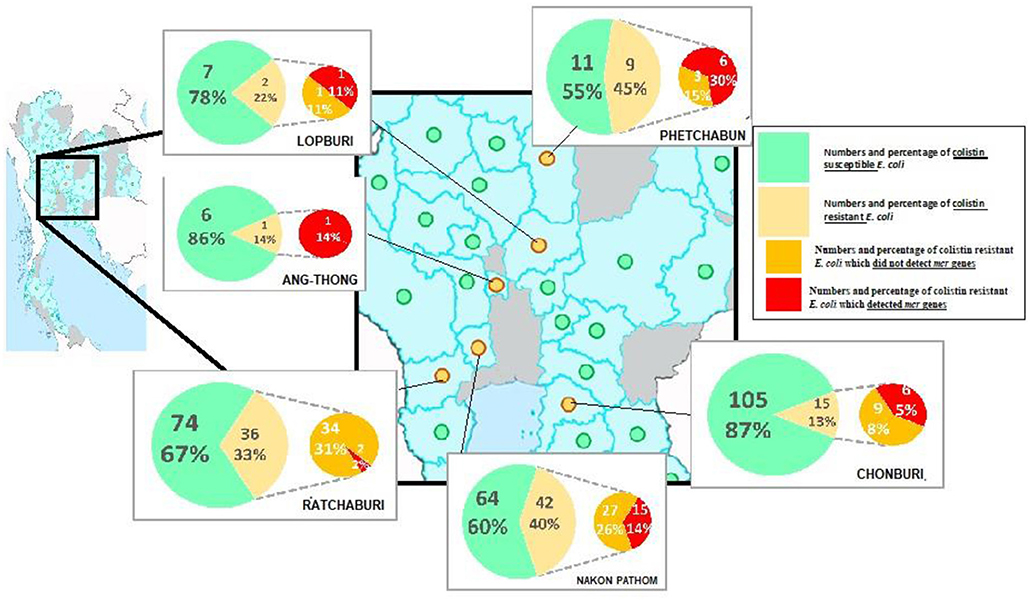
Figure 1. Geographical distribution of either colistin resistant or susceptible E. coli from the surveyed contracted pig farms in Thailand.
All 31 MCRPE were multidrug resistant (Figure 2), with all being resistant to AMX, AMP, PIP, and TET, and over 50% were resistant to CEX, INN, XNL, GEN, ENR, C, and the SXT combination. No pan-drug resistance was detected among the MCRPE isolates. ESBL was found in 32.3% (10/31) mcr-1 positive isolates. A total of 26 antibiogram patterns were recorded for 31 MCRPE isolates. Forty-eight percent (15/31) of these isolates were MDR with resistance to six antimicrobial groups (Table 1).
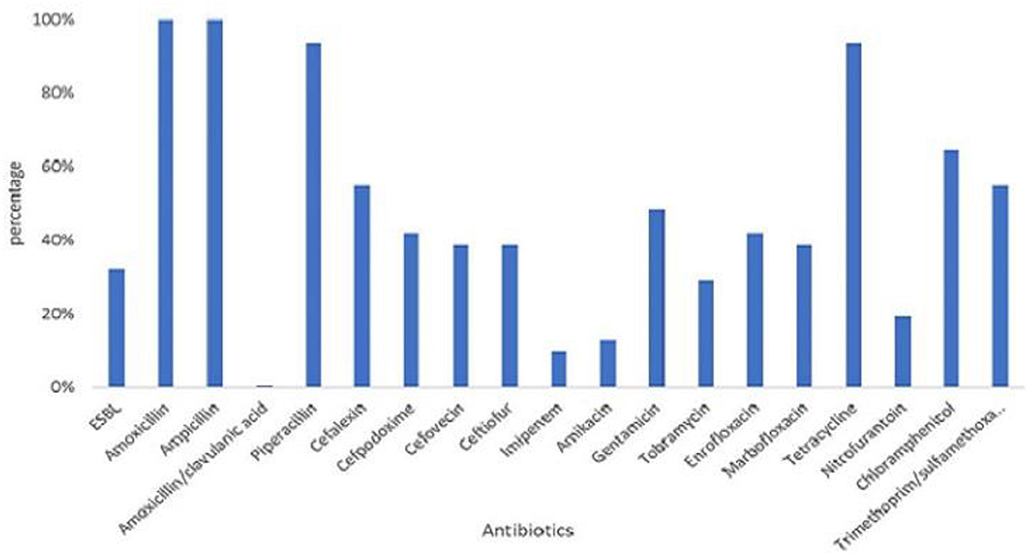
Figure 2. Distribution of resistant rates against 18 antimicrobials and presence of extended- spectrum beta-lactamase (ESBL) characteristic among 105 colistin-resistant E. coli isolated from contracted pig farms in Thailand.
Most isolates were from phylogenetic group A (51.6%), followed by group B1 (29%) and groups E (12.9%), B2 (3.2%), and F (3.2%) (Figure 3).
The predominantly found plasmid replicons were of the IncF and IncFIB replicon types at 80.6 and 61.3%, respectively. Plasmid replicon types L/M, W, Y, A/C, T, and K were not detected in this study (Figure 4). The other replicon types were found at low prevalence rates among the MCRPE isolates, with IncX, IncB/O, and IncHI1 being present at the lowest percentages (3.2%).
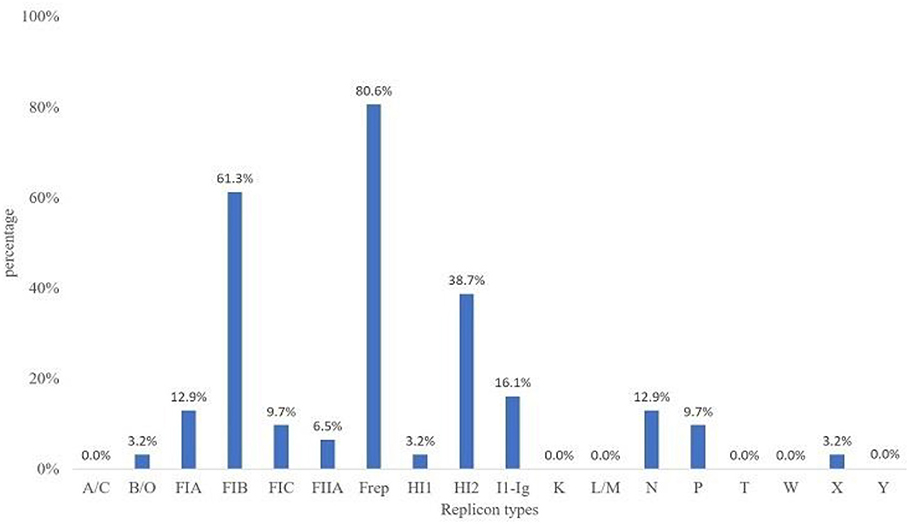
Figure 4. Plasmid replicon types detected among 31 MCRPE isolates in contracted pig farms in Thailand.
The virulent genes representing ETEC or EHEC were found in 18 out of 31 (58.1%) MCRPE isolates (Table 2). The ETEC strains possessed the StaP and Stb enterotoxin-encoding genes as the most frequent pathotype, and one strain (from Phetchabun province) showed a hybrid ETEC–EHEC genotype.
The association between antimicrobial susceptibility and pathogenicity of the 31 MCRPE isolates was analyzed by Fischer's exact test (Table 3). No association between pathogenicity and resistance to the six antibiotic groups was found (fluoroquinolones, sulfonamides, tetracyclines, nitrofurazones, phenicols, and aminoglycosides) (p = 0.28, 1.00, 1.00, 1.00, 1.00, and 0.15, respectively).
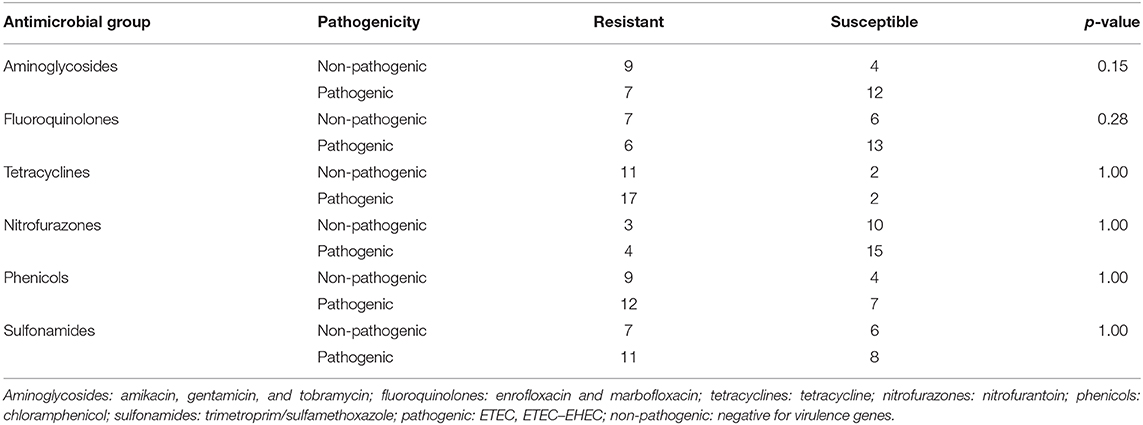
Table 3. Relation analysis between MCRPE resistance to the other six antimicrobial groups and their pathogenicity.
This national-scale study of contract-farmed pigs in Thailand confirmed the existence of colistin-resistant E. coli containing mcr genes and that they showed diversity in their phylogenetic group, replicon type, antibiogram, ESBL trait, and pathogenic potential. All recruited contracted pig farms had strict historical data and management records that can be traced back as an essential inclusion criteria. The sample collection criteria were set up and executed by the farm workers under the authority of veterinarians. In this study, MALDI-TOF MS was used for the identification and confirmation of bacteria strains. This technique has emerged as a powerful technique for the identification of microorganisms with an overall 95% accuracy at the species level. The main advantage of MADLI-TOF is being able to identify bacterial species directly from the culture plates as fast as 1 to 15 min in a few simple steps (12).
According to mPCR, our results indicated the lower resistance rate of mcr-1 (4.4% or 31/696) when compared with a previous report from healthy pigs in China (21%) (20). This study covered all parts of Thailand where high-intensity pig farming is done. Unfortunately, all the historical data could not be analyzed due to the company's policy. However, the positive areas were distributed in the western, central, and eastern parts within a radius of about 300 km. The distributions of colistin-resistant E. coli were higher (15–30%) in Nakhon Pathom, Ratchaburi, Chonburi, Lopburi, and Phetchabun provinces. These provinces reported to have a huge number of pig farms and total number of pigs. Colistin was legally use in pig feeds for prophylactic purposes in Thailand until March 2018. The high percentage of MCRPE isolates in certain provinces might come from prolonged cumulative selective pressure from their history of colistin usage in pig feeds. To the best of our knowledge, this is the first report of mcr-1 gene in E. coli isolates from pigs in Thailand. Interestingly, three of the mcr-1-positive isolates also co-expressed mcr-3. These results could highlight the awareness of the distribution of mcr genes and for the national policy of livestock immigration. The mcr-1 genes have been widely shown to be distributed in Asia, Europe, Africa, and America and primarily due to the consequence of long-term colistin application in animals (25). The mcr-3 gene was first reported in China in 2017 (26) and the prevalence and spread of the mcr-3 gene in Thailand should be carefully monitored from now on.
According to phylogenetic grouping, the majority of the isolates in our study were in phylogroups A or B1, predominantly related with commensal strains (27). On the other hand, for the virulent E. coli groups, phylogroup D was not detected in the current study and there was a low frequency of phylogroup B2. Several studies have reported that phylogroups B2 and D were associated with intestinal and extraintestinal pathogenic E. coli as well as MDR strains (28, 29). Nonetheless, even commensal E. coli from various phylogroups have been reported to harbor pathogenicity islands that can serve as integration sites for virulence and/or AMR determinants (30) and so may facilitate in converting commensal strains to pathogens.
With respect to plasmid replicon typing, the IncFIB and F plasmids were the most commonly found replicon types in this study. They are narrow host-range-type plasmids, which have been reported in worldwide members of the Enterobacteriaceae family, associated with various antimicrobial-resistant genes (31). The mcr-1 and mcr-3 genes were previously described on the IncI, IncHI2, and IncX4 plasmids (32). A variety of replicon types were found in the MCRPE isolates in this study, which suggest that the mcr genes can locate and/or transfer to different plasmid types. This is in accordance with a previous report that the mcr-1 genes and ESBL could be co-transferred by more than one type of conjugative plasmid, which might alleviate their effective dissemination among bacteria (33).
The antibiogram profiles characterized among the MCRPE isolates revealed that MDR was a common phenotype in this study. E. coli resistance to beta-lactam and the tetracycline antibiotic groups was very common in Thailand, and aminoglycoside and fluoroquinolone resistance was found to be varied in farm management such as using antibiotic for prophylactic or treatment purposes (21). The MDR traits among mcr-1-positive E. coli have been reported frequently in pigs due to the usage of antibiotics in the production cycle (34). Interestingly, ESBLs were found at a high prevalence among the MCRPE isolates of this study, which might due to co-selection under selective pressure (33). Moreover, E. coli plasmids that harbor co-localization of mcr-1 and blaCTX−M genes and/or mcr-1 and blaNDM−5 genes have been reported previously (35). Genomic characterization should be performed to resolve the reason for this apparent correlation.
The presence of the Ltb, Stb, StaP, Stx2e, and K88 virulence genes in MCRPE isolates indicated that they also had the potential to cause an infection. Thus, healthy pigs could be an important reservoir of colistin-resistant ETEC. Interestingly, one MCRPE isolate was found to be an ETEC–EHEC hybrid strain. E. coli with highly virulent hybrid pathotype strains had been reported previously both in animals and human diarrhea patients (36). Since many of the virulence genes of E. coli are carried on mobile genetic elements, the genetic combination of these MGE resulted in the emergence of STEC/ETEC hybrid strains in multiple events (37). The recent finding of a clone of sequence type (ST) 95 showing extreme drug resistance with a high virulence potential underscores the need to monitor new and emerging trends in antibiotic resistance development in this important global lineage (38). On the other hand, aminoglycoside- and fluoroquinolone-resistant E. coli seemed to have a lower probability to act as an ETEC pathotype in this study. Pathogenic E. coli tends to be more susceptible to many antimicrobials (39). However, the mechanism is still not elucidated and clonal typing should be included for a more convincing analysis.
In conclusion, a low carriage rate of mcr-1 and mcr-3 co-positive E. coli was detected in large-scale contract pig farms in Thailand. The MCRPE isolates showed MDR E. coli and most of the isolates contained virulence genes representing an ETEC pathotype. These data provide an insight into the occurrence of colistin resistance among E. coli in healthy pig carriages and their characteristics, in terms of virulence genes and antibiograms. However, genomic characterization of mcr genes found in Thailand is required for further study.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethical review and approval was not required for the animal study because all fecal samples were submitted from veterinarians in pig industrial field to the veterinary diagnostic laboratory as the annual surveillance. We asked the permission to use these sort of samples which did not directly collect the feces by our team. However, the biohazard execution control was approved by the Institutional Biosafety Committee of the Faculty of Veterinary Science, Chulalongkorn University (IBC 1731021).
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
The present study was supported financially by the Royal Golden Jubilee Ph.D. (RGJPHD) program, Agricultural Research Development Agency: ARDA (CRP6205031110), the CHE-TRF Senior Research Fund (RTA6280013), Thailand Science Research and Innovation, Pathogen Bank, Faculty of Veterinary Science, Chulalongkorn University, Bangkok, Thailand, and Chulalongkorn Academic Advancement into its 2nd Century Project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2020.582899/full#supplementary-material
1. Organization WH. WHO Global Strategy for Containment of Antimicrobial Resistance. Geneva: World Health Organization (2001).
2. Kaspar H. Results of the antimicrobial agent susceptibility study raised in a representative, cross-sectional monitoring study on a national basis. Int J Med Microbiol. (2006) 296(Suppl.41):69–79. doi: 10.1016/j.ijmm.2006.01.063
3. European Food Safety Authority European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2014. EFSA J. (2016) 14:4380. doi: 10.2903/j.efsa.2016.4380
4. Duggett NA, Randall LP, Horton RA, Lemma F, Kirchner M, Nunez-Garcia J, et al. Molecular epidemiology of isolates with multiple MCR plasmids from a pig farm in Great Britain: the effects of colistin withdrawal in the short and long term. J Antimicrob Chemother. (2018) 73:3025–33. doi: 10.1093/jac/dky292
5. AbuOun M, Stubberfield EJ, Duggett NA, Kirchner M, Dormer L, Nunez-Garcia J, et al. mcr-1 and mcr-2 (mcr-6.1) variant genes identified in Moraxella species isolated from pigs in Great Britain from 2014 to 2015. J Antimicrob Chemother. (2018) 73:2904. doi: 10.1093/jac/dky272
6. Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. (2018) 73:1791–5. doi: 10.1093/jac/dky111
7. Wang X, Wang Y, Zhou Y, Li J, Yin W, Wang S, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. (2018) 7:122. doi: 10.1038/s41426-018-0124-z
8. Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible salmonella enterica serotype typhimurium isolate. mBio. (2019) 10:e00853–19. doi: 10.1128/mBio.00853-19
9. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Euro Surveill. (2016) 21:30155. doi: 10.2807/1560-7917.ES.2016.21.9.30155
10. Lugsomya K, Yindee J, Niyomtham W, Tribuddharat C, Tummaruk P, Hampson DJ, et al. Antimicrobial resistance in commensal Escherichia coli isolated from pigs and pork derived from farms either routinely using or not using in-feed antimicrobials. Microb Drug Resist. (2018) 24:1054–66. doi: 10.1089/mdr.2018.0154
11. Lay KK, Koowattananukul C, Chansong N, Chuanchuen R. Antimicrobial resistance, virulence, and phylogenetic characteristics of Escherichia coli isolates from clinically healthy swine. Foodborne Pathog Dis. (2012) 9:992–1001. doi: 10.1089/fpd.2012.1175
12. Singhal N, Kumar M, Kanaujia PK, Virdi JS. MALDI-TOF mass spectrometry: an emerging technology for microbial identification and diagnosis. Front Microbiol. (2015) 6:791. doi: 10.3389/fmicb.2015.00791
13. Clinical M100 LSIJCs. Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: Clinical and Laboratory Standards Institute (2017).
14. Rebelo AR, Bortolaia V, Kjeldgaard JS, Pedersen SK, Leekitcharoenphon P, Hansen IM, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. (2018) 23:672. doi: 10.2807/1560-7917.ES.2018.23.6.17-00672
15. Lugsomya K, Chanchaithong P, Tribudharat C, Thanawanh N, Niyomtham W, Prapasarakul N. Prevalence of plasmid-mediated colistin resistance genes (mcr-1) in commensal Escherichia coli from fattening pigs in Thailand. Thai J Vet Med. (2016) 46:2016.
17. Food and Drug Administration. Microbiology Data for Systemic Antibacterial Drugs — Development, Analysis, and Presentation Guidance for Industry. Washington, DC: Food and Drug Administration Center for Drug Evaluation and Research (CDER) (2018).
18. EUCAST. The European Committee on Antimicrobial Susceptibility Testing: Breakpoint Tables for Interpretation of MICs. (2018). Available online at: http://www.eucast.org
19. Dhara M, Disha P, Sachin P, Manisha J, Seema B, Vegad MJNJMR. Comparison of various methods for the detection of extended spectrum beta-lactamase in Klebsiella pneumoniae isolated from neonatal intensive care unit. Natl J Med Res. (2012) 2:348–53.
20. Clermont O, Christenson JK, Denamur E, Gordon DM. The Clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. (2013) 5:58–65. doi: 10.1111/1758-2229.12019
21. Lugsomya K, Chatsuwan T, Niyomtham W, Tummaruk P, Hampson DJ, Prapasarakul NJMDR. Routine prophylactic antimicrobial use is associated with increased phenotypic and genotypic resistance in commensal Escherichia coli isolates recovered from healthy fattening pigs on farms in Thailand. Microb Drug Resist. (2018) 24:213–23. doi: 10.1089/mdr.2017.0042
22. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. (2005) 63:219–28. doi: 10.1016/j.mimet.2005.03.018
23. Casey TA, Bosworth BT. Design and evaluation of a multiplex polymerase chain reaction assay for the simultaneous identification of genes for nine different virulence factors associated with Escherichia coli that cause diarrhea and edema disease in swine. J Vet Diagn Invest. (2009) 21:25–30. doi: 10.1177/104063870902100104
24. Prapasarakul N, Tummaruk P, Niyomtum W, Tripipat T, Serichantalergs O. Virulence genes and antimicrobial susceptibilities of hemolytic and nonhemolytic Escherichia coli isolated from post-weaning piglets in central Thailand. J Vet Med Sci. (2010) 72:1603–8. doi: 10.1292/jvms.10-0124
25. Elbediwi M, Li Y, Paudyal N, Pan H, Li X, Xie S, et al. Global burden of colistin-resistant bacteria: mobilized colistin resistance genes study (1980-2018). Microorganisms. (2019) 7:461. doi: 10.3390/microorganisms7100461
26. Yin W, Li H, Shen Y, Liu Z, Wang S, Shen Z, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. (2017) 8:e00543–17. doi: 10.1128/mBio.00543-17
27. Yilmaz ES, Aslantas O. Phylogenetic group/subgroups distributions, virulence factors, and antimicrobial susceptibility of Escherichia coli Strains from urinary tract infections in Hatay. Rev Soc Bras Med Trop. (2020) 53:e20190429. doi: 10.1590/0037-8682-0429-2019
28. Sarshar M, Scribano D, Marazzato M, Ambrosi C, Aprea MR, Aleandri M, et al. Genetic diversity, phylogroup distribution and virulence gene profile of PKS positive Escherichia coli colonizing human intestinal polyps. Microb Pathog. (2017) 112:274–8. doi: 10.1016/j.micpath.2017.10.009
29. Iranpour D, Hassanpour M, Ansari H, Tajbakhsh S, Khamisipour G, Najafi A. Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. Biomed Res Int. (2015) 2015:846219. doi: 10.1155/2015/846219
30. Raimondi S, Righini L, Candeliere F, Musmeci E, Bonvicini F, Gentilomi G, et al. Antibiotic resistance, virulence factors, phenotyping, and genotyping of E. coli isolated from the feces of healthy subjects. Microorganisms. (2019) 7:251. doi: 10.3390/microorganisms7080251
31. Johnson TJ, Wannemuehler YM, Johnson SJ, Logue CM, White DG, Doetkott C, et al. Plasmid replicon typing of commensal and pathogenic Escherichia coli isolates. Appl Environ Microbiol. (2007) 73:1976–83. doi: 10.1128/AEM.02171-06
32. Kieffer N, Nordmann P, Moreno AM, Zanolli Moreno L, Chaby R, Breton A, et al. Genetic and functional characterization of an MCR-3-like enzyme-producing Escherichia coli isolate recovered from Swine in Brazil. Antimicrob Agents Chemother. (2018) 62:e00278–18. doi: 10.1128/AAC.00278-18
33. Wu C, Wang Y, Shi X, Wang S, Ren H, Shen Z, et al. Rapid rise of the ESBL and mcr-1 genes in Escherichia coli of chicken origin in China, 2008–2014. Emerg Microbes Infect. (2018) 7:30. doi: 10.1038/s41426-018-0033-1
34. Rhouma M, Theriault W, Rabhi N, Duchaine C, Quessy S, Fravalo P. First identification of mcr-1/mcr-2 genes in the fecal microbiota of Canadian commercial pigs during the growing and finishing period. Vet Med. (2019) 10:65–7. doi: 10.2147/VMRR.S202331
35. Rhouma M, Letellier A. Extended-spectrum β-lactamases, carbapenemases and the mcr-1 gene: is there a historical link? Int J Antimicr Agents. (2017) 49:269–71. doi: 10.1016/j.ijantimicag.2016.11.026
36. Leonard SR, Mammel MK, Rasko DA, Lacher DW. Hybrid shiga toxin-producing and enterotoxigenic Escherichia sp. cryptic lineage 1 strain 7v harbors a hybrid plasmid. Appl Environ Microbiol. (2016) 82:4309–19. doi: 10.1128/AEM.01129-16
37. Prager R, Fruth A, Busch U, Tietze E. Comparative analysis of virulence genes, genetic diversity, and phylogeny of Shiga toxin 2g and heat-stable enterotoxin STIa encoding Escherichia coli isolates from humans, animals, and environmental sources. Int J Med Microbiol. (2011) 301:181–91. doi: 10.1016/j.ijmm.2010.06.003
38. Forde BM, Zowawi HM, Harris PNA, Roberts L, Ibrahim E, Shaikh N, et al. Discovery of mcr-1-mediated colistin resistance in a highly virulent Escherichia coli Lineage. mSphere. (2018) 3:e00486–18. doi: 10.1128/mSphere.00486-18
Keywords: colistin resistance, Escherichia coli, mcr genes, pigs, virulent factor
Citation: Khine NO, Lugsomya K, Kaewgun B, Honhanrob L, Pairojrit P, Jermprasert S and Prapasarakul N (2020) Multidrug Resistance and Virulence Factors of Escherichia coli Harboring Plasmid-Mediated Colistin Resistance: mcr-1 and mcr-3 Genes in Contracted Pig Farms in Thailand. Front. Vet. Sci. 7:582899. doi: 10.3389/fvets.2020.582899
Received: 13 July 2020; Accepted: 02 October 2020;
Published: 10 November 2020.
Edited by:
Marina Spinu, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaReviewed by:
Sidharath Dev Thakur, Chaudhary Sarwan Kumar Himachal Pradesh Krishi Vishvavidyalaya, IndiaCopyright © 2020 Khine, Lugsomya, Kaewgun, Honhanrob, Pairojrit, Jermprasert and Prapasarakul. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuvee Prapasarakul, bnV2ZWUucEBjaHVsYS5hYy50aA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.