- 1College of Veterinary Medicine, Yangzhou University, Yangzhou, China
- 2Jiangsu Co-innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, Yangzhou, China
- 3Joint International Research Laboratory of Agriculture and Agri-Product Safety, The Ministry of Education of China, Yangzhou, China
- 4College of Agriculture and Forestry Science, Linyi University, Linyi, China
Bovine mastitis is a prevalent disease that causes serious economic problems globally in the dairy industry. Staphylococcus aureus is an important pathogen of bovine mastitis. This study was conducted to characterize S. aureus isolates from clinical bovine mastitis cases in large-scale dairy herds in China. S. aureus was isolated from 624 clinical mastitis cases and confirmed by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS). In total, 62 S. aureus isolates were obtained. Cluster analysis, genetic diversity, quantification of biofilm formation, antimicrobial resistance, and detection of virulence genes were performed on these isolates of S. aureus. Eight isolates harbored the mecA gene and were sensitive to oxacillin. MALDI-TOF MS cluster analysis revealed that the 62 isolates were divided into three major clusters (I, II, III) and eight main groups (A–H) at the distance level of 700. The agr II was the most prevalent (56.5%). The 62 S. aureus isolates were assigned to seven spa types. The most common spa type was t529(58.1%), followed by t2196 (14.5%), t518 (14.5%), t571(6.5%), t034 (3.2%), t2734 (1.6%), and t730 (1.6%). Five STs were identified from seven representative isolates as follows: ST630/CC8, ST97/CC97, ST50, ST398, and ST705. All isolates had the ability to form biofilm. Antimicrobial resistance was most frequently observed to ciprofloxacin (29%), followed by penicillin (24.2%), and streptomycin (9.6%). All isolates harbored the fnbA, clfB (100%), icaA, and icaD genes. This study provides the basis for the development of bovine mastitis prevention program on large-scale dairy farms.
Introduction
Bovine mastitis, inflammation of the mammary gland, is predominantly induced by intramammary bacterial infection, and causes serious economic losses in the global dairy industry (1). Staphylococcus aureus (S. aureus) is one of the most important and common pathogenic microorganisms in bovine mastitis (2). S. aureus carries many virulence factors, such as hemolysins (Hla and Hlb), toxic shock syndrome toxin-1 (Tsst-1), leukocidin, fibronectin binding proteins (FnbA and FnbB), and clumping factors (ClfA and ClfB), which facilitate adhesion of S. aureus to the host extracellular matrix components, damaging host cells and impeding the immune system (3). S. aureus has developed antimicrobial resistance due to selective pressures from the indiscriminate use of antimicrobial agents (4), and multidrug-resistant (MDR) strains have emerged including methicillin-resistant S. aureus (MRSA). Furthermore, biofilm formation helps S. aureus survive in the pressure of antimicrobial agents and evade the host immune response, which can give rise to a persistent infection (5). Thus, mastitis caused by S. aureus has a low cure rate in cattle.
Conventional methods used for the epidemiological studies of S. aureus include pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), polymorphism of protein A gene (spa typing), and accessory gene regulator typing (agr typing). Utilization of MALDI-TOF MS for identification and classification of species of the genus Staphylococcus has been evaluated in both human and veterinary medicine (6, 7). However, few studies have assessed the value of MALDI-TOF MS as an epidemiological typing tool in bovine mastitis. The MALDI-TOF MS fingerprinting approach could extend phenotypic and genotypic approaches, allowing for more detailed classification of S. aureus.
In China, large-scale dairy farms have developed rapidly in recent years and have become the mainstay of raw milk production. Although mastitis control programs have achieved considerable progress, the incidence of clinical mastitis remains high in large dairy herds in China (8). S. aureus infection can sometimes result in acute and clinical mastitis with changes in milk composition, leading to milk being discarded and even culling of cows. If appropriate control strategies are not taken in time to prevent the transmission of S. aureus in herds, immeasurable losses can occur on large-scale dairy farms. It is thus necessary to monitor the incidence of clinical mastitis caused by S. aureus on large-scale dairy farms. Moreover, data analysis regarding the epidemiology of S. aureus can provide references for developing scientific prevention and control programs. Currently, there is a lack of relevant data available for large-scale farms. The objective of this study was to provide epidemiological information on S. aureus isolates from large-scale Chinese dairy herds by examining cluster analysis, genetic diversity, biofilm formation, antimicrobial resistance, and virulence genes.
Materials and Methods
Samples
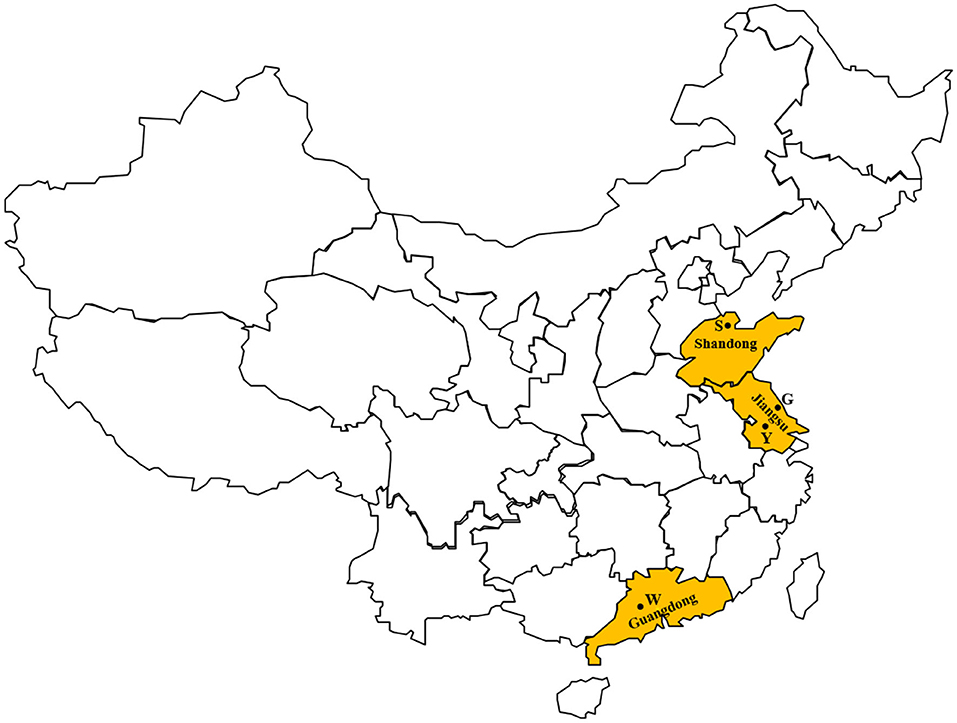
A total of 624 milk samples from cases of clinical bovine mastitis were collected aseptically from four farms between May 2016 and August 2017. These milk samples were collected from all the clinical mastitis cases during the investigation. Mastitis was initially identified by farm staffs at milking time, and was confirmed by a veterinarian. The four farms were in three provinces of China, including Shandong (n = 1), Jiangsu (n = 2), and Guangdong (n = 1) (Figure 1). These provinces are located in the eastern coastal areas, and have distinctly different climates. Shandong province has a cool and dry climate, while it is warm and humid in Guangdong province. Jiangsu province is in a transitional zone between temperate and subtropical zones, and has a mild climate. According to a report of the Dairy Association of China, the average cow population of Chinese farms was 166 in 2019. In a previous investigation, farms in China with a cow population more than 500 were defined as large-scale farms (8). In this study, there were at least 1,000 lactating Holstein-Friesian cows on the farms, hence these were large-scale dairy farms. Related information on the four farms is shown in Table 1.
Isolation and Identification of S. aureus
Milk samples (100 μL) were inoculated on Baird-Parker agar (BPA) supplemented with 5% egg yolk and tellurite (Hope Biotech Co., Ltd, Qingdao, China), and cultured at 37°C for 24 h. S. aureus isolates were initially identified by morphology and biochemical methods (colony morphology, Gram staining, and catalase testing), then were confirmed by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany). MRSA strains were confirmed by targeting the mecA gene and oxacillin disc diffusion test.
Cluster Analyses of S. aureus Isolates
Cluster analyses of S. aureus isolates were conducted using MALDI Biotyper OC version 4.0.19 (Bruker Daltonics, Bremen, Germany).
Molecular Typing
The agr types I–IV were determined by multiplex PCR assay as described previously (9). Briefly, a 25 μL reaction mixture was prepared containing 12.5 μL 2 × EasyTaq PCR SuperMIX (TransGen Biotech Co., Ltd, Beijing, China), 2 μL DNA, 1 μL each of forward and reverse primers (10 μmoL), and 8.5 μL ultrapure water. The thermal cycling program comprised an initial denaturation at 94°C for 3 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s, and a final extension at 72°C for 5 min. The polymorphic X region of the spa gene was amplified by the method described previously (1). MLST was performed as described by Enright et al. and the seven housekeeping genes were amplified by PCR (10). PCR amplicons were submitted to Tsingke Biological Technology Co., Ltd (Nanjing, China) for sequencing. The spa types were determined using the spa-server (http://spa.ridom.de/;) (11). The allelic profile of each strain was identified and assigned to the respective sequence type (ST) using the PubMLST database (https://pubmlst.org).
Biofilm Formation
Quantification of biofilm was performed by spectrophotometry in microplates using crystal violet staining as described previously (12). Isolates were classified into the following categories: no biofilm producer, weak, moderate, or strong biofilm producer according to a previous method (13).
Antimicrobial Susceptibility Tests
Antimicrobial susceptibility of S. aureus isolates was determined by the agar disk diffusion method according to the guideline of the Clinical Laboratory Standards Institute (14). Antimicrobial agents often used on farms to treat bovine mastitis were selected and included the following: penicillin (P, 10 U), cephalexin (CEP, 30 μg), ceftiofur (EFT, 30 μg), gentamicin (CN, 10 μg), streptomycin (STR, 10 μg), clindamycin (DA, 2 μg), ciprofloxacin (CIP, 5 μg), doxycycline (DX, 30 μg), tetracycline (TE, 30 μg), amikacin (AK, 30 μg), and kanamycin (KAN, 30 μg). All antimicrobial agents were purchased from Hangzhou Microbial Reagent Co., Ltd., except ceftiofur (Oxoid). MDR isolates were defined as showing resistance to three or more antimicrobial agents (4).
Detection of Virulence Determinants
Virulence genes, including lukM, fnbA, fnbB, clfA, clfB, hlα, hlβ, icaA, icaD, pvl, bap, and tsst-1 were detected by PCR as described previously (15–21). Positive and negative controls were included in all PCRs.
Statistical Analysis
Statistical analysis was performed with SPSS Statistics 22.0 software (IBM, USA). Chi-square tests were used to analyze the association between multidrug resistance and biofilm formation ability. A p-value of < 0.05 was deemed to be statistically significant.
Results
Incidence of S. aureus and MRSA
The incidence of clinical mastitis was 14.8, 5.5, 1.1, and 1.3% on farms S, Y, G, and W, respectively. From the milk samples, 62 S. aureus isolates were obtained, including 18 isolates in 301 Shandong samples, 39 isolates in 183 Jiangsu samples, and 5 isolates in 140 Guangdong samples. The proportion of clinical mastitis caused by S. aureus was 6, 14.5, 24.2, and 3.6% on farm S, Y, G, and W, respectively. Eight isolates of S. aureus from farm Y harbored the mecA gene and were sensitive to oxacillin, hence were classed as oxacillin-susceptible and mecA-positive strains (OS-MRSA).
Cluster Analyses of S. aureus Isolates
A dendrogram was generated based on the protein spectral fingerprints of the 62 S. aureus isolates (Figure 2). This MALDI-TOF MS cluster analysis showed that the 62 isolates were divided into three major clusters (I, II, III) and eight main groups (A–H) at the distance level of 700. The majority of isolates (83.9%, 52/62) were classified into cluster III. Cluster I and II contained 1 and 9 isolates, respectively. In groups F and H, the isolates obtained from different farms were classified into sub-clades at a close relative distance (<200). Eight mecA-positive S. aureus strains were discriminated from the other 54 methicillin-sensitive S. aureus (MSSA) and were grouped in clusters I and II.
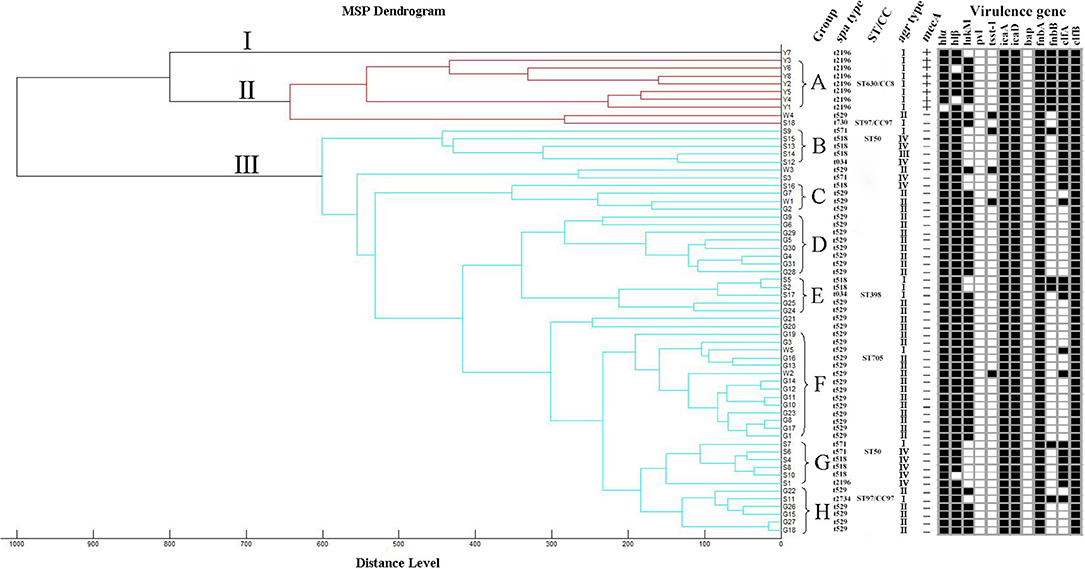
Figure 2. Dendrogram and spa types, ST/CC, agr types, mecA gene, and virulence genes. MSP dendrogram generated by the standard MALDI Biotyper MSP creation method for 62 S. aureus isolates. The scale below the dendrogram indicates the relative distance. Letters (A–H) represent 8 groups. The detection of virulence genes was summarized by a heat map. Black squares denote that the studied genes were detected. White squares denote that those isolates lack these genes.
Molecular Typing
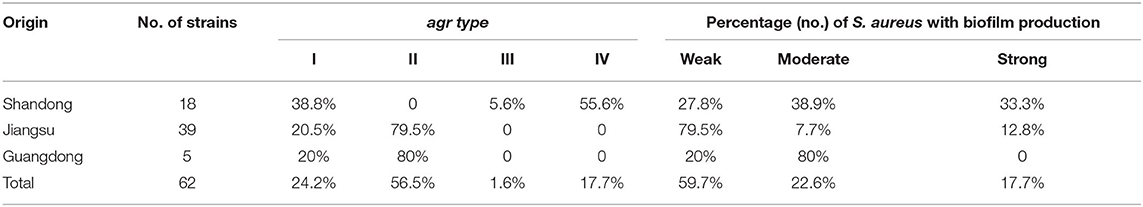
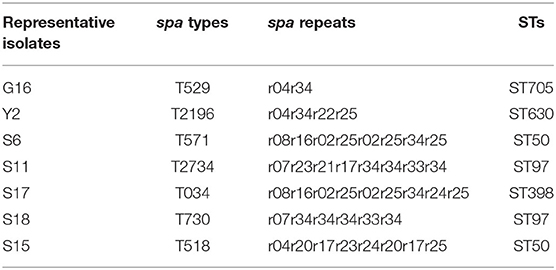
The agr alleles were successfully identified in the 62 isolates by multiplex PCR. As shown in Table 2, agr II was most prevalent (56.5%; 35/62), followed by agr I (24.2%; 15/62), agr IV (17.7%; 11/62), and agr III (1.6%; 1/62). The most common agr type in S. aureus isolated from Shandong herd was agr IV, whereas, the agr II was predominant in S. aureus isolates obtained from herds in Jiangsu and Guangdong provinces. The 62 S. aureus isolates were also assigned to seven spa types. The predominant spa type was t529 (58.1%; 36/62), followed by t2196 (14.5%; 9/62), t518 (14.5%; 9/62), t571 (6.5%; 4/62), t034 (3.2%; 2/62), t2734 (1.6%, 1/62), and t730 (1.6%, 1/62) (Figure 2). The spa type t529 was found in S. aureus isolated from farms G and W. Six spa types (t2196, t571, t2734, t034, t730, and t518) were found in S. aureus isolated from farm S. Seven representative isolates that belonged to different spa types were then subjected to MLST. Five sequence types (STs) were identified as follows: ST630/CC8, ST97/CC97, ST50, ST398, and ST705. The corresponding relationship between STs and spa types is shown in Table 3.
Biofilm Formation
All isolates had the ability to form biofilms. The rates of weak, moderate, and strong biofilm producers were 59.7% (37/62), 22.6% (14/62), and 17.7% (11/62), respectively (Table 2). Among the S. aureus isolates obtained from the Shandong herd, 27.8% (5/18) were weak biofilm producers, 38.9% (7/18) were moderate biofilm producers, and 33.3% (6/18) were strong biofilm producers. Most isolates of S. aureus from the Jiangsu herds showed a weak biofilm-forming ability, whereas, isolates obtained from the Guangdong herd were predominantly moderate biofilm producers.
Antimicrobial Susceptibility
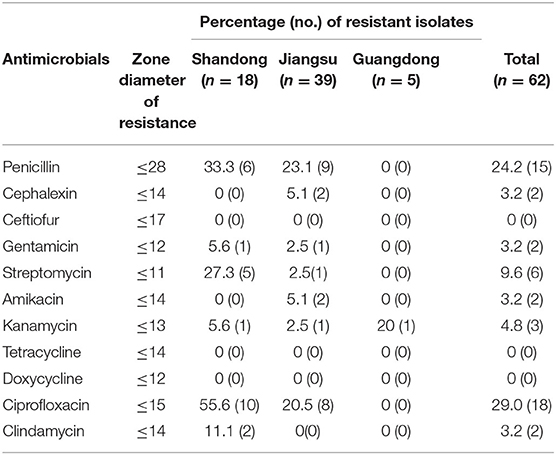
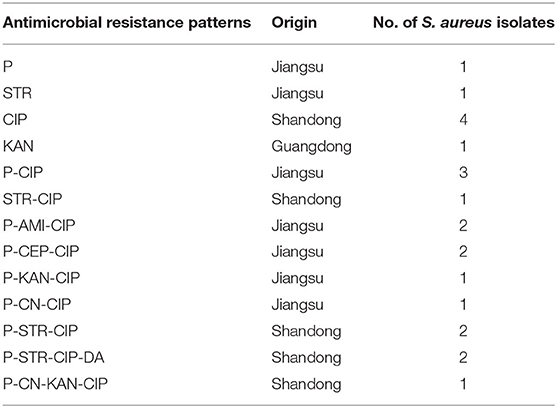
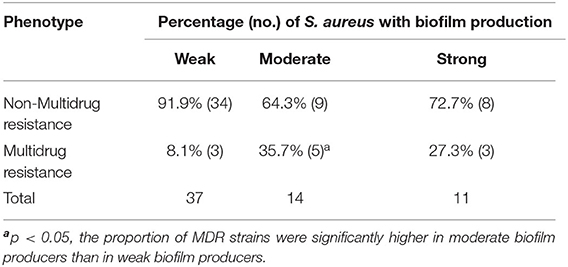
Antimicrobial resistance was most frequently observed to ciprofloxacin (29%,18/62), followed by penicillin (24.2%, 15/62), streptomycin (9.6%, 6/62), kanamycin (3.2%, 2/62), cephalexin (3.2%, 2/62), gentamicin (3.2%, 2/62), amikacin (3.2%, 2/62), and clindamycin (3.2%, 2/62). All isolates were sensitive to ceftiofur, tetracycline, and doxycycline (Table 4). Thirteen resistance patterns were identified and 11 isolates (17.7%, 11/62) were characterized as multidrug resistant (MDR) (Table 5). All MDR isolates were from the Jiangsu and Shandong herds. The relationship between prevalence of MDR strains and biofilm formation ability of the S. aureus isolates was further analyzed (Table 6). The proportion of MDR isolates was significantly higher in moderate biofilm producers than in weak biofilm producers (p < 0.05).
Detection of Virulence Genes
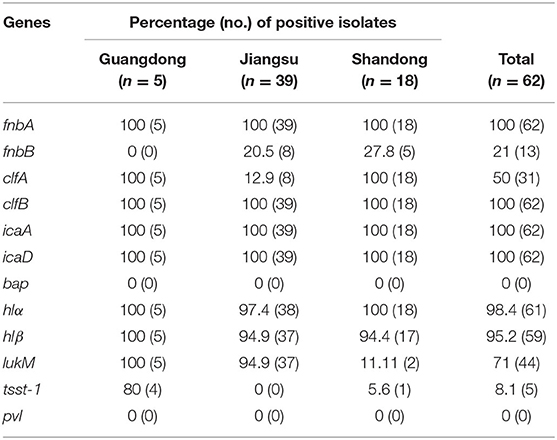
The 12 virulence genes examined in this study were distributed with varying frequencies among the S. aureus isolates (Table 7). The genes fnbA, clfB, icaA, and icaD were detected in all isolates. Nearly all isolates also harbored the hlα (98.4%) and hlβ (95.2%) genes. The lukM, clfA, fnbB, and tsst-1 genes were detected in 71% (44/62), 50% (31/62), 21% (13/62), and 8.1% (5/62) of isolates, respectively. The genes pvl or bap were not detected in any of the isolates. Furthermore, 10 different virulence gene patterns were observed among the S. aureus isolates (Table 8). The most frequent number of virulence genes per isolate was 7 and this number was detected in all isolates.
Discussion
Mastitis is an important economic disease restricting the development of the dairy industry. Rapid and accurate identification of pathogens is crucial for the development of targeted prevention and control strategies for mastitis, especially on large-scale farms. MALDI-TOF MS is a rapid and reliable technique for the accurate detection of various microorganisms (22). In this study, 62 S. aureus isolates were identified in milk samples from clinical bovine mastitis cases and were subjected to cluster analysis by MALDI-TOF MS. This is the first time that this technique has been applied to detect and analyze bovine mastitis pathogens in China. The incidence of clinical mastitis caused by S. aureus was 9.9% in this investigation, similar to the results of Gao et al. (8). In contrast, a lower incidence (2.8%) of S. aureus bovine mastitis was reported in large dairy herds in Wisconsin, USA (23). The emergence of MRSA not only challenges the treatment of bovine mastitis, but also poses a threat to human health through food chain or other ways. In this study, eight of the 62 isolates were OS-MRSA. A high prevalence of OS-MRSA has been reported among S. aureus of bovine mastitis origin in China (24). Molecular typing indicated that eight OS-MRSA isolates belonged to t2196-agr I and the genotype pattern of the representative MRSA isolate was ST630-CC8-t2196-agr I. Previous studies revealed that the spa types of OS-MRSA from different regions were diverse. The spa types t267, t1234, t324, and t121 were found in OS-MRSA isolates from different countries (24, 25).
The diverse genetic backgrounds of the 62 isolates were indicated by agr typing, spa typing, and MLST. The predominant agr type in this study was agr II. This was in contrast to previous reports, in which agr I was the dominant type in S. aureus isolated from bovine mastitis cases (26, 27). Strains of S. aureus belonging to agr type I were reported to invade epithelial cells more efficiently than strains classified into other agr types (28). This is more likely to result in antibiotic treatment failure. A certain proportion of S. aureus isolates from different farms in the current study were classified as agr I type. In terms of spa types, seven recognized spa types were identified among the 62 isolates. The most common spa type was t529, consistent with a previously report in which t529 was the dominant spa type in Switzerland. Isolates shown to belong to agr II has only a single spa type, however, there were seven or four spa types among the isolates belonging to agr I or agr IV. Seven representative isolates were selected from different spa types for MLST typing. ST97 and ST50 each corresponded to two spa types (ST97-t2734 and ST97-t730, and ST50-t518 and ST50-t571, respectively). Another isolate was identified as ST398, which was common livestock-associated MRSA type (29). The ST398 was originally reported in swine farmers and often appeared in Europe and America countries (30). Furthermore, ST398 MRSA was shown to correspond to several distinct spa types (t011, t034, t108, and t1451) and was resistant to many non-β-lactam antibiotics (31). In this study, an isolate of ST398-t034 was MSSA and was sensitive to the tested antibiotics. To further understand the circulation of S. aureus, cluster analysis based on MALDI-TOF MS spectra was performed. In group F, the isolates obtained from different farms were classified into sub-clades at a close relative distance, which indicated that there were some relationships in the circulation of S. aureus between the different herds. The development of the dairy industry in China has led to many small farms bring eliminated or choosing to expand, resulting in cows being transferred between different regions or farms. This transfer of animals provides opportunities for S. aureus to spread between different herds.
Information concerning the antimicrobial resistance of pathogens is crucial for selecting effective antibiotic therapies. In this study, antimicrobial resistance to ciprofloxacin was most frequently observed. Previous studies reported that antimicrobial resistance to ciprofloxacin ranged from 29.6 to 53.4% in S. aureus isolated from bovine mastitis in China (29, 32, 33). Conversely, ciprofloxacin-resistant S. aureus is rarely detected in other countries (34, 35). Fluoroquinolone antibiotics are regarded as extremely important antibiotics for human use by the World Health Organization (36). Consequently, special care should be taken regarding the use of these antibiotics in livestock. S. aureus was found to be the most common penicillin-resistant pathogen in bovine mastitis (37). In this investigation, penicillin resistance was the second most prevalent antimicrobial resistance phenotype. The proportion of clinical mastitis caused by S. aureus varies greatly (from 3.6 to 24.2%) on the studied farms. The high proportion of S. aureus in Jiangsu herds was speculated to be related to the use of benzylpenicillin, nafcillin sodium, and dihydrostreptomycin sulfate in dry cows. Because some strains of S. aureus isolated from the Jiangsu herds were resistant to penicillin and streptomycin. The emergence of MDR S. aureus has become a growing public health concern. In this study, 54.5% (6/11) of MDR S. aureus isolates belong to spa type t2196-agr I. Staphylococcal biofilms can enhance the resistance of S. aureus to antibiotics (38). In this investigation, the percentage of MDR isolates was significantly higher in moderate biofilm producers than in weak biofilm producers. In contrast, Zhang et al. (39) reported that there was no significant association between MDR and biofilm formation ability of S. aureus isolated from pork production samples.
Adhesion were essential for S. aureus to invade host cells and evade immune responses (40). Specific genetic changes in spa, clfA, fnbA, as well as a loss of fnbB can lead to a Staphaurex-negative phenotype of S. aureus (41). In this study, all S. aureus isolates harbored the fnbA and clfB genes. This was congruent with previous reports, in which fnbA and clfB were detected in all S. aureus isolates from cases of bovine mastitis (21, 29). In contrast, only 21% of isolates harbored the fnbB gene, which was much lower than that of previous reports (40, 42). Similarly, Gogoi et al. (43) reported that the fnbB gene was only detected in 1.3% of S. aureus isolates in Australia. Biofilm contributes to the development of antimicrobial resistance in S. aureus. All isolates in the current study had the ability to form biofilm. Biofilm-related genes (icaA and icaD) were detected in all isolates, and none of the isolates harbored biofilm associated protein (bap) gene. This was in agreement with previous reports, in which the genes icaA and icaD were frequently detected, but the bap gene was low incidence or not detected in S. aureus of bovine mastitis origin (12, 29, 40). The high prevalence of hlα and hlβ in this study was similar to previous reports in South Africa (100%) and in other regions of China (94.3 and 97.1%, respectively) (26, 42). Furthermore, 16 hlb-positive isolates failed to produce β-hemolysin though Christie, Atkins, and Munch-Petersen (CAMP) tests (data not shown). The reason for this may be associated with the sak gene encoding staphylokinase, which can be integrated into the hlb gene by phage and causes S. aureus to fail to produce β-hemolysin (44). S. aureus can acquire two phage-encoded leukocidins, Panton-Valentine leukocidin (PVL) and LukMF. None of the tested isolates in this study harbored the pvl genes, and this was in agreement with previous investigations, where the pvl gene were often absent in S. aureus isolated from bovine mastitis (45, 46). The bicomponent leukocidin LukMF has high leukotoxic activity to bovine polymorphonuclear cells (47) and the level of LukM in milk is associated with the severity of mastitis during the course of infection (48). The genes encoding this leukocidin are predominantly found in S. aureus isolated from ruminants with mastitis (49). The high prevalence of the lukM gene in the current study was in agreement with other reports (50, 51). Another toxin, Tsst-1 causes toxic shock syndrome in humans by hyperactivation the host immune response (52). In this study, five S. aureus isolates harbored the tsst-1 gene. It is imperative for the protection of public health to continuously monitor the epidemiology of such super antigenic toxin genes.
In conclusion, this study presents the first insights into the characterization of S. aureus isolated from clinical mastitis cases on large-scale dairy farms in China. The findings suggest that selecting effective antibiotics is important to reduce the incidence of S. aureus mastitis in dry cows. Moreover, it is necessary to strictly implement inspection and quarantine procedures to prevent the transmission of S. aureus among different farms. Knowledge generated in this study will contribute to improvements in prevention and control strategies to minimize the risk of bovine mastitis and associated losses on the large-scale dairy farms.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the departmental committee of the College of Veterinary Medicine, Yangzhou University.
Author Contributions
HW participated in the design of this study. KL, LT, LF, JuL, and CB collected the samples. KL and LT performed the experiments. KL analyzed the data and wrote the paper. JiL, GZ, LC, and XM contributed to the preparation of the manuscript. All authors read and approved the final version of the manuscript.
Funding
The investigation was supported by the Natural Science Foundation of Jiangsu Province (No. BK20160062), National Natural Science Foundation of China (No. 31802254), Earmarked fund for Jiangsu Agricultural Industry Technology System [No. JATS (2019)461], Outstanding Young Backbone Teacher Foundation of Yangzhou University, Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and Top-notch Academic Programs Project of Jiangsu Higher Education Institutions (TAPP).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Käppeli N, Morach M, Corti S, Eicher C, Stephan R, Johler S. Staphylococcus aureus related to bovine mastitis in Switzerland: clonal diversity, virulence gene profiles, and antimicrobial resistance of isolates collected throughout 2017. J Dairy Sci. (2019) 102:3274–81. doi: 10.3168/jds.2018-15317
2. Monistero V, Graber HU, Pollera C, Cremonesi P, Castiglioni B, Bottini E, et al. Staphylococcus aureus isolates from bovine mastitis in eight countries: genotypes, detection of genes encoding different toxins and other virulence genes. Toxins. (2018) 10:247–69. doi: 10.3390/toxins10060247
3. Watkins KE, Unnikrishnan M. Evasion of host defenses by intracellular Staphylococcus aureus. Adv Appl Microbiol. (2020) 112:105–41. doi: 10.1016/bs.aambs.2020.05.001
4. Kroning IS, Iglesias MA, Mendonca KS, Lopes GV, Silva WP. Presence of classical enterotoxin genes, agr typing, antimicrobial resistance, and genetic diversity of Staphylococcus aureus from milk of cows with mastitis in Southern Brazil. J Food Prot. (2018) 81:738–42. doi: 10.4315/0362-028X.JFP-17-436
5. Aslantas O, Demir C. Investigation of the antibiotic resistance and biofilm-forming ability of Staphylococcus aureus from subclinical bovine mastitis cases. J Dairy Sci. (2016) 99:8607–13. doi: 10.3168/jds.2016-11310
6. Manukumar HM, Umesha S. MALDI-TOF-MS based identification and molecular characterization of food associated methicillin-resistant Staphylococcus aureus. Sci Rep. (2017) 7:11414–30. doi: 10.1038/s41598-017-11597-z
7. Nonnemann B, Lyhs U, Svennesen L, Kristensen KA, Klaas IC, Pedersen K. Bovine mastitis bacteria resolved by MALDI-TOF mass spectrometry. J Dairy Sci. (2019) 102:2515–24. doi: 10.3168/jds.2018-15424
8. Gao J, Barkema HW, Zhang L, Liu G, Deng Z, Cai L, et al. Incidence of clinical mastitis and distribution of pathogens on large Chinese dairy farms. J Dairy Sci. (2017) 100:4797–806. doi: 10.3168/jds.2016-12334
9. Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl Environ Microb. (2003) 69:18–23. doi: 10.1128/AEM.69.1.18-23.2003
10. Enright MC, Day NPJ, Davies C, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillinresistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. (2000) 38:1008–15. doi: 10.1128/JCM.38.3.1008-1015.2000
11. Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, et al. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol. (2003) 41:5442–8. doi: 10.1128/JCM.41.12.5442-5448.2003
12. Marques VF, Motta CC, Soares BD, Melo DA, Coelho SM, Coelho ID, et al. Biofilm production and beta-lactamic resistance in Brazilian Staphylococcus aureus isolates from bovine mastitis. Braz J Microbiol. (2017) 48:118–24. doi: 10.1016/j.bjm.2016.10.001
13. Stepanović S, Vuković D, Hola V, Bonaventure GD, Djukic S, Cirkovic I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilmproduction by staphylococci. Apmis. (2007) 115:891–9. doi: 10.1111/j.1600-0463.2007.apm_630.x
14. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Document M100–S23. Clinical and Laboratory Standard Institute (CLSI), Wayne, PA (2013).
15. Lovseth A, Loncarevic S, Berdal KG. Modified multiplex PCR method for detection of pyrogenic exotoxin genes in staphylococcal isolates. J Clin Microbiol. (2004) 42:3869–72. doi: 10.1128/JCM.42.8.3869-3872.2004
16. Felipe V, Morgante CA, Somale PS, Varroni F, Zingaretti ML, Bachetti RA, et al. Evaluation of the biofilm forming ability and its associated genes in Staphylococcus species isolates from bovine mastitis in Argentinean dairy farms. Microb Pathog. (2017) 104:278–86. doi: 10.1016/j.micpath.2017.01.047
17. Xu J, Tan X, Zhang X, Xia X, Sun H. The diversities of staphylococcal species, virulence and antibiotic resistance genes in the subclinical mastitis milk from a single Chinese cow herd. Microb Pathog. (2015) 88:29–38. doi: 10.1016/j.micpath.2015.08.004
18. Kalorey DR, Shanmugam Y, Kurkure NV, Chousalkar KK, Barbuddhe SB. PCR-based detection of genes encoding virulence determinants in Staphylococcus aureus from bovine subclinical mastitis cases. J Vet Sci. (2007) 8:151–4. doi: 10.4142/jvs.2007.8.2.151
19. Simojoki H, Hyvonen P, Plumed Ferrer C, Taponen S, Pyorala S. Is the biofilm formation and slime producing ability of coagulase-negative staphylococci associated with the persistence and severity of intramammary infection? Vet Microbiol. (2012) 158:344–52. doi: 10.1016/j.vetmic.2012.02.031
20. Jarraud S. Relationships between Staphylococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect Immun. (2002) 70:631–41. doi: 10.1128/IAI.70.2.631-641.2002
21. Pereyra EA, Picech F, Renna MS, Baravalle C, Andreotti CS, Russi R, et al. Detection of Staphylococcus aureus adhesion and biofilm-producing genes and their expression during internalization in bovine mammary epithelial cells. Vet Microbiol. (2016) 183:69–77. doi: 10.1016/j.vetmic.2015.12.002
22. Ueda O, Tanaka S, Nagasawa Z, Hanaki H, Shobuike T, Miyamoto H. Development of a novel matrix-assisted laser desorption/ionization time-of-flight mass spectrum (MALDI-TOF-MS)-based typing method to identify meticillin-resistant Staphylococcus aureus clones. J Hosp Infect. (2015) 90:147–55. doi: 10.1016/j.jhin.2014.11.025
23. Oliveira L, Hulland C, Ruegg PL. Characterization of clinical mastitis occurring in cows on 50 large dairy herds in Wisconsin. J Dairy Sci. (2013) 96:7538–49. doi: 10.3168/jds.2012-6078
24. Pu W, Su Y, Li J, Li C, Yang Z, Deng H, et al. High incidence of oxacillin-susceptible mecA-positive Staphylococcus aureus (OS-MRSA) associated with bovine mastitis in China. PLoS ONE. (2014) 9:e88134. doi: 10.1371/journal.pone.0088134
25. Haran KP, Godden SM, Boxrud D, Jawahir S, Bender JB, Sreevatsan S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farms. J Clin Microbiol. (2012) 50:688–95. doi: 10.1128/JCM.05214-11
26. Schmidt T, Kock MM, Ehlers MM. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in south African dairy herds: genetic diversity and inter-species host transmission. Front Microbiol. (2017) 8:511. doi: 10.3389/fmicb.2017.00511
27. Magro G, Biffani S, Minozzi G, Ehricht R, Monecke S, Luini M, et al. Virulence genes of S. aureus from dairy cow mastitis and contagiousness risk. Toxins. (2017) 9:195–207. doi: 10.3390/toxins9060195
28. Buzzola FR, Alvarez LP, Tuchscherr LP, Barbagelata MS, Lattar SM, Calvinho L, et al. Differential abilities of capsulated and noncapsulated Staphylococcus aureus isolates from diverse agr groups to invade mammary epithelial cells. Infect Immun. (2007) 75:886–91. doi: 10.1128/IAI.01215-06
29. Wang W, Lin X, Jiang T, Peng Z, Xu J, Yi L, et al. Prevalence and characterization of Staphylococcus aureus cultured from raw milk taken from dairy cows with mastitis in Beijing, China. Front Microbiol. (2018) 9:1123. doi: 10.3389/fmicb.2018.01123
30. Smith TC, Pearson N. The emergence of Staphylococcus aureus ST398. Vector Borne Zoonotic Dis. (2011) 11:327–39. doi: 10.1089/vbz.2010.0072
31. Raji MA, Garaween G, Ehricht R, Monecke S, Shibl AM, Senok A. Genetic characterization of Staphylococcus aureus isolated from retail meat in Riyadh, Saudi Arabia. Front Microbiol. (2016) 7:911. doi: 10.3389/fmicb.2016.00911
32. Liu H, Li S, Meng L, Dong L, Zhao S, Lan X, et al. Prevalence, antimicrobial susceptibility, and molecular characterization of Staphylococcus aureus isolated from dairy herds in northern China. J Dairy Sci. (2017) 100:8796–803. doi: 10.3168/jds.2017-13370
33. Zhang L, Li Y, Bao H, Wei R, Zhou Y, Zhang H, et al. Population structure and antimicrobial profile of Staphylococcus aureus strains associated with bovine mastitis in China. Microb Pathog. (2016) 97:103–9. doi: 10.1016/j.micpath.2016.06.005
34. Bhat AM, Soodan JS, Singh R, Dhobi IA, Hussain T, Dar MY, et al. Incidence of bovine clinical mastitis in Jammu region and antibiogram of isolated pathogens. Vet World. (2017) 10:984–9. doi: 10.14202/vetworld.2017.984-989
35. McMillan K, Moore SC, McAuley CM, Fegan N, Fox EM. Characterization of Staphylococcus aureus isolates from raw milk sources in Victoria, Australia. BMC Microbiol. (2016) 16:169–81. doi: 10.1186/s12866-016-0789-1
36. WHO. Critically Important Antimicrobials for Human Medicine –5th rev. Geneva: World Health Organization (2017).
37. Cheng J, Qu W, Barkema HW, Nobrega DB, Gao J, Liu G, et al. Antimicrobial resistance profiles of 5 common bovine mastitis pathogens in large Chinese dairy herds. J Dairy Sci. (2019) 102:2416–26. doi: 10.3168/jds.2018-15135
38. Schilcher K, Horswill AR. Staphylococcal biofilm development: structure, regulation, and treatment strategies. Microbiol Mol Biol R. (2020) 84:e00026–19. doi: 10.1128/MMBR.00026-19
39. Zhang Y, Xu D, Shi L, Cai R, Li C, Yan H. Association between agr type, virulence factors, biofilm formation and antibiotic resistance of Staphylococcus aureus isolates from pork production. Front Microbiol. (2018) 9:1876. doi: 10.3389/fmicb.2018.01876
40. Zaatout N, Ayachi A, Kecha M, Kadlec K. Identification of staphylococci causing mastitis in dairy cattle from Algeria and characterization of Staphylococcus aureus. J Appl Microbiol. (2019) 127:1305–14. doi: 10.1111/jam.14402
41. Stutz K, Stephan R, Tasara T. SpA, ClfA, and FnbA genetic variations lead to Staphaurex test-negative phenotypes in bovine mastitis Staphylococcus aureus isolates. J Clin Microbiol. (2011) 49:638–46. doi: 10.1128/JCM.01148-10
42. Wang D, Zhang L, Zhou X, He Y, Yong C, Shen M, et al. Antimicrobial susceptibility, virulence genes, and randomly amplified polymorphic DNA analysis of Staphylococcus aureus recovered from bovine mastitis in Ningxia, China. J Dairy Sci. (2016) 99:9560–9. doi: 10.3168/jds.2016-11625
43. Gogoi-Tiwari J, Waryah CB, Eto KY, Tau M, Wells K, Costantino P, et al. Relative distribution of virulence-associated factors among Australian bovine Staphylococcus aureus isolates: potential relevance to development of an effective bovine mastitis vaccine. Virulence. (2015) 6:419–23. doi: 10.1080/21505594.2015.1043508
44. Goerke C, Wirtz C, Fluckiger U, Wolz C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol Microbiol. (2006) 61:1673–85. doi: 10.1111/j.1365-2958.2006.05354.x
45. Srednik ME, Usongo V, Lepine S, Janvier X, Archambault M, Gentilini ER. Characterisation of Staphylococcus aureus strains isolated from mastitis bovine milk in Argentina. J Dairy Res. (2018) 85:57–63. doi: 10.1017/S0022029917000851
46. Zhang L, Gao J, Barkema HW, Ali T, Liu G, Deng Y, et al. Virulence gene profiles: alpha-hemolysin and clonal diversity in Staphylococcus aureus isolates from bovine clinical mastitis in China. BMC Vet Res. (2018) 14:63–75. doi: 10.1186/s12917-018-1374-7
47. Schlotter K, Ehricht R, Hotzel H, Monecke S, Pfeffer M, Donat K. Leukocidin genes lukF-P83 and lukM are associated with Taphylococcus aureus clonal complexes 151, 479 and 133 isolated from bovine udder infections in Thuringia, Germany. Vet Res. (2012) 43:42. doi: 10.1186/1297-9716-43-42
48. Vrieling M, Boerhout EM, van Wigcheren GF, Koymans KJ, Mols-Vorstermans TG, de Haas CJ, et al. LukMF' is the major secreted leukocidin of bovine Staphylococcus aureus and is produced in vivo during bovine mastitis. Sci Rep. (2016) 6:37759. doi: 10.1038/srep37759
49. Alonzo F III, Torres VJ. The bicomponent pore-forming leucocidins of Staphylococcus aureus. Microbiol Mol Biol Rev. (2014) 78:199–230. doi: 10.1128/MMBR.00055-13
50. Li T, Lu H, Wang X, Gao Q, Dai Y, Shang J, et al. Molecular characteristics of Staphylococcus aureus causing bovine mastitis between 2014 and 2015. Front Cell Infect Microbiol. (2017) 7:127–37. doi: 10.3389/fcimb.2017.00127
51. Ben Said M, Abbassi MS, Bianchini V, Sghaier S, Cremonesi P, Romano A, et al. Genetic characterization and antimicrobial resistance of Staphylococcus aureus isolated from bovine milk in Tunisia. Lett Appl Microbiol. (2016) 63:473–81. doi: 10.1111/lam.12672
Keywords: Staphylococcus aureus, bovine mastitis, MALDI TOF MS, molecular typing, virulence gene, antimicrobial resistance (AMR)
Citation: Liu K, Tao L, Li J, Fang L, Cui L, Li J, Meng X, Zhu G, Bi C and Wang H (2020) Characterization of Staphylococcus aureus Isolates From Cases of Clinical Bovine Mastitis on Large-Scale Chinese Dairy Farms. Front. Vet. Sci. 7:580129. doi: 10.3389/fvets.2020.580129
Received: 04 July 2020; Accepted: 12 November 2020;
Published: 07 December 2020.
Edited by:
Alejandra Victoria Capozzo, Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), ArgentinaReviewed by:
László Ózsvári, University of Veterinary Medicine Budapest, HungaryStefan Monecke, Leibniz Institute of Photonic Technology (IPHT), Germany
Copyright © 2020 Liu, Tao, Li, Fang, Cui, Li, Meng, Zhu, Bi and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heng Wang, c2RhdWxlbGxvd0AxNjMuY29t
†These authors have contributed equally to the work
 Kangjun Liu
Kangjun Liu Luyao Tao
Luyao Tao Jianji Li
Jianji Li Li Fang
Li Fang Luying Cui
Luying Cui Jun Li
Jun Li Xia Meng
Xia Meng Guoqiang Zhu
Guoqiang Zhu Chongliang Bi
Chongliang Bi Heng Wang
Heng Wang